Abstract
Repeated alcohol consumption leads to the development of tolerance, simply defined as an acquired resistance to the physiological and behavioral effects of the drug. This tolerance allows increased alcohol consumption, which over time leads to physical dependence and possibly addiction1–3. Previous studies showed that Drosophila develop ethanol tolerance with kinetics of acquisition and dissipation that mimic those seen in mammals. This tolerance requires the catecholamine octopamine, the functional analog of mammalian noradrenaline4. Here we describe a novel gene, hangover, required for normal development of ethanol tolerance. hangover flies are also defective in responses to environmental stressors, such as heat and the free radical-generating agent paraquat. Using genetic epistasis tests we show that ethanol tolerance in Drosophila relies on two distinct molecular pathways, a cellular stress pathway defined by hangover and a parallel pathway requiring octopamine. hangover encodes a large nuclear zinc-finger protein suggesting a role in nucleic acid binding. There is growing recognition that stress, at the cellular and systemic levels, contributes to drug- and addiction-related behaviors in mammals. Our studies suggest that this role may be conserved in evolution.
When flies are exposed to ethanol vapor they become hyperactive, uncoordinated, and eventually sedated. These effects of ethanol cause loss of postural control, which can be readily quantified in the inebriometer5. Naïve wild-type flies emerge from the inebriometer with a mean elution time (MET) of ~20 minutes at standard ethanol vapor concentrations6,7. A single exposure to ethanol in the inebriometer leads to the development of tolerance; flies reintroduced into the apparatus 4 hours after their initial exposure elute with a MET of ~28 minutes4 (Fig. 1a). This acquired resistance, or tolerance, correlates with an increase in the absorbed ethanol levels needed to induce loss of postural control, and is measured as the % increase (~35–40% at standard ethanol concentrations) in MET between the first and second ethanol exposures4.
Figure 1.
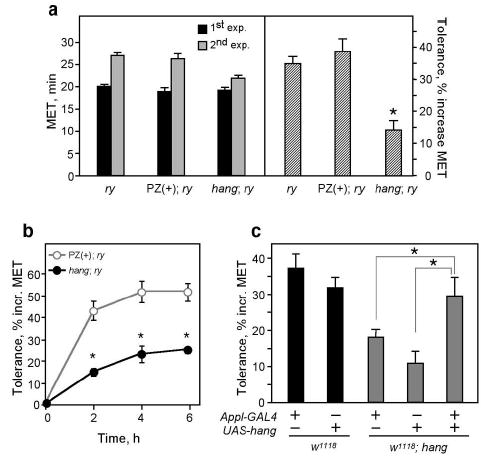
hangAE10 is an ethanol tolerance mutant.
a. Left panel: Mean elution times (METs) from the inebriometer of naïve (black bars) and ethanol pre-exposed (gray bars) flies. Right panel: Tolerance development, expressed as a % increase in MET between the two exposures. hangAE10 mutants (hang;ry) show significantly reduced tolerance compared to two controls, (ry) and (PZ(+);ry). n=18 experiments, *p<0.0001. In all figures, error bars correspond to the standard error of the mean (SEM).
b. hangAE10 flies are defective in a chronic tolerance paradigm, in which flies are exposed to ethanol in the inebriometer four times at 2-hour intervals. n=8 experiments, *p<0.001.
c. The hangAE10 tolerance defect can be rescued by expression of a UAS-hang transgene in the nervous system under the control of Appl-GAL4. Mutant w1118,hang flies carrying either transgene alone show reduced tolerance similar to that of hangAE10 flies, while flies carrying both transgenes show normal tolerance. n=5–10 experiments, *p<0.005.
To identify molecules and pathways involved in tolerance development, we carried out a screen for P element-induced mutants with aberrant tolerance (see Methods). Because the degree of tolerance is proportional to the length of initial ethanol exposure4, we limited our screen to strains that reacted normally to their first ethanol exposure. One mutant strain, AE10, that showed a normal initial MET, but a reduced ability to develop tolerance (14±3% compared to 35±2% for controls), was named hangover (hang)(Fig. 1a). The tolerance defect in hangAE10 is not simply due to a change in the rate of tolerance acquisition, as the mutant flies were also impaired when tested in a paradigm that induces maximal tolerance, through several consecutive ethanol exposures4 (Fig. 1b). hang flies show normal ethanol absorption and metabolism (Suppl. Fig. 1a); their phenotype is therefore not due to altered drug pharmacokinetics.
The P-element in hangAE10 is inserted in the predicted open reading frame of a novel gene, CG32575 (http://www.fruitfly.org/; Fig. 2a), encoding a protein with 15 nucleic acid-binding zinc-finger domains, two of which are of the U1 subclass found in RNA-binding proteins8 (Fig. 2b). The P-element insertion causes the mutant phenotype, as precise excision of the transposon causes reversion to the wild-type phenotype (Suppl. Fig. 1b). hang is expressed ubiquitously in the nervous system (Suppl. Fig. 2a,b) and HANG protein is localized to the nuclei of neurons (Fig.2c and Suppl. Fig. 2c). hangAE10 appears to be a null allele, as the ~ 7 kb mRNA encoded by hang (Fig. 2d) and the HANG protein (Fig. 2c) are undetectable in the mutant. Consistent with a role for hang in the nervous system, expression of a UAS-hang transgene under the control of the pan-neuronal Appl-GAL4 driver9 restores normal ethanol, tolerance to hangAE10 flies (Fig. 1c).
Figure 2.
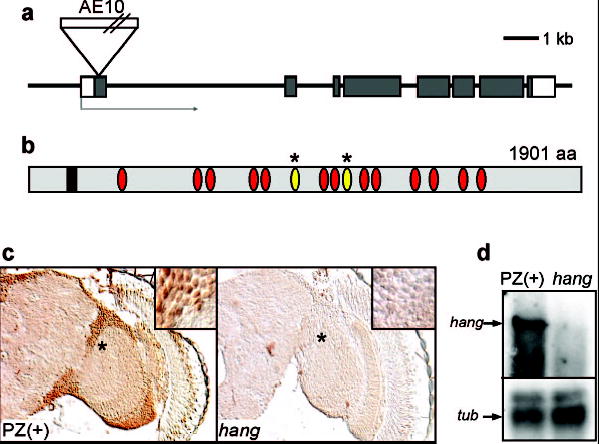
The hangAE10 mutation disrupts a gene encoding a novel zinc-finger protein. a. Genomic map of the hang locus (CG32575). The 8 exons are shown as boxes; gray shading indicates protein-coding regions. The position of the P-element insertion AE-10 is shown.
b. Conceptual translation predicts a protein of 1901 amino acids (http://www.fruitfly.org/). The positions of an EF hand (box) and zinc-finger domains (ovals) are indicated; the U1-like zinc-fingers are marked with asterisks.
c. HANG protein is expressed broadly in the nervous system (left panel) and localized to nuclei of neurons (see also Suppl. Fig. 2). Expression is undetectable in the hangAE10 mutant (right panel).
d. Northern blot analysis shows that expression of the full length hang transcript (~7Kb in length, upper arrow) is absent (or severely reduced) in the hangAE10 mutant compared to the PZ(+) control strain. The lower panel (tub) shows expression of Tubulin 84B, used as a control.
Ethanol, at high concentrations, is known to induce cellular responses similar to those elicited by heat shock10,11. Indeed, ethanol exposure in adult flies induces the expression of the heat shock protein Hsp70 (data not shown). To assess if these cellular stress responses may mediate ethanol tolerance development, we asked if a heat pulse could mimic the effects of ethanol pre-exposure. Heat exposure of control flies (37°C for 30 minutes) led to a 46±4% increase in MET (compared to untreated flies) when measured in the inebriometer 4 hours later. This heat-ethanol cross-tolerance is substantially reduced in hangAE10 flies (26±4%, Fig. 3a). The fact that hangAE10 flies are deficient in both forms of tolerance leads us to conclude that the cellular changes induced by ethanol and heat overlap. However, hangAE10 flies retain some ability to develop tolerance (Fig. 1a, Fig. 3a,b), suggesting that other pathways are also involved.
Figure 3.
