Abstract
For nuclear entry of large nucleoprotein complexes, it is thought that one key nuclear localization signal (NLS) of a protein component becomes exposed to mediate importin recognition. We show that the nuclear entry of simian virus 40 involves a dynamic interplay between two distinct interiorly situated capsid NLSs, the Vp1 NLS and the Vp3 NLS, and the selective exposure and importin recognition of the Vp3 NLS. The Vp3 NLS-null mutants assembled normally into virion-like particles (VLP) in mutant DNA-transfected cells. When used to infect a new host, the null VLP entered the cell normally but was impaired in viral DNA nuclear entry due to a lack of recognition by the importin α2/β heterodimer, leading to reduced viability. Both Vp3 and Vp1 NLSs directed importin interaction in vitro, but the Vp1 NLS, which overlaps the Vp1 DNA binding domain, did not bind importins in the presence of DNA. The results suggest that certain canonical NLSs within a nucleoprotein complex, such as the Vp1 NLS, can be masked from functioning by binding to the nucleic acid component and that the availability of an NLS that is not masked and can become exposed for importin binding, such as the Vp3 NLS, is a general feature of the nuclear entry of the nucleoprotein complexes, including those of other animal viruses.
Nucleocytoplasmic trafficking of macromolecules, including protein multimers and multiprotein complexes with nucleic acid, is important for their functioning in the nucleus of eukaryotic cells. A major site of exchange is the nuclear pore complex (NPC) on the nuclear envelope. Much has been learned about the structure of the NPC, the canonical nuclear localization signals (NLSs) that interact with cytoplasmic import receptors, and the mechanism of receptor-mediated import of proteins through the NPC (2, 3, 11, 37, 38, 49). For the nuclear import of a nucleoprotein complex, the individual component proteins often harbor multiple canonical NLSs, one of which may function to promote the nuclear entry of the complex (40, 41). For example, the spliceosomal small nuclear RNAs and a set of Sm core proteins form a complex and enter the nucleus as ribonucleoprotein particles (see references in reference 54). The genomes of certain eukaryotic viruses are also targeted to the nucleus as nucleoprotein complexes.
For diverse groups of animal viruses, nuclear entry of the viral genome after invading the host cell is a prerequisite for viral genome expression and replication and is thus essential for initiating the viral life cycle (25, 53). The incoming virion may progressively uncoat its protein components, and the viral genome may pass through the NPC in association with a few virion proteins, as has been suggested previously for nuclear entry of the adenovirus (14). It is expected that an elaborate interplay of multiple and canonical NLSs regulates the nuclear entry of viral genomes. An in vitro approach using digitonin-permeabilized cells with intact nuclei has identified the viral proteins (22), their respective NLSs (8), and cellular import receptors (44) that mediate the nuclear entry. Genetic study has identified the NLS of human immunodeficiency virus (HIV) integrase as contributing to nuclear entry of the preintegration complex (5), while other studies indicate the role of other virion proteins in its nuclear entry (6, 46). Modification of the flanking amino acids of the NLS by phosphorylation has also been proposed for its functioning in the nuclear entry of hepatitis B virus core particle (23). Understanding the molecular mechanism that regulates the nuclear entry of nucleoprotein complexes should illuminate the requirements for crucial protein motifs that function in the nuclear entry.
Although nuclear import mechanisms of proteins in isolation are well known (e.g., references 11 and 37), little is known about the sequence of events that enable nuclear entry of a nucleoprotein complex that contains multiple proteins. Most of them carry resident NLSs that may not be on the surface of the complex or particle. Three questions need to be addressed in elucidating how a large nucleoprotein complex enters the nucleus. First, which of the NLSs of different proteins is the critical NLS and is recognized by an NLS receptor such as the importin (karyopherin) α/β heterodimer? Conversely, which of the NLSs, if any, is excluded for this function? Since an NLS can spatially overlap with a protein's nucleic acid binding motif, sharing some or all of the basic residues (27), binding of the protein to the nucleic acid could mask the NLS's function, as has been described previously for the NLSs of nucleic acid binding proteins (28, 30, 42, 50). Second, when the NLS is concealed in a multimolecular particle, does the particle undergo a conformational change to expose the signal so that importin can interact with it? Finally, given that the diameter of certain nucleoprotein complexes exceeds an upper limit of 26 nm for the passage of a nondeformable cargo (10) through the NPC, how can such a large complex pass through the NPC?
Simian virus 40 (SV40) offers a unique system for addressing these central questions. Besides its double-stranded, circular DNA molecule, each SV40 particle contains 360 copies of the major capsid protein Vp1, 72 copies of the minor capsid proteins Vp2 and Vp3, and about 200 copies of histones, all harboring distinct resident NLSs (4; see also references in reference 25) that are in the virion's interior (7, 33). SV40 enters the nucleus through the NPC, and its entry is inhibited by wheat germ agglutinin, antinucleoporin antibody, or depletion of ATP (25). Furthermore, the SV40 DNA in complex with empty capsids, but not with histones, enters the nucleus, and the presence of antibody to Vp3 in the cytoplasm effectively blocks the nuclear entry of the virion (39). Thus, capsid proteins direct viral DNA nuclear entry. The results also suggest that a structural alteration of the infecting virion occurs prior to its nuclear entry. The resident NLSs and DNA binding domains (DBD) for all capsid proteins have been mapped (see references in reference 31). Vp2 and Vp3, identical except for Vp2's additional N-terminal segment, share the carboxyl-terminal NLS and DBD and will hereafter be collectively referred to as Vp3.
For SV40, the capsid proteins' NLSs may function at two stages of the viral life cycle—in the nuclear targeting of the infecting viral genome and in the nuclear entry of the newly synthesized capsid proteins. In the latter event, the interaction between Vp1 and Vp3 while still in the cytoplasm allows the nuclear entry of an NLS-defective Vp3 when Vp1 with a functional NLS is present (19). We term this phenotypic NLS complementation. When such complementation occurs, virion-like particles (VLP) could form efficiently in the nucleus during a single cycle of infection initiated by mutant DNA transfection. It is then conceivable that such VLP, internalized normally into a new host, would not deliver viral genome to the nucleus, thus compromising the initiation of a new infection cycle. This reduced infectivity may be due to the failure of the VLP's mutant signal to interact with nuclear import receptors in the cytoplasm.
We used a genetic approach to show that nuclear entry of SV40 DNA is mediated by interaction of the Vp3 NLS with the importin α2/β heterodimer. Following the particle's entry into the cell, the VLP with intact Vp1 NLSs and defective null mutant Vp3 NLSs is impaired in delivering the viral DNA to the nucleus. Unlike its wild-type counterpart, the internalized null mutant VLP is not recognized by importin α2/β. Thus, the Vp3 NLS, but not the Vp1 NLS, is the NLS of the infecting SV40.
MATERIALS AND METHODS
Plasmids.
The Vp3 residue number is used for common amino acids of Vp2 and Vp3 displayed in the single-letter code. Sequences of oligonucleotide primers and linkers used in this study can be obtained upon request.
Nonoverlapping (NO) viral plasmids NO-pSV40 and NO-pSV40 KTKRK (NO-pSV40202T) have been described previously (19). NO-pSV40 KTKTK and NO-pSV40 NNNGN221 were constructed as described previously (19). NO-pSV40 SRBSM, which is as viable as the previously described NO-pSV40 and harbors nonnatural restriction sites for SalI, PstI, XbaI, and RsrII at the coding sequence for Vp3 residues 3 and 4, 96 and 97, 164 to 166, and 197 to 199, respectively, was constructed from NO-pSV40 BSM (32). The Vp3 NLS mutants NO-pSV40 KGAGK, NO-pSV40 KGAGA, NO-pSV40 AGAGA (Null AG), and NO-pSV40 NNNGN (Null NG) were made by replacing the RsrII-to-XbaI fragment of NO-pSV40 SRBSM with the respective mutant linker fragments. The pSV-Vp3 plasmids were made by replacing the XbaI-to-SacI Vp1 segment of the respective NO-pSV40 plasmids with the XbaI-BamHI-SacI linker. In NO-pSV40ΔNcoI the SV40 nucleotide 37-to-560 sequences between two NcoI sites are deleted.
Plasmids for expressing dihydrofolate reductase (DHFR) fused with various Vp3 open reading frames (ORFs) were constructed as follows. First, three fragments encoding Vp3 residues 3 to 164, 165 to 194, and 195 to 234 were sequentially inserted into pBlueScript II(+). (i) In pBS195-234, the Vp3(195-234) coding fragment whose sequence is optimized for bacterial codon usage was made by ligating four sets of double-stranded oligonucleotide linkers and inserted into pBlueScript II(+) via NotI-XhoI sites. (ii) Similarly oligonucleotide linkers encoding Vp3(165-194) were inserted through XbaI and RsrII sites of pBS195-234 to make pBS165-234. (iii) Finally, two PCR fragments, one encoding Vp3(3-96) and the other encoding Vp3(96-164), were ligated through the nonnatural unique PstI site at the 96th residue and inserted into pBS165-234 via SalI-XbaI sites, generating pBS3-234. pBS3-234 and its construction intermediates contain unique restriction sites, SalI, AvrII, PstI, XmaI, and RsrII, at Vp3 residues 3 and 4, 55 and 56, 96 and 97, 164 to 166, and 197 to 199, respectively, and the coding sequence for Vp3(165-234) residues is optimized for bacterial expression.
The Vp3 ORFs were then inserted, in frame, downstream of the DHFR coding sequence in pQE-16 (Qiagen). pQEVp3195-234 was made by inserting, via BglII-HindIII sites, the ORF in pBS195-234 that had been amplified with a sense primer which incorporates a unique BglII site and a hexahistidine tag and an antisense primer which incorporates a unique HindIII site. pQEVp3165-234 and pQEVp33-234 were constructed by inserting into pQEVp3195-234 the NotI-to-RsrII fragment from pBS165-234 and pBS3-234, respectively. pQEVp3222-234 was constructed by replacing the NotI-to-HindIII fragment of pQEVp3195-234 with a linker encoding Vp3(222-234) amino acids. In pQEVp3165-193, Vp3(194-234) residues were deleted from pQEVp3165-234 by HindIII digestion and self-ligation. For pQEVp3165-206, a stop codon was introduced at the 207th amino acid in pQEVp3165-234.
Recombinant protein preparation.
Preparation of Vp1ΔC58 protein containing mostly the Vp1 pentamer has been described previously (31). The histidine-tagged DHFR and the series of DHFR-Vp3 ORF fusion proteins were expressed in XL-1 Blue cells carrying the pQE-16 derivatives harboring the respective Vp3 ORFs. Induced proteins were purified under denaturing conditions with Talon beads (Clontech). Glutathione S-transferase (GST)-importin α and GST-importin β were prepared as reported previously (17, 18).
Subcellular localization and viability assay.
Immunocytochemistry of viral proteins, preparation of viral DNAs, and preparation of lysates containing virion or VLP were described previously (31). The amount of DNase I-resistant virion- or VLP-packaged viral DNA in the transfected cell lysate was estimated as described previously (32). Each transfected cell lysate was adjusted to 1 μg of DNase I-resistant viral DNA per ml and assayed for PFU per milliliter. The value of DNase I-resistant viral DNA provides a reliable measure to predict PFU of virus preparation; wild-type transfected cell lysate, purified virus, and purified virus mixed with untransfected cell lysate gave the same PFU per milliliter when the volume of the preparation was adjusted to contain the same amount of DNase I-resistant viral DNA per milliliter (data not shown).
VLP analysis.
Analyses of the transfected cell lysate for viral DNA replication and encapsidation and for VLP formation were described previously (32). For the capsid composition of VLP, proteins in an aliquot of pooled fractions were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting with respective antibodies as described previously (34).
Virion cell entry and nuclear entry.
For all experiments described here, TC-7 cells were infected with approximately 103 particles per cell and incubated at 4°C for 1 h and then at 37°C for designated time periods. For virion cell entry, cells in two 100-mm-diameter culture dishes were infected and were harvested either by trypsin treatment or by scraping off, followed by extraction of low-molecular-weight DNA (15). For nuclear entry of viral DNA, cells harvested by trypsin treatment were resuspended in buffer H (10 mM HEPES [pH 7.5], 10 mM KCl, 0.5 mM MgCl2, 0.1 mM dithiothreitol, 1× PIM [10 μg of aprotinin/ml, 1 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, and 10 μg of pepstatin/ml]), left on ice for 30 min, separated into the cytoplasmic and the nuclear fractions by Dounce homogenization, and centrifuged at 700 × g for 1 min at 4°C. The supernatant was designated as the cytoplasmic fraction. The pellet was washed twice with buffer H containing 0.5% Nonidet P-40 and resuspended in 10 mM Tris-HCl, pH 8.0, which was designated as the nuclear fraction. Low-molecular-weight DNA was extracted, digested with BamHI, and resolved in an agarose gel. Viral DNA was detected by Southern blotting with 32P-labeled SV40 DNA as a probe.
Analysis of the internalized particles by immunoprecipitation.
The cytoplasmic fraction, prepared at 6 h postinfection (hpi) as described above with buffer K (10 mM HEPES [pH 7.4], 10 mM KOAc, 0.5 mM MgOAc, 1× PIM) instead of buffer H, was cleared by centrifugation at 12,000 × g at 4°C for 10 min. Fifty microliters of the fraction from 2.5 × 105 cells was mixed with either rabbit anti-Vp1 antiserum, affinity-purified rabbit anti-Vp3 antibody, a mixture of mouse anti-importin α (anti-RchI; Transduction Laboratories) and anti-importin β (anti-karyopherin β; Transduction Laboratories) antibodies, or rabbit anti-mouse immunoglobulin G (IgG) antiserum and incubated on ice for 2 h, followed by another 2-h incubation along with supplementation with 300 μl of buffer B (10 mM HEPES [pH 6.8], 150 mM KOAc, 2 mM MgOAc, 0.5 mM CaCl2, 0.1% Tween 20, 0.1% Casamino Acids, 5 mM glutathione, 1× PIM) containing either 15 μl of protein A beads (Amersham-Pharmacia) or the same amount of protein A beads bound with rabbit anti-mouse IgG. After extensive washing, the immunoprecipitates were mixed with 20 pg of standard DNA (NO-pSV40ΔNcoI), and DNA was extracted and resuspended in 20 μl of 10 mM Tris-HCl (pH 8.0). Viral DNAs (1 μl) were detected by semiquantitative PCR with a set of primers that hybridize to SV40 nucleotide sequences at positions 4517 to 4542 (sense) and at positions 1464 to 1487 (antisense) (Herculase DNA polymerase [Stratagene] for 25 cycles of 92°C for 30 s, 65°C for 30 s, and 72°C for 2 min). The PCR products resolved on the agarose gel were detected by Southern blotting with 32P-labeled SV40 DNA. The radioactive signals were measured by phosphorimaging (Molecular Dynamics). The amount of amplified products is proportional to the template DNAs, ranging from 0.1 to 100 pg (data not shown).
In vitro importin binding.
A hundred picomoles of each purified DHFR-Vp3 or DHFR-His protein was bound to Talon beads and reacted with 67 nM (each) 125I-labeled GST-importins in buffer B supplemented with 5 mM iodoacetic acid (B/I). After incubation for 30 min at room temperature, the beads were washed extensively. Bound proteins were released from the beads by denaturation in SDS sample buffer, resolved by SDS-PAGE, and detected by either Coomassie blue staining or autoradiography.
