Abstract
Although anti-human immunodeficiency virus type 1 (HIV-1) therapy has prolonged the lives of patients, drug resistance is a significant problem. Of particular concern are mutations that cause cross-resistance to a particular class of drugs. Among the mutations that cause resistance to several nucleoside analogs are the insertion of amino acids in the fingers subdomain of HIV-1 reverse transcriptase (RT) at positions 69 and 70. These insertions are usually associated with changes in the flanking amino acids and with a change to F or Y at position 215. We have proposed that the T215F/Y mutation makes the binding of ATP to HIV-1 RT more effective, which increases the excision of 3-azido-3′-deoxythymidine-5′-monophosphate (AZTMP) in vitro and increases zidovudine (AZT) resistance in vivo. Although the mechanism of AZT resistance involves enhanced excision, resistance to 3TC involves a block to incorporation of the analog. We measured the effects of fingers insertion mutations on the misincorporation and excision of several nucleoside analogs. RT variants with the amino acid insertions in the fingers and T215Y have a decreased level of misincorporation of ddATP and 3TCTP. These mutants also have the ability to excise AZTMP by ATP-dependent pyrophosphorylysis. However, unlike the classic AZT resistance mutations (M41L/D67N/K70R/T215Y or F/K219E or Q), the combination of the amino acid insertions in the fingers and the T215Y mutation allows efficient excision of ddTMP and d4TMP, even when relatively high levels of deoxynucleoside triphosphates are present in the reaction. Although the dideoxynucleoside analogs of other nucleosides were excised more slowly than AZTMP, ddTMP, and d4TMP, the mutants with the fingers insertion and T215Y excised all of the nucleoside analogs that were tested more efficiently than wild-type RT or a mutant RT carrying the classical AZT resistance mutations. In the ternary complex (RT/template-primer/dNTP), the presence of the bound dNTP prevents the end of the primer from gaining access to the nucleotide binding site (N site) where excision occurs. Gel shift analysis showed that the amino acid insertions in the fingers destabilized the ternary complex compared to wild-type HIV-1 RT. If the ternary complex is unstable, the end of the primer can gain access to the N site and excision can occur. This could explain the enhanced excision of the nucleoside analogs.
Nucleoside analogs are widely used in anti-human immunodeficiency virus type 1 (HIV-1) therapy, usually in combination with protease inhibitors and/or nonnucleoside reverse transcriptase (RT) inhibitors. The nucleoside analogs used in HIV-1 therapy lack the 3′ OH group found in normal deoxynucleoside triphosphates (dNTPs). The triphosphate forms of the nucleoside analogs are incorporated into viral DNA by HIV-1 RT and once incorporated act as chain terminators, blocking viral DNA synthesis. Therapy with nucleoside analogs selects for resistant viruses; the resistant viruses have mutations in RT. Each of the different nucleoside analogs selects for a particular set of resistance mutations; however, there is often cross-resistance to other nucleoside analogs. The drug-resistant RTs must retain the ability to incorporate normal dNTPs reasonably well, yet have an enhanced ability to discriminate against the nucleoside analogs. This enhanced discrimination can either occur at the incorporation step or, alternatively, the enzyme can selectively excise the analog after it has been incorporated. Resistance to the analog 3TC, for example, involves an enhanced discrimination at the incorporation step and usually arises by either an M184I or M184V mutation in HIV-1 RT; both of these mutations selectively interfere with the incorporation of 3TCTP. The underlying mechanism is steric hindrance; a β-branched amino acid at position 184 clashes with the oxathiolane ring of 3TCTP (11, 23). In contrast, resistance to zidovudine (AZT) involving the M41L, D67N, K70R, T215Y/F, and K219Q/E mutations involves the enhanced excision of 3′-azido-3′-deoxythymidine-5′-monophosphate (AZTMP) by pyrophosphorolysis (3, 5, 7, 16-18). If the pyrophosphate donor in an excision reaction is pyrophosphate itself, then the excision reaction is simply the polymerase reaction run in reverse, leading to the release of a nucleoside analog triphosphate. Because the underlying enzymatic mechanism is the same, mutations that decrease polymerase activity should also decrease pyrophosphate-mediated pyrophosphorylysis. However, there is evidence to suggest that the pyrophosphate donor in the excision reaction is ATP, with the β and γ phosphates of ATP playing the role of pyrophosphate in the excision reaction, which leads to the release of a dinucleotide tetraphosphate. Because the polymerase and excision reactions are related, the efficiency of the excision reaction with ATP is influenced by the level of polymerase activity of HIV-1 RT. However, because the products of the forward polymerase reaction (addition of dNMP to the end of the primer and release of pyrophosphate) differ from the substrates for the reverse excision reaction (ATP and a template-primer), there are mutations that preferentially affect the excision reaction. For example, the T215Y mutation enhances the binding of ATP (but not pyrophosphate) to the HIV-1 RT. Therefore, these are mutations which increase excision not by increasing the polymerase activity of HIV-1 RT but rather by increasing the probability that an ATP will be bound to RT and available for the excision reaction (7, 8).
The mutations that confer AZT resistance (M41L, D67N, K70R, T215Y/F, and K219Q/E) selectively enhance AZTMP excision; the excision of other nucleotide analogs is increased to a much smaller extent. This selective excision appears to be inherent in the interactions of the AZTMP-terminated primer with the structure of HIV-1 RT and the incoming dNTP (7, 8). The AZTMP-terminated primer is readily accommodated in the nucleotide binding site (N site) of HIV-1 RT; however, if the end of the primer is moved (by translocation) to the priming site (P site), there is a potential steric clash between the azido group and D185 and/or one of the active site metals, which causes a displacement in the position of the AZTMP-terminated primer when it resides in the P site compared to a normal, dNMP-terminated primer (G. Sarafianos, unpublished observations). This alteration in the position of the AZTMP-terminated primer interferes with the binding of the incoming dNTP at the N site and with the formation of a closed ternary complex. If a stable closed complex does not form, the AZTMP-terminated primer has good access to the N site, where the AZTMP can be excised. In contrast, a dideoxy-terminated primer is efficiently translocated to the P site, and the incoming dNTP is bound at the N site, forming a stable closed complex and blocking the access of a dideoxy-terminated primer to the N site. This is the reason that AZT resistance is selective for AZT: dideoxy nucleotides are excised from the end of a primer much more slowly than AZTMP because in the presence of dNTPs, they have relatively poor access to the N site (7, 8).
As antiretroviral therapy has become more complex, mutations with multinucleoside resistance profiles have emerged (for a review, see reference 24). Among these are RT variants with insertions in the fingers subdomain between amino acids 67 and 70 (2, 4, 9, 10, 13, 21, 26). The insertion mutations are usually associated with changes in the amino acids flanking the insertion as well as an alteration to Y or F at residue T215. By themselves, some of the fingers insertion mutations appear (at least in vitro) to provide low-level resistance by interfering with the incorporation of ddITP and ddATP (5). The fingers insertions also seem able to cause enhanced excision of thymidine analogs, including AZT (14, 15). However, some of the published excision data are difficult to interpret, in part because the data were obtained with enzymes that contain, in addition to an insertion in the fingers domain, other mutations, including the suite of mutations that are normally associated with enhanced AZT excision or complex suites of mutations present in clinical isolates. Taken together, the published data suggest that some of these additional mutations are important for drug resistance and for the excision reaction; however, it is difficult to try to understand the role(s) of the various mutations from the available data. We have revisited the problem with a relatively simple set of site-directed mutants. These mutants were based on the T69K70→S69SSR70 and T69K70→S69SGR70 mutants that were originally isolated from a patient treated with ddI and hydroxyurea (HU) (9). The T69K70→S69SSR70 mutant appeared first but was replaced by the T69K70→S69SGR70 mutant, suggesting that in the patient undergoing ddI/HU dual therapy, the T69K70→S69SGR70 variant had some growth advantage over the T69K70→S69SSR70 variant. In vitro analysis of these variants suggested that the T69K70→S69SGR70 mutant had some kinetic advantages with normal dNTPs relative to the T69K70→S69SSR70 variant (5). Here we present data which show that the insertion mutations can interfere with the incorporation of certain triphosphate analogs, in particular 3TCTP. In addition, the insertion mutations, in concert with T215Y, a mutation directly associated with ATP binding, can lead to enhanced excision of AZTMP, ddTMP, and d4TMP.
MATERIALS AND METHODS
Preparation of HIV-1 RT.
The open reading frames encoding wild-type HIV-1 RT and each of the RT mutants were cloned into a plasmid containing the HIV-1 PR open reading frame as previously described (6). The plasmid is based on the expression vector pT5m and was introduced into the Escherichia coli strain BL21(DE3)(pLysE). After induction with isopropyl-β-d-thiogalactopyranoside, the plasmid expresses both the p66 form of HIV-1 RT (either wild-type or a mutant) and HIV-1 PR. Approximately 50% of the overexpressed p66 RT is converted to the p51 form by HIV-1 PR, and p66/p51 heterodimers accumulate in E. coli. The p66/p51 heterodimers were purified by metal chelate chromatography (6).
Polymerase assays.
The polymerase assays were done as previously described (6) and were done in duplicate. For each sample, 0.25 μg of single-stranded M13mp18 DNA (New England Biolabs) was hybridized to 0.5 μl of 1.0-optical density/ml −47 sequencing primer (New England Biolabs). The template-primer was suspended in a solution containing 100.0 μl of 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg of bovine serum albumin (BSA)/ml, 10.0 mM 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 2.0 mM dithiothreitol (DTT), 10.0 μM each of dATP, dTTP, and dGTP, 5.0 μM dCTP, 2.0 μM [α-32P]dCTP, and the indicated concentration of inhibitor. Extension was initiated by the addition of 1.0 μg of wild-type RT or RT variant. The mixture was incubated for 30 min at 37o, and then the reaction was halted by the addition of 3 ml of ice-cold 10% trichloroacetic acid. Precipitated DNA was collected by suction filtration through Whatman GF/C glass filters. The amount of incorporated radioactivity was determined by liquid scintillation counting.
Processivity and primer extension assays.
The processivity and primer extension assays have been previously described (6) and were done in duplicate. In brief, for each sample to be assayed, 0.5 μl of 1.0-optical density/ml −47 sequencing primer (New England Biolabs) was 5′ end labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to single-stranded M13mp18 DNA (1.0 μl of a 0.25-μg/μl DNA stock for each sample to be assayed) by heating and slow cooling. The labeled template-primer was resuspended in a solution containing 25 mM Tris (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 100.0 μg of BSA/ml, 10.0 mM CHAPS, and 2.0 mM DTT. One microgram of wild-type RT or RT variant was added to each tube and allowed to bind the labeled template primer for 2 min. Extension was initiated by the addition of dNTPs to a final concentration of 10.0 μM each (for the primer extension assay) or 10.0 μM dNTPs plus 0.5 U of poly(rC) · oligo(dG)/ml (for the processivity assay). The addition of the poly(rC) · oligo(dG) “trap” limits the extension of the labeled primer by HIV-1 RT by binding the RT after it disassociates from the labeled template-primer.
Primer block excision and extension assays.
The primer in these assays is complementary to the HIV-1 primer binding site (PBS) sequence (5′ GTCCCTGTTCGGGCGCCA 3′). The primer was 5′ end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. After purification, the labeled primer was annealed to a fivefold excess of template oligonucleotide that is based on sequence from the U5-PBS region of the HIV-1 genome (5′ AGTCAGTGTGGACAATCTCTAGCAATGGCGCCCGAACAGGGACTTGAAAGCGAAAGTAAA 3′) by heating and slow cooling. The position in italics is normally an A in the pNL 4-3 sequence. It was changed to a C to alter a run of A residues. After the primer is annealed to the template, the underlined A residue will be the first base of the template strand after the double-stranded region. To block the primer, the template-primer was suspended in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, 10.0 mM CHAPS, and 10.0 μM of either 3′-azido 3′-deoxythymidine 5′-triphosphate (AZTTP) (Moravek Biochemicals), 2′,3′ dideoxythymidine 5′-triphosphate (ddTTP) (Boehringer Mannheim), or other analog. Wild-type RT (1.0 μg) was added to the labeled template-primer, and the reactions were allowed to proceed at 370 for 30 min and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume isopropanol, followed by an ethanol precipitation. The blocked template-primer was then resuspended in 25 mM Tris-Cl (pH 8.0), 75 mM KCl, 16.0 mM MgCl2, 2.0 mM DTT, 100 μg of BSA/ml, and 10.0 mM CHAPS. The level of MgCl2 was increased to 16.0 mM from 8.0 mM to ensure that the addition of ATP or sodium pyrophosphate did not bind all of the magnesium ions. The concentration of template-primer was 0.15 nM. Depending upon the experiment (see the figure legends for details), the reaction buffer was supplemented with various amounts of dNTPs, nucleoside analogs (AZTTP or ddTTP), and a pyrophosphate donor (ATP or sodium pyrophosphate). Wild-type RT (1.0 μg) or variant RT was added to each reaction, with a final reaction volume of 50 μl; the approximate concentration of enzyme is 200 nM. The reactions were allowed to proceed for 10 min at 37°C and then halted by phenol-chloroform extraction. The samples were precipitated by the addition of one volume isopropanol, fractionated by electrophoresis on a 15.0% polyacrylamide gel, and autoradiographed. The total amount of template-primer (blocked and unextended plus deblocked and extended) and the amount of full-length product were determined using a PhosphorImager.
