Abstract
A key early event in the replication of herpes simplex virus 1 (HSV-1) is the localization of infected-cell protein no. 0 (ICP0) in nuclear structures knows as ND10 or promyelocytic leukemia oncogenic domains (PODs). This is followed by dispersal of ND10 constituents such as the promyelocytic leukemia protein (PML), CREB-binding protein (CBP), and Daxx. Numerous experiments have shown that this dispersal is mediated by ICP0. PML is thought to be the organizing structural component of ND10. To determine whether the virus targets PML because it is inimical to viral replication, telomerase-immortalized human foreskin fibroblasts and HEp-2 cells were transduced with wild-type baculovirus or a baculovirus expressing the Mr 69,000 form of PML. The transduced cultures were examined for expression and localization of PML in mock-infected and HSV-1-infected cells. The results obtained from studies of cells overexpressing PML were as follows. (i) Transduced cells accumulate large amounts of unmodified and SUMO-I-modified PML. (ii) Mock-infected cells exhibited enlarged ND10 structures containing CBP and Daxx in addition to PML. (iii) In infected cells, ICP0 colocalized with PML in ND10 early in infection, but the two proteins did not overlap or were juxtaposed in orderly structures. (iv) The enlarged ND10 structures remained intact at least until 12 h after infection and retained CBP and Daxx in addition to PML. (v) Overexpression of PML had no effect on the accumulation of viral proteins representative of α, β, or γ groups and had no effect on the accumulation of infectious virus in cells infected with wild-type virus or a mutant (R7910) from which the α0 genes had been deleted. These results indicate the following: (i) PML overexpressed in transduced cells cannot be differentiated from endogenous PML with respect to sumoylation and localization in ND10 structures. (ii) PML does not affect viral replication or the changes in the localization of ICP0 through infection. (iii) Disaggregation of ND10 structures is not an obligatory event essential for viral replication.
One of the early hallmarks of herpes simplex virus 1 (HSV-1) infection is structural changes in infected cells associated with the function of infected-cell protein no. 0 (ICP0) (reviewed in reference 51). Specifically, ICP0 made immediately after infection is transported to structures known as nuclear bodies, promyelocytic leukemia oncogenic domains, Kr bodies, or ND10 (reviewed in references 25, 36, and 41). In this report we designate these bodies ND10 based largely on the preeminence of this term in virological literature. Within hours after the localization of the ICP0 protein in these structures, the contents of ND10 become dispersed (16, 39, 40). This dispersal has been linked specifically to ICP0 inasmuch as these structures are also affected in cells transfected with the ICP0 gene in the absence of other viral genes (44, 46). Following dispersal on ND10, ICP0 is translocated to the cytoplasm (27, 35). The major constituent of ND10 structures is the promyelocytic leukemia protein (PML). Although ND10 structures contain additional components such as SP100, SP140, ISG20, SUMO-I, CREB-binding protein (CBP), pRB, Daxx, BAX, HAUSP, P53, and nascent RNA (reviewed in references 10, 25, 30, 36, 41, and 67), PML is responsible for the proper localization of all other ND10-associated proteins since they are all dispersed in PML−/− cells (26, 65). The dispersal of ND10 is accompanied by proteasome-dependent degradation of sumoylated PML (7, 15, 44). One conclusion of the existing literature is that degradation of ND10 is essential for the translocation of ICP0 from the nucleus into the cytoplasm since in cells treated with proteasomal inhibitors ND10 remains intact and translocation does not ensue (7, 15). The argument that HSV-1 targets PML for degradation because it is inimical to viral replication is supported by the evidence that PML has been reported to have an antiviral activity during infection with different viruses such as vesicular stomatitis virus, adenovirus, and influenza virus A (8, 9). Additional evidence that PML may play an inhibitory role in viral replication is based on the known functions of PML (reviewed in references 38, 41, and 50).
Briefly, PML is a RING finger, B-box, coiled-coil protein; it contains a characteristic C3HC4 zinc (Ring) finger and two additional cysteine-rich regions (B boxes) followed by a predicted coiled-coil region (for reviews see references 3, 4, and 52). PML has been implicated in a significant number of cellular process including transcription (10, 22, 30, 32, 55, 56, 64, 66), cell proliferation, senescence, tumorigenesis, apoptosis, resistance to virus infection (38, 50), and hormone signaling on the following bases. (i) In vitro experiments have shown that PML can inhibit transformation induced by neu (c-erb2, ERBB2), Ha-ras, mutant p53, or Ha-ras plus c-myc (33, 42, 43). (ii) PML is involved in the regulation of p53-dependent senescence upon oncogenic transformation (17, 48). (iii) PML−/− mice develop more tumors than wild-type mice do when challenged with carcinogens (60). (iv) PML−/− mice and PML−/− cells grown in vitro are protected from caspase-dependent apoptotic signals (60, 61). PML is also implicated in caspase-independent apoptosis involving the activation and relocalization of BAX or Daxx into the nuclear bodies (49, 54, 67). (v) PML is involved in the activation of CREB-binding protein (CBP), suggesting an involvement in hormone signaling (10, 30). (vi) Expression of nuclear body proteins PML, ISG120, SP140, and SP100 and proteasome components is increased in response to interferon (IFN) (19-21, 23, 31). The PML promoter contains functional elements responsive to IFN-α/β and IFN-γ (53). Thus PML has been reported to be the primary target of IFN. In addition to HSV-1, other viruses, notably cytomegalovirus (CMV) (1) and adenoviruses (9), cause drastic changes to ND10s and induce their disruption or the relocalization of their constituents.
In essence PML and the ND10 structures have emerged as components of the host defense system designed to thwart viral infection, and, conversely, they are the targets of viral functions designed to block the cell from blocking viral replication. The involvement of ICP0 in this process became more significant with the accruing evidence that ICP0 mediates the destruction of several cellular proteins and acts as a multiple ubiquitin ligase (5, 13, 24, 57). To define better the role of PML, we transduced human cells with a baculovirus expressing PML. We report that (i) the induced PML is sumoylated and aggregates with CBP and Daxx in enlarged ND10 structures. (ii) Late in HSV-1 infection, the induced PML, CBP, and Daxx are retained in ND10 structures. (iii) ICP0 is distributed similarly in treated and untreated cells. (iv) Finally, the overexpression of PML has no effect on the accumulation of infectious virus or of proteins representative of various kinetic classes of HSV-1.
MATERIALS AND METHODS
Cells and viruses.
Telomerase-transformed human embryonic foreskin (pHF) fibroblasts were obtained from Thomas Shenk (Princeton University, Princeton, N.J.). HEp-2, U-2 Os (human osteosarcoma cells), and Vero cells were obtained from the American Type Culture Collection (ATCC). pHF, HEp-2, and Vero cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10 (pHF) or 5% (HEp-2 and Vero) fetal bovine serum (FBS). U-2 Os cells (HTB-96) were grown in McCoy medium (GIBCO-BRL) supplemented with 10% FBS. HSV-1 strain F [HSV-1(F)], a limited-passage isolate, is the prototype HSV-1 strain used in this laboratory (12). The construction and phenotypic properties of recombinant virus R7910, lacking both copies of the α0, were described elsewhere (28).
Construction of recombinant baculovirus.
The construction of the pAcCMV baculovirus transfer vector, which contains the human CMV immediate-early promoter/enhancer sequences, was described elsewhere (68). PML cDNA was excised from a simian virus 40-PML plasmid encoding the Mr 69,000 form of PML (kindly provided by K. Borden, Mount Sinai School of Medicine, New York, N.Y.) with NcoI and inserted into the NcoI site of pAcSG2 (PharMingen, San Diego, Calif.) to generate the pAc-PML baculovirus transfer vector. The XhoI-EcoRI fragment from pAcCMV containing the human CMV immediate-early promoter/enhancer sequences was inserted into the XhoI-EcoRI site of pAc-PML to generate the pAcCMV-PML baculovirus transfer vector. Baculovirus encoding PML (Bac-PML) was generated by cotransfecting Sf9 cells with BaculoGold linearized baculovirus DNA with the pAcCMV-PML transfer vector according to the manufacturer's instructions (PharMingen). Bac-PML and the wild-type baculovirus (PharMingen) were propagated in Sf9 cells grown in 150-cm3 flasks in TNM-FH insect medium (PharMingen). Virus stocks were prepared and titered as described by the manufacturer. In mammalian cells infected with Bac-PML, the expression of PML is directed by the CMV promoter.
Infection of cells.
HEp-2 cell grown in a 25-cm2 flask were exposed to 20 PFU of Bac-PML per cell or mock infected and incubated at 37°C for 2 h in TNM-FH insect cell medium. The inoculum was replaced with fresh DMEM containing 10% FBS and 5 mM sodium butyrate. Expression of PML was verified 24 h after baculovirus infection. In coinfection experiments, pHF and HEp-2 cells were exposed to 20 PFU of wild-type baculovirus or Bac-PML per cell and incubated at 37°C for 2 h in TNM-FH insect cell medium. The inoculum was replaced with fresh DMEM containing 10% FBS and 10 (pHF) or 5 mM (HEp-2) sodium butyrate for 10 h. The cells were then exposed to 5 PFU of HSV-1(F) or 1 PFU of R7910 recombinant virus per cell in mixture 199 (Gibco) supplemented with 1% calf serum (199V). At 2 h after exposure to the virus, the inocula were replaced with fresh DMEM containing 10% FBS and 10 mM sodium butyrate. The cells were harvested at various times after herpesvirus infection as indicated in Results.
Virus titration.
pHF or Hep-2 cells (4 × 106) seeded on a 25-cm2 flask were infected with 20 PFU of wild-type or Bac-PML baculovirus for 2 h at 37°C. The inocula were replaced by fresh DMEM containing 10% FBS and either 10 (pHF) or 5 mM (HEp-2) sodium butyrate. After 10 h of incubation, the cells were exposed to HSV-1(F) or the R7910 mutant for 2 h. At 2 or 24 h after infection, the cells were rinsed three time with 199V and then harvested in 1.5 ml of mixture 199V, frozen and thawed twice, sonicated, and titrated in Vero [(HSV-1(F)] or U-2 Os (R7910 mutant) cell cultures.
Immunoblotting of electrophoretically separated cell lysates.
Cells grown in 25-cm2 flasks were infected as described in Results. At various intervals the medium was replaced with ice-cold phosphate-buffered saline (PBS; 0.15 M NaCl, 0.01 M Na2HPO4, 0.01 M NaH2PO4, pH 7.4) and the cultures were stored on wet ice for 10 min and then harvested by scraping. After centrifugation the cell pellet was rinsed and the cells were pelleted again by centrifugation in 1 ml of PBS and then lysed in a 1:3 mixture of buffer I (5% sodium dodecyl sulfate [SDS], 0.15 M Tris-HCl [pH 6.8], 30% glycerol) and buffer II (25 mM Tris-HCl [pH 8.3], 50 mM NaCl, 0.5% NP-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitors). The lysate was sonicated briefly, centrifuged for 10 min at 13,000 rpm at 4°C (Biofuge pico, 3325 Heraeus rotor; Sorvall, Norwalk, Conn.). The supernatant fluid was transferred into a tube containing [1/4] volume of 4× disruption buffer (8% SDS, 200 mM Tris [pH 6.8], 12% sucrose, 20% β-mercaptoethanol, and bromophenol blue). The extracts were boiled for 5 min, and 50 μg of protein per lane was subjected to electrophoresis on an SDS-10% N,N′-diallyltardiamide-acrylamide gel and then transferred to nitrocellulose membranes. The membranes were blocked for 1 h with PBS containing 5% nonfat dry milk and 0.05% Tween 20, rinsed, and reacted overnight at 4°C with the appropriate primary antibody in PBS containing 1% bovine serum albumin (BSA) and 0.05% Tween 20. Mouse monoclonal antibodies to PML (Santa Cruz Biotechnology, Santa Cruz, Calif.), SUMO-I (Santa Cruz Biotechnology), actin (Sigma), ICP4, and ICP0 (H1083) were used at 1:500, 1:400, 1:200, 1:1,000, and 1:1,000 dilutions, respectively. The rabbit polyclonal antibodies against PML (Santa Cruz Biotechnology), ICP22, the product of UL38, and thymidine kinase (TK) were used at 1:500 dilutions. The rabbit polyclonal antibodies against SUMO-I (Santa Cruz Biotechnology) and ICP0 (28) were used at 1:200 and 1:1,000 dilutions, respectively. The membrane was rinsed and then reacted with secondary antibodies. Goat anti-mouse and goat anti-rabbit peroxidase-conjugated antibodies (Sigma) and goat anti-mouse and goat anti-rabbit alkaline phosphatase-conjugated antibodies (Bio-Rad, Hercules, Calif.) were used at 1:1,000, 1:1,000, 1:3,000, and 1:3,000 dilutions, respectively. All rinses were done in PBS and 0.05% Tween 20. Immunoblots were developed either with alkaline phosphatase or through enhanced chemiluminescence detection according to the instructions of the manufacturer (ECL and RPN2209; Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom).
Preparation of pHF fibroblast cultures for immunofluorescence.
pHF cells were seeded onto glass slides (Cell-line, Newfield, N.J.) at 104 cells per well. One day after the seeding the cultures were exposed to 40 PFU of Bac-PML or wild-type baculovirus per cell and incubated at 37°C for 2 h in TNM-FH insect medium. Then the inoculum was replaced with fresh DMEM containing 10% FBS and 10 mM sodium butyrate. Cells were fixed at 15 h after infection in ice-cold methanol. In coinfection experiments, after exposure to baculovirus for 2 h and incubation for an additional 10 h in DMEM containing 10% FBS and 10 mM sodium butyrate, the cells were exposed to 20 PFU of HSV-1(F) per cell for 2 h in 199V. The inocula were replaced with fresh DMEM containing 10% FBS and 10 mM sodium butyrate. Cells were fixed at the times indicated in Results after infection with HSV-1(F).
Immunofluorescence analyses.
At the times indicated in Results, the slide cultures were rinsed five times in DMEM, fixed with cold methanol for 2 h, and blocked for 1 h in PBS containing 1% BSA and 20% normal human serum. The primary antibodies were diluted in PBS containing 1% BSA and 10% normal human serum. The mouse monoclonal antibodies to PML (Santa Cruz Biotechnology) and glycoprotein D (gD) were used at 1:400 and 1:1,000 dilutions, respectively. The rabbit polyclonal antibodies to ICP0 (28), CBP, and Daxx (Santa Cruz Biotechnology) were used at dilutions of 1:1,000, 1:400, and 1:400, respectively. After reaction overnight at 4°C, the slide cultures were rinsed at least five times with PBS and exposed to the secondary antibodies in PBS containing 1% BSA and 10% normal human serum. Goat anti-rabbit and goat anti-mouse secondary antibodies coupled with Texas red (Molecular Probes, Eugene, Oreg.) were used at a 1:400 dilution, and goat anti-rabbit and goat anti-mouse antibodies coupled with fluorescein isothiocyanate (FITC; Sigma) were used at 1:160 and 1:64 dilutions, respectively. After 1 h of reaction with the secondary antibody, the slide cultures were again rinsed at least five times with PBS and mounted in 90% glycerol containing 1 mg of 1-4-phenylenediamine per ml (Aldrich Chemical Co., Milwaukee, Wis.). The slides were examined in Zeiss confocal microscope. Digitized images of the fluorescent antibody-stained cells were taken with a Zeiss camera (AxioCam) or with confocal laser technology (Zeiss) and were acquired with software provided by Zeiss.
Electron microscopy.
pHF cells were harvested 15 h after infection with Bac-PML and 3 h after infection with HSV-1(F), rinsed, and fixed in 4% paraformaldehyde-0.1% gluteraldehyde in PBS for 1 to 2 h at 4°C. The fixed cells were then pelleted by centrifugation, rinsed, resuspended in PBS, dehydrated, infiltrated, and embedded at low-temperature (E. Kellenberger, E. Carlemaln, W. Villiger, J. Roth, and R. M. Garavito, Law denaturation embedding for electron microscopy of thin sections, Chemische Werke Lowi GMbH, Waldkraiburg, Germany, 1980) in Lowicryl K4M (Polysciences, Inc., Warrington, Pa.) by the technique of progressive lowering of temperature, as described in detail elsewhere (6). Sections 80 to 90 nm thick were cut on a Sorvall MT-2 ultramicrotome with a diamond knife (Diatome, Fort Washington, Pa.) and collected onto 300 mesh nickel grids (Ted Pella Inc., Redding Calif.). The procedure for immunoelectron microscopy has been reported elsewhere (2). Briefly, grids were floated on microdrops (25 to 50 μl) for the following procedures, except where exhaustive rinsing was involved. Sections collected on grids were quenched with 0.5 M ammonium chloride, pH 5.5, for 1 h, and then transferred directly for blocking with 10% normal goat sera (PBS, 1% BSA, 1% Triton X-100) for 15 min at room temperature. Grids prepared in this manner were then incubated on microdrops of dilutions (e.g., 1:5 to 1:100 in PBS-1% BSA-1% Triton X-100) of primary antibodies and reacted overnight in a humidified chamber at 4°C. Following incubation, grids were rinsed briefly over a series of 7 microdrops of PBS containing 1% BSA and 1% Triton X-100 at room temperature. Reaction mixtures with secondary antibodies conjugated with colloidal gold and diluted 1:75 (PBS, 1% BSA, 1% Triton X-100) were incubated for 1 h in a humidified chamber at room temperature. The secondary antibodies used were a goat anti-rabbit antibody conjugated with 5-nm colloidal gold (GFAR-5) and a goat anti-mouse antibody conjugated with 5- (GFAM-5) or 15-nm (GMHL-15) colloidal gold (Ted Pella). The grids were rinsed exhaustively in three successive 50-ml beakers of distilled water, air dried at room temperature, and very lightly stained with 2% aqueous uranyl acetate for 10 min to enhance the visualization of colloidal gold and then viewed in a Elmiskop 201 (Siemens) electron microscope. Photography was done with Kodak OS-163 film and the plates shown are without image processing.