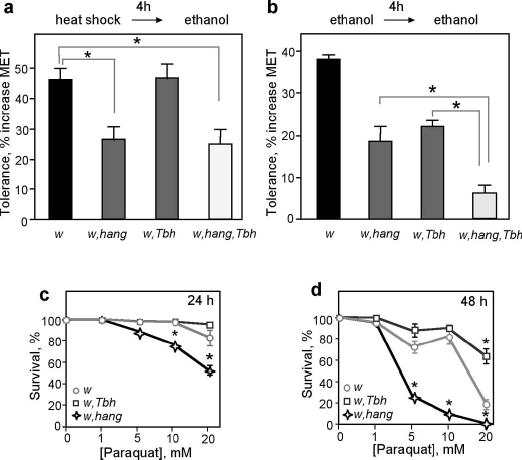
hangAE10 shows impaired heat-ethanol cross-tolerance. a. Heat-ethanol cross-tolerance: Control flies (w) and flies lacking octopamine (w,T□h) show normal cross-tolerance, while hangAE10 mutants (w,hang) and the double mutant (w,T□h,hang) display strongly reduced cross-tolerance. n=8–10 experiments, *p<0.0001.
b. Ethanol tolerance, while reduced in hangAE10 and T□h flies (significant difference with control not indicated in panel), is essentially absent in the T□h,hang double mutant flies. n=4–6 experiments, *p<0.001.
c, d. hangAE10 flies show increased sensitivity to paraquat. Panel c and d depict survival data obtained after 24 and 48 hours, respectively. n=5 experiments, *p<0.001.
We had found previously that flies lacking the neuromodulator octopamine, due to a mutation in the gene encoding Tyramine β Hydroxylase (Tbh)12, show a reduction in ethanol tolerance similar to that seen with hangAE10 flies4 (22±5% compared to 34±7% for control flies, Fig. 3b). To test if the residual tolerance seen with hangAE10 mutant flies may be mediated by octopaminergic systems (and vice versa), we tested flies lacking both octopamine and the hang gene product. We find that ethanol tolerance is almost completely abolished in hangAE10,Tbh double mutant flies (6±2% compared to 34±7% for control flies, Fig. 3b). Because both mutations cause complete loss of gene function (ref. 12 and Fig. 2), these data suggest that the development of ethanol tolerance relies on two parallel molecular pathways, one involving octopaminergic systems and the other the hang gene. The octopamine pathway is specific to ethanol tolerance, as Tbh flies develop normal heat-ethanol cross-tolerance (Fig. 3a). The hang pathway, on the other hand, involves cellular stress responses that are shared with those induced by heat. Finally, the fact that some heat-ethanol cross-tolerance remains in hang and Tbh,hang double mutant flies (Fig. 3b) suggests that heat treatment engages a third unidentified pathway that can induce ethanol resistance.
To test if hangAE10 flies are more generally deficient in their ability to deal with environmental stressors, we exposed the mutant flies to paraquat, an agent that induces the formation of reactive oxygen species (ROS) and that has been used to test for oxidative-stress responses13. When fed paraquat-containing food, hangAE10 flies show a dose- and time-dependent reduction in survival rate as compared to control flies (Fig. 3c, d). This reduced survival was not seen with Tbh flies, which appear to be even slightly resistant to the effects of the drug (Fig. 3c, d). The generation of endogenous ROS is believed to contribute to organismal aging14. We find that hangAE10, but not Tbh flies, show a reduced life span (Suppl. Fig. 3a, b), which may be caused by an impaired ability to cope with ROS. Mutations that cause neurodegeneration often show reduced lifespan15. This does not appear to be the case in hangAE10 flies, as their overall brain structure is normal, even in old flies (Suppl. Fig. 3c). This suggests a more direct involvement of hang in the functional response to environmental stressors. hang expression was not altered by ethanol exposure (data not shown). However, the predicted hang protein contains, in addition to multiple zinc-fingers, a calcium-binding EF hand (Fig. 2b), suggesting that hang function may be regulated by calcium. As calcium levels are sensitive to ethanol, ROS, and heat exposure10,16–18, this provides a potential mechanism by which these environmental stressors may regulate HANG function.
In summary, development of ethanol tolerance in Drosophila engages two systems that function in parallel, one involving a cellular stress pathway defined by the hang gene and the other involving octopaminergic systems. Octopamine has recently been implicated in the formation of appetitive (sugar-reinforced) memories in Drosophila19. The contribution of learned behaviors and stress, at the cellular and systemic levels, to drug- and addiction-related behaviors in mammals is increasingly being recognized20,21. Our studies in Drosophila suggest that these pathways are conserved allowing their analysis in this genetically tractable model organism.
Methods
Fly stocks and genetics
We generated and tested 404 homozygous viable X-linked lines carrying PZ(ry+)-element insertions22 for their ability to develop ethanol tolerance. Lines were generated6 and tested as described before4. Two lines with altered ethanol tolerance were isolated, one of them the insertion line hangAE10. To insure that the phenotype was not caused by chromosomal alterations unlinked to the PZ(ry+)-element, hangAE10 mutants were crossed for 5 generations to ry506 flies and re-tested for tolerance; hangAE10 flies retained their ethanol tolerance defect. The PZ(+) control line displays normal ethanol sensitivity and tolerance as measured in the inebriometer6,4. Excisions of the hangAE10 PZ(ry+)-insertion were generated as described before6. Four independent excision stocks were tested for ethanol tolerance and analyzed by PCR and DNA sequencing; a perfect correlation between precise excision of the P element and phenotype reversion was observed (Suppl. Fig. 1b).
The T□hnM18,hangAE10double mutant stock was generated by recombination as both genes reside on the X chromosome. Stocks that displayed female sterility, associated with the T□hnM18 mutation 12, were tested for β-galatosidase expression to determine the presence of the hangAE10 PZ(ry+)-insertion. A fertile (T□H+) w1118, hangAE10 strain was also generated as a control. Two independent recombinant strains (w1118, T□hnM18, hangAE10) were tested for ethanol tolerance with identical results.
Behavioral analyses
For all behavioral tests flies were generated as described before6,4. Approximately 100 3-4 day old males were tested in the inebriometer (MET1)6,7. After the initial exposure, flies were collected in vials and allowed to recovered in a humidified incubator at 25°C. The second exposure in the inebriometer (MET2) was initiated exactly 4hrs after the start of the first exposure. Tolerance was calculated as described before4 (MET2- MET1/MET1 x 100). Ethanol absorption was quantified as described before in extracts prepared from adult flies6,7. Significance was established using either Student’s paired t-tests assuming equal variance or one-way ANOVAs with Newman-Keuls post hoc tests.
For heat shock-ethanol cross-tolerance experiments, flies were incubated in a vial at 37°C for 30 min in a water bath. After a recovery period of 3 hrs and 30 min at 25°C in a humidified incubator, these flies were exposed to ethanol in the inebriometer (METhs+). Tolerance was calculated with respect to METhs−, the MET of flies that were not heat-treated (METhs+- METhs−/METhs− x 100).
To measure sensitivity to paraquat groups of 20, 3-4 day old male flies were transferred into a glass vial containing a filter paper saturated with aqueous solutions of paraquat (0, 1, 5, 10 and 20 mM) in 5% sucrose. Flies were kept in an incubator with 70% humidity at 25°C. Every 12 hrs dead flies were counted and live flies were transferred into new vials.
Molecular Biology
The genomic DNA flanking the hangAE10 PZ(ry+) insertion was isolated by plasmid rescue. Comparisons with the published genome sequence of Drosophila revealed that the insertion was located in the first exon of CG32575, a finding that was confirmed by PCR. Several cDNA clones (GH14331, LD34334 and GH19829) were sequenced. The GH14331 cDNA is approximately 7kb long and its sequence corresponds to the predicted structure of CG32575 (http://www.fruitfly.org/). The sequence of GH14331 does not differ from the genomic sequence, but differs from annotated transcripts CG32575RA and RB. The full-length cDNA of GH14331 was cloned into pUAST23 to generate the UAS-hang transgene. For Northern blots, 1μg polyA+ RNA from PZ(+) and hangAE10 was used. A 3.7 Kb EcoRI/KpnI fragment from the GH14331 cDNA clone, spanning from exon 1 (3’ of the insertion site) to exon 5, was used as a hybridization probe. A probe that recognizes Tubulin 84B mRNA (nucleotides 10–340; http://flybase.bio.indiana.edu/) was used as a control.
Histology
To generate antibodies to the HANG protein, a His-tagged antigen was made. The sequence coding for amino acids 62-319 of HANG were cloned into the pET28-b vector (Novagene). Protein was purified from E. coli extracts using a nickel column as indicated by manufacturers, and injected into rabbits. For HANG immunohistochemistry, flies were fixed for 2.5 hrs in 5% paraformaldehyde at 4°C, washed and infiltrated with a 25% sucrose solution overnight at 4°C. Whole flies were sectioned into 10 μm slices using a cryostat. Sections were washed twice for 5 min in PBT (PBS plus 0.3% Triton X-100) and preincubated with 10% normal goat serum in PBT for 2 hrs at room temperature. Incubation with anti-HANG serum (1:5000 in PBT plus 10% NGS) was carried out overnight at 4°C. After two 5-minute washes with PBT, signal detection was carried out using the Vectastain® ABC elite kit with a peroxidase-coupled mouse IgG antibody.