The Vp1ΔC58 binding with importin α/β was performed in buffer B/I containing 100 nM (each) GST-importin α/β and 200 nM Vp1ΔC58 in the absence or the presence of a 326-bp DNA fragment (31) with various concentrations from none to 1 μM. The Vp1-importin complex, retrieved by Talon beads, was washed and detected by Coomassie blue staining as described above. Similarly, the Vp3 and importin interaction in the presence of DNA was measured by using the Talon beads bound with 200 pmol of DHFR-VP3165-234 protein, 100 nM (each) GST-importin α and GST-importin β, and the DNA in buffer B/I.
Interaction of recombinant Vp3 with endogenous importin α was examined by using an uninfected cytoplasmic fraction prepared as described above. As for association of recombinant Vp3 and endogenous importin β, cells were extracted as described previously (52). The fraction from 4 × 107 cells was diluted in 1 ml with buffer B/I for importin α or the lysis buffer plus 200 mM NaCl for importin β and was incubated for 2 h at 4°C with Talon beads bound with respective DHFR-Vp3 proteins. After extensive washing the bead-bound proteins were screened for importins by Western blotting with respective importin antibodies followed by enhanced chemiluminescence (West Pico; Pierce).
RESULTS
Viability reduction of Vp3 NLS mutants.
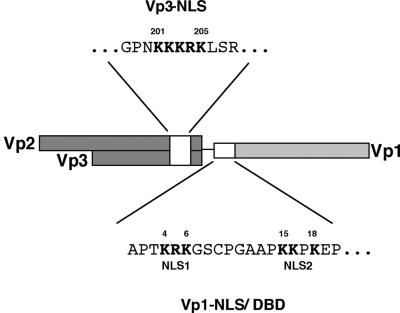
Our previous work has established the importance of capsid proteins for the nuclear entry of SV40 DNA (39). The protein domain responsible for the virion's nuclear entry may be the resident NLS of either Vp1 (Vp1 NLS, Fig. 1) or Vp3 (Vp3 NLS, Fig. 1), both of these, or an unidentified capsid protein domain. Studies of the bipartite Vp1 NLS have identified the two clusters of basic residues, NLS1 and NLS2, which precisely overlap the DNA binding activity (31). A mutant of NLS1 is nonviable, and the mutant Vp1 cannot enter the nucleus; hence, no VLP was made (31). By contrast, the nuclear localization defect of a mutant NLS2 is complemented by coexpression of Vp3, and yet the mutant shows severe defects in viral DNA packaging (31). Since few mutant VLP are formed, the NLS2's function in virion nuclear entry could not be distinguished from that in nuclear virion assembly. These results led us to focus on the role of the Vp3 NLS in nuclear entry of viral DNA. We introduced a series of Vp3 NLS mutations to the viable, nonoverlapping SV40 (NO-SV40) genome (Fig. 1) to assess whether mutants with defective Vp3 NLSs give rise to a mutant VLP with reduced infectivity.
FIG. 1.
A map for NLSs of NO-SV40 Vp1 and Vp3. Locations for Vp1 and Vp3 NLS coding regions (white boxes) as well as their respective amino acid sequences in single letters are shown. The amino acid numbers of the first and last residues for the respective NLSs are given, and the basic amino acids of the NLS are highlighted in boldface.
The nuclear accumulation of all capsid proteins, including the NLS-defective Vp3, is a prerequisite for mutant VLP formation in the nucleus. To verify whether coexpression of NLS-intact Vp1 complements the nuclear localization defect of Vp3, subcellular localization of NLS-defective Vp3 was examined by expressing the mutant Vp3 from either the SV-Vp3 viral variants that lack the Vp1 coding region or NO-SV40 variants in which all viral protein coding regions are present. Confirming our earlier studies (19), all mutant Vp3s mostly localized to the cytoplasm in the absence of Vp1, but they localized to the nucleus as effectively as wild-type Vp3 when Vp1 was coexpressed (Table 1).
TABLE 1.
Viability and the proteins' subcellular localization of Vp3 NLS mutantsa
| DNA | Vp3 NLS | Protein localization
|
Particle infectivity (PFU/ml) | |
|---|---|---|---|---|
| Vp3 | Vp1 | |||
| 201205 | ||||
| NO-SV40 KKKRK (wild type)b | GPNKKKRKLSRG | Nuc | Nuc | 1.0 × 108 |
| NO-SV40 KTKRK (202T) | ----T------- | Nucc | Nucc | 9.1 × 107 |
| NO-SV40 KTKTK | ----T-T----- | Nuc | Nuc | 9.5 × 107 |
| NO-SV40 KGAGK | ----GAG----- | Nuc | Nuc | 2.5 × 107 |
| NO-SV40 KGAGA | ----GAGA---- | Nuc | Nuc | 2.1 × 107 |
| NO-SV40 AGAGA (Null AG) | ---AGAGA---- | Nuc | Nuc | 1.1 × 106 |
| NO-SV40 NNNGN (Null NG) | ---NNNGN---- | Nuc | Nuc | 1.0 × 106 |
| NO-SV40 NNNGN221d | ---NNNGN---- | Nuc | Nuc | 1.0 × 104 |
| SV-Vp3 KKKRK | ------------ | Nuc | ||
| SV-Vp3 KTKKK | ----T------- | Cyt | ||
| SV-Vp3 KTKTK | ----T-T----- | Cyt | ||
| SV-Vp3 NNNGN | ---NNNGN---- | Cyt | ||
Subcellular localization of capsid proteins was designated either nuclear (Nuc) or cytoplasmic (Cyt). Amino acid sequence, including the Vp3 NLS (boldfaced and underlined), is given in single letters. Particle infectivity is given as the number of PFU originating from 1 μg of DNase I-resistant viral DNA in 1 ml of viral lysate.
Prepared from NO-pSV40 SRBSM.
Results are from reference 19.
Mutation of alanine, the 221st residue in Vp3, to threonine.
Mutant NO-SV40 DNA was then transfected into the cells, and DNA that had entered the nucleus was allowed to initiate a single infection cycle. Provided that the mutation did not affect virion assembly in the nucleus, virion or VLP should have formed in the transfected cells. The mutant VLP's ability to initiate the next infection cycle should depend on whether the mutation compromises cell entry, nuclear entry, or both and would indicate the viability of the mutant. First we harvested cells transfected with the mutant DNA to prepare lysates. The resulting lysates, titrated for DNase I-resistant DNA, were used as the source of mutant VLP to examine its ability to form plaques (Table 1). Single or double point mutations did not affect viability, while triple (KGAGK) and quadruple (KGAGA) mutations reduced viability fivefold. A hundredfold reduction in viability was observed for null mutants, Null AG and Null NG, in which all basic residues of the NLS were changed to AGAGA and to NNNGN, respectively. The Null NG mutation with an additional alteration of alanine 221 to threonine (NNNGN221T) reduced viability by 4 log units. Clearly, null mutations of the Vp3 NLS exert significant impact on viral viability.
Null mutants can replicate and form VLP.
Reduced viability of the null mutants could arise either when VLP production is reduced in the transfected cells or when the VLP is normally formed but is compromised in initiating the next cycle of infection. We examined the mutants' ability to produce VLP by measuring the extent of viral DNA replication and packaging and verifying VLP formation. Low-molecular-weight DNA was extracted from the lysates, and replicated viral DNA was detected by Southern blotting. Figure 2a (upper panel) shows a representative set of results. The amounts of replicated DNA in both mutant and wild-type lysates differed by less than twofold (data not shown). The proportion of viral DNA packaged into protective structures (i.e., virion or VLP) was then examined by measuring DNase I-resistant viral DNA (Fig. 2a, lower panel). The amount of DNase I-resistant mutant DNA was no less than 60% of total replicated viral DNA, comparable to that of the wild type (Fig. 2a, upper and lower panels).
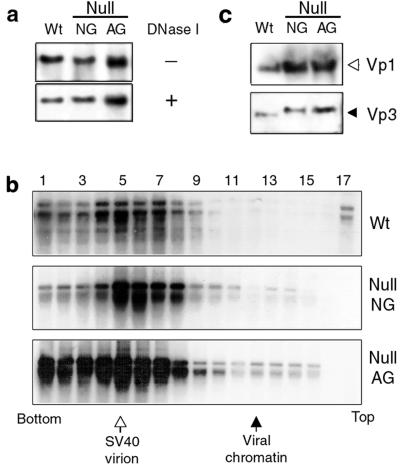
FIG. 2.
Normal DNA replication and VLP formation of null mutants. Cell lysate prepared following DNA transfection with wild-type (Wt), Null NG, or Null AG viral DNA was used for the analysis. (a) Viral DNA replication and DNA encapsidation. An aliquot of transfected cell lysate was treated either with (+) or without (−) DNase I, and the extracted DNA was doubly digested with DpnI and BamHI. The DNA was resolved in an agarose gel and screened for viral DNA. (b) VLP formation. The DNA transfected-cell lysates were fractionated by sedimentation through 5 to 32% sucrose gradients, and viral DNA in the respective fractions was detected by Southern blotting. Locations for SV40 virion (open arrow) and 75S viral chromatin (closed arrow) were determined by parallel sedimentation of purified SV40 or nuclear lysate of [3H]thymidine-labeled SV40-infected cells (51). (c) Incorporation of capsid proteins into VLP. Vp1 and Vp3 in an aliquot of pooled fractions 4 to 7 in panel b were detected by Western blotting.
To determine if viral DNA was packaged in a particle form, the DNase I-treated cell lysate was sedimented through a sucrose gradient and fractions were analyzed for viral DNA. In both wild-type and mutant lysates, viral DNA appeared in fractions 4 through 7, positions for mature virion particles (Fig. 2b), indicating that mutant VLP similar in size to that of wild-type virions was formed in the transfected cells. The higher DNA signal in the fractions for Null AG than in that for either the wild type or Null NG reflects the relative amounts of the DNAs in the set of transfections (Fig. 2a).
Incorporation of mutant Vp3 proteins into VLP was confirmed by detecting Vp3 proteins as well as Vp1 in the VLP peak fractions (Fig. 2c). Higher protein signals were found in the Null NG and Null AG samples than in the wild-type sample, paralleling the relative levels of viral DNAs found in the corresponding fractions (Fig. 2b). The ratio of Vp1 to Vp3 in the fractions was constant (data not shown). Thus, mutant Vp3 was normally incorporated into the VLP. Migration of both mutant Vp3s was slower than that of wild-type Vp3, probably due to alterations of all five basic residues to neutral residues. Thus, once the transfected null mutant DNA reaches the nucleus, the mutant DNA replicates normally and is packaged into VLP indistinguishable from the virion in size and composition. We conclude that null mutants are not impaired in VLP production.
Null VLP can enter a new host cell but is defective in nuclear delivery of its DNA.
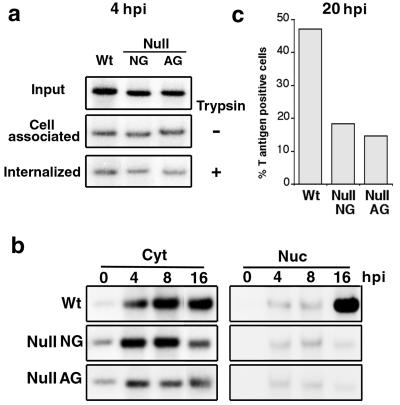
Since null VLP is produced normally, we asked if the cell entry process—cell surface binding and internalization—is impaired in null VLP infection. To distinguish the efficacy of VLP binding to cell surface from the VLP internalization, VLP-infected cells were harvested either by scraping off the cells or by trypsin treatment. The latter removes surface-bound particles from which the amount of internalized viral DNA can be determined. Total cell-associated viral DNA was measured from the cells collected without trypsin treatment. At 4 hpi, the amount of cell-associated (Fig. 3a, trypsin −) and internalized (Fig. 3a, trypsin +) viral DNA showed no difference between the mutant and wild-type infection, and approximately 12 to 15% and 5 to 6% of the input viral DNAs were cell associated and internalized, respectively, indicating that the mutant is not blocked in cell entry.
FIG. 3.
Detection of viral DNA in subcellular fractions. (a) Cells infected with the VLP lysate were harvested at 4 hpi either by scraping off (trypsin −, cell associated) or by trypsin treatment (trypsin +, internalized). Low-molecular-weight DNA was extracted and screened for viral DNA by Southern blotting. “Input” represents 1/50 of the viral DNA in total VLP lysate. Each lane of the cell-associated or internalized DNA panel was loaded with 1/20 of the DNA samples. (b) VLP-infected cells were harvested and separated into subcellular fractions at 0, 4, 8, or 16 hpi. Viral DNA in each fraction was detected as for panel a. One twenty-fourth of the cytoplasmic (Cyt) or 1/8 of the nuclear (Nuc) fraction was used for the analysis. (c) VLP-infected cells were fixed at 20 hpi and examined immunocytochemically for T-positive cells. Percent T-positive cells indicates the proportion of cells with visible T antigen in the 2,000 examined cells. Wt, wild type.
We then studied whether the internalized viral DNA enters the nucleus. The intracellular viral DNA was monitored at different hours postinfection, and the amount of full-length viral DNA accumulated in either the cytoplasmic or the nuclear fraction of infected cells was measured. As expected, an increase in the cytoplasmic viral DNA was seen between 0 and 4 hpi in all samples (Fig. 3b). The amount of cytoplasmic, full-length viral DNA was unchanged at the later time points. In the nuclear fraction, a slight increase in the wild-type viral DNA was noted from 4 hpi, and the onset of viral DNA replication was evident at 16 hpi. In a separate experiment, we observed evidence of viral DNA replication as early as 12 hpi (data not shown).
In contrast, although a slight increase of mutant DNA was similarly noted at 4 hpi, the level of mutant DNA was unchanged from 4 to16 hpi, indicating that mutant DNA did not replicate. Whether the lack of DNA replication reflects a lack of delivery of biologically active viral DNA to the nucleus was tested by examining T-antigen-positive cells, a hallmark of the nuclear entry of viral DNA. At 20 hpi, nearly 50% of the cells were T antigen positive in wild-type particle-infected cells, whereas only 15 to 17% were positive in the mutant VLP-infected cells (Fig. 3c). This result is consistent with the nuclear accumulation defect of the mutant viral DNA; thus, the mutant VLP can enter the cytoplasm normally but is compromised in delivering its DNA to the nucleus.
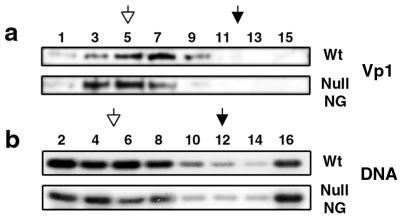
Two possible scenarios could account for the observed defect: (i) premature disassembly of the internalized null VLP, generating capsid-free viral DNA that enters the nucleus poorly (39), or (ii) poor recognition of the mutant NLS of the internalized particle by the nuclear import machinery. To test whether disassembly occurred, the cytoplasmic fraction prepared at 4 hpi was sedimented through sucrose gradients, and the profiles for viral DNAs and Vp1 in fractions were compared. The sedimentation profile of Null NG Vp1 was similar to that of the virion, but that of wild-type Vp1 was slightly shifted toward the top (Fig. 4a). The distribution of internalized, particle-derived DNA in the gradient, however, was similar between the wild type and the mutant: 49% of wild-type DNA and 46% of mutant DNA were found in the fractions expected for virions (Fig. 4b). Thirteen percent of wild-type viral DNA and 19% of mutant DNA were also present at the top of the gradient (fraction 16), and the rest were distributed in other fractions. If viral DNA in fraction 16 represents the dissociated DNA, the mutant particles did not dissociate to any larger extent than wild-type particles did. It is more likely, therefore, that mutants' defect in the nuclear delivery of viral DNA is due to poor recognition of the internalized mutant particles by the cellular import machinery.
FIG. 4.