RESULTS
PML encoded in baculoviruses is expressed and sumoylated.
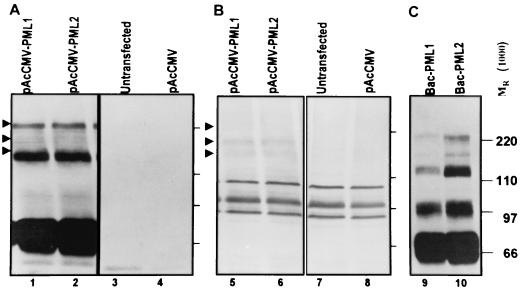
As described in Materials and Methods, the cDNA encoding the 69-kDa form of human PML driven by the CMV immediate-early promoter was cloned into baculovirus transfer vector pAcCMV to generate pAcCMV-PML. To verify that the pAcCMV-PML-encoded 69-kDa form of PML is sumoylated, HEp-2 cells were transfected with pAcCMV-PML or empty vector pAcCMV. The cells were harvested 48 h after transfection, lysed, subjected to electrophoresis on an SDS-denaturing 10% polyacrylamide gel, transferred to a nitrocellulose membrane, and reacted with a rabbit polyclonal antibody against PML. As shown in Fig. 1A, the transfected cells accumulated a protein with an apparent Mr of 69,000 and higher-molecular-weight forms that reacted with the anti-PML antibody (Fig. 1A). The same membrane was reacted with a mouse monoclonal antibody to SUMO-I (Fig. 1B). Some of the higher-molecular-weight forms of PML correspond to SUMO-I-modified PML since the corresponding bands could be also detected with the SUMO-I antibody only in cells transfected with pAcCMV-PML (compare Fig. 1A and C).
FIG. 1.
PML cloned in a plasmid or baculovirus is expressed and modified by SUMO-I. (A and B) HEp-2 cells were transfected with pAcCMV (lanes 4 and 8) or pAcCMV-PML (lanes 1, 2, 5, and 6) or were untransfected (lanes 3 and 7). After 48 h of incubation the cells were harvested, solubilized, subjected to electrophoresis in denaturing gels, and reacted with a mouse monoclonal antibody to PML (A) or with a rabbit polyclonal antibody to SUMO-I (B). (C) Lysates of cells infected with two different preparations of Bac-PML (lanes 9 and 10) were solubilized, subjected to electrophoresis in denaturing gels, and reacted with a mouse monoclonal antibody to PML. Because of the large amount of PML expressed in these cells, the exposure to the antibody was very short, too short to detect endogenous PML in panel A, lanes 3 and 4.
The pAcCMV-PML vector was used to generate baculovirus Bac-PML after cotransfection of Sf9 insect cells with the baculoGold linearized DNA as described in Materials and Methods. To verify that the Bac-CMV baculovirus is also able to express PML in mammalian cells, HEp-2 cells in a 25-cm2 flask were mock infected or exposed to 20 PFU of Bac-PML per cell for 2 h and then incubated at 37°C. The cells harvested 24 h after exposure to baculoviruses were processed as described above. As shown in Fig. 1C, the cells exposed to Bac-PML expressed large amounts of PML and more slowly migrating forms corresponding in electrophoretic mobility to sumoylated PML. Because of very high levels of PML produced in these cells, the reaction time with the anti-PML antibody was very short, too short for detection of endogenous PML in lanes 3 and 4 of Fig. 1A.
Overexpression of PML causes the formation of large ND10 structures.
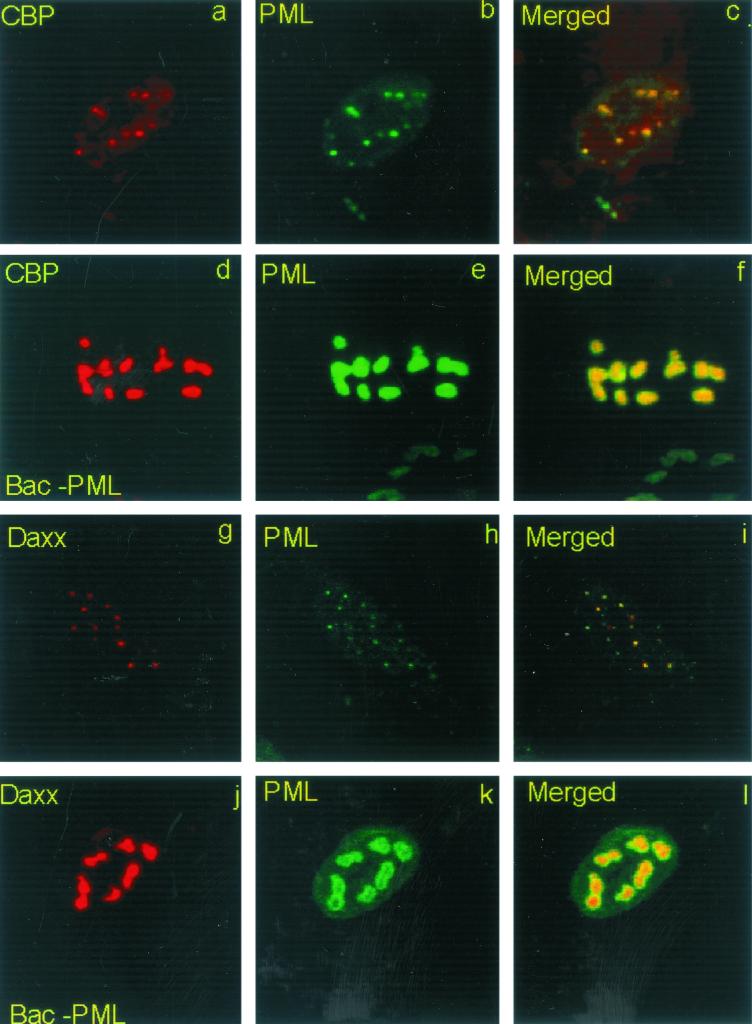
The objective of this series of experiments was to verify by immunofluorescence microscopy that the exogenous PML, encoded by the Bac-PML baculovirus, was correctly localized in ND10 structures of cells not infected with HSV-1. To localize ND10 structures, we used antibodies to two known ND10 components, Daxx and CBP (10, 30, 67). As detailed in Materials and Methods, replicate slide cultures seeded with 104 pHF cells were exposed to 40 PFU of wild-type or Bac-PML baculovirus per cell for 2 h. After 13 h of additional incubation, the cells were fixed in ice-cold methanol for 2 h and then reacted first with a mouse monoclonal antibody to PML and a rabbit polyclonal antibody to Daxx or CBP and then with a goat anti-mouse antibody coupled with FITC and a goat anti-rabbit antibody coupled with Texas red. The results, shown in Fig. 2, were as follows. (i) In mock-infected cells, PML, CBP, and Daxx colocalized in structures defined in the literature as ND10 (Fig. 2a to c and g to i). (ii) In cells exposed to Bac-PML, PML aggregated in large nuclear punctate structures more prominent than those present in cells exposed to wild-type baculovirus (Fig. 2, compare b and h to e and k). (iii) CBP and Daxx colocalized with the exogenous PML in the large nuclear punctate structures (Fig. 2d to f and j to l). We conclude from these two series of experiments that the PML gene carried by Bac-PML was expressed, that PML product was sumoylated, and that the exogenous PML correctly colocalized with CBP and the Daxx protein in enlarged ND10 structures.
FIG. 2.
