Abstract
Synthetic small interfering RNAs (siRNAs) have been shown to induce the degradation of specific mRNA targets in human cells by inducing RNA interference (RNAi). Here, we demonstrate that siRNA duplexes targeted against the essential Tat and Rev regulatory proteins encoded by human immunodeficiency virus type 1 (HIV-1) can specifically block Tat and Rev expression and function. More importantly, we show that these same siRNAs can effectively inhibit HIV-1 gene expression and replication in cell cultures, including those of human T-cell lines and primary lymphocytes. These observations demonstrate that RNAi can effectively block virus replication in human cells and raise the possibility that RNAi could provide an important innate protective response, particularly against viruses that express double-stranded RNAs as part of their replication cycle.
The phenomenon now generally termed RNA interference (RNAi) is an evolutionarily conserved process whereby the expression or introduction of double-stranded RNA (dsRNA) sequences results in the specific posttranscriptional inactivation of genes which are complementary to the dsRNA sequence used (reviewed in reference 37). In several organisms, including plants, nematodes, and fruit flies, RNAi can be used to inactivate a specific host gene subsequent to microinjection or transfection of the appropriate dsRNA (16, 21, 39). However, in mammalian cells, dsRNAs consisting of more than 30 nucleotides (nt) induce not only RNAi but also a set of responses, including activation of the interferon response, that collectively result in a global, nonspecific inhibition of host cell mRNA expression (32, 36).
Efforts to resolve the mechanisms underlying RNAi in the plant and drosophila systems revealed that longer dsRNAs were processed by a host RNase, termed dicer, into ∼21-nt dsRNAs, called small interfering RNAs (siRNAs), that contain 2-nt 3′ overhangs (14, 19-21, 25, 26, 28, 43). These siRNAs are key mediators of RNAi and are thought to serve as guide RNAs for a large protein complex, termed the RNA-induced silencing complex, that is essential for target mRNA degradation (22). Importantly, it soon became apparent that direct introduction of synthetic siRNAs could also induce RNAi not only in drosophila cells but also in human cells (8, 14, 15). Subsequently, transfection of synthetic siRNAs has been used to selectively block the expression of a small but rapidly growing list of human genes and to determine the phenotypic consequences of this inhibition (1, 11, 18, 23, 25, 33).
RNAi is believed to have evolved as a host defense mechanism directed at transposable elements and infecting viruses (12, 27, 34, 38). We therefore wondered if RNAi might provide an effective defense against a pathogenic human virus. Here, we describe siRNAs targeted against the essential human immunodeficiency virus type 1 (HIV-1) regulatory proteins Tat and Rev (13) and show that these siRNAs can effectively and specifically block Tat or Rev expression and function in transfected 293T cells. More importantly, we show that these same siRNAs can prevent the productive infection of both immortalized and primary human T cells by HIV-1 in culture. These data provide evidence in favor of the hypothesis that RNAi can be used to selectively block viral gene expression, and hence replication, in human cells.
MATERIALS AND METHODS
Plasmid constructs.
The mammalian expression plasmids pcTat, pcRev, pBC12/CMV, pBC12/CMV/β-Gal, pcRex, and pbTat (5, 31) and the indicator constructs pDM128/PL, pDM128/RRE, pDM128/RxRE, pTAR/CAT, and pbTAR/CAT (5, 24, 40) have been described in detail elsewhere. In addition, the HIV-1 proviral clones pNL-ADA and pNL-luc-ADA and plasmids expressing CD4 (pCMV5/CD4) and CCR5 (pCMV/CCR5) have also been described (41).
The plasmid pHIV/Tat, expressing the Tat protein under the control of the HIV-1 long terminal repeat (LTR) was constructed by PCR amplification of the tat cDNA sequence contained within pcTat (40). Oligonucleotide primers were designed to introduce HindIII and BamHI sites so that the amplified Tat sequence could be ligated into HindIII-BamHI-digested pHIV/TAR/CAT (40).
RNA interference.
Twenty-one nucleotide RNA duplexes with 2-nt (2′-deoxy)- thymidine 3′ overhangs directed against nucleotides 5886 to 5909 and 8462 to 8485 of the respective HIV-1 tat and rev coding sequences were obtained from Dharmacon Research, Inc. (Lafayette, Colo.). RNAi in 293T cells was performed essentially as described previously (14). Briefly, 2 × 105 cells were plated in a 24-well plate 1 day prior to transfection. Annealed siRNA duplexes (0.1 μM) and the mammalian expression and indicator constructs denoted in the relevant figure legends were cotransfected with Lipofectamine 2000 (Invitrogen) according to the manufacturers' instructions. Tat and Rev expression was assayed by Western analysis and by biological activity as previously described (4, 31, 40).