Stability of internalized particles. The cytoplasmic fraction prepared at 4 hpi was fractionated through a sucrose gradient as described for Fig. 2b. (a) Vp1 in odd-numbered fractions was detected immunologically as described for Fig. 2c. (b) Viral DNA in even-numbered fractions was detected by semiquantitative PCR, generating a 2.2-kbp fragment, followed by Southern blotting with 32P-labeled SV40 DNA as a probe. Wt, wild type.
Both the Vp1 NLS and the Vp3 NLS define importin α2/β binding in vitro.
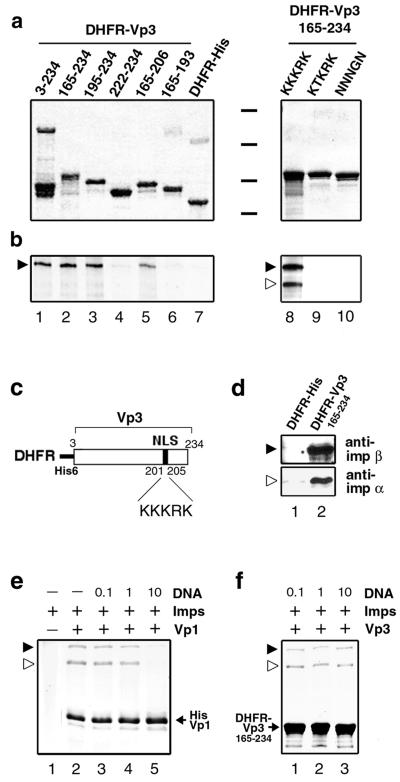
The importin α/β heterodimer recognizes proteins that harbor canonical NLSs and mediates their nuclear import (1, 13, 18). We first tested the interaction of the importin α2/β heterodimer, hereafter referred to as importin α/β, with either Vp3 or Vp1 (Fig. 5). A series of Vp3 fragments fused with the polyhistidine-tagged DHFR (Fig. 5c) were reacted with a combination of either unlabeled GST-importin α and 125I-labeled GST-importin β or both 125I-labeled GST-importins. The bound proteins were resolved in an SDS-polyacrylamide gel and probed for their mass (Fig. 5a) and importin binding (Fig. 5b). Binding of importin α/β was observed with the nearly full-length recombinant Vp3 protein (Fig. 5a and b, lane 1, 3-234) and proteins harboring Vp3 fragments 165 to 234 (lane 2), 195 to 234 (lane 3), and 165 to 206 (lane 5) but not with proteins harboring Vp3 fragments 222 to 234 (lane 4) and 165 to 193 (lane 6) or DHFR-His alone (lane 7).
FIG. 5.
In vitro binding of Vps with importins α2 and β. (a and b) An equimolar amount of DHFR-Vp3 fusion proteins immobilized onto beads was individually incubated with either a mixture of cold GST-importin α and 125I-labeled GST-importin β (lanes 1 through 7) or 125I-labeled GST-importin α and 125I-labeled GST-importin β (lanes 8 through 10). The bead-bound proteins were analyzed by SDS-PAGE followed by Coomassie blue staining (a) and autoradiography (b). Black bars between the two subpanels in panel a indicate positions for molecular mass markers of 97.4, 66, 46, and 30 kDa. Locations for GST-importin α (open arrowhead) and GST-importin β (filled arrowhead) are marked. (c) Schematic diagram of recombinant DHFR-Vp33-234 protein. (d) DHFR-Vp3 can recognize endogenous importin α/β. The beads bound with either DHFR-His (lane 1) or DHFR-Vp3165-234 (lane 2) were mixed with the uninfected TC-7 cell lysate. The cellular proteins bound to the beads were probed with either anti-importin α (anti-imp α) or anti-importin β (anti-imp β) antibody by Western blotting. (e) The presence of DNA blocks importin α/β-Vp1 interaction. GST-importin α/β (Imps) was incubated either with or without histidine-tagged Vp1ΔC58 (Vp1) in the presence or absence of the DNA fragment (DNA). The molecular ratio of DNA to importins in each reaction mixture is marked on the respective lane. The Vp1ΔC58-importin complex was recovered with Talon beads and resolved by SDS-PAGE, and the bead-bound proteins were visualized by Coomassie blue staining. (f) Importin α/β binding with Vp3 is not blocked by DNA. Interaction of GST-importin α and β and bead-bound DHFR-VP3165-234 in the presence of DNA was examined as for panel d.
The results show that amino acids 194 to 206, which include the Vp3 NLS (amino acids 198 to 206), are necessary for importin binding. As expected, mutations of the NLS in the DHFR-Vp3165-234 background—either a single mutation, KTKRK (lane 9), or the Null NG mutation (lane 10)—eliminated the importin binding, but the wild-type counterpart fully bound to the importins (lane 8).
Interaction of the Vp3 NLS with the endogenous cellular importin α/β complex was also confirmed (Fig. 5d). The extracts from uninfected TC-7 cells were incubated with resin immobilized with either DHFR-Vp3165-234 or DHFR-His, and the importins bound to the fusion proteins were detected by Western blotting with the respective importin antibodies. Importin α and importin β were retrieved by the resin that contained DHFR-Vp3165-234 (lane 2, top and bottom panels), whereas very little binding of either importin to DHFR-His was observed (lane 1). These results show that the Vp3 NLS defines the importin interaction and can bind to endogenous importins.
We next tested whether the Vp1 NLS binds importin α/β. Polyhistidine-tagged Vp1 with a truncation of the carboxyl-terminal 58 residues (31) bound importin α/β (Fig. 5e, lane 2) with no visible importin binding to resin alone (lane 1). The binding is specific to the Vp1 NLS, as mutant NLS proteins showed negligible binding to importins (data not shown). When an increasing concentration of the short DNA fragment was added to the binding reaction mixture (lanes 3, 4, and 5), importin-Vp1 binding was eliminated in the presence of a 10-fold excess of DNA over the amount of importin α/β (lane 5). These observations are consistent with the functional overlap of the Vp1 NLS with the Vp1 DBD (31) and show that the Vp1 NLS and/or DBD cannot interact with importin α/β if it engages in DNA binding. In contrast, the Vp3 NLS can still bind importins in the presence of increasing concentrations of DNA (Fig. 5f, lanes 1, 2, and 3).
Null mutant proteins of internalized VLP were not recognized by endogenous importins.
Findings from in vitro binding studies imply that Vp1 NLSs may remain associated with viral DNA and not be accessible for importin binding, whereas the Vp3 NLS may not remain concealed inside the particles. If Vp3 NLSs of the internalized VLP could become exposed, the cellular importin α/β heterodimer would bind the NLS. The binding of importin to a particle would depend on the integrity of the particle's Vp3 NLSs, and the presence of such an importin-particle complex could be shown in the immunocomplex resulting from immunoprecipitation of lysate with anti-importin antibody. The immunocomplexes should contain coprecipitated viral DNAs derived from the VLP, and the amounts of the DNAs are expected to be proportional to the extent of interaction of the signals on VLP with importin. If the null mutant proteins were defective in importin interaction, very little DNA would be coimmunoprecipitated. It is also expected that if the Vp3 NLSs become exposed, the otherwise internal Vp3 epitopes could become reactive to anti-Vp3 antibody. Immunoprecipitation by anti-Vp3 should provide a measure for the extent of exposure of the internal Vp3 epitope on the mutant VLP relative to that of the wild type, even if their mutant Vp3 NLSs are not visible to cytoplasmic importins. By comparison with the anti-Vp1-immunoprecipitated viral DNA, the anti-Vp3-precipitated DNA would indicate the proportion of viral DNA-Vps complex with exposed internal Vp3 epitopes.
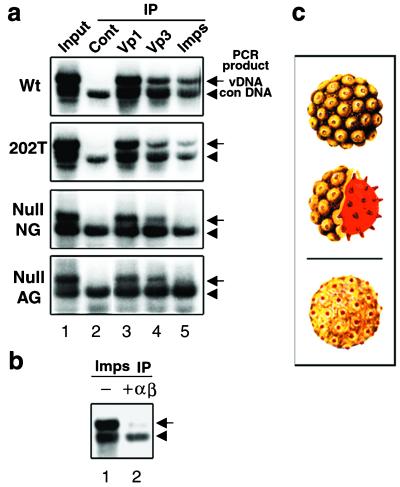
Cytoplasmic lysates prepared from cells infected with either wild-type, KTKRK (202T), Null NG, or Null AG particles were reacted with control antibody, anti-Vp1 serum, anti-Vp3 antibody, or a mixture of anti-importin α and β antibodies. The immunocomplexes were prepared with immunoaffinity beads, mixed with a constant amount of standard DNA, and purified for DNAs. The viral DNAs were then detected by semiquantitative PCR that generated 2.2- or 1.7-kbp fragments from the viral or standard DNA templates, respectively (Fig. 6a).
FIG. 6.
Coimmunoprecipitation of viral DNA by antibodies to capsids or cellular importins. (a) The cytoplasmic fraction from respective VLP-infected cells was immunoprecipitated with anti-mouse IgG (Cont), anti-Vp1 (Vp1), anti-Vp3 (Vp3), or a mixture of anti-importin α and anti-importin β antibodies (Imps). The respective immunocomplexes were mixed with a standard DNA, and their DNA was extracted and subjected to semiquantitative PCR for viral DNA. The amplification of the NO-SV40 genome generates the 2.2-kbp fragment (arrow, vDNA) and the standard DNA, the 1.7-kbp PCR fragment (arrowhead, con DNA). In “Input” (lane 1), 10 μl of each cytoplasmic fraction was mixed with the standard DNA, and the extracted DNAs were subjected to PCR. The total viral DNA in the respective cytoplasmic fractions was estimated as 20, 20, 15, or 10 pg for each 50 μl of the wild-type (Wt), KTKRK (202T), Null NG, or Null AG particle-infected cytoplasm, respectively. (b) The wild-type-infected cytoplasmic fraction was immunoreacted with anti-importin in the absence (−) or presence (+αβ) of recombinant GST-importins α and β to which a fivefold excess over the concentration of endogenous importins was added. Viral DNA in each immunoprecipitate was detected as for panel a. (c) Illustrations of an SV40 virion and an internalized particle. Shown are an intact SV40 particle with some of the 72 Vp1 pentamers (top panel, yellow) and a cross section revealing the internal core composed of minor capsid proteins Vp2 and Vp3 and a minichromosome (middle panel, red). A possible alteration of the internalized particle exposing a portion of internal minor capsid proteins on the surface is also shown (bottom panel).
From the total viral DNA present in the respective cytoplasmic lysates (Fig. 6a, lane 1), no DNA was coimmunoprecipitated by control antibody (lane 2). Anti-Vp1 precipitation yielded a significant proportion of viral DNAs for all samples (lane 3). Amounts of viral DNA precipitated by anti-Vp3 were also similar for all samples (lane 4), though less DNA was precipitated than in those by anti-Vp1. The reduced level of coimmunoprecipitated DNA could reflect the condition either that the concentration of the surface Vp3 epitope was low relative to the Vp1 epitope or that the Vp3 epitope was not exposed in all internalized particles. Nonetheless, coprecipitation of viral DNA with anti-Vp3 indicates that internal Vp3s become exposed once the particles enter the cytoplasm.
Striking differences were observed in the amount of DNA coprecipitated with anti-importins (Fig. 6a, lane 5). The 202T mutant DNA was coprecipitated to a similar extent as the wild-type DNA, indicating that mutant Vp3 as well as wild-type Vp3 was exposed and recognized by the importins. This observation contrasts with that seen in the in vitro binding assay (Fig. 5, lane 9): DHFR-VP3165-234 KTKRK, whose mutation is identical to that in the mutant 202T VLP, did not bind importins (see Discussion). On the other hand, both null mutant DNAs were coprecipitated at much reduced levels, indicating that the mutant Vp3s of internalized null VLPs were defective in interacting with cytoplasmic importins.
That the reduced level of the viral DNAs found in anti-importin immunocomplexes was due to a decrease in specific interaction with importin was tested by supplying excess recombinant importin α and β to the cytoplasmic lysate of wild-type virion-infected cells (Fig. 6b). The level of viral DNA that could coprecipitate with anti-importin antibodies seen in lane 1 was reduced (lane 2). We also observed the reduction in the level of the viral DNA when the infected lysate was supplemented with either excess importin α or excess importin β (data not shown).
We noted that Vp1 proteins of null mutant VLP were not reactive to anti-importin. If any Vp1 NLSs had been exposed on internalized VLP, the cellular importin would have recognized the signals. In that case, viral DNA precipitation by anti-importins would not have been affected by the null mutations in Vp3. Thus, these results show that the internalized particles did not expose the Vp1 NLS, consistent with the inhibition of importin binding in the presence of DNA in vitro (Fig. 5d) and the dual function of the NLS-DBD signal (31). We conclude that the Vp3 NLS, but not the Vp1 NLS, of the particle becomes visible for importin recognition once particles enter the cytoplasm. Cellular importin α/β does not recognize null mutant Vp3. The interaction of importins and Vp3 NLSs in the particle mediates the nuclear entry of infecting SV40.
DISCUSSION
In this study, we show that the nuclear entry of a nucleoprotein complex involves a dynamic interplay among distinct resident NLSs residing in the particle. The nuclear entry signal of the infecting SV40, which is a multisubunit nucleoprotein complex, was identified genetically and biochemically as the NLS of the minor structural Vp3 protein. The Vp3 NLS-null mutants, in which all the basic residues of the resident signal were altered, replicated and assembled into VLP normally, but the viability of the mutants was compromised. The mutant VLP, which had 72 defective Vp3 NLSs and 360 normal Vp1 NLSs, was able to enter the cell, and the extent of structural alteration that occurred in the cytoplasm resembled that of wild-type particles. Both the Vp1 NLS and the Vp3 NLS determine in vitro importin α/β interaction, but the importins did not bind the Vp1 NLS in the presence of DNA. In the cytoplasm of particle-infected cells, Vp1 NLSs of both particles were not recognized by importins and were therefore not exposed on the particles. In contrast, the interior Vp3 became at least partly exposed, and the Vp3 NLS of the wild-type infecting particle, but not the null-NLS Vp3 of mutant VLPs, was recognized by the importin α/β heterodimer. Consequently, nuclear entry of null mutant DNA was impaired, leading to reduced T-antigen expression and viability. These results show that Vp3 NLSs of SV40 mediate importin interaction and, as such, direct the nuclear entry of the viral DNA. They also suggest that a specific feature of a key and functional NLS for nuclear entry of a nucleoprotein complex is its ability to avoid masking by nucleic acid binding, given the basic properties of the canonical NLS.
Exposure of the interior Vp3 NLS in the internalized particle is a key step.
In SV40 virions, both the Vp1 NLS and the Vp3 NLS are interior and inside the capsid (7, 33), as are histone NLSs (4). Before any of them can function as the virion's nuclear targeting signal, they must be exposed so that the import machinery in the cytoplasm can recognize them. Histone NLSs are likely to be inaccessible in the minichromosome, and Vp1 NLSs remain bound to viral DNA within the particle, as this experiment shows. Thus, Vp3 NLSs could serve as the virion's NLSs if they become exposed through alteration by partial virion disassembly or a conformational change of capsid.