Overexpression of PML results in the formation of large nuclear structures (ND10) containing CBP and Daxx. pHF fibroblast slide cultures were infected with Bac-PML (d to f and j to l) or wild-type baculovirus (a to c and g to i) as described in Materials and Methods. The cells were fixed and reacted with a mouse monoclonal antibody to PML (b, c, e, f, h, i, k, and l) or a rabbit polyclonal antibody to CBP (a, c, d, and f) or Daxx (g, i, j, and l). The secondary antibodies were anti-mouse immunoglobulin G (IgG) conjugated to fluorescein isothiocyanate and an anti-rabbit IgG conjugated to Texas red. Left and middle columns, single-color images captured separately; right column, merged images. The yellow visualized in the overlaid images represents colocalization of PML and CBP or PML and Daxx. The images were captured with a Zeiss camera (AxioCam) and software provided by Zeiss. The digitized images were not modified subsequent to capture.
The localization of ICP0 is not dependent on the localization of PML.
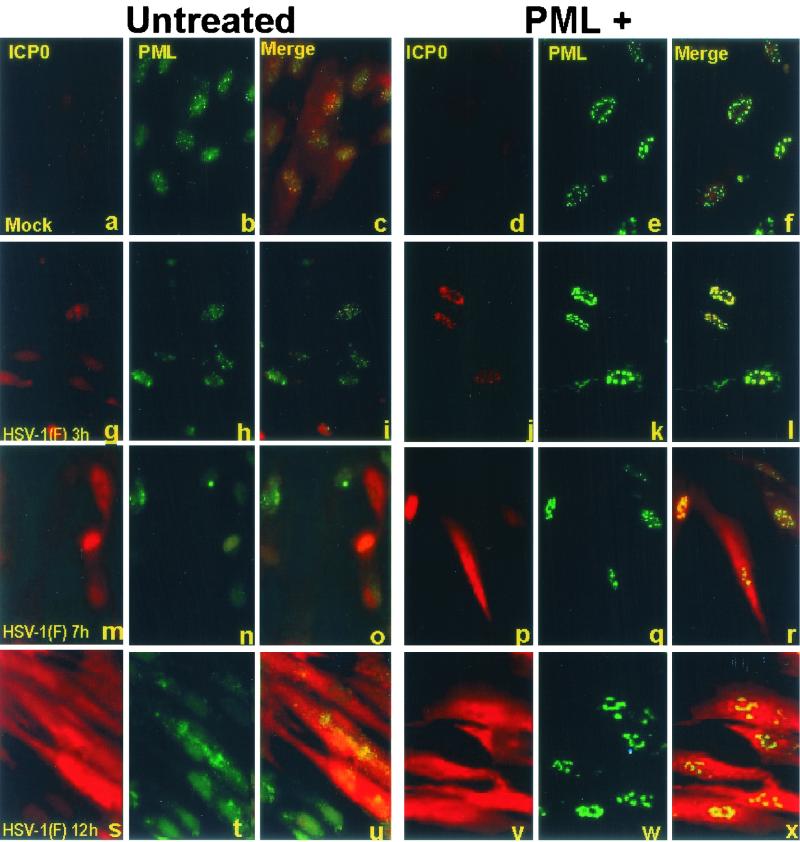
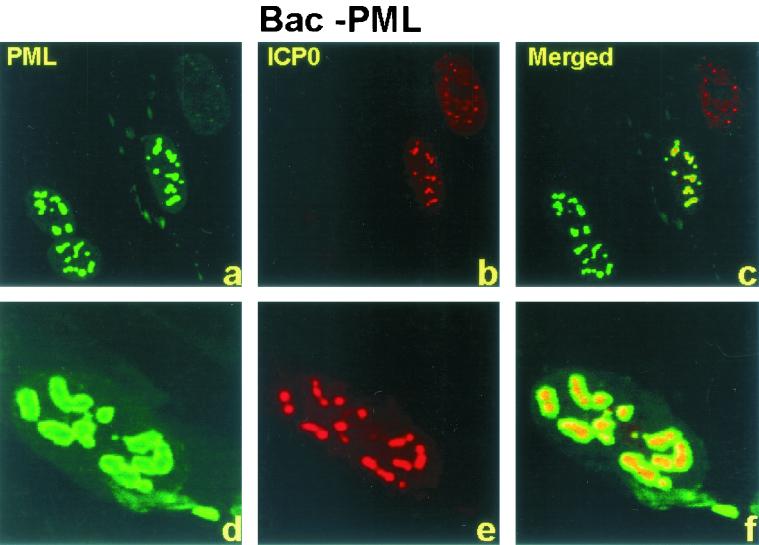
Numerous reports from this and other laboratories have shown that ICP0 expressed by open reading frames introduced into cells by infection or transfection initially colocalizes with ND10 structures and then causes the disappearance of sumoylated PML and the dispersal of ND10 components in a proteasome-dependent manner (7, 15, 16, 39, 40, 44). Thus addition of a proteasome inhibitor, such lactacystin or MG132, prior to or early in infection blocks the dispersal of ND10 components (7, 15). In these cells ICP0 remains associated with ND10 for the duration of infection, suggesting that the dispersion of ND10 is required for the translocation of ICP0 from the nucleus to the cytoplasm (27, 35). The purpose of this series of experiments was therefore twofold. The first objective was to determine the fate of the enlarged ND10 structures after infection with wild-type HSV-1. The second objective was to determine whether the expression and translocation of ICP0 was affected by overexpression of PML. In this series of experiments coverslip cultures of pHF cells were exposed to 40 PFU of wild-type or Bac-PML baculovirus per cell for 2 h. After 10 h of additional incubation, the cells were either mock infected or exposed to 20 PFU of HSV-1(F) per cell for 2 h. Replicate cultures were fixed and reacted with the antibody at 3, 7, or 12 after HSV-1 infection as described in Materials and Methods and in the preceding section. The results (Fig. 3) were as follows. (i) In pHF cells exposed to wild-type baculovirus and infected with HSV-1(F), ICP0 localized in nuclear punctate structures at 3 h after infection. ICP0 was then detected as diffuse fluorescence in both the nucleus and cytoplasm between 3 and 7 h after infection and was primarily cytoplasmic at 12 after infection (Fig. 3g, m, and s). (ii) In cells overexpressing PML, at 3 h after infection, ICP0 appeared to colocalize with PML in the large punctate structures seen in Fig. 2 (Fig. 3j to l and 4a to c). At higher magnification, however, the colocalization appeared to be imperfect. In essence, ICP0 appeared to form dense aggregates surrounded by a halo of fluorescent PML (Fig. 4d to f). (iv) At late times after infection ICP0 increased in amount and was dispersed throughout the cell (Fig. 3p to r and v to x). In contrast PML remained totally contained in the enlarged punctate structures (Fig. 3k, q, and w) throughout the experimental interval (3 to 12 h after infection). Because of the large amounts of ICP0 present at late times after infection, it was not possible to ascertain definitively whether any ICP0 remained associated with PML in the enlarged punctate structures. The appearance of the punctate structures suggested that they did not contain amounts of ICP0 greater than the surrounding cellular compartments.
FIG. 3.
Overexpressed PML remains associated with enlarged ND10 structures but does not affect the localization of ICP0 in the course of infection of pHF fibroblasts. pHF fibroblast slide cultures were infected with Bac-PML (d to f, j to l, p to r, and v to x) or wild-type baculovirus (a to c, g to i, m to o, and s to u) and then either mock infected (a to f) or infected with HSV-1(F) (g to x) as described in Materials and Methods. The cells were fixed at the indicated times and reacted with a mouse monoclonal antibody to PML and a rabbit polyclonal antibody to ICP0 as indicated at the top of each column. The secondary antibodies were anti-rabbit immunoglobulin G (IgG) conjugated to Texas red and an anti-mouse IgG conjugated to FITC. The images were captured as described in the legend to Fig. 2 and were not modified subsequent to capture.
FIG. 4.
ICP0 colocalizes, but does not overlap, with PML. pHF fibroblast slide cultures were infected with Bac-PML and HSV-1(F) at 3 h after HSV-1 infection and were treated as indicated in the legend for Fig. 2. The images were captured with a Zeiss confocal microscope with the aid of software provided by the manufacturer and were not modified subsequent to capture.
We conclude from this series of experiments that in pHF fibroblasts exogenous PMLs were localized throughout infection in punctate structures similar to those seen in uninfected cells throughout infection and that the localization of ICP0 was independent of and not hindered by the presence or localization of exogenously produced PMLs.