For Jurkat T cells, annealed Tat and Rev siRNA duplexes (12 μM concentration) were mixed with 1.5 × 107 cells in 0.5 ml of OPTI-Mem I reduced serum medium (Invitrogen) and subjected to electroporation in a 0.4-cm-gap cuvette at 300 mV and 960 μF with a Bio-Rad gene pulser. The electroporated cells were immediately transferred to 15 ml of RPMI 1640-10% fetal calf serum and grown overnight in the absence of antibiotics. Inhibition of viral replication was assayed by enzyme-linked immunosorbent assay (ELISA) for p24 Gag protein production, as described below.
Peripheral blood mononuclear cells (PBMC) from healthy HIV-1-negative donors were isolated by Ficoll-Hypaque gradient centrifugation. Cells, 108/ml, were frozen at −150°C in human AB serum until required. Thawed cells were resuspended in AIM V medium (Invitrogen) and supplemented with 10% human AB serum (Sigma) and 10 U of interleukin-2 (R&D Systems) per ml as described previously (41). Nonadherent cells were stimulated with 3 μg of phytohemagglutinin (Sigma) per ml for 2 days prior to electroporation as described above.
Cell maintenance and preparation of viral stocks.
Human 293T cells were maintained as described previously (4), while Jurkat T cells were maintained in RPMI 1640 (Invitrogen) supplemented with 10% fetal calf serum. Fresh viral stocks were prepared by transfection of 293T cells with 1 μg of pNL-ADA or pNL-luc-ADA (41) with FuGENE 6 reagent (Roche). After 48 h, the cells were washed once with phosphate-buffered saline (PBS), and newly produced virions were harvested over 3 h in 0.5 ml of fresh medium. Viral supernatants were clarified by passage through a 0.4-μm syringe filter. HIV-1 stocks pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) were prepared in a similar manner, except that 100 ng of pHIT/G (17) was cotransfected with 1 μg of pNL-ADA with FuGENE 6 reagent (Roche).
Viral replication assays.
293T cells were cotransfected with 100 ng of pCMV5/CD4, 500 ng of pCMV/CCR5, and siRNA oligonucleotides, as denoted in the relevant figure legends. After 48 h, CD4+ CCR5+ 293T cells were infected with 50 ng of p24 antigen of the NL-ADA or NL-luc-ADA virus (41). The following day, the cells were washed extensively with PBS and overlaid with fresh growth medium. Supernatants were harvested 48 h later and p24 Gag antigen production was quantified by ELISA (NEN). In the case of Jurkat T cells, cells were harvested 24 h after electroporation and counted. Cells (106) were then infected with 50 ng of p24 antigen of VSV-G-pseudotyped NL-ADA virus in a 24-well dish as described above. Progeny virus production was measured 48 h later by p24 Gag ELISA. Similarly, PBMC were also infected 24 h after electroporation with 50 ng of p24 of the CCR5-tropic virus NL-ADA, and progeny virus production was measured 48 h later.
Viral gene expression was measured in 293T cells with a luciferase reporter virus (10). After 48 h, the cells were washed with PBS and resuspended in 200 μl of cell lysis buffer (Promega) and luciferase activity was determined as previously described (41).
RNA, DNA, and Western analyses.
All nucleic acid blotting and hybridization techniques were performed essentially as described previously (35). Total RNA was isolated from infected 293T cells with a QIAGEN RNeasy minikit in accordance with the manufacturers' instructions. RNA (15 μg) was separated on a 1% agarose gel containing formaldehyde and transferred to a Hybond-N membrane (Amersham Pharmacia Biotech). RNA was fixed by UV cross-linking in a Stratalinker (Stratagene), and HIV-1 RNAs were detected with a 32P-labeled random-primed DNA probe derived from pNL-ADA by PCR amplification with oligonucleotide primers directed towards the HIV-1 U3 region (nucleotides 9078 to 9609).
Total DNA was extracted from infected 293T cells with a QIAGEN DNeasy tissue kit in accordance with the manufacturers' instructions. Samples were treated with 400 μg of RNase A to remove contaminating RNA. DNA (10 μg) was dried in a speed vacuum, denatured in 2 μl of 0.2 M NaOH for 15 min at 37°C, and spotted onto a Hybond-N membrane. After UV cross-linking, the membrane was probed with the HIV-1-specific 32P-labeled probe as described above or with a random-primed β-globin cDNA probe.
Western analysis of protein expression in transfected 293T cells was performed with rabbit polyclonal antisera directed against the HIV-1 Tat or Rev protein or against human Tap/NXF1, as previously described (9, 31). All antibodies were used at a dilution of 1:1,000.