Our finding that the interior Vp3 becomes visible to anti-Vp3 once the virion enters the cytoplasm (Fig. 6a) is consistent with previous data showing that the nuclear entry of an infecting virus can be blocked if anti-Vp3 is present in the cytoplasm (39). They imply that the virion is at least partially altered once it enters the cytoplasm. The Vp3 NLS's exposure in the cytoplasm is essential for the binding of wild-type NLSs to importins. The exposure might be caused by disassembly of the particle or an alteration that maintains the shape and composition of proteins housing the viral genome. The latter possibility, a plausible change in the particle harboring partially exposed Vp3 (Fig. 6c, lower illustration) from SV40 virions (upper two illustrations), is consistent with our results. Fractionating the viral protein complexes by size and probing them with anti-importin would help identify the composition of virion proteins that associate with endogenous importins and clarify the molecular nature of the internalized particles bearing the NLS. How these interior proteins become exposed on the surface and how such structural alteration accommodates the virion's entry through the NPC, which is often smaller than the particle, remain unresolved.
Vp1 NLSs present in the internalized mutant VLP were apparently not recognized by importin α/β (Fig. 6a, lane 5). The NLSs may be masked due to their interaction with the SV40 minichromosome and thus be unavailable for importin binding. In support of this notion, the presence of DNA inhibited importin binding to Vp1 but not to Vp3 in vitro (Fig. 5e and f). This result does agree with the virion's structure in that the amino-terminal Vp1 residues are disordered and are proximal to the virion core (33). Thus, the Vp1 NLSs, not generally exposed on the internalized particle, are unlikely to play a role in nuclear entry.
Partially defective multiple Vp3 NLSs on the internalized particle are functional for importin recognition.
Our finding that Vp3 NLSs of a 202T mutant VLP can recognize importins in the cytoplasm (Fig. 6a) provides in vivo evidence for compensation of partly defective multiple NLSs occurring in the nuclear entry of the genomes. VLP bearing multiple defective Vp3 NLSs with single or double mutations were fully viable (Table 1) despite the inability of the single substitution mutant protein to bind importins in vitro (Fig. 5a and b). A close proximity of multiple NLSs on a VLP could allow the suboptimal signals to cumulatively function as an intact NLS. The local concentration of the KTKRK 202T Vp3 NLS in the 202T VLP is high, approximately 30 mM, and could allow the NLSs to be proximal to each other and effectively function as an NLS. The concentration of the DHFR-Vp3165-234 KTKRK attached to the affinity beads, about 10 μM, would be far less effective at achieving such a condition. Our results agree with earlier reports that (i) a nonnuclear protein coupled to suboptimal, multiple NLSs can accumulate in the nucleus (29, 48) and (ii) Vp2 capsid protein of minute virus of mice is imported as homo-oligomers, and single or double point mutations of the NLS do not influence the protein's karyophilic property (35). The presence of two NLS-binding sites 28 Å apart in the importin α molecule could allow the simultaneous binding of the two weak NLSs, leading to stable binding by importins to a protein harboring such NLSs (9, 26).
Viability of Vp3 NLS mutants.
Our results here show a role of the Vp3 NLS in the nuclear entry of infecting SV40 via an NPC-mediated route. The findings that the null mutants retained partial viability and that a small proportion of cells became positive for T antigen upon mutant VLP infection indicate that in such cells DNA had entered the nucleus. They also suggest that the protein domains yet to be defined could play an auxiliary role in the process. Two explanations are possible: (i) amino acids upstream and/or downstream of the Vp3 NLS could function cooperatively in VLP nuclear import or (ii) other capsid domains exposed on the surface of the SV40 virion cooperatively function in the VLP nuclear entry. The fact that viability was further reduced by 2 log units after additional replacement of Vp3 alanine 221 with threonine within the Null NG background (Table 1) supports the first possibility and agrees with our interpretation that Vp3 NLSs in concert with the flanking amino acids are responsible for the virion's nuclear entry. Importin binding of DHFR-Vp3165-234 having the A221T mutation was similar to that observed for the wild-type DHFR-Vp3165-234 protein (Fig. 5, lane 8, data not shown). The flanking amino acids could trigger covalent modifications of the region, thereby preventing NLSs from functioning (21). For example, phosphorylation of the upstream amino acids down-regulates the NLS recognition of the NF-ATc transcription factor leading to the cytoplasmic localization of the NF-ATc (55). An opposite effect is observed for the phosphorylation of amino acids upstream of the T-antigen NLS in vitro (16) and in vivo (20).
Alternatively, other non-NPC-mediated nuclear entry routes could provide auxiliary access to the nucleus. Upon infection with 10,000 or more particles per cell, SV40 accumulates in the endoplasmic reticulum (ER) (24, 36) or in the syntaxin 17-positive smooth ER (45). The particles that entered the lumen of the ER could traverse to the perinuclear space of the nuclear envelope and then penetrate its inner membrane to the nucleus. The biological activity accompanying such particles has not been shown so far. Perhaps obtaining and analyzing mutant VLP that is defective in ER targeting would be instrumental in clarifying the role of ER in virion nuclear targeting.
Cellular importins for SV40 nuclear entry.
Our study has shown that the nuclear entry of infecting SV40 involves importin α2/β and follows a common import pathway for a number of nuclear proteins. In vitro nuclear import assays have been instrumental in determining the role of importins in viral genome nuclear entry. For example, (i) influenza virus nucleoproteins target viral ribonucleoprotein particles to the nucleus through their interaction with importin α1 (NPI1) or α2 (RchI) (44), (ii) hepatitis delta antigen (HDag) of hepatitis delta virus associating with viral RNA can bind importin α2 and mediate the nuclear import of the RNA/HDag complex (8), (iii) NPC binding of the hepatitis B virus capsid is mediated by the interaction of the viral core protein's NLSs and importin α2/importin β (23), and (iv) the nucleocapsid of herpes simplex virus type 1 whose envelope has been removed can bind NPC, implying a role for importin β as the import receptor for herpes simplex virus (43).
In our study, the importin α2 (RchI/PTAC53)/importin β heterodimer, both components of which are abundant in the cell (12, 47), was identified as the mediator of SV40 virion entry. Other importin α subtypes could participate in SV40 nuclear entry, and further study may clarify whether a certain importin α subtype is preferred over others for the SV40 nuclear entry. Furthermore, other cellular import receptors, such as the importin β ortholog (11) or a structural component(s) of NPC, may participate in recognizing the internalized particles and thus contribute to the entry of the infecting DNA. Further analysis employing a combination of genetic and biochemical approaches as shown in this study may unveil roles of other import receptors that are critical for the nuclear entry of the infecting virion. Findings on the nuclear entry of the SV40 genome would help illuminate the nuclear entry mechanisms of many different nucleoprotein complexes, including many viruses that use the nucleus for genome replication.
Acknowledgments
We are grateful to Y. Yoneda and N. Imamoto for their gift of pGEX-2T PTAC58 and PTAC97 and for their suggestions for isolating recombinant importins. We thank N. Ishii for construction of NO-pSV40 KTKTK; C. Fernandez, J. Chan, and A. Nakamura for technical help in the steps of subcloning various mutant DNAs; D. Mayer for helpful discussion of subcellular fractionation; T. Benjamin, M. Moore, M. Rosbash, and Y. Yoneda for suggestions and discussions; and P. Li and T. Kasamatsu for invaluable assistance in the preparation of the manuscript.
This work was supported by NIH grant CA50574 and by a grant from the UCLA Academic Senate.
REFERENCES
- 1.Adam, E. J. H., and S. A. Adam. 1994. Identification of cytosolic factors required for nuclear location sequence-mediated binding to the nuclear envelope. J. Cell Biol. 125:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adam, S. A. 1999. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol. 11:402-406. [DOI] [PubMed] [Google Scholar]
- 3.Azuma, Y., and M. Dasso. 2000. The role of Ran in nuclear function. Curr. Opin. Cell Biol. 12:302-307. [DOI] [PubMed] [Google Scholar]
- 4.Baake, M., D. Doenecke, and W. Albig. 2001. Characterisation of nuclear localisation signals of the four human core histones. J. Cell. Biochem. 81:333-346. [PubMed] [Google Scholar]
- 5.Bouyac-Bertoia, M., J. D. Dvorin, R. A. M. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 6.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chou, H.-C., T.-Y. Hsieh, G.-T. Sheu, and M. M. C. Lai. 1998. Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J. Virol. 72:3684-3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conti, E., M. Uy, L. Leighton, G. Blobel, and J. Kuriyan. 1998. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin α. Cell 94:193-204. [DOI] [PubMed] [Google Scholar]
- 10.Dworetzky, S. I., R. E. Lanford, and C. M. Feldherr. 1988. The effects of variations in the number and sequence of targeting signals on nuclear uptake. J. Cell Biol. 107:1279-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Görlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 12.Görlich, D., S. Prehn, R. A. Laskey, and E. Hartmann. 1994. Isolation of a protein that is essential for the first step of nuclear protein import. Cell 79:767-778. [DOI] [PubMed] [Google Scholar]
- 13.Görlich, D., F. Vogel, A. D. Mills, E. Hartmann, and R. A. Laskey. 1995. Distinct functions for the two importin subunits in nuclear protein import. Nature 377:246-248. [DOI] [PubMed] [Google Scholar]
- 14.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 15.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 16.Hubner, S., C.-Y. Xiao, and D. A. Jans. 1997. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J. Biol. Chem. 272:17191-17195. [DOI] [PubMed] [Google Scholar]
- 17.Imamoto, N., T. Shimamoto, S. Kose, T. Takao, T. Tachibana, M. Matsubae, T. Sekimoto, Y. Shimonishi, and Y. Yoneda. 1995. The nuclear targeting complex binds to nuclear pores after association with a karyophile. FEBS Lett. 368:415-419. [DOI] [PubMed] [Google Scholar]
- 18.Imamoto, N., T. Shimamoto, T. Takao, T. Tachibana, S. Kose, M. Matsubae, T. Sekimoto, Y. Shimonishi, and Y. Yoneda. 1995. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 14:3617-3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ishii, N., A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu. 1994. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J. Virol. 68:8209-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jans, D. A., M. J. Ackermann, J. R. Bischoff, D. H. Beach, and R. Peters. 1991. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV-40 T antigen proteins. J. Cell Biol. 115:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jans, D. A., C.-Y. Xiao, and M. H. C. Lam. 2000. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays 22:532-544. [DOI] [PubMed] [Google Scholar]
- 22.Kann, M., A. Bischof, and W. H. Gerlich. 1997. In vitro model for the nuclear transport of the hepadnavirus genome. J. Virol. 71:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kartenbeck, J., H. Stukenbrok, and A. Helenius. 1989. Endocytosis of simian virus 40 into the endoplasmic reticulum. J. Cell Biol. 109:2721-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kasamatsu, H., and A. Nakanishi. 1998. How do the animal viruses get to the nucleus? Annu. Rev. Microbiol. 52:627-686. [DOI] [PubMed] [Google Scholar]
- 26.Kobe, B. 1999. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin α. Nat. Struct. Biol. 6:388-397. [DOI] [PubMed] [Google Scholar]
- 27.LaCasse, E. C., and Y. A. Lefebvre. 1995. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 23:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, M.-C., R.-I. Lin, and W.-Y. Tarn. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA 98:10154-10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanford, R. E., R. G. White, R. G. Dunham, and P. Kanda. 1988. Effect of basic and nonbasic amino acid substitutions on transport induced by simian virus 40 T-antigen synthetic peptide nuclear transport signals. Mol. Cell. Biol. 8:2722-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, D. C. Y., and J. D. Aitchison. 1999. Kap104p-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J. Biol. Chem. 274:29031-29037. [DOI] [PubMed] [Google Scholar]
- 31.Li, P. P., A. Nakanishi, D. Shum, P. C.-K. Sun, A. M. Salazer, C. F. Fernandez, S.-W. Chan, and H. Kasamatsu. 2001. Simian virus 40 Vp1 DNA-binding domain is functionally separable from the overlapping nuclear localization signal and is required for effective virion formation and full viability. J. Virol. 75:7321-7329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, P. P., A. Nakanishi, M. A. Tran, A. M. Salazar, R. C. Liddington, and H. Kasamatsu. 2000. Role of simian virus 40 Vp1 cysteines in virion infectivity. J. Virol. 74:11388-11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liddington, R. C., Y. Yan, J. Moulai, R. Sahli, T. L. Benjamin, and S. C. Harrison. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354:278-284. [DOI] [PubMed] [Google Scholar]
- 34.Lin, W., T. Hata, and H. Kasamatsu. 1984. Subcellular distribution of viral structural proteins during simian virus 40 infection. J. Virol. 50:363-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombardo, E., J. C. Ramirez, M. Agbandje-McKenna, and J. M. Almendral. 2000. A beta-stranded motif drives capsid oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maul, G. G., G. Rovera, A. Vorbrodt, and J. Abramczuk. 1978. Membrane fusion as a mechanism of simian virus 40 entry into different cellular compartments. J. Virol. 28:936-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 38.Moroianu. J. 1999. Nuclear import and export pathways. J. Cell. Biochem. Suppl. 32/33:76-83. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi, A., J. Clever, M. Yamada, P. L. Li, and H. Kasamatsu. 1996. Association with capsid proteins promotes nuclear targeting of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 93:96-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-685. [DOI] [PubMed] [Google Scholar]
- 41.Nakielny, S., U. Fischer, W. M. Michael, and G. Dreyfuss. 1997. RNA transport. Annu. Rev. Neurosci. 20:269-301. [DOI] [PubMed] [Google Scholar]
- 42.Nelson, L. M., R. C. Rose, L. LeRoux, C. Lane, K. Bruya, and J. Moroianu. 2000. Nuclear import and DNA binding of human papillomavirus type 45 L1 capsid protein. J. Cell. Biochem. 79:225-238. [PubMed] [Google Scholar]
- 43.Ojala, P. M., B. Sodeik, M. W. Ebersold, U. Kutay, and A. Helenius. 2000. Herpes simplex virus type 1 entry into host cells: reconstitution of capsid binding and uncoating at the nuclear pore complex in vitro. Mol. Cell. Biol. 20:4922-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Neill, R. E., R. Jaskunas, G. Blobel, P. Palese, and J. Moroianu. 1995. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem. 270:22701-22704. [DOI] [PubMed] [Google Scholar]
- 45.Pelkmans, L., J. Kartenbeck, and A. Helenius. 2001. Caveolar endocytosis of simian virus 40 reveals a new two-step vesicular-transport pathway to the ER. Nat. Cell Biol. 3:473-483. [DOI] [PubMed] [Google Scholar]
- 46.Popov, S., M. Rexach, G. Zybarth, N. Reiling, M.-A. Lee, L. Ratner, C. M. Lane, M. S. Moore, G. Blobel, and M. Bukrinsky. 1998. Viral protein R regulates nuclear import of the HIV-1 pre-integration complex. EMBO J. 17:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ribbeck, K., G. Lipowsky, H. M. Kent, M. Stewart, and D. Görlich. 1998. NTF2 mediates nuclear import of Ran. EMBO J. 17:6587-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts, B. L., W. D. Richardson, and A. E. Smith. 1987. The effect of protein context on nuclear location signal function. Cell 50:465-475. [DOI] [PubMed] [Google Scholar]
- 49.Ryan, K. J., and S. R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361-371. [DOI] [PubMed] [Google Scholar]
- 50.Senger, B., G. Simos, F. R. Bischoff, A. Podtelejnikov, M. Mann, and E. Hurt. 1998. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 17:2196-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spence, S. L., and J. M. Pipas. 1994. Simian virus 40 large T antigen host range domain functions in virion assembly. J. Virol. 68:4227-4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tiganis, T., A. J. Flint, S. A. Adam, and N. K. Tonks. 1997. Association of the T-cell protein tyrosine phosphatase with nuclear import factor p97. J. Biol. Chem. 272:21548-21557. [DOI] [PubMed] [Google Scholar]
- 53.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 54.Will, C. L., and R. Lührmann. 2001. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13:290-301. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, J., F. Shibasaki, R. Price, J.-C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]