Supplementary Material
Acknowledgments
We thank R. Threlkeld for the Northern blot, N. Funk and I. Schwenkert for production of rescue constructs and HANG antibody, H.Saumweber and G.Krohne for gifts of antibodies, B. Poeck, A. Corl, A. Rothenfluh, D. Guarnieri, F. Wolf, and C. Kenyon for comments on the manuscript. Work was supported by NIH/NIAAA (UH), ABMRF (UH), the Sandler Foundation (UH), the Wheeler Center (HS, UH), and the German Science Foundation DFG 656 (HS).
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
References
- 1.Tabakoff B, Cornell N, Hoffman PL. Annals of Emergency Medicine. 1986;15:1005–12. doi: 10.1016/s0196-0644(86)80119-6. [DOI] [PubMed] [Google Scholar]
- 2.Lê, A. D. & Mayer, J. M. in Pharmacological effects of ethanol on the nervous system (eds. Deitrich, R. A. & Erwin, V. G.) 251–268 (CRC Press, Boca Raton, 1996).
- 3.Fadda F, Rossetti ZL. Prog Neurobiol. 1998;56:385–431. doi: 10.1016/s0301-0082(98)00032-x. [DOI] [PubMed] [Google Scholar]
- 4.Scholz H, Ramond J, Singh CM, Heberlein U. Neuron. 2000;28:261–271. doi: 10.1016/s0896-6273(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 5.Weber KE, Diggins LT. Genetics. 1990;125:585–97. doi: 10.1093/genetics/125.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moore MS, et al. Cell. 1998;93:997–1007. doi: 10.1016/s0092-8674(00)81205-2. [DOI] [PubMed] [Google Scholar]
- 7.Singh CM, Heberlein U. Alcoholism: Clinical and Experimental Research. 2000;24:1127–1136. [PubMed] [Google Scholar]
- 8.Legrain P, Choulika A. Embo J. 1990;9:2775–81. doi: 10.1002/j.1460-2075.1990.tb07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torroja, L., Chu, H., Kotovsky, I. & White, K. Curr Biol 9, 489–92 (1999). [DOI] [PubMed]
- 10.Wilke N, Sganga M, Barhite S, Miles MF. Exs. 1994;71:49–59. doi: 10.1007/978-3-0348-7330-7_6. [DOI] [PubMed] [Google Scholar]
- 11.Piper PW. Fems Microbiology Letters. 1995;134:121–7. doi: 10.1111/j.1574-6968.1995.tb07925.x. [DOI] [PubMed] [Google Scholar]
- 12.Monastirioti M, Linn CEJ, White K. The Journal of Neuroscience. 1996;16:3900–3911. doi: 10.1523/JNEUROSCI.16-12-03900.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arking R, Buck S, Berrios A, Dwyer S, Baker GT., 3rd Dev Genet. 1991;12:362–70. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- 14.Finkel T, Holbrook NJ. Nature. 2000;408:239–47. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 15.Min KT, Benzer S. Curr Biol. 1997;7:885–8. doi: 10.1016/s0960-9822(06)00378-2. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki YJ, Forman HJ, Sevanian A. Free Radic Biol Med. 1997;22:269–85. doi: 10.1016/s0891-5849(96)00275-4. [DOI] [PubMed] [Google Scholar]
- 17.Walter HJ, Messing RO. Neurochem Int. 1999;35:95–101. doi: 10.1016/s0197-0186(99)00050-9. [DOI] [PubMed] [Google Scholar]
- 18.Sun AY, et al. Alcohol Clin Exp Res. 2001;25:237S–243S. doi: 10.1097/00000374-200105051-00038. [DOI] [PubMed] [Google Scholar]
- 19.Schwaerzel M, et al. J Neurosci. 2003;23:10495–502. doi: 10.1523/JNEUROSCI.23-33-10495.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hyman SE, Malenka RC. Nat Rev Neurosci. 2001;2:695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- 21.Nestler EJ. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 22.Mlodzik M, Hiromi Y. Methods Neurosci. 1992;9:397–414. [Google Scholar]
- 23.Brand AH, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