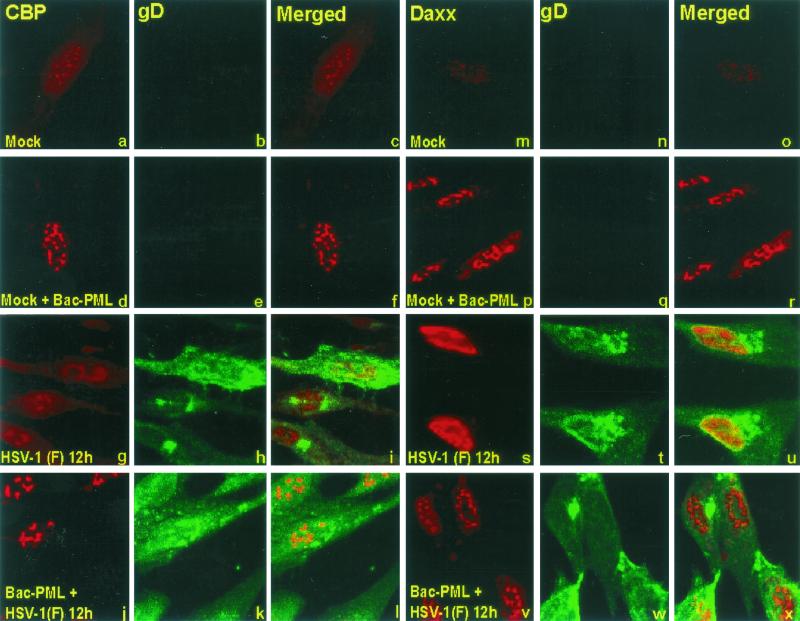
Overexpression of PML blocks the dispersal of CBP and Daxx induced by HSV-1.
As shown in Fig. 1, overexpression of PML by Bac-PML led to the enlargement of ND10 structures, which contain CBP and Daxx in addition to PML. In wild-type virus-infected cells, the components of ND10 are dispersed in an ICP0- and proteasome-dependent manner. The results shown in Fig. 3 and 4 indicated that, in pHF cells transduced with Bac-PML, PML remained associated with the enlarged punctate bodies throughout the viral replicative cycle. To verify that these structures contain other components of ND10, the experiment described above was repeated except that the transduced, infected slide cultures were probed with a polyclonal rabbit antibody against CBP or Daxx. In addition, to visualize more clearly the nuclei of infected cells, the cells were also reacted with monoclonal antibody anti-gD. This antibody was chosen because gD accumulates largely in cytoplasmic membranes and would not interfere with the visualization of ND10 or its components. The procedures were as described in Materials and Methods. The results (Fig. 5) were as follows. (i) As expected, in control cultures both CBP and Daxx localized in small punctate structures in nuclei of mock-infected cells. In this and other experiments, CBP and Daxx also formed a diffuse layer throughout the cell (Fig. 5a to c and m to o). In cells exposed to Bac-PML, the punctate structures were significantly larger and, moreover, the diffuse material seen in control cells was less apparent (Fig. 5d to f and p to r). (ii) In cultures of cells fixed 12 h after infection, CBP and especially Daxx appeared to be dispersed from the punctate structures (Fig. 5g to i and s to u). (iii) In cells exposed to Bac-PML and fixed 12 h after infection with HSV-1(F) both CBP and Daxx were retained in the enlarged punctate bodies (Fig. 5j to l and v to x). Except for the expression of gD, the appearance of the nuclei was very similar to that of nuclei of uninfected cells overexpressing PML (compare Fig. 5d to f and p to r with j to l and v to x). We conclude from these experiments that, in pHF fibroblasts overexpressing PML, the components of ND10 structures assayed in these experiments are not dispersed as late as 12 h after infection with wild-type virus.
FIG. 5.
CBP and Daxx colocalize with PML in enlarged ND10 structures throughout the replicative cycle of HSV-1. pHF fibroblast slide cultures were exposed to Bac-PML (d to f, j to l, p to r, and v to x) or wild-type baculovirus (a to c, g to i, m to o, and s to u). The cultures were then mock infected or infected with HSV-1(F) as indicated. At 12 h after mock infection or infection with HSV-1(F) the cells were fixed and reacted with the antibody to CBP (conjugated to Texas red), gD (conjugated to FITC), or Daxx (conjugated to Texas red) as shown at the top. The procedures were as described in Materials and Methods. Single-color images captured separately (first, second, fourth and fifth columns from the left) and merged images (third and sixth columns) are shown. The images were captured with a Zeiss confocal microscope with the aid of software provided by the manufacturer and were not modified subsequent to capture.
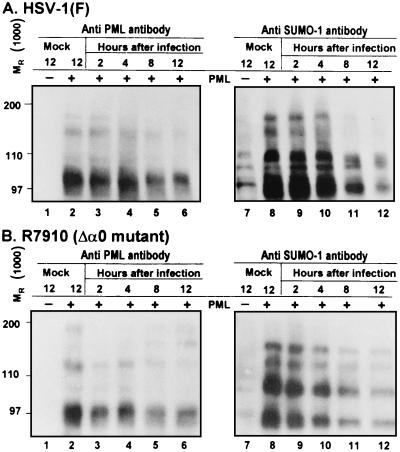
SUMO-I-modified proteins accumulate late in infection in cells transduced with PML.
The correct modification of PML by SUMO-I has been reported to be important for its localization in ND10 structures and for the formation of mature ND10 (11, 29, 45, 65). ICP0 has been shown to induce the disappearance of sumoylated PML and the dispersion of ND10 (7, 15, 16, 39, 40, 44). In this report we have shown that that overexpression of PML led to the persistence of PML, Daxx, and CBP in enlarged ND10 structures late in HSV-1 infection. To test if sumoylated protein persisted late in infection, pHF fibroblasts grown in a 25-cm2 flask, were exposed to 20 PFU of Bac-PML or wild-type baculovirus for 2 h. After 10 additional hours of incubation at 37°C, the cells were exposed for 2 h either to HSV-1(F) (Fig. 6A) or the ICP0-null mutant, R7910 (Fig. 6B). The cells were harvested at 2, 4, 8, or 12 h later, processed as described in Materials and Methods, and reacted with a mouse monoclonal antibody to PML and a rabbit polyclonal antibody to SUMO-I. The results were as follows. (i) The amounts of sumoylated proteins decreased during the course of the infection both in wild-type and ICP0-null mutant virus-infected cells. The disappearance of sumoylated proteins appeared to be more dramatic in cells infected with wild-type virus than in cells infected with the R7910 mutant. (ii) There was at least as much sumoylated protein in cells transduced with Bac-PML at late times after HSV-1 infection as in mock-infected cells. We conclude that sumoylated proteins persist throughout the course infection.
FIG. 6.
In cells overexpressing PML, sumoylated proteins decrease with time but are detectable late in infection. Replicate cultures of pHF fibroblasts were exposed to wild-type baculovirus (PML −) or baculovirus expressing PML. After 10 h of incubation, the cells were mock infected or exposed to 5 PFU of HSV-1(F) (A) or 1 PFU of the R7910 (α0) mutant (B) per cell. The cultures were harvested at 2, 4, 8, or 12 h after infection, solubilized, and subjected to electrophoresis in denaturing polyacrylamide gels, and the products were transferred to a nitrocellulose sheet and reacted with a monoclonal antibody to PML (left) or a rabbit polyclonal antibody to SUMO-I (right).
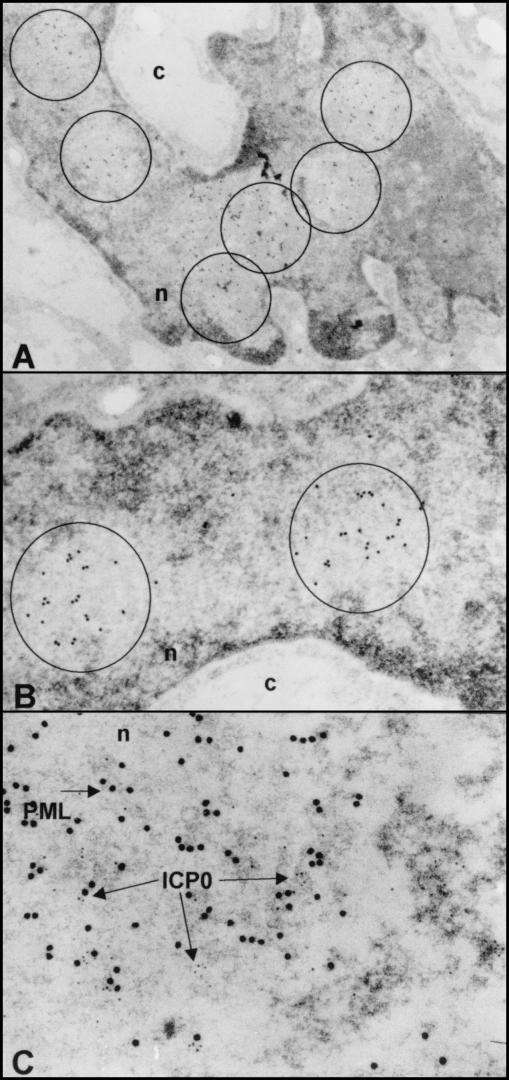
Electron-microscopic studies of enlarged ND10 structures in cells exposed to Bac-PML and infected with HSV-1.
The objective of this series of experiments was to examine the ND10 structures by electron microscopy in order to determine whether PML and ICP0 form topologically identifiable aggregates. The procedures were as described in Materials and Methods. Briefly, the cells were harvested at 3 h after infection with HSV-1(F). Thin sections were reacted with antibodies to PML or PML and ICP0 and then with secondary antibodies conjugated with 15-nm gold particles for PML and 5-nm gold particles for ICP0. Examinations of thin sections showed that the 15-nm gold particles were present in roughly circular electron-translucent areas of this section. Some thin sections contain numerous such electron-translucent areas (Fig. 7A). The 3-h time point was selected to insure that ICP0 colocalized predominantly with ND10 structures. As shown in Fig. 7C, the 5-nm gold particles were present in clusters but were also singly dispersed throughout the ND10 structures. There was no apparent clustering of ICP0 with PML. ICP0 is also seen in the electron-dense material immediately to the right of the electron-translucent area. The PML (15-nm gold particles) at the lower right portion of Fig. 7C may represent a portion of another ND10 structure. We conclude that the electron-microscopic examination of cells overexpressing PML and showing enlarged ND10 structures does not show unequivocal aggregation of ICP0 with PML in organized structures.
FIG.7.
Electron photomicrographs of enlarged ND10 structures overexpressing PML and infected with HSV-1(F). pHF fibroblasts were harvested 3 h after infection with HSV-1(F) and processed for immunoelectron microscopy as described in Materials and Methods. Thin sections of cells were labeled with a mouse monoclonal antibody to PML (Santa Cruz Biotechnology) and then reacted with a goat anti-mouse immunoglobulin G (IgG) antibody conjugated with 15-nm colloidal gold (GMHL-15) as a secondary antibody (Ted Pella). (A) Low magnification of a cell showing multiple ND10 structures delineated by circles and containing multiple 15-nm gold particles. (B) Higher magnification showing individual ND10 structures delineated by the circles and containing 15-nm gold particles indicating the presence of PML. (C) Thin sections of cells were labeled simultaneously with a mouse monoclonal antibody to PML and a rabbit antibody to ICP0 and then reacted with a goat anti-mouse IgG antibody conjugated with 15-nm colloidal gold as a secondary antibody and a goat anti-rabbit IgG antibody conjugated with 5-nm colloidal gold (GFAR-5). This high magnification of a single ND10 structure shows the presence of both ICP0 (5-nm) and PML (15-nm) particles.
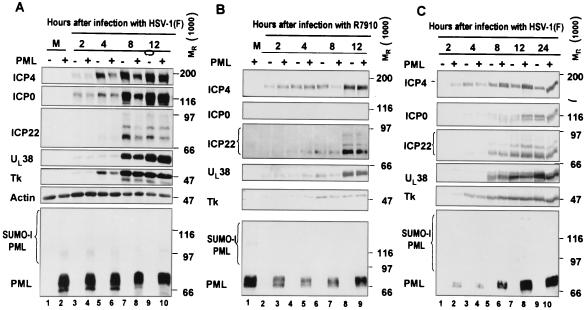
Overexpression of PML does not have any effect the accumulation of viral proteins expressed by wild-type virus HSV-1(F) or the α0-null mutant, R7910.
The objectives of this series of experiments were to determine the effect of overexpression of PML on the accumulation of viral proteins representative of α (e.g., ICP0, ICP4, and ICP22), β (TK), and γ (UL38) groups. The procedures were as described in Materials and Methods. In the first series of experiments (Fig. 8A and B), pHF fibroblasts grown in a 25-cm2 flask were exposed to 20 PFU of Bac-PML or wild-type baculovirus for 2 h at 37°C and incubated in fresh DMEM containing 10% FBS and 10 mM sodium butyrate for 10 additional hours. Then cells were exposed to HSV-1(F) (Fig. 8A) or R7910 (Fig. 8B) for 2 h. The cells were harvested at 2, 4, 8, or 12 h after the end of the 2-h adsorption interval and processed as described in Materials and Methods and the legend to Fig. 8. The experiment was repeated with HEp-2 cells and HSV-1(F) (Fig. 8C). The results, shown in Fig. 8, were as follows. (i) There were no significant differences in the accumulation of tested viral proteins throughout the 12-h interval after infection between control cells and HEp-2 cells overexpressing PML (Fig. 8C). The slightly decreased amounts of ICP0 and ICP4 at 4 h after infection of pHF cells with wild-type virus were not discernible at 12 h, and no apparent differences in the accumulations of UL38 and TK were noted (Fig. 8A). It is of interest that the slowest-migrating form of overexpressed PML (Fig. 8A, lanes 2, 4, and 6) decreased in relative amounts at 8 h after infection and disappeared entirely at 12 h after infection. (ii) In pHF fibroblasts infected with the α0-null mutant, R7910, there was a decrease in the amounts of accumulated ICP4 at 8 h, but not at 12 h, after infection of cells overexpressing PML. This observation was reproduced in different experiments. Also, we noted the disappearance of the fast-migrating form of PML (Fig. 8B, compare lanes 1 and 3 with 5, 7, and 9). We conclude that overexpression of PML had no appreciable effect on the expression of proteins encoded by wild-type or ICP0-null mutant virus.
FIG. 8.
Overexpression of PML does not affect the expression of viral proteins. Replicate cultures of pHF fibroblasts were exposed to wild-type baculovirus (PML −) or baculovirus expressing PML (PML +). At 10 h later, the cells were mock infected or exposed to 5 PFU of HSV-1(F) (A) or 1 PFU of the R7910 (α0-null) mutant (B) per cell. The cells were harvested at 2, 4, 8, or 10 h after the 2-h adsorption interval, solubilized, subjected to electrophoresis in denaturing gels, and reacted with antibodies as described in Materials and Methods. (C) Same procedures as for panels A and B with HEp-2 cells.
Overexpression of PML does not repress replication of HSV-1(F) or the R7910 mutant.
Inasmuch as overexpression of PML had no significant effects on the accumulation of viral proteins, it was of interest to determine whether overexpression of the protein interferes with viral replication. In this series of experiments replicate cultures of pHF or HEp-2 cells were exposed to 20 PFU of wild-type baculovirus or Bac-PML for 2 h at 37°C. The inoculum was then replaced with fresh medium and incubation continued for another 10 h. At that time the cells were infected with 5 PFU of HSV-1(F) or 1 PFU of R7910 per cell. After 2 h of adsorption, the cells were rinsed and incubated in fresh medium for either 2 or 24 h at 37°C. At 2 and 12 h after infection cells were harvested and titered in Vero cells [HSV-1(F)] or U-2 Os (R7910 mutant virus) as reported elsewhere (62). The procedures were as described in Materials and Methods. The 2-h time point measured the amount of virus that attached to but did not penetrate the cells and represents the background amounts of the virus. The results, summarized in Table 1, indicate that overexpression of PML had no effect on the accumulation of infectious virus. As expected, the α0-null mutant, R7910, replicated less well than its parent virus, HSV-1(F), in both pHF and HEp-2 cells. In other experiments, not shown, we measured the effect of overexpression of PML on cells exposed to 1 PFU of HSV-1(F) per cell with similar results (data not shown)
TABLE 1.
Effect of overexpression of PML on the replication of wild-type and Δα0 mutant viruses
| Virus | Cell line | Baculovirus | Virus yield (PFU) at indicated hours after infection
|
|
|---|---|---|---|---|
| 2 | 24 | |||
| HSV-1(F) | pHF | WTa | 1.92 × 104 | 2.5 × 107 |
| Bac-PML | 2.63 × 104 | 1.3 × 107 | ||
| HEp-2 | WT | 3.5 × 105 | 7 × 108 | |
| Bac-PML | 4 × 105 | 5.5 × 108 | ||
| R7910 | pHF | WT | 1.47 × 105 | 6.8 × 106 |
| Bac-PML | 1.48 × 105 | 7 × 106 | ||
| HEp-2 | WT | 1.35 × 105 | 1.5 × 107 | |
| Bac-PML | 1.1 × 105 | 9 × 106 | ||
WT, wild type.
DISCUSSION
A significant fraction of the functions encoded by HSV-1 concern themselves with blocking potential cellular responses to infection. Thus HSV-1 blocks cellular protein synthesis, cellular RNA synthesis, the processing of cellular mRNA, activation of cellular pathways that lead to programmed cell death, presentation of antigenic peptides to the immune system on the surfaces of infected cells, and activation of IFN pathways that mediate shutoff of protein synthesis (for a review see reference 51). The evidence that HSV-1 targets for destruction the ND10 structures suggested the possibility that these structures are components of the cellular armamentarium evolved to combat viral infections. Several studies have shown that SUMO-I PML is a targeted for proteasome-dependent disappearance mediated by ICP0, and indeed the functions attributed to PML listed in the introduction suggest that this protein may pose a threat to viral replication, and hence the virus could be expected to evolve the means to block it (7, 15, 44). The rationale of the studies described in this report is that, if PML expresses antiviral activity, then overexpression of PML would be expected to affect viral replication as reflected in decreased accumulation of viral proteins and diminished viral yields. This effect would be expected to be magnified in cells infected with an α0-negative mutant.
The experimental design employed in these studies was to transduce cells with a baculovirus expressing PML. The evidence presented in this report suggests that the product of the open reading frame cloned into the baculovirus is an authentic PML. Specifically, the electrophoretic mobility of the protein was the expected molecular weight of authentic PML, the protein was sumoylated, and, in addition, it accumulated in structures containing CBP and Daxx proteins known to be contained in ND10 structures (10, 30, 67). The key conclusion of the studies presented here is that overexpression of PML had no effect on viral replication or the accumulation of viral proteins in cells infected with either wild-type virus or virus lacking the ICP0 gene.
The published literature indicates that ICP0 targets sumoylated proteins for destruction (7, 15, 44). The results presented in this report show the accumulation of sumoylated protein bands in cells overexpressing PML. The results also show that, at middle times after infection, the amounts of sumoylated proteins decrease (Fig. 6). However, while the decrease was more dramatic in wild-type virus-infected cells than in cells infected with the α0 mutant, R7910, we noted that there was a decrease in both ICP0− and in wild-type virus-infected cells. It is also noteworthy that, at the end of the assay interval, i.e., 12 h after infection, the levels of sumoylated proteins were significantly higher than those in mock-infected cells (Fig. 6).
The salient features of the results obtained in this report were as follows. (i) The sizes of the ND10 structures were not fixed but rather appeared to be dependent on the amounts of PML expressed in both infected and uninfected cells. These observations are consistent with the reports that PML is a determinant of the assembly of ND10 (26, 65). (ii) The ND10 structures contained in cells transduced with Bac-PML were stable throughout the 12 h of infection with wild-type virus, in sharp contrast to the fate of ND10 structures in cells transduced with wild-type baculovirus. However, the failure of ND10 structures to disperse had no effect on the replication or viral gene expression of wild-type virus or of the R7910 (ICP0-null) mutant (Fig. 8 and Table 1). The implication of this observation is that the degradation on the ND10 structures is not essential for viral replication. (iii) The increase in the size of the ND10 structures permitted a higher resolution of the interaction between ICP0 and the PML. The results presented in this report do not support the hypothesis that PML and ICP0 interact. The results of confocal microscopy suggest that ICP0 and PML are contained in the same structures but do not fully overlap (Fig. 4). The immunoelectron-microscopic experiments illustrated in Fig. 7 do not support the hypothesis that PML and ICP0 form orderly structures in which the two proteins are juxtaposed. (iv) In untreated wild-type virus-infected cells, ICP0 mediates the dispersal of ND10 components. ICP0 then becomes dispersed throughout the nucleus and ultimately becomes translocated to the cytoplasm. Exposure of cells to proteasomal inhibitors early in infection blocks the dispersal of ND10 components and retains ICP0 in the nucleus. The results presented here indicate that the compartmentalization of ICP0 in the infected cells is totally independent of the stability of the ND10 structures (Fig. 3).
The results presented in this report raise several questions. The first is that, in the absence of a specific interaction between PML and ICP0, what causes the translocation of the newly made ICP0 to the ND10 structures? We can exclude cyclin D3 since the D199A mutant, which does not bind cyclin D3, also localizes with the ND10 structures early in infection (59). One interesting alternative is that ICP0 interacts early with one subunit of the proteasomes that aggregate in or near the ND10 structures (29, 37). This hypothesis is consistent with the observation that ICP0 colocalizes with components of the ubiquitin-proteasome system at the early stages of infection (13, 57).
The second relevant question relates to the role of ICP0 in mediating the degradation of SUMO-I-modified PML and the dispersal of ND10 components. Is SUMO-I-modified PML the intended target or is it affected because it shares properties with an as yet unknown target of ICP0? ICP0 mediates the degradation of several seemingly unrelated proteins. These include centromeric proteins A and C, DNA-dependent protein kinase SP100, etc. (7, 14, 34, 47). If PML is the intended target of ICP0, one possible explanation for the failure of the overexpressed PML to inhibit HSV-1 replication is that the virus blocks PML-mediated repression by several mechanism. One such alternative mechanism is posttranslational modification of the protein. It is noteworthy for example, that in pHF fibroblasts infected with wild-type or R7910 mutant virus the fast-migrating form of PML present in mock-infected cells and in infected cells early in infection disappears at later times after infection (Fig. 7). An alternative hypothesis is that the ubiquitin-conjugating enzymes conscripted by the ubiquitin ligase function of ICP0 regulate the degradation of several proteins including that of PML. One notable example of such an association is related to the observation that ICP0 mediates the stabilization of both cyclin D3 and cyclin D1 (58, 59). While cyclin D3 interacts with ICP0 and has been shown to play a role in the phenotypic properties of ICP0, no such an interaction has been shown for cyclin D1 (59). However, ICP0 appears to target for destruction UbcH3 (cdc34), the ubiquitin-conjugating enzyme know to regulate the turnover of D cyclins (18, 24, 63). In this instance the stabilization of cyclin D1 reflects the viral requirement to block the turnover of cyclin D3.
In essence, we have demonstrated that viral replication does not depend on or require the destruction of ND10. The true function of ICP0 in the ND10 structures remains to be determined.
Acknowledgments
We thank K. Border for making the plasmid encoding PML available to us.
P.L. was a recipient of a grant from “la fondation Philippe (France/USA)” and of a postdoctoral fellowship from The University of Chicago Committee on Cancer Biology. These studies were aided by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860) and the U.S. Public Health Service.
REFERENCES
- 1.Ahn, J. H., E. J. Brignole III, and G. S. Hayward. 1998. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol. Cell. Biol. 18:4899-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., R. J. Jacob, L. Simmerman, and B. Roizman. 1995. The herpes simplex virus 1 UL11 proteins are associated with cytoplasmic and nuclear membranes and with nuclear bodies of infected cells. J. Virol. 69:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden, K. L. 1998. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem. Cell Biol. 76:351-358. [DOI] [PubMed] [Google Scholar]
- 4.Borden, K. L., M. N. Boddy, J. Lally, N. J. O'Reilly, S. Martin, K. Howe, E. Solomon, and P. S. Freemont. 1995. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 14:1532-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlemaln, E., W. Villiger, and R. M. Garavito. 1982. Resin development for electron microscopy and analysis of embedding at low temperature. J. Microsc. 126:123-143. [DOI] [PubMed] [Google Scholar]
- 7.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 8.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. Koken, and H. de The. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 10.Doucas, V., M. Tini, D. A. Egan, and R. M. Evans. 1999. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc. Natl. Acad. Sci. USA 96:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duprez, E., A. J. Saurin, J. M. Desterro, V. Lallemand-Breitenbach, K. Howe, M. N. Boddy, E. Solomon, H. de The, R. T. Hay, and P. S. Freemont. 1999. SUMO-1 modification of the acute promyelocytic leukaemia protein PML: implications for nuclear localisation. J. Cell Sci. 112:381-393. [DOI] [PubMed] [Google Scholar]
- 12.Ejercito, P. M., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behaviour of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferbeyre, G., E. de Stanchina, E. Querido, N. Baptiste, C. Prives, and S. W. Lowe. 2000. PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14:2015-2027. [PMC free article] [PubMed] [Google Scholar]
- 18.Ganiatsas, S., R. Dow, A. Thompson, B. Schulman, and D. Germain. 2001. A splice variant of Skp2 is retained in the cytoplasm and fails to direct cyclin D1 ubiquitination in the uterine cancer cell line SK-UT. Oncogene 20:3641-3650. [DOI] [PubMed] [Google Scholar]
- 19.Gongora, C., G. David, L. Pintard, C. Tissot, T. D. Hua, A. Dejean, and N. Mechti. 1997. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 272:19457-19463. [DOI] [PubMed] [Google Scholar]
- 20.Grotzinger, T., K. Jensen, and H. Will. 1996. The interferon (IFN)-stimulated gene Sp100 promoter contains an IFN-gamma activation site and an imperfect IFN-stimulated response element which mediate type I IFN inducibility. J. Biol. Chem. 271:25253-25260. [DOI] [PubMed] [Google Scholar]
- 21.Grotzinger, T., T. Sternsdorf, K. Jensen, and H. Will. 1996. Interferon-modulated expression of genes encoding the nuclear-dot-associated proteins Sp100 and promyelocytic leukemia protein (PML). Eur. J. Biochem. 238:554-560. [DOI] [PubMed] [Google Scholar]
- 22.Guiochon-Mantel, A., J. F. Savouret, F. Quignon, K. Delabre, E. Milgrom, and H. De The. 1995. Effect of PML and PML-RAR on the transactivation properties and subcellular distribution of steroid hormone receptors. Mol. Endocrinol. 9:1791-1803. [DOI] [PubMed] [Google Scholar]
- 23.Guldner, H. H., C. Szostecki, T. Grotzinger, and H. Will. 1992. IFN enhance expression of Sp100, an autoantigen in primary biliary cirrhosis. J. Immunol. 149:4067-4073. [PubMed] [Google Scholar]
- 24.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodges, M., C. Tissot, K. Howe, D. Grimwade, and P. S. Freemont. 1998. Structure, organization, and dynamics of promyelocytic leukemia protein nuclear bodies. Am. J. Hum. Genet. 63:297-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishov, A. M., A. G. Sotnikov, D. Negorev, O. V. Vladimirova, N. Neff, T. Kamitani, E. T. Yeh, J. F. Strauss III, and G. G. Maul. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 147:221-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lallemand-Breitenbach, V., J. Zhu, F. Puvion, M. Koken, N. Honore, A. Doubeikovsky, E. Duprez, P. P. Pandolfi, E. Puvion, P. Freemont, and H. de The. 2001. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J. Exp. Med. 193:1361-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LaMorte, V. J., J. A. Dyck, R. L. Ochs, and R. M. Evans. 1998. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc. Natl. Acad. Sci. USA 95:4991-4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lavau, C., A. Marchio, M. Fagioli, J. Jansen, B. Falini, P. Lebon, F. Grosveld, P. P. Pandolfi, P. G. Pelicci, and A. Dejean. 1995. The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11:871-876. [PubMed] [Google Scholar]
- 32.Li, H., C. Leo, J. Zhu, X. Wu, J. O'Neil, E. J. Park, and J. D. Chen. 2000. Sequestration and inhibition of Daxx-mediated transcriptional repression by PML. Mol. Cell. Biol. 20:1784-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J. H., Z. M. Mu, and K. S. Chang. 1995. PML suppresses oncogenic transformation of NIH/3T3 cells by activated neu. J. Exp. Med. 181:1965-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 35.Lopez, P., C. Van Sant, and B. Roizman. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matera, A. G. 1999. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9:302-309. [DOI] [PubMed] [Google Scholar]
- 37.Mattsson, K., K. Pokrovskaja, C. Kiss, G. Klein, and L. Szekely. 2001. Proteins associated with the promyelocytic leukemia gene product (PML)-containing nuclear body move to the nucleolus upon inhibition of proteasome-dependent protein degradation. Proc. Natl. Acad. Sci. USA 98:1012-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maul, G. G. 1998. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays 20:660-667. [DOI] [PubMed] [Google Scholar]
- 39.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 40.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 41.Maul, G. G., D. Negorev, P. Bell, and A. M. Ishov. 2000. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J. Struct. Biol. 129:278-287. [DOI] [PubMed] [Google Scholar]
- 42.Mu, Z. M., K. V. Chin, J. H. Liu, G. Lozano, and K. S. Chang. 1994. PML, a growth suppressor disrupted in acute promyelocytic leukemia. Mol. Cell. Biol. 14:6858-6867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mu, Z. M., X. F. Le, A. B. Glassman, and K. S. Chang. 1996. The biologic function of PML and its role in acute promyelocytic leukemia. Leuk Lymphoma 23:277-285. [DOI] [PubMed] [Google Scholar]
- 44.Muller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 49.Quignon, F., F. De Bels, M. Koken, J. Feunteun, J. C. Ameisen, and H. de The. 1998. PML induces a novel caspase-independent death process. Nat. Genet. 20:259-265. [DOI] [PubMed] [Google Scholar]
- 50.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 51.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams & Wilkins, Philadelphia, Pa.
- 52.Saurin, A. J., K. L. Borden, M. N. Boddy, and P. S. Freemont. 1996. Does this have a familiar RING? Trends Biochem. Sci. 21:208-214. [PubMed] [Google Scholar]
- 53.Stadler, M., M. K. Chelbi-Alix, M. H. Koken, L. Venturini, C. Lee, A. Saib, F. Quignon, L. Pelicano, M. C. Guillemin, C. Schindler, et al. 1995. Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11:2565-2573. [PubMed] [Google Scholar]
- 54.Torii, S., D. A. Egan, R. A. Evans, and J. C. Reed. 1999. Human Daxx regulates Fas-induced apoptosis from nuclear PML oncogenic domains (PODs). EMBO J. 18:6037-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallian, S., K. V. Chin, and K. S. Chang. 1998. The promyelocytic leukemia protein interacts with Sp1 and inhibits its transactivation of the epidermal growth factor receptor promoter. Mol. Cell. Biol. 18:7147-7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallian, S., J. A. Gaken, E. B. Gingold, T. Kouzarides, K. S. Chang, and F. Farzaneh. 1998. Modulation of Fos-mediated AP-1 transcription by the promyelocytic leukemia protein. Oncogene 16:2843-2853. [DOI] [PubMed] [Google Scholar]
- 57.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. USA 96:8184-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICPO. J. Virol. 75:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, Z. G., L. Delva, M. Gaboli, R. Rivi, M. Giorgio, C. Cordon-Cardo, F. Grosveld, and P. P. Pandolfi. 1998. Role of PML in cell growth and the retinoic acid pathway. Science 279:1547-1551. [DOI] [PubMed] [Google Scholar]
- 61.Wang, Z. G., D. Ruggero, S. Ronchetti, S. Zhong, M. Gaboli, R. Rivi, and P. P. Pandolfi. 1998. PML is essential for multiple apoptotic pathways. Nat. Genet. 20:266-272. [DOI] [PubMed] [Google Scholar]
- 62.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu, Z. K., J. L. Gervais, and H. Zhang. 1998. Human CUL-1 associates with the SKP1/SKP2 complex and regulates p21CIP1/WAF1 and cyclin D proteins. Proc. Natl. Acad. Sci. USA 95:11324-11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhong, S., L. Delva, C. Rachez, C. Cenciarelli, D. Gandini, H. Zhang, S. Kalantry, L. P. Freedman, and P. P. Pandolfi. 1999. A RA-dependent, tumour-growth suppressive transcription complex is the target of the PML-RARα and T18 oncoproteins. Nat. Genet. 23:287-295. [DOI] [PubMed] [Google Scholar]
- 65.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 66.Zhong, S., P. Salomoni, and P. P. Pandolfi. 2000. The transcriptional role of PML and the nuclear body. Nat. Cell Biol. 2:E85-E90. [DOI] [PubMed] [Google Scholar]
- 67.Zhong, S., P. Salomoni, S. Ronchetti, A. Guo, D. Ruggero, and P. P. Pandolfi. 2000. Promyelocytic leukemia protein (PML) and Daxx participate in a novel nuclear pathway for apoptosis. J. Exp. Med. 191:631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou, G., V. Galvan, G. Campadelli-Fiume, and B. Roizman. 2000. Glycoprotein D or J delivered in trans blocks apoptosis in SK-N-SH cells induced by a herpes simplex virus 1 mutant lacking intact genes expressing both glycoproteins. J. Virol. 74:11782-11791. [DOI] [PMC free article] [PubMed] [Google Scholar]