RESULTS
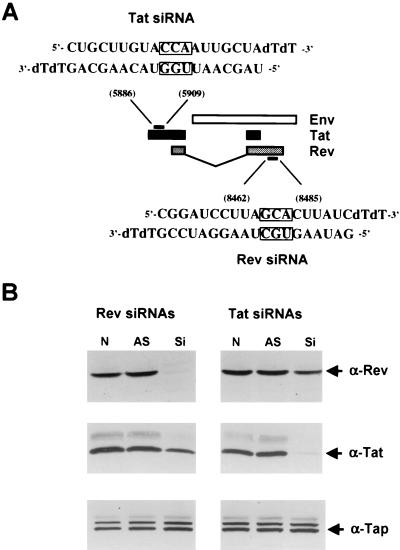
In the context of the HIV-1 genome, the viral Tat and Rev proteins are encoded by two partially overlapping exons separated by an intron consisting largely of envelope coding sequences (Fig. 1A). Previously, Elbashir et al. (14) demonstrated that synthetic 21-nt siRNA duplexes, bearing 2-nt 3′ overhangs, can function as effective inhibitors of mRNA expression in transfected human cells. In addition, these researchers suggested that the 2-nt overhang should ideally consist of uridine or, in the interest of stability, deoxythymidine. Based on these observations, we therefore searched the nonoverlapping parts of the Tat and Rev coding sequences for appropriate siRNA targets. The location of the sequences in the HIV-1 Tat and Rev open reading frame that were chosen as targets and the structure of the synthetic siRNAs that were used in this analysis are shown in Fig. 1A.
FIG. 1.
(A) Schematic representation of part of the HIV-1 genome. HIV-1 tat and rev exons are depicted with black and grey boxes, respectively, while the env open reading frame is shown as a white box (not to scale). The structure and target coordinates of the siRNAs specific for tat and rev are depicted. Boxed residues were mutated by transversion for use as control duplexes in some experiments. (B) siRNAs directed against tat and rev specifically inhibit Tat and Rev protein expression. Human 293T cells were cotransfected with 50 ng of both the pcTat and pcRev expression plasmid in the absence of RNA oligonucleotides (N) or in the presence of 0.1 μM antisense oligonucleotides (AS) or 0.1 μM siRNA duplexes (Si). Cell lysates were prepared 60 h posttransfection, and protein expression was assayed by Western analysis with polyclonal antiTat and antiRev antisera. Western blots were also probed with antibodies directed against the endogenous TAP/NXF-1 protein, which here served as an internal control for both loading and specificity.
Specific inhibition of HIV-1 Tat and Rev expression and function by RNAi.
To test whether siRNAs that were targeted to these short Tat and Rev sequence elements would specifically block Tat or Rev protein expression, we cotransfected 293T cells with the Tat expression construct pcTat and the Rev expression plasmid pcRev (31). These cDNA expression plasmids contain exclusively the coding sequence of Tat or Rev (Fig. 1A), and therefore each should be subject to inhibition by only one of the siRNAs. Cells were also cotransfected with one of the double-stranded siRNAs or, as controls, with only the relevant antisense RNA strand or with nothing.
As shown in Fig. 1B, Western analysis revealed that the Tat-specific siRNA blocked Tat protein expression but did not significantly affect expression of an irrelevant cellular protein termed Tap. Similarly, the Rev-specific siRNA also selectively blocked expression of the Rev protein. In neither case did the antisense RNA oligonucleotide alone exert any effect. In some experiments, we did notice a slight (≤20%) effect of the Rev-specific siRNA on Tat protein expression or of the Tat-specific siRNA on Rev expression. This may reflect a weak nonspecific inhibitory effect or, more trivially, may suggest that the double-stranded siRNA, unlike the single-stranded antisense RNA, slightly reduced transfection efficiency in this experiment. Nevertheless, it is clear that these siRNAs can indeed selectively block expression of the HIV-1 Tat or Rev protein in transfected 293T cells.
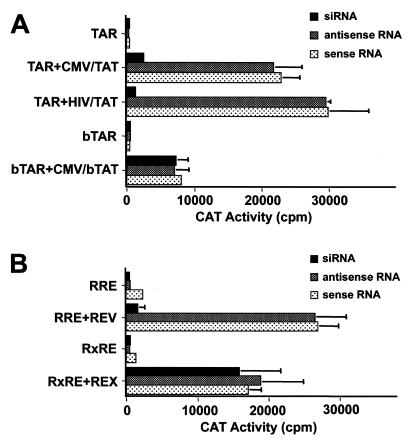
We next wished to determine whether these siRNAs would also block Tat or Rev function. The HIV-1 Tat protein serves as a specific transcriptional transactivator of gene expression directed by the HIV-1 LTR promoter after binding to a specific target site termed TAR (reviewed in reference 13). As shown in Fig. 2A, we confirmed that the HIV-1 Tat protein expressed under the control of either the cytomegalovirus (CMV) immediate early promoter or the HIV-1 LTR itself can strongly activate chloramphenicol acetyltransferase (CAT) activity directed by the HIV-1 LTR promoter in transfected 293T cells. As shown in Fig. 2A, this activation was potently inhibited by cotransfection of the specific siRNA but not by cotransfection of either the sense or antisense RNA strand alone. This inhibition is specific, since activation by the bovine immunodeficiency virus (BIV) Tat protein (bTat) of an HIV-1 LTR promoter that contains the BIV TAR element in place of the HIV-1 TAR element was not affected (Fig. 2A). Importantly, while the bTat protein is very similar to HIV-1 Tat in its mechanism of action, the HIV-1 Tat coding sequence indicated in Fig. 1A is not conserved in bTat (5).
FIG. 2.
(A) siRNAs directed against HIV-1 tat inhibit HIV-1 Tat but not BIV Tat function. Human 293T cells were transfected with 100 ng of pTAR/CAT or pbTAR/CAT, 50 ng of the internal control plasmid pBC12/CMV/β-Gal, and 0.1 ng of the indicated effector plasmid. siRNA oligonucleotides or siRNA duplexes were cotransfected with the reporter and expression plasmids as described in Materials and Methods. Cells were harvested 48 h after transfection, and induced CAT activities were determined. The data shown represent averages for three experiments with the standard deviations indicated by error bars. (B) siRNAs directed against HIV-1 rev specifically inhibit HIV-1 Rev but not HTLV Rex function. 293T cells were transfected with 25 ng of the pDM128/RRE or pDM128/RxRE reporter plasmid, 50 ng of pBC12/CMV/β-Gal, and 1 ng of the indicated effector plasmid. siRNA oligonucleotides or siRNA duplexes were cotransfected with the reporter and expression plasmids as described in Materials and Methods. Cells were harvested 48 h after transfection, and induced CAT and β-galactosidase activities were determined. The data shown represent averages for three experiments with the standard deviations indicated by error bars.
As shown in Fig. 2B, we demonstrated that the siRNA targeted to the HIV-1 Rev protein can also specifically block HIV-1 Rev function in transfected 293T cells. This experiment utilizes an indicator construct termed pDM128/RRE that contains an mRNA in which the cat gene and the HIV-1 Rev response element (RRE) are sequestered within an intron (4, 24). This RNA is normally unable to exit the cell nucleus but is exported and expressed in the presence of the HIV-1 Rev nuclear RNA export factor. As shown in Fig. 2B, this induction is blocked by the Rev-specific siRNA but not by either the sense or antisense RNA strand alone. This inhibition is specific, since the Rev siRNA did not inhibit expression of the unspliced cat mRNA encoded by the similar indicator plasmid pDM128/RxRE, which contains the human T-cell leukemia virus (HTLV) Rex response element in place of the RRE, in the presence of the functionally analogous but unrelated HTLV Rex protein (4).
Inhibition of HIV-1 gene expression and replication by RNAi.
Although the human cell line 293T is not normally infectible by HIV-1, 293T cells can support HIV-1 infection and replication if they are programmed to express the CD4 receptor and the CCR5 coreceptor for HIV-1 (41). We have previously described the pNL-ADA and pNL-luc-ADA HIV-1 proviral expression plasmids. The NL-ADA virus is a fully replication-competent HIV-1 that was derived from the NL4-3 HIV-1 isolate by substitution of the NL4-3 env gene with an env gene derived from the CCR5-tropic HIV-1 isolate ADA (41, 42). NL-luc-ADA is similar to NL-ADA except that the HIV-1 nef gene has been replaced by the luc indicator gene (10). Therefore, infection of cells by NL-luc-ADA can be readily monitored by measurement of the level of luciferase expression.
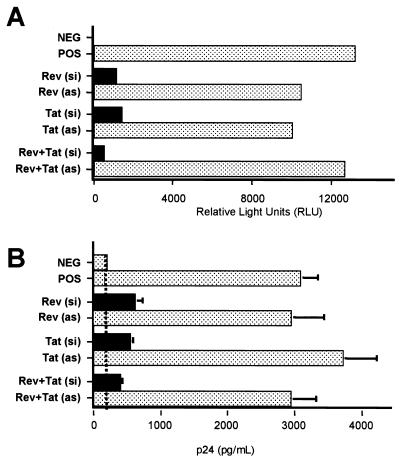
We initially determined the effect of the siRNAs on the level of viral gene expression by infecting 293T cells transfected with plasmids encoding CD4 and CCR5, as well as with RNA oligonucleotides, with the NL-luc-ADA indicator virus. As shown in Fig. 3A, both the Tat siRNA and the Rev siRNA, and particularly the combination of both, were able to effectively inhibit expression of the virally encoded luc indicator gene in these HIV-1-infected cells. In contrast, the antisense RNA oligonucleotides alone had no specific effect.
FIG. 3.
Inhibition of HIV-1 replication in human cells by RNAi. 293T cells were transfected with CD4 and CCR5 expression plasmids and siRNA duplexes (si) or antisense RNAs (as) (0.1 μM each). After 48 h, CD4+ CCR5+ cells were infected overnight with 50 ng of p24 antigen of the luciferase reporter virus NL-luc-ADA or the wild-type virus NL-ADA. Viral gene expression was determined by luciferase activity after 48 h of infection (A), while viral replication was measured by p24 Gag antigen production 60 h posttransfection (B). 293T cells lacking CD4 and CCR5 served as the negative control (NEG), while CD4+ CCR5+ 293T cells infected in the absence of siRNAs served as the positive control (POS). A low level of carryover of the virus stock used to infect the cells explains the background level of p24 Gag seen in panel B (vertical dashed line).
We next examined whether the siRNAs would be able to inhibit the release of progeny HIV-1 virion particles from infected 293T cells. As shown in Fig. 3B, the two siRNAs acting either alone or in combination were indeed able to effectively inhibit the production of progeny HIV-1 virions. Again, the antisense RNA oligonucleotides had little or no effect. We therefore conclude that these siRNAs are able to effectively inhibit HIV-1 gene expression and replication in infected CD4+ CCR5+ 293T cells.
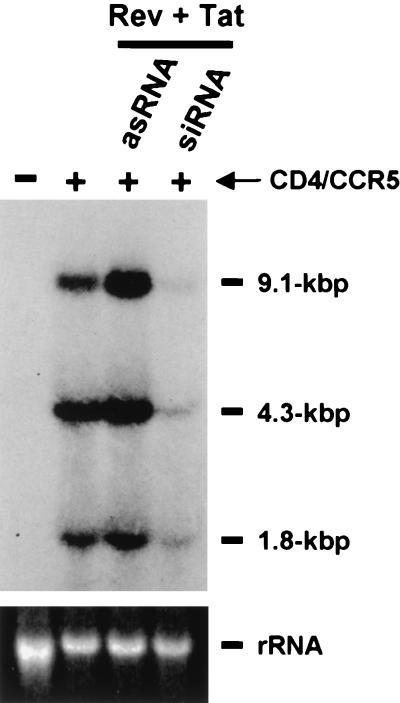
RNAi is predicted to induce the selective degradation of mRNAs containing the appropriate target sequence (37), and we therefore wished to confirm that the siRNAs were indeed reducing the steady-state expression of HIV-1 mRNAs in the infected 293T cells. Although the siRNAs used were targeted to the tat and rev genes of HIV-1, which are encoded by the small, multiply spliced class of HIV-1 mRNAs, it is predicted that the siRNAs would also reduce the expression of the unspliced and singly spliced class of mRNAs contained in HIV-1. This could be achieved either directly, given that many of these mRNAs retain tat and rev sequences within their 3′ untranslated region, or indirectly, by blocking their production by interfering with Tat and/or Rev synthesis. As shown in Fig. 4, we indeed observed a marked reduction in the level of expression of all three classes of HIV-1 mRNAs in the infected CD4+ CCR5+ cells that had been transfected with both siRNAs relative to levels in infected cells that were transfected with only one RNA strand or cells that were mock transfected.
FIG. 4.
Analysis of HIV-1 RNA expression in human cells reveals that tat and rev siRNAs can reduce the expression of all three classes of HIV mRNA. Human 293T cells were cotransfected with siRNA duplexes (si) or with antisense RNAs (as) (0.1 μM each) and the CD4 and CCR5 expression plasmids as indicated at the top of the lanes. CD4+ CCR5+ cells were then infected overnight with 50 ng of p24 antigen. After 48 h of infection, total RNA was extracted from infected 293T cells and subjected to Northern analysis. The approximate sizes of the three classes of HIV-1 transcripts are indicated on the right. 18S rRNA, stained with ethidium bromide, is shown at the bottom of the figure as a loading control.
While the result shown in Fig. 4 is consistent with the hypothesis that the siRNAs are blocking HIV-1 gene expression and replication by inducing the degradation of viral mRNA species, it is also possible that the siRNAs could act to block infection directly by inducing the degradation of the RNA genome present in the infecting HIV-1 virion prior to reverse transcription. To examine this possibility, we quantified the level of HIV-1 proviral DNA production in HIV-1-infected CD4+ CCR5+ 293T cells by Southern dot blot analysis (Fig. 5). The endogenous β-globin gene was used as a loading control.
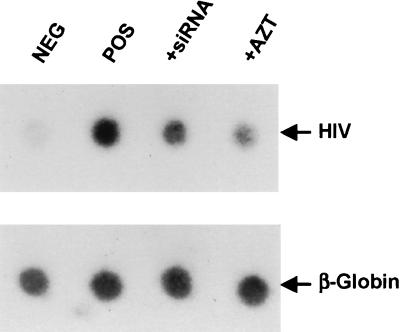
FIG. 5.
siRNAs directed against tat and rev can initiate the degradation of HIV-1 genomic RNA prior to reverse transcription. Human 293T cells were transfected with siRNA duplexes (si) or with antisense RNAs (as). CD4+ CCR5+ cells were infected overnight with 50 ng of p24 antigen. After 48 h, total DNA was extracted from infected 293T cells, denatured in 0.2 M NaOH, spotted onto a nylon membrane, and probed with a 32P-labeled HIV-1-specific probe as described in Materials and Methods. As a control for loading, equivalent amounts of DNA were probed with a 32P-labeled β-globin probe. 293T cells treated with 0.1 μM AZT, the reverse transcriptase inhibitor (2), are also shown (+AZT). DNA isolated from mock-infected 293T cells served as the negative control (NEG), while CD4+ CCR5+ 293T cells infected in the absence of siRNAs served as the positive control (POS).
As shown in Fig. 5, we were indeed able to readily detect HIV-1 proviral DNA in HIV-1-infected CD4+ CCR5+ 293T cells but not in 293T cells lacking CD4 and CCR5 that were incubated with the same infectious virus dose. The level of proviral DNA production was reduced but not totally eliminated in a CD4+ CCR5+ culture treated with the reverse transcriptase inhibitor azidodeoxythymidine (AZT) (2), thus arguing that the bulk of the observed signal does represent newly synthesized viral DNA. Interestingly, treatment of infected cells with the siRNAs also modestly reduced the level of HIV-1 proviral DNA detected. While this result suggests that siRNAs can indeed inhibit HIV-1 infection to some degree, presumably by inducing the degradation of the viral RNA genome prior to completion of reverse transcription, the effect illustrated in Fig. 5 does not appear sufficient to explain the dramatic drop in viral gene expression induced by these siRNAs in HIV-1-infected CD4+ CCR5+ 293T cells (Fig. 3A).
Inhibition of HIV-1 replication in T cells by RNAi.
All of the experiments reported thus far, including the HIV-1 infection experiments, have used human 293T cells as an experimental system. 293T cells are fully competent to support HIV-1 infection when engineered to express the appropriate receptors, and they are readily transfected with synthetic siRNAs. However, these cells may not be representative of the tissues, including CD4+ T cells, that are infected by HIV-1 in vivo.
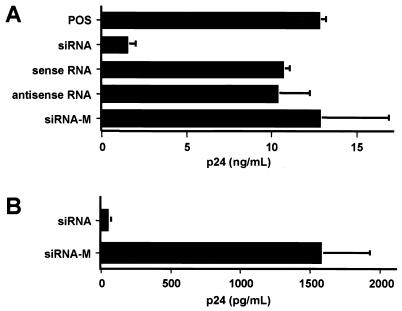
To examine whether RNAi could also inhibit HIV-1 replication in human T cells, we first determined the effect of the siRNAs characterized above on HIV-1 replication in the Jurkat human CD4+ T-cell line. As shown in Fig. 6A, we indeed observed a significant inhibition in progeny virus release from HIV-1-infected Jurkat T cells that had been transfected with the wild-type Tat and Rev siRNAs relative to T cells that were transfected with either RNA strand alone or with control siRNAs bearing a 3-nt missense mutation (Fig. 1A). Importantly, introduction of such central mismatches has been previously reported to block RNAi (15). Next, we asked whether RNAi would inhibit HIV-1 replication in human peripheral blood lymphocytes. In fact, relative to mutant forms of the same siRNA duplexes, the wild-type Tat and Rev siRNAs produced a dramatic drop in the release of progeny virions from HIV-1-infected peripheral blood lymphocytes (Fig. 6B). Therefore, we conclude that RNAi is functional in cells of lymphoid origin and that RNAi can be used to effectively inhibit HIV-1 replication in human T cells in culture.
FIG. 6.
siRNA inhibition of HIV-1 replication in T cells. (A) Jurkat T cells were transfected with a 12 μM concentration of each of the indicated siRNA duplex or single-stranded RNA oligonucleotides. Mock-transfected Jurkat T cells served as the positive control (POS), while the two 3-nt mismatch siRNAs shown in Fig. 1A served as the control (siRNA-M). After 24 h, transfected cells were infected overnight with VSV-G-pseudotyped NL-ADA. Viral production was determined by measurement of p24 Gag production at 48 h after infection. The data shown represent averages for three experiments with the standard deviations indicated by error bars. (B) Human PBMCs were transfected with the wild-type or mutant forms of the Tat and Rev siRNAs by electroporation and were infected with the R5-tropic NL-ADA strain 24 h later. Progeny virus production was measured 48 h after infection by p24 ELISA.
DISCUSSION
The clearance of viral infections from mammalian hosts normally requires pathogen-specific immune responses, such as neutralizing antibodies and cytotoxic T lymphocytes. However, target-nonspecific or innate defenses can also play an important protective role, particularly in the early stages of the infection prior to the clonal expansion of lymphocytes that are able to mount target-specific responses. The most important and best understood innate response to many viral infections is that mounted by the interferon system (reviewed in reference 36). This complex system is designed to block the spread of viruses within the infected host by blocking the production of progeny virions. Importantly, activation of the interferon response can also profoundly inhibit the synthesis of host cell proteins and may even result in the death of the infected cell.
One potent activator of the interferon system is dsRNA molecules that are larger than ∼30 nt (32). Mammalian cells therefore differ from the cells of lower animal species, such as nematodes and fruit flies, which respond to introduced dsRNA molecules primarily by inducing RNAi (16, 21). Indeed, for a long time it remained unclear whether mammalian cells could even mount an RNAi response, given the profound physiological changes that are induced upon activation of the interferon response by longer dsRNAs (7).
The demonstration that synthetic siRNAs of ∼21 nt can induce the specific inhibition of target mRNA expression in human cells without inducing the interferon response (8, 14) has raised the issue of whether RNAi might also play an important role in antiviral defense in mammalian, and particularly human, cells, as indeed has been previously demonstrated in plant cells (12, 34). In this report, we have taken a first step towards addressing this issue by demonstrating that RNAi induced by transfection of synthetic siRNAs can indeed effectively protect human cells, including primary T cells, from the pathogenic retrovirus HIV-1 (Fig. 3 through 6). RNAi was able to potently and specifically inhibit HIV-1 gene expression and replication by inhibiting viral infection (Fig. 5) and by reducing viral mRNA expression (Fig. 4). These data therefore indicate that RNAi has the potential to protect human cells against viral infection, and to prevent virus spread, independently of the normally overlying interferon response.
Clearly, this observation raises a number of important questions, including the following. (i) Is RNAi normally activated by viruses that produce dsRNAs as part of their replication cycle? Such viruses include all RNA viruses as well as some DNA viruses, such as adenovirus. If so, then one would predict that virus-specific siRNAs might be produced in infected cells. (ii) Does induction of RNAi exert an inhibitory effect on virus replication and spread? This question could be addressed in cell culture by using synthetic siRNAs to block proteins, such as dicer, that are known to be essential for de novo siRNA production (21, 25). (iii) Have viruses evolved mechanisms to specifically inhibit RNAi, in addition to the mechanisms that many viruses have evolved to overcome the inhibitory effect of the interferon response on their replication (36)? Ample precedent for such viral defense mechanisms exists in the case of plants (reviewed in reference 29). (iv) Could RNAi be used to protect humans or animals against viral infection? The recent demonstration that biologically active siRNAs can be transcribed from DNA templates (6, 44) suggests that it might be possible to protect human or animal cells against specific virus infections by constitutively expressing siRNAs that are specific for conserved viral sequence elements, as indeed has been previously reported for plants (30). In the case of economically important animal species, it might eventually prove feasible to construct transgenic animals that constitutively express siRNAs that are targeted against important viral pathogens, a potentially powerful variation on the previously proposed concept of “intracellular immunization” (3). In effect, this could render the animal entirely nonpermissive for the targeted virus. Clearly, the potential role of RNAi in antiviral defense is an issue that merits considerable future attention.
Acknowledgments
This research was funded by the Howard Hughes Medical Institute.
We thank Michael Morse for generously providing the PBMCs used in the experiments illustrated in Fig. 6B and Mario Stevenson for helpful discussions.
REFERENCES
- 1.Ancellin, N., C. Colmont, J. Su, Q. Li, N. Mittereder, S. S. Chae, S. Steffansson, G. Liau, and T. Hla.2002. Extracellular export of sphingosine kinase-1 enzyme. Sphingosine 1-phosphate generation and the induction of angiogenic vascular maturation. J. Biol. Chem. 277:6667-6675. [DOI] [PubMed] [Google Scholar]
- 2.Arts, E. J., M. E. Quiñones-Mateu, J. L. Albright, J.-P. Marois, C. Hough, Z. Gu, and M. A. Wainberg.1998. 3′-Azido-3′-deoxythymidine (AZT) mediates cross-resistance to nucleoside analogs in the case of AZT-resistant human immunodeficiency virus type 1 variants. J. Virol. 72:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baltimore, D.1988. Intracellular immunization. Nature 335:395-396. [DOI] [PubMed] [Google Scholar]
- 4.Bogerd, H. P., A. Echarri, T. M. Ross, and B. R. Cullen.1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627-8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, P. D. Bieniasz, and B. R. Cullen.2000. Functional differences between human and bovine immunodeficiency virus Tat transcription factors. J. Virol. 74:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami.2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Caplen, N. J., J. Fleenor, A. Fire, and R. A. Morgan.2000. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene 252:95-105. [DOI] [PubMed] [Google Scholar]
- 8.Caplen, N. J., S. Parrish, F. Imani, A. Fire, and R. A. Morgan.2001. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98:9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coburn, G. A., H. L. Wiegand, Y. Kang, D. N. Ho, M. M. Georgiadis, and B. R. Cullen.2001. Using viral species specificity to define a critical protein/RNA interaction surface. Genes Dev. 15:1194-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. I., B. K. Chen, S. Choe, and N. R. Landau.1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 11.Cortez, D., S. Guntuku, J. Qin, and S. J. Elledge.2001. ATR and ATRIP: partners in checkpoint signaling. Science 294:1713-1716. [DOI] [PubMed] [Google Scholar]
- 12.Covey, S. N., N. S. Al-Kaff, A. Langara, and D. S. Turner.1997. Plants combat infection by gene silencing. Nature 385:781-782. [Google Scholar]
- 13.Cullen, B. R.1998. HIV-1 auxiliary proteins: making connections in a dying cell. Cell 93:685-692. [DOI] [PubMed] [Google Scholar]
- 14.Elbashir, S. M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494-498. [DOI] [PubMed] [Google Scholar]
- 15.Elbashir, S. M., J. Martinez, A. Patkaniowska, W. Lendeckel, and T. Tuschl.2001. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 20:6877-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello.1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 17.Fouchier, R. A. M., B. E. Meyer, J. H. M. Simon, U. Fischer, and M. H. Malim.1997. HIV-1 infection of non-dividing cells: evidence that the amino-terminal basic region of the viral matrix protein is important for Gag processing but not for post-entry nuclear import. EMBO J. 16:4531-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, Z. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Côté, R. L. Rich, D. G. Myszka, and W. I. Sundquist.2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 19.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello.2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 20.Hamilton, A. J., and D. C. Baulcombe.1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286:950-952. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, S. M., E. Bernstein, D. Beach, and G. J. Hannon.2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404:293-295. [DOI] [PubMed] [Google Scholar]
- 22.Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi, and G. J. Hannon.2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293:1146-1150. [DOI] [PubMed] [Google Scholar]
- 23.Harborth, J., S. M. Elbashir, K. Bechert, T. Tuschl, and K. Webe.2001. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 114:4557-4565. [DOI] [PubMed] [Google Scholar]
- 24.Hope, T. J., X. Huang, D. McDonald, and T. G. Parslow.1990. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc. Natl. Acad. Sci. USA 87:7787-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutvágner, G., J. McLachlan, A. E. Pasquinelli, É. Bálint, T. Tuschl, and P. D. Zamore.2001. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 26.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk.2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ketting, R. F., T. H. Haverkamp, H. G. van Luenen, and R. H. Plasterk.1999. Mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner syndrome helicase and RNaseD. Cell 99:133-141. [DOI] [PubMed] [Google Scholar]
- 28.Knight, S. W., and B. L. Bass.2001. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in C. elegans. Science 293:2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, W. X., and S. W. Ding.2001. Viral suppressors of RNA silencing. Curr. Opin. Biotechnol. 12:150-154. [DOI] [PubMed] [Google Scholar]
- 30.Lindbo, J. A., L. Silva-Rosales, W. M. Proebsting, and W. G. Dougherty.1993. Induction of a highly specific antiviral state in transgenic plants: implications for regulation of gene expression and virus resistance. Plant Cell 5:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malim, M. H., J. Hauber, S.-Y. Le, J. V. Maizel, and B. R. Cullen.1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 32.Manche, L., S. R. Green, C. Schmedt, and M. B. Mathews.1992. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol. Cell. Biol. 12:5238-5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martins, L. M., I. Iaccarino, T. Tenev, S. Gschmeissner, N. F. Totty, N. R. Lemoine, J. Savopoulos, C. W. Gray, C. L. Creasy, C. Dingwall, and J. Downward.2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277:439-444. [DOI] [PubMed] [Google Scholar]
- 34.Ratcliff, F., B. D. Harrison, and D. C. Baulcombe.1997. A similarity between viral defense and gene silencing in plants. Science 276:1558-1560. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. W. Russell.2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 36.Sen, G. C.2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 37.Sharp, P. A.2001. RNA interference-2001. Genes Dev. 15:485-490. [DOI] [PubMed] [Google Scholar]
- 38.Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok, L. Timmons, A. Fire, and C. C. Mello.1999. The rde-1 gene, RNA interference and transposon silencing in C. elegans. Cell 99:123-132. [DOI] [PubMed] [Google Scholar]
- 39.Tenllado, F., and J. R. Díaz-Ruíz.2001. Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 75:12288-12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiley, L. S., S. J. Madore, M. H. Malim, and B. R. Cullen.1992. The VP16 transcription activation domain is functional when targeted to a promoter-proximal RNA sequence. Genes Dev. 6:2077-2087. [DOI] [PubMed] [Google Scholar]
- 41.Tokunaga, K., M. L. Greenberg, M. A. Morse, R. I. Cumming, H. K. Lyerly, and B. R. Cullen.2001. Molecular basis for cell tropism of CXCR4-dependent human immunodeficiency virus type 1 isolates. J. Virol. 75:6776-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Westervelt, P., H. E. Gendelman, and L. Ratner.1991. Identification of a determinant within the human immunodeficiency virus 1 surface envelope glycoprotein critical for productive infection of primary monocytes. Proc. Natl. Acad. Sci. USA 88:3097-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zamore, P. D., T. Tuschl, P. A. Sharp, and D. P. Bartel.2000. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101:25-33. [DOI] [PubMed] [Google Scholar]
- 44.Zeng, Y., E. J. Wagner, and B. R. Cullen. 2002. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell, in press.Cell 9:1327-1333. [DOI] [PubMed] [Google Scholar]