Abstract
The complete nucleotide sequences of genomic RNA1 (9,407 nucleotides [nt]) and RNA2 (8,223 nt) of Sweet potato chlorotic stunt virus (SPCSV; genus Crinivirus, family Closteroviridae) were determined, revealing that SPCSV possesses the second largest identified positive-strand single-stranded RNA genome among plant viruses after Citrus tristeza virus. RNA1 contains two overlapping open reading frames (ORFs) that encode the replication module, consisting of the putative papain-like cysteine proteinase, methyltransferase, helicase, and polymerase domains. RNA2 contains the Closteroviridae hallmark gene array represented by a heat shock protein homologue (Hsp70h), a protein of 50 to 60 kDa depending on the virus, the major coat protein, and a divergent copy of the coat protein. This grouping resembles the genome organization of Lettuce infectious yellows virus (LIYV), the only other crinivirus for which the whole genomic sequence is available. However, in striking contrast to LIYV, the two genomic RNAs of SPCSV contained nearly identical 208-nt-long 3′ terminal sequences, and the ORF for a putative small hydrophobic protein present in LIYV RNA2 was found at a novel position in SPCSV RNA1. Furthermore, unlike any other plant or animal virus, SPCSV carried an ORF for a putative RNase III-like protein (ORF2 on RNA1). Several subgenomic RNAs (sgRNAs) were detected in SPCSV-infected plants, indicating that the sgRNAs formed from RNA1 accumulated earlier in infection than those of RNA2. The 5′ ends of seven sgRNAs were cloned and sequenced by an approach that provided compelling evidence that the sgRNAs are capped in infected plants, a novel finding for members of the Closteroviridae.
Sweet potato chlorotic stunt virus (SPCSV; previously also known as sweet potato sunken vein virus) belongs to the genus Crinivirus in the family Closteroviridae and is a pathogen of sweet potato (Ipomoea batatas L.) (23, 35, 61). It is a required component for the manifestation of several severe viral diseases caused by coinfection with other viruses (9, 13, 23). For example, sweet potato virus disease is caused by coinfection with SPCSV and Sweet potato feathery mottle virus (genus Potyvirus) and is the major disease of sweet potato in Africa and perhaps worldwide (8).
Viruses of the family Closteroviridae are phloem limited and transmitted in a semipersistent manner by specific homopteran vectors (1, 31). For instance, SPCSV is phloem limited in sweet potato plants (35) and is transmitted by whiteflies (10, 56). In the flexuous filamentous virions, the viral genome of Closteroviridae is encapsidated by two types of coat protein (CP) arranged in a polar configuration (3, 21, 58). These viruses have the largest and the most complex positive-strand single-stranded RNA (ssRNA) genomes of plant viruses. They always contain the domains for a papain-like proteinase (P-Pro), methyltransferase, and helicase (open reading frame 1a [ORF1a]) and an RNA-dependent RNA polymerase (RdRp) (ORF 1b).
The Closteroviridae hallmark gene array consists of ORFs for a putative small hydrophobic protein (SHP), a heat shock protein homologue (Hsp70h), a protein of 50 to 60 kDa depending on the virus, the CP, and the minor CP (CPm), which are, like most proteins in Closteroviridae, translated from subgenomic RNAs (sgRNAs) (2, 20, 24, 30, 33, 34, 37, 41, 43, 65). The genomic sequence of SPCSV is unknown except for partial sequences of Hsp70h and CP gene sequences from a few isolates (4, 28, 61).
The family Closteroviridae contains two genera. The genus Closterovirus encompasses all viruses with a monopartite genome, whereas the genus Crinivirus consists of viruses with a bipartite genome (59). The genomic sequences of five closteroviruses (2, 30, 33, 43, 65) and the partial genomic sequences of four other closteroviruses (20, 24, 34, 41) indicate a diverse genus, prompting calls for revision of the classification (31). In contrast, diversity among the criniviruses is largely unknown, since the only reported crinivirus genome sequence available is that of Lettuce infectious yellows virus (LIYV) (37).
The rapidly increasing number of viruses in the genus Crinivirus (reviewed in reference 62) and the very significant negative impact of SPCSV on sweet potato production prompted us to characterize the complete sequence of the SPCSV genome. After Citrus tristeza virus (CTV), SPCSV appears to have the second largest genome of all plant viruses containing a positive-strand ssRNA genome. Analyses of the two genomic RNAs revealed several new features compared to the genome of LIYV, such as the presence of a unique ORF that putatively encodes a protein belonging to the RNase III family, the novel genomic position of the small hydrophobic protein characteristic of Closteroviridae, and identical 3′ regions with predicted stable secondary structures on RNA1 and RNA2. Our study also revealed seven sgRNAs as well as evidence that these sgRNAs contain 5′ caps. Differential accumulation of sgRNAs at different stages of infection in the leaves of Ipomoea setosa was observed.
MATERIALS AND METHODS
Plant material and virus isolate.
The SPCSV isolate used in this study belongs to the serotype East Africa 2, as determined with anti-CP monoclonal antibodies according to the methods described by Alicai et al. (4). It was isolated by whitefly transmission from a sweet potato virus disease-affected sweet potato plant (Mpigi district, Uganda) (35) and maintained in the sweet potato cultivar Tanzania. The highly susceptