Abstract
Double-stranded RNA (dsRNA) viruses conceal their genome from the host to avoid triggering unfavorable cellular responses. The crystal structure of the core of one such virus, bluetongue virus, reveals an outer surface festooned with dsRNA. This may represent a deliberate strategy to sequester dsRNA released from damaged particles to prevent host cell shutoff.
When double-stranded RNA (dsRNA) is released into the cytoplasm of a mammalian cell, it elicits host cell defense mechanisms that can modify host cell translation control and lead to an apoptotic response. Presumably to avoid shutdown of the host cell, the replication strategy of dsRNA viruses, including the members of the family Reoviridae, is such that genomic dsRNA is never released into an infected cell (5, 13). For example, bluetongue virus (BTV), the type species of the genus Orbivirus, produces capped mRNA internally within an enclosed core particle, releasing only single-stranded RNA into the cytoplasm of the infected cell. Some of these viruses, such as Orthoreovirus, possess viral proteins with an affinity for dsRNA. It has been proposed that this provides an effective mechanism for soaking up any available dsRNA, acting as a further line of defense against the release of genomic material into the cell in the event of rupture of virus particles (1, 8, 10).
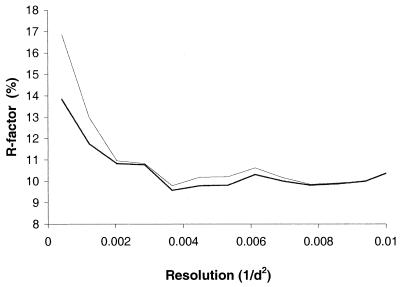
We have determined the atomic structure of the BTV core (BTV type 1 South Africa [BTV-1 SA]) and learned something of the internal organization of the dsRNA genome segments and transcription complex, along with the routes of entry and exit for substrate and product molecules (4, 6, 7). However, during this analysis, we noted that our model for the crystal structure was in unexpectedly poor agreement with our observed diffraction data at low resolution (Fig. 1). In order to investigate the reason for this, we calculated a difference electron density map between our best model for the virus particle and the experimental data, including data only to 10-Å Bragg spacings (Table 1) (experimental details and further statistics are given in reference 4). This low-resolution map had no symmetry imposed on it additional to that imposed by the formation of the crystal lattice and showed essentially no features within the viral core, indicating that the model for the structure of the protein and internal RNA was, as expected, consistent with the data. However, to our surprise, there were beautifully defined ropes of electron density festooning the outer surface of the virus particles (Fig. 2). These were the only significant features in the difference map and were present at a height of, on average, 4σ, demonstrating the exquisite power of difference Fourier syntheses. This electron density could be readily interpreted in terms of A-form dsRNA with the major and minor grooves clearly delineated (Fig. 2). Fitting A-form RNA to the electron density (bent and kinked where necessary) indicated that the festoons of RNA comprised three distinct segments of 412, 276, and 265 bp, respectively. The coordinates (code 1h1k) and structure factors have been deposited with the Protein Data Bank. The net result of this was a virus core in the crystal that was intimately associated with some 2,000 bp of dsRNA (equivalent to approximately 10% of the 19.2 kbp of the complete viral genome) which adhere to its surface via predominantly electrostatic interactions. Two of the three ropes of dsRNA are passed from one particle to another within the crystal lattice. Thus, the crystal lattice is held together by a combination of RNA-protein and protein-protein interactions. The crystal-packing interactions that are mediated by particle-particle contacts total some 10,000 Å2 of surface area per half particle (calculated by AREAIMOL [3] with a probe of 3-Å radius; the crystallographic asymmetric unit contains half a particle). In contrast, the interactions mediated by RNA-protein interactions total 42,000 Å2 per half particle, suggesting that RNA acts as a mesh, helping to hold the crystals together. This presumably explains the observation (2) that the BTV core crystals grow over a period of several months at 29°C, suggesting that the decay of some particles to release dsRNA gene segments may well be a rate-limiting stage in crystal growth. The RNA lattice may also explain the ability of the crystals to survive transfer from a crystallization mother liquor containing 11 to 16% ammonium sulfate and 25% ethylene glycol to a 0.1 M Tris-HCl buffer containing 9 mM magnesium chloride (4).
FIG. 1.
Improvement in the agreement between observed diffraction data and data calculated from an atomic model, with and without external RNA. The R-factor between the data and the initial model missing external RNA is plotted, with respect to resolution, as a thin line; the R-factor between the data and the improved model is plotted as a thick line.
TABLE 1.
Data collection and processing statisticsa
| No. of crystals | No. of images | Resolution limits (Å) | Total no. of reflections | No. of unique reflections | Complete- ness (%) | I/σIa | Rmerge (%)b |
|---|---|---|---|---|---|---|---|
| 587 | 1,045 | 105.0-10.0 | 490,727 | 251,202 | 94.7 | 18.5 | 10.0 |
I/σI, mean signal-to-noise ratio for the diffraction data.
Rmerge, ΣjΣh(|Ij,h − 〈Ih〉)|/ ΣjΣh(〈Ih〉), where h is the unique reflection index, Ij,h is intensity of symmetry-related reflection, and 〈Ih〉 is the mean intensity.
FIG. 2.
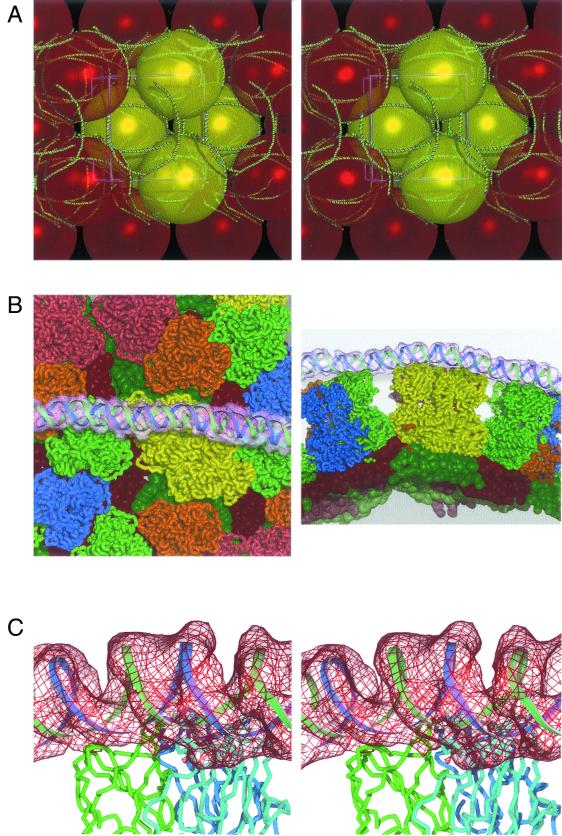
Ropes of RNA within BTV core crystals. (A) Stereo figure showing the meshing together of the BTV core particles in the crystals by the long ropes of external dsRNA. The model for the dsRNA is drawn as a green-and-red worm; the core particles are shown to scale as colored yellow and red spheres. (B) Two orthogonal views of the interactions of the parts of one RNA rope with the BTV core. The RNA model and electron density are shown (the density is rendered semitransparent). The VP7 trimers are colored as described in reference 7, with the underlying VP3 molecules shown in slightly darker colors (again, colored as in reference 7). (C) A stereo image of the fit of the dsRNA model, with a portion of the difference electron density map in red, showing the typical disposition of the RNA on the top surface of a VP7 trimer. The strands of the RNA model are shown as green and blue ribbons, and the three polypeptide chains of VP7 are drawn in green, cyan, and blue.
The T=13 symmetry of the outer layer of the core means that any potential binding site on VP7(T13) is presented 780 times on the surface of the particle. In the crystal, only 78 sites are occupied (39 in the half-virus that represents the crystallographic asymmetric unit). We found that these independent binding sites make repeated use of polar residues; out of 2,419 residues that contact the RNA, 1,606 belong to the set Asn 164, Asn 200, Gln 202, Gln 239, and Asn 240. These residues are conserved across BTVs and reside on the outer surface of the VP7(T13) trimers. Further evidence for the specificity of the core-dsRNA interactions is shown in Fig. 3, which presents the trajectories of the 39 independent instances of RNA binding. Over half of the interactions (24 of 39) are strikingly similar and involve contact with two subunits of the trimer. The remaining binding events are more diverse but tend to involve interactions with, predominantly, a single VP7 subunit. These rarer interactions may nevertheless be classified into three distinct binding modes (Fig. 3). The presence of 780 subunits of VP7(T13) on the outside of the BTV core provides immense scope for the amplification of binding strength by avidity effects. This is facilitated by the arrangement of the BTV trimers on the T=13 surface lattice, which presents the most favored dsRNA binding sites appropriately aligned, zipper-like, along great circles of the core surface. Most of the minor binding modes are generated as contacts interspersed between these driving interactions. If we sum the surface area of interaction along the longest piece of interacting RNA, we find that the individually rather modest contacts produce a total contact area of some 17,000 Å2 which, by analogy to protein-protein interactions, would correspond to an extremely robust interaction (9).
FIG. 3.
Binding modes. The 39 crystallographically independent interactions between the dsRNA and VP7 trimers are displayed superimposed, with the threefold degenerate trimer orientation chosen so as to best align the RNA trajectories. The upper panel illustrates the RNA colored according to binding mode. The principal mode (24 members, lower bundle) is colored green-yellow to cyan. The next-most populous mode (8 members) is colored yellow (top). Two minor intermediate modes (3 and 4 members) are colored red and orange, respectively. The lower panel is identical except that the RNA is translucent, revealing the structure of the underlying VP7 trimer.
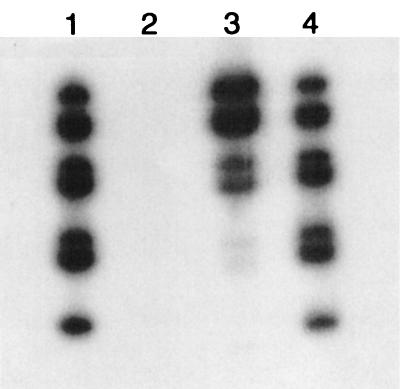
To investigate this core-RNA interaction in solution, BTV-1 SA dsRNA was labeled with [32P]cytidine bis-diphosphate (pCp) and RNA ligase as described previously (12), a method which labels each of the genome segments in equimolar amounts. Labeled dsRNA was separated from unincorporated label by using Microspin S-200 high-resolution columns (Amersham Pharmacia) as recommended by the manufacturer. Purified BTV core particles (1 mg) were incubated with labeled dsRNA (1.25 μg of labeled RNA ≈ 320,000 cpm) in 100 μl of 0.1 M Tris-HCl (pH 8.0)-9 mM MgCl2 for 20 min at 30°. The sample was then layered on a 5.5-ml cushion of 40% (wt/vol) sucrose in an SW50 rotor and centrifuged for 2 h at 40,000 rpm (108,000 × g) at 20°C. Under these conditions, approximately 1% of the labeled dsRNA was pelleted with the core particles, although in a “minus cores” control tube, less than 0.02% of the label was detected at the bottom of the tube. In initial sucrose gradient experiments, addition of the labeled dsRNA had prevented the core particles from banding in 10 to 50% (wt/vol) sucrose gradients, suggesting that they had aggregated and had pelleted at a faster rate. The pelleted cores (through the 40% [wt/vol] sucrose cushion) were resuspended in electrophoresis sample buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 4). Labeled RNA bands were detected in the pellet containing the BTV cores but not at the bottom of the minus cores control tube. The amount of RNA that pelleted with the cores was considerably less than one molecule per core particle, and the individual segments did not bind in an equimolar manner but rather in amounts that appeared to show a direct correlation with the size of the RNA segment (Fig. 4). These observations suggest that relatively few binding sites are available or that binding is a slow process. Under the experimental conditions used, the larger dsRNA molecules may simply represent a larger target for core particle binding, suggesting a lack of nucleotide sequence specificity in the binding mechanism. Further experiments will be required to see if there is any binding specificity for RNA over DNA molecules or for single-stranded RNA over dsRNA molecules and if the amount of RNA bound to the cores increases with longer incubation times. Nevertheless, the direct association of dsRNA with BTV core particles in solution suggests that our crystallographic observations may reflect a general property of the BTV core particles.
FIG. 4.
Autoradiograph of a sodium dodecyl sulfate-11% polyacrylamide electrophoresis gel that was used to analyze interactions between BTV-1 SA core particles and [32P]pCp-labeled BTV-1 SA dsRNA. dsRNA binding was detected by pelleting core particles through a 40% (wt/vol) sucrose cushion. Lanes 1 and 4 are positive controls and show that, although genome segments 2 and 3, 5 and 6, and 8 and 9 were not well separated, all 10 segments were present in the dsRNA starting material and were labeled in equimolar amounts. Lane 2 shows analysis of the pellet from a sample of the labeled dsRNA that was centrifuged without BTV core particles, showing the absence of detectable RNA bands. Lane 3 shows analysis of the pellet when cores were incubated with the labeled dsRNA prior to centrifugation, demonstrating that there is an association between the dsRNA and core particles. In addition, although all of the dsRNAs are present in the pellet (including small amounts of segment 10), it is clear that there is a higher ratio of binding of the larger segments of dsRNA to cores than that of the smaller segments.
The intimate interaction of dsRNA with the BTV core is likely to be biologically useful to the virus. The inadvertent release of dsRNA in an infected cell would be disastrous for an ongoing infection, which, in insect cells, may persist for several weeks (11), and our analysis suggests that the BTV core particles themselves scour infected cells of dsRNA released from damaged particles.
Acknowledgments
We thank disease security officers S. Williams and D. Goodridge, R. Esnouf and J. Dong for computing support, and R. Bryan for software. We also thank the ESRF and the EMBL Outstation, Grenoble, France, for assistance in data collection and financial support for travel (grant contract HPRI-CT-1999-00022) and, in particular, the staff of ID14.
This work was supported by the Biotechnology and Biological Sciences Research Council and Medical Research Council and the European community (grant contract Bio4 CT 97-2364). P.P.C.M. and J.N.B. are supported by the Department for Environment, Food and Rural Affairs; J.M.G. is supported by the Royal Society, and D.I.S. is supported by the Medical Research Council.
REFERENCES
- 1.Bergeron, J., T. Mabrouk, S. Garzon, and G. Lemay. 1998. Characterization of the thermosensitive ts453 reovirus mutant: increased dsRNA binding of sigma 3 protein correlates with interferon resistance. Virology 246:199-210. [DOI] [PubMed] [Google Scholar]
- 2.Burroughs, J. N., J. M. Grimes, P. P. C. Mertens, and D. I. Stuart. 1995. Crystallization and preliminary X-ray analysis of the core particle of bluetongue virus. Virology 210:217-220. [DOI] [PubMed] [Google Scholar]
- 3.Collaborative Computational Project, No. 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Diprose, J. M., J. N. Burroughs, G. C. Sutton, A. Goldsmith, P. Gouet, R. Malby, I. Overton, S. Zientara, P. P. C. Mertens, D. I. Stuart, and J. M. Grimes. 2001. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 20:7229-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eaton, B. T., A. D. Hyatt, and S. M. Brookes. 1990. The replication of bluetongue virus. Curr. Top. Microbiol. Immunol. 162:89-118. [DOI] [PubMed] [Google Scholar]
- 6.Gouet, P., J. M. Diprose, J. M. Grimes, R. Malby, J. N. Burroughs, S. Zientara, D. I. Stuart, and P. P. C. Mertens. 1999. The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell 97:481-490. [DOI] [PubMed] [Google Scholar]
- 7.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. C. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs, B. L., and J. O. Langland. 1998. Reovirus sigma 3 protein: dsRNA binding and inhibition of RNA-activated protein kinase. Curr. Top. Microbiol. Immunol. 233:185-196. [DOI] [PubMed] [Google Scholar]
- 9.Janin, J., S. Miller, and C. Chothia. 1988. Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol. 204:155-164. [DOI] [PubMed] [Google Scholar]
- 10.Martínez-Costas, J., C. González-López, V. N. Vakharia, and J. Benavente. 2000. Possible involvement of the double-stranded RNA-binding core protein σA in the resistance of avian reovirus to interferon. J. Virol. 74:1124-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mellor, P. S. 1990. The replication of bluetongue virus in Culicoides vectors. Curr. Top. Microbiol. Immunol. 162:143-161. [DOI] [PubMed] [Google Scholar]
- 12.Mertens, P. P., S. Pedley, J. Cowley, and J. N. Burroughs. 1987. A comparison of six different bluetongue virus isolates by cross-hybridization of the dsRNA genome segments. Virology 161:438-447. [DOI] [PubMed] [Google Scholar]
- 13.Zarbl, H., and S. Millward. 1983. The Reovirus multiplication cycle, p. 107-196. In W. Joklik (ed.), The Reoviridae. Plenum Press, New York, N.Y.