The assay for the M13-templated reaction (see Fig. 8) was similar to the assay described above except that the −47 M13 sequencing primer was 5′ labeled. The −47 primer was annealed to single-strand M13mp18 DNA. Unlike the assays described above, the labeled −47 plus M13mp18 template-primer was not blocked by the addition of a nucleoside analog before the excision assay. The labeled template-primer was extended in the presence of 100.0 μM concentrations of each dNTP, 50.0 μM AZTTP, and the indicated amount of ATP. The samples were treated as described above.
Gel shift assay.
This assay was done as previously described with some modifications (11). For each sample, 2.4 × 10−5 nmol of primer identical to the PBS sequence of HIV-1 (5′ GTC CCT GTT CGG GCG CCA 3′) was end labeled with [γ-32P]ATP and T4 polynucleotide kinase. The labeled primer was separated from unincorporated ATP by passage through a Centri-Sep column (Princeton Separations) and then annealed to a threefold excess of template (5′ AGT CAG TGT GGA CAA TCT CTA GCA ATG GCG CCC GAA CAG GGA CTT GAA 3′) by heating and slow cooling. The underlined adenosine base in the template sequence is the first base after the 3′ end of the primer in the annealed template-primer.
The end of the primer strand was blocked by the addition of ddTTP using 1.0 μg of wild-type RT in 1× RT buffer (25 mM Tris-Cl [pH 8.0], 75 mM KCl, 8.0 mM MgCl2, 2.0 mM DTT, 100-μg/ml BSA, 10.0 mM CHAPS, 10.0 μM 2′,3′-dideoxythymidine-5′-triphosphate [ddTTP] [Boehringer Mannheim]). The reaction was allowed to proceed at 370 for 30 min and then halted by phenol-chloroform extraction. Excess ddTTP was removed by passage over a Centri-Sep column. The samples were then precipitated by the addition of one volume of isopropanol followed by an ethanol wash.
For each sample, wild-type or mutant HIV-1 RT (10.0 nM final concentration) was allowed to bind to the labeled, blocked template-primer described above (1.5 nM final concentration) in a solution containing 40.0 mM Tris (pH 8.0), 20.0 mM MgCl2, 60.0 mM KCl, 1.0 mM DTT, 100.0-μg/ml BSA, 10.0 mM CHAPS, 2.5% glycerol, and the indicated amount of dTTP for 5 min at room temperature. The bound complex will be either a binary (RT plus template-primer) or a ternary (RT plus template-primer plus dTTP) complex. To decrease the amount of binary complex, additional KCl was added (to a final concentration of 100 mM) as was an unlabeled chase substrate (poly[rC] · oligo[dG]) to a final concentration of 0.8 U/ml. The mixtures were incubated at 37o for an additional 5 min, and then 4.0 μl of loading buffer (Novex) was added to each reaction. The samples were fractionated by electrophoresis on a 6.0% polyacrylamide DNA retardation gel (Novex). The amount of template-primer in the bound form and the amount free was determined by using a PhosphorImager.
RESULTS
Polymerase activity of the fingers insertion mutants of HIV-1 RT.
As described in the introduction, in an in vitro excision assay the ability of HIV-1 RT to remove a chain terminator and then complete the synthesis of the DNA is related to the polymerase activity. In general, enzymes that have greater polymerase activity should have greater excision activity. For this reason we began by comparing the overall polymerase activity and processivity of wild-type HIV-1 RT and four mutant enzymes. AZT-21 carries five mutations associated with enhanced excision of AZTMP (M41L, D67N, K70R, T215Y, and K219Q). The RT variant T69K70→S69SGR70, which we have designated SSGR, has a two-amino-acid insertion between positions 69 and 70 of the fingers subdomain and has changes in the flanking amino acids. SSGR/T215Y has, in addition to the changes in the fingers, a change at position 215 associated with enhanced ATP binding. SSSR/T215Y, while having the same flanking amino acid changes, has a different insertion in the fingers (T69K70→S69SSR70). It also contains the T215Y mutation. In a simple polymerase assay using an M13 template, the mutant enzymes were all slightly less active than wild-type HIV-1 RT. AZT-21 was the least active (72% of wild-type), followed by SSGR/T215Y (74%), SSGR (90%), and SSSR/T215Y (92%).
The magnitude of the effect of the incorporation of a chain terminator depends on the average chain length produced by the polymerase (20). Therefore, the enzymes were also tested for processivity and primer extension. As expected, all of the mutant enzymes showed a modest decrease in processivity compared to wild-type HIV-1 RT (Fig. 1). The decrease in primer extension was most noticeable for AZT-21 (Fig. 1). These data show that all of the mutant enzymes were reasonably active in both the in vitro processivity and polymerase assays. Similar results were obtained in a low dNTP extension assay (data not shown). Taken together, the data from the polymerase assays show that the mutant enzymes are sufficiently similar to the wild-type enzyme to allow a direct comparison of the resistance data. The data also show that any increase in excision activity for the mutant RTs is not due to an increase in polymerase activity.
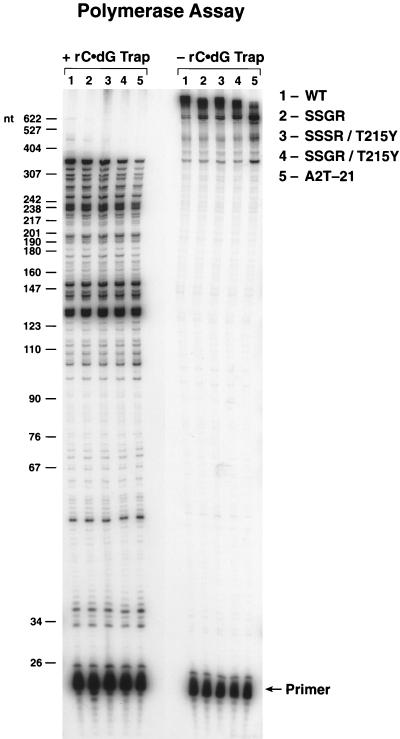
FIG. 1.
Processivity and primer extension of wild-type HIV-1 RT and the RT mutants. The addition of poly(rC) · oligo(dG) traps the HIV-1 RT after it dissociates from the labeled template-primer. This limits RT to one round of extension of the labeled primer and provides a measure of processivity (left side of figure). Without the trap, HIV-1 RT can undergo multiple rounds of primer extension (right side of figure). The reactions were run for 10 min, and the reaction products were fractionated on a 6% polyacrylamide gel. The products were visualized by autoradiography. The scale on the left indicates the sizes of the products, in nucleotides.
Inhibition of polymerization by nucleoside analogs.
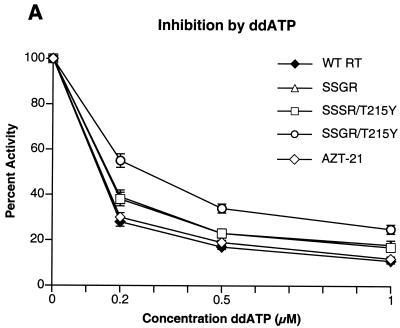
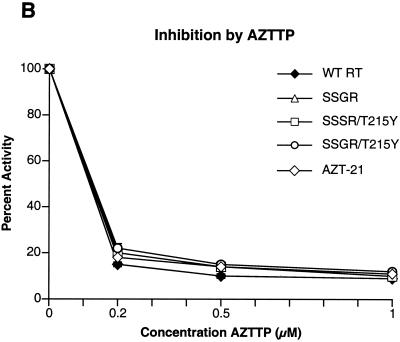
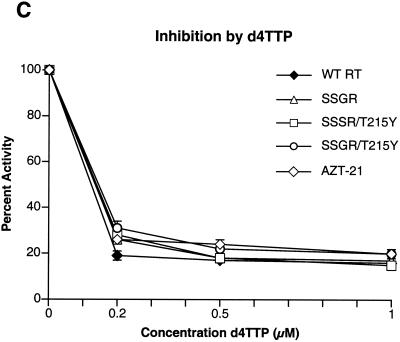
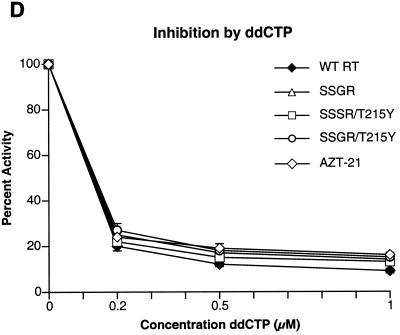
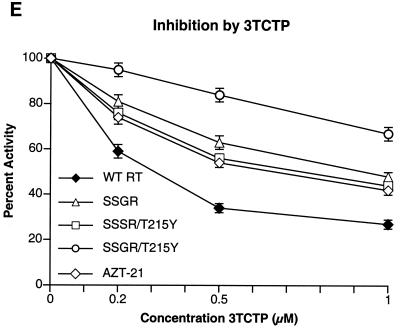
To determine if any of the mutations interfered with the incorporation of nucleoside analog triphosphates, inhibition assays were done with AZTTP, ddATP, ddCTP, 3TCTP, and d4TTP, which are the triphosphate forms of nucleoside analogs approved for HIV-1 therapy. There was little difference in the ability of AZTTP to inhibit wild-type HIV-1 RT and the four mutant enzymes (Fig. 2A). With ddATP, only the SSGR/T215Y mutant appeared to have a modest level of resistance (Fig. 2B); the mutant enzymes do not appear to differ markedly from the wild-type enzyme in their ability to incorporate d4TTP or ddCTP (Fig. 2C and D). Interestingly, all of the mutant enzymes, including AZT-21, showed a significant resistance to 3TCTP; it would appear that the presence of the T215Y mutation enhances the resistance of the SSGR mutant (Fig. 2E). All the simple polymerase inhibition assays were done without a pyrophosphate donor (the dNTP level was relatively low), and 3TCTP is poorly excised by either wild-type or mutant HIV-1 RT (see Fig. 5C), so it is unlikely that the resistance of the mutant enzymes to inhibition by 3TCTP is due to excision (see Discussion). We have previously reported that the mutations in AZT-21 cause modest levels of resistance to 3TCTP in vitro (7, 8); moreover, AZT resistance mutations have been shown to confer a low level resistance to 3TC in viral replication assays (19).
FIG. 2.
Inhibition of polymerase activity by various nucleoside analogs. To simplify the comparisons, the activity of each of the enzymes in the absence of an inhibitor was considered 100%. Polymerase activity in the presence of an inhibitor was normalized to this value. Various concentrations of the analogs were added to polymerization assays containing a −47 primer annealed to a M13mp18 DNA template (see Materials and Methods). After 30 min, the reactions were stopped by the addition of 10% trichloroacetic acid, and the newly synthesized DNA was collected on Whatman GF/C filters. All reactions were done in duplicate. SSGR was plotted for all panels; however data overlap may obscure the data points. Panel A shows the effects of adding ddATP to the polymerization reactions, while panel B shows the effects of AZTTP. Panel C shows the effects of d4TTP; panel D is for ddCTP; panel E is for 3TCTP.
FIG. 5.
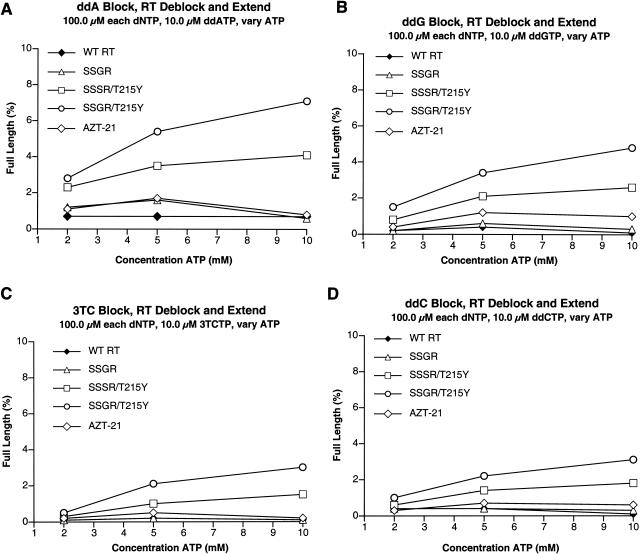
Comparison of the abilities of the various RTs to excise other nucleoside analogs. Note the change in the scale of the percent full length (y axis). All reactions were carried out in duplicate. Reactions run with ddAMP (A), ddGMP (B), 3TCMP (C), or ddCMP (D) as the primer blocking group are shown.
Excision of nucleoside analogs.
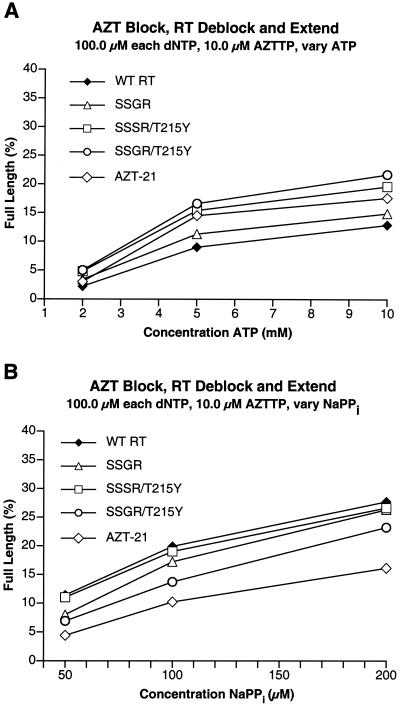
We compared the abilities of wild-type HIV-1 RT, AZT-21, SSGR, SSSR/T215Y, and SSGR/T215Y to excise AZTMP in the presence of high levels of dNTPs and either ATP or pyrophosphate. To minimize any effects of polymerase activity and/or processivity on the excision reaction, we used a relatively short template extension (25 nucleotides). The reactions contained the triphosphate form of the nucleoside analog, which allowed multiple blocking and excision reactions to occur. We have shown that the efficiency of the excision reaction can vary at different positions along a template (8). Multiple excision reactions will tend to average out the efficiency of events at individual sites and will also mimic the situation in a patient where multiple blocks will occur along the viral template. As previously described (7, 8), in the presence of ATP, AZT-21 shows enhanced excision of AZTMP compared to wild-type RT (Fig. 3A). The SSGR mutant was more efficient in carrying out AZTMP excision than wild-type HIV-1 RT but less efficient than AZT-21 (Fig. 3A). However, the SSSR/T215Y mutant was more efficient in excising AZTMP than AZT-21, and the SSGR/T215Y mutant was better than SSSR/T215Y (Fig. 3A). In the presence of pyrophosphate, wild-type HIV-1 RT was the most efficient at AZTMP excision (Fig. 3B). The ability of the enzymes to excise AZTMP with pyrophosphate appears to be related to their relative activities in simple polymerase assays (see Discussion).
FIG. 3.
Comparison of the abilities of the various HIV-1 RTs (wild-type and mutant) to excise AZTMP from the end of the primer and extend the primer in the presence of AZTTP, high concentrations of dNTPs, and various concentrations of a pyrophosphate donor. The primer is fully blocked with AZTMP at the beginning of the reaction (see Materials and Methods). A short (25-nucleotide-long) template extension provides additional opportunity for the enzymes to incorporate AZTTP and excise AZTMP. This will tend to average out the events at individual sites. The assay measures the percentage of the primer that is completely extended by each enzyme at the various pyrophosphate donor concentrations. All assays were done in duplicate. Panel A shows the results obtained when the pyrophosphate donor is ATP; Panel B shows the results when sodium pyrophosphate (NaPPi) is the pyrophosphate donor.
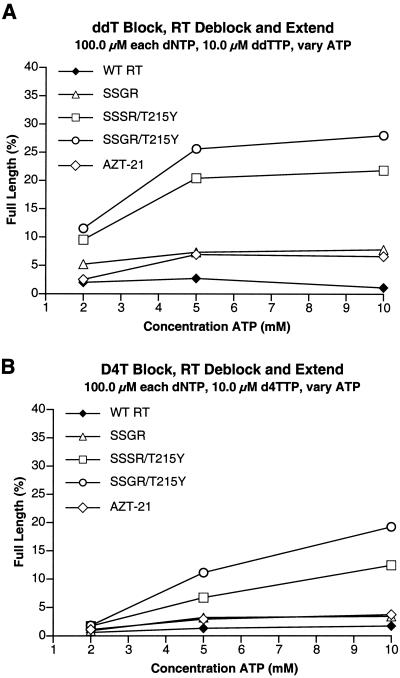
There were more dramatic differences in the abilities of the mutant enzymes to excise other thymidine analogs in the presence of ATP. As has already been discussed, previous studies (7, 8, 16-18) showed that a sufficiently high level of dNTPs will block the excision of dideoxy nucleotide analogs with either wild-type RT or an RT containing the classic AZT-resistance mutations (M41L, D67N, K70R, T215F/Y, and K219Q/E). The data obtained with d4TTP and ddTTP were, in general, quite similar. As expected, the wild-type enzyme is relatively inefficient at excising either ddTMP or d4TMP in the presence of high levels of dNTPs (Fig. 4A and B). The AZT-21 and SSGR mutants were slightly better than wild-type RT, but still quite inefficient. However, both the SSSR/T215Y and the SSGR/T215Y mutants can excise either ddTMP or d4TMP reasonably well when ATP is the pyrophosphate donor; the SSGR/T215Y enzyme is more efficient than SSSR/T215Y (Fig. 4A and B). Although the relative excision efficiencies of the enzymes were similar for ddTMP and d4TMP, the absolute level of excision/extension was higher for ddTMP than for d4TMP, suggesting that d4TMP is somehow more difficult to excise.
FIG. 4.
Comparison of the abilities of the various RTs to excise other thymidine analogs. The assay conditions are similar to those described in the legend to Fig. 3 except that instead of an AZTMP-terminated primer, the primer was terminated with either ddTMP or d4TMP, and in the reactions, a 10 μM concentration of the triphosphate form of the indicated nucleoside analog was used instead of AZTTP. ATP was the pyrophosphate donor in all reactions, which were run in duplicate. Panel A shows the results of reactions run with ddTMP as the blocking group on the primer. Panel B corresponds to reactions run with d4TMP as the blocking group on the primer.
Since SSSR/T215Y and SSGR/T215Y were able to excise ddTMP and d4TMP from the end of the primer even in the presence of high dNTP concentrations, we examined the abilities of the enzymes to excise other nucleoside analogs from the 3′ end of the primer. Because ddATP is the active metabolite of ddI (1) and the SSSR and SSGR insertions were selected by treatment with ddI/HU, we first tested the various RTs for their ability to excise ddAMP from the primer. Although the excision of ddAMP was less efficient than the excision of ddTMP or d4TMP for all of the enzymes (Fig. 5A), the relative excision efficiencies of the various enzymes were similar for ddTMP, d4TMP, and ddAMP. The insertion mutants with T215Y (in particular SSGR/T215Y) were more efficient than wild-type RT, SSGR, or AZT-21 (Fig. 5A). We also tested the abilities of the various enzymes to excise ddGMP (Fig. 5B), 3TCMP (Fig. 5C), and ddCMP (Fig. 5D). All were excised poorly; however, the SSGR/T215Y mutant was always the most efficient, and SSSR/T215Y always the next best at excising the various nucleoside analogs (Fig. 5B to D). Taken together, these data suggest two things. First, enhanced ATP binding (the presence of T215Y) is important for the fingers insertions to carry out efficient excision. Second, the fingers insertion somehow allows the ddTMP, d4TMP, and ddAMP terminated primers better access to the N site, where excision takes place, even in the presence of high dNTP concentrations. One possibility is that the fingers mutations decrease the stability of the closed complex. We used gel shift assays to test this idea.
Stability of a closed complex with a ddTMP terminated primer.
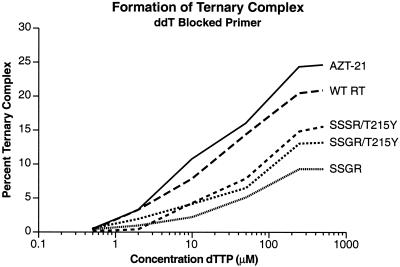
The stability of a closed complex formed with a ddTMP-terminated primer was measured using a gel shift assay (11, 25). In this assay, the ternary complex (RT/DNA/dNTP) was assembled in a low-salt environment, but before the complexes were fractionated on the gel, a large amount of salt was added to disrupt any binary (RT/DNA) complexes (11). The ternary (closed) complex is more stable than the binary (RT/DNA) complex and can withstand higher salt levels. In this assay, the AZT-21 mutant formed the most stable closed complex (Fig. 6). The wild-type complex was also relatively stable. The fingers insertions clearly destabilized the closed complex. The closed complex appeared to be less stable for SSGR/T215Y, which had the highest level of nucleoside analog excision, than for SSSR/T215Y, which was next in stability. The closed complex for SSGR was less stable than those for the SSSR/T215Y or SSGR/T215Y variants, suggesting that the presence of T215Y may stabilize the closed complex (Fig. 6). Since the AZT-21 mutations include T215Y, it may be that it is the T215Y mutation that contributes to the enhanced stability of the closed complex formed with the AZT-21 enzyme.
FIG. 6.
Detection of the ternary complex by gel electrophoresis. As described in Materials and Methods, a 5′-end-labeled primer was annealed to a template by heating and slow cooling. The end of the primer strand was then blocked by the addition of ddTMP. The blocked template-primer was then purified. For each sample, wild-type or mutant RT was allowed to bind to labeled ddTMP blocked template-primer in 1× RT binding buffer plus the indicated concentration of dTTP (0 to 500 μM) for 5 min at room temperature. The salt concentration was then increased to 100 mM KCl, and an unlabeled chase substrate, poly(rC) · oligo(dG), was added. The reactions were incubated at 37o for 5 min and then fractionated on a 6% Novex polyacrylamide DNA retardation gel. The percentage of gel-shifted complex was determined by a PhosphorImager.
DISCUSSION
The AZT-21 mutations cause high-level resistance to AZT but not other nucleoside analogs. As expected, in an appropriate in vitro assay, the AZT-21 mutant excises AZTMP more efficiently than wild-type HIV-1 RT. The enhanced excision is selective for AZTMP; AZT-21 is not much more efficient than wild-type HIV-1 RT at excising other nucleoside analogs. This is the expected result. We believe that the primary role of the AZT-21 mutations is to enhance the binding of ATP, which serves as the pyrophosphate donor in the excision reaction. What determines the propensity of both wild-type HIV-1 RT and the AZT-21 mutant to selectively excise AZTMP is the steric conflict that occurs when the AZTMP-terminated primer is translocated from the N site to the P site. This decreases the stability of the closed ternary complex with an AZTMP-terminated primer. As a consequence, the AZTMP-terminated primer has better access to the N site, where it can be excised. A dideoxy-terminated primer can form a relatively stable closed complex; because the complex is more stable, the end of the primer has little access to the N site and the excision of a ddNMP from the end of a primer is low.
In contrast, the fingers insertions lead to the enhanced excision of AZTMP, ddTMP, d4TMP, and ddAMP. The excision reaction is still ATP dependent; the T215Y mutation, which we believe is directly involved in ATP binding, strongly enhances the excision reaction carried out by mutant RTs carrying the fingers insertion. These data suggest that the fingers insertion somehow increases the ability of the 3′ end of a primer to access the N site. The gel shift experiments suggest that this increased access is due, at least in part, to the fact that the fingers insertions decrease the stability of the closed (or ternary) complex. This, in turn, will increase the access of the end of the primer to the N site. This type of model predicts that the fingers insertion would have a much greater effect on the ability of the enzyme to excise d4TMP and ddTMP (which have restricted access to the N site) than on the excision of AZTMP (which already has good access to the N site); the data match the model.
We tested several other nucleoside analogs and found that while the relative abilities of the various RTs to excise the different analogs was quite similar (SSGR/T215Y showed greater ability than SSSR/T215Y, which showed greater ability than SSGR, which showed approximately the same ability as AZT-21, which showed greater ability than WT RT), there were dramatic differences in the overall efficiencies of excision. For the dideoxy analogs, ddTMP and d4TMP were excised more readily than ddAMP, which in turn was excised more readily than ddGMP. ddCMP and 3TCMP were excised quite poorly. The assay we used involved both the incorporation of the triphosphate form of the nucleoside analog and the excision of the monophosphate from the 3′ end of the primer, so it is theoretically possible that the ability of the enzymes to incorporate the inhibitor could influence the outcome of the assay. However, with the exception of ddATP and 3TCTP, the inhibition curves (polymerization in the absence of ATP) were sufficiently similar that it is unlikely that differential incorporation of the analogs affected the excision data significantly. This means that both the nature of the base and modifications on the deoxyribose ring can affect the efficiency of excision. Presumably, this reflects the ability of the different analogs to bind in a position appropriate for excision.
As described in the introduction, a patient who had the T215Y mutation at the baseline time point was treated with ddI/HU. The SSSR/T215Y variant appeared first and then was replaced with the SSGR/T215Y mutant (9). This would suggest that the SSGR/T215Y variant is better able to replicate, at least in this patient undergoing ddI/HU treatment, than the SSSR/T215Y variant. Previous analyses indicated that in the absence of the T215Y mutation, the SSGR variant had some advantages in in vitro polymerase assays relative to the SSSR variant (5). Based on the assays reported here, it would also appear that the SSGR/T215Y mutant is better able to excise AZTMP, d4TMP, and the various dideoxy analogs than is the SSSR/T215Y variant. The closed complex is also less stable for the SSGR/T215Y variant than for the SSSR/T215Y mutant, which could explain the differences in excision efficiency for these two mutants.
These observations have implications for normal polymerization. The fact that mutations that decrease the stability of the closed complex have only a modest effect on polymerization suggests that for the wild-type enzyme, the stability of the closed complex is greater than what is necessary for reasonably efficient polymerization. This is not an unreasonable idea; during normal polymerization the closed state is transient. During polymerization, the chemistry step, with the concomitant release of the pyrophosphate, destabilizes the closed complex, allowing translocation to occur. As a consequence, the stability of the closed state is actually a much more important issue for determining the access of the end of the primer to the N site in the excision reaction than it is for normal polymerization.
In addition to affecting the excision of nucleoside analogs after they have been incorporated, the fingers insertion mutations also have an effect on the incorporation of some nucleoside analogs, in particular on the incorporation of 3TCTP. Resistance to 3TCTP usually involves a β-branched amino acid at position 184 in HIV-1 RT. However, the M184A mutation (which has not yet been isolated from patients) provides significant resistance to 3TCTP in vitro (6). The equivalent mutation in murine leukemia virus RT confers resistance to 3TC in vivo and to 3TCTP in vitro (6, 12). We have suggested that despite the fact that alanine has no β-branch, changes in the active site (either in the protein itself, in the position of the nucleic acid, or both) still lead to some sort of steric clash which interferes with the incorporation of 3TCTP. We believe that the fingers insertions cause something similar to occur. This would mean that the fingers insertions are altering the active site directly or indirectly. This type of proposal is supported by the data which show that the fingers insertions affect the stability of the closed complex. The data also show that the T215Y mutation can influence the ability of the enzyme to incorporate 3TCTP and that this mutation affects the stability of the closed complex. These are more surprising observations. The fingers insertions are located in the β3/β4 loop. This loop closes down directly onto the dNTP binding site. It is easy to imagine how the insertion of the two amino acids in the fingers at positions 69 and 70, together with the changes in the flanking amino acids (T69K70→S69SSR70 or T69K70→S69SGR70), could affect the interactions of the β3/β4 loop and the polymerization substrates. In contrast, the T215Y mutation is relatively distant from the polymerase active site. However, the T215Y mutation affects both the resistance to 3TCTP (at the level of incorporation) and the stability of the closed complex, both of which suggest that the T215Y mutation has some effect on the structure at or near the polymerase active site. It was previously reported that the mutation T215F caused alterations in the structure of the polymerase active site (22). However, the claim is based on an HIV-1 RT structure that did not include a template-primer and included a nonnucleoside inhibitor bound to the protein. It is unclear whether the reported changes in the structure would have any relevance to dNTP binding or 3TC resistance.
Our results also raise the question of what level of resistance to misincorporation of a nucleoside analog is sufficient to cause resistance in vivo. The level is probably different for each nucleoside analog. The AZT resistance mutations that comprise AZT-21 cause moderate resistance to 3TCTP in vitro and have been reported to cause low-level resistance to 3TC in vivo; however, 3TC treatment selects for M184I/V, not the suite of mutations found in AZT-21. A number of factors contribute to which mutation(s) is actually selected in vivo, including effects on viral fitness, number of nucleotide and/or amino acid changes needed to obtain resistance, and the fold change in susceptibility.
The data also show that in the case of HIV-1 RT, a particular mutant enzyme can cause both a decrease in the ability of HIV-1 RT to incorporate the triphosphate form of certain nucleoside analogs and an increase in the excision of other analogs, which makes both the task of understanding the exact molecular mechanism(s) that engenders drug resistance and the design of more effective inhibitors quite difficult challenges.
Acknowledgments
We thank Pat Clark for preparing purified wild-type and mutant RT and Hilda Marusiodis for help in preparing the manuscript.
Research in S.H.H.'s laboratory was supported by the National Cancer Institute and NIGMS. Research in E.A.'s laboratory was supported by grants AI 27690 and GM 55609 from the National Institutes of Health.
REFERENCES
- 1.Ahluwalia, G., D. A. Cooney, H. Mitsuya, A. Fridland, K. P. Flora, Z. Hao, M. Dalal, S. Broder, and D. G. Johns. 1987. Initial studies on the cellular pharmacology of 2′,3′-dideoxyinosine, an inhibitor of HIV infectivity. Biochem. Pharmacol. 36:3797-3800. [DOI] [PubMed] [Google Scholar]
- 2.Andréoletti, L., L. Weiss, A. Si-Mohamed, C. Piketty, T. Prazuck, G. Calamy, J.-E. Malkin, M. Matta, F.-X. Mbopi-Kéou, F. Clavel, M. D. Kazatchkine, and L. Bélec. 2002. Multidrug-resistant HIV-1 RNA and proviral DNA variants harboring new dipeptide insertions in the reverse transcriptase pol gene. J. Acquir. Immune Defic. Syndr. 29:102-104. [DOI] [PubMed] [Google Scholar]
- 3.Arion, D., N. Kaushik, S. McCormick, G. Borkow, and M. A. Parniak. 1998. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry 37:15908-15917. [DOI] [PubMed] [Google Scholar]
- 4.Bonfanti, P., I. Faggion, S. L. S. Catamancio, M. Violin, C. Balotta, and S. Rusconi. 2000. Response to antiretroviral therapy in a patient with an uncommon codon 69 insertion in the human immunodeficiency virus type 1 reverse transcriptase. Antimicrob. Agents Chemother. 44:1767-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer, P. L., J. Lisziewicz, F. Lori, and S. H. Hughes. 1999. Analysis of amino acid insertion mutations in the fingers subdomain of HIV-1 reverse transcriptase. J. Mol. Biol. 286:995-1008. [DOI] [PubMed] [Google Scholar]
- 6.Boyer, P. L., H.-Q. Gao, P. K. Clark, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. YADD mutants of human immunodeficiency virus type 1 and Moloney murine leukemia virus reverse transcriptase are resistant to lamivudine triphosphate (3TCTP) in vitro. J. Virol. 75:6321-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2001. Selective excision of AZTMP by drug-resistant human immunodeficiency virus reverse transcriptase. J. Virol. 75:4832-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, P. L., S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2002. The M184V mutation reduces the selective excision of zidovudine 5′-monophosphate (AZTMP) by the reverse transcriptase of human immunodeficiency virus type 1. J. Virol. 76:3248-3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Antoni, A., A. Foli, J. Lisziewicz, and F. Lori. 1997. Mutations in the pol gene of human immunodeficiency virus type 1 in infected patients receiving didanosine and hydroxyurea combination therapy. J. Infect. Dis. 176:899-903. [DOI] [PubMed] [Google Scholar]
- 10.de Jong, J. J., J. Goudsmit, V. V. Lukashov, M. E. Hillebrand, E. Baan, R. Huismans, S. A. Danner, J. H. ten Veen, F. de Wolf, and S. Jurriaans. 1999. Insertion of two amino acids combined with changes in reverse transcriptase containing tyrosine-215 of HIV-1 resistant to multiple nucleoside analogs. AIDS 13:75-80. [DOI] [PubMed] [Google Scholar]
- 11.Gao, H.-Q., P. L. Boyer, S. G. Sarafianos, E. Arnold, and S. H. Hughes. 2000. The role of steric hindrance in 3TC resistance of human immunodeficiency virus type-1 reverse transcriptase. J. Mol. Biol. 300:403-418. [DOI] [PubMed] [Google Scholar]
- 12.Halvas, E. K., E. S. Svarovskaia, E. O. Freed, and V. K. Pathak. 2000. Wild-type YMDD mutant murine leukemia virus reverse transcriptases are resistant to 2′,3′-dideoxy-3′-thiacytidine. J. Virol. 74:6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larder, B. A., S. Bloor, S. D. Kemp, K. Hertogs, R. L. Desmet, V. Miller, M. Sturmer, S. Staszewski, J. Ren, D. K. Stammers, D. I. Stuart, and R. Pauwels. 1999. A family of insertion mutants between codons 67 and 70 of human immunodeficiency virus type 1 reverse transcriptase confer multinucleoside analog resistance. Antimicrob. Agents Chemother. 43:1961-1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lennerstrand, J., K. Hertogs, D. K. Stammers, and B. A. Larder. 2001. Correlation between viral resistance to zidovudine and resistance at the reverse transcriptase level for a panel of human immunodeficiency virus type 1 mutants. J. Virol. 75:7202-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mas, A., M. Parera, C. Briones, V. Soriano, M. A. Martínez, E. Domingo, and L. Menéndez-Arias. 2000. Role of a dipeptide insertion between codons 69 and 70 of HIV-1 reverse transcriptase in the mechanism of AZT resistance. EMBO J. 19:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyer, P. R., S. E. Matsurra, A. G. So, and W. A. Scott. 1998. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc. Natl. Acad. Sci. USA 95:13471-13476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meyer, P. R., S. E. Matsurra, A. M. Mian, A. G. So, and W. A. Scott. 1999. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol. Cell 4:35-43. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, P. R., S. E. Matsurra, R. F. Schinazi, A. G. So, and W. A. Scott. 2000. Differential removal of thymidine nucleotide analogues from blocked DNA chains by human immunodeficiency virus reverse transcriptase in the presence of physiological concentrations of 2′-deoxynucleoside triphosphates. Antimicrob. Agents Chemother. 44:3465-3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quan, Y., L. Rong, C. Liang, and M. A. Wainberg. 1999. Reverse transcriptase inhibitors can selectively block synthesis of differently sized viral DNA transcripts in cells acutely infected with human immunodeficiency virus type 1. J. Virol. 73:6700-6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rakik, A., M. Ait-Khaled, P. Griffin, D. A. Thomas, M. Tisdale, and J.-P. Kleim. 1999. A novel genotype encoding a single amino acid insertion and five other substitutions between residues 64 and 74 of the HIV-1 reverse transcriptase confers high-level cross-resistance to nucleoside reverse transcriptase inhibitors. J. Aquir. Immune Defic. Syndr. 22:139-145. [DOI] [PubMed] [Google Scholar]
- 22.Ren, J., R. M. Esnouf, A. L. Hopkins, E. Y. Jones, I. Kirby, J. Keeling, C. K. Ross, B. A. Larder, D. I. Stuart, and D. K. Stammers. 1998. 3′-azido-3′-deoxythymidine drug resistance mutations in HIV-1 reverse transcriptase can induce long range conformational changes. Proc. Natl. Acad. Sci. USA 95:9518-9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sarafianos, S., K. Das, A. D. Clark, Jr., J. Ding, P. L. Boyer, S. H. Hughes, and E. Arnold. 1999. 3TC (lamivudine) resistance in HIV-1 reverse transcriptase involves steric hindrance with β-branched amino acids. Proc. Natl. Acad. Sci. USA 96:10027-10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schinazi, R. F., B. A. Larder, and J. W. Mellors. 2000. Mutations in retroviral genes associated with drug resistance: 2000-2001 update. Int. Antivir. News 8:65-91. [Google Scholar]
- 25.Tong, W., C.-D. Lu, S. K. Sharma, S. Matsuura, A. G. So, and W. A. Scott. 1997. Nucleotide-induced stable complex formation by HIV-1 reverse transcriptase. Biochemistry 36:5749-5757. [DOI] [PubMed] [Google Scholar]
- 26.Winters, M. A., K. L. Coolley, Y. A. Girard, D. J. Levee, H. Hamdan, R. W. Schafer, D. A. Katzenstein, and T. C. Merigan. 1998. A 6-basepair insert in the reverse transcriptase gene of human immunodeficiency virus type 1 confers resistance to multiple nucleoside inhibitors. J. Clin. Investig. 102:1769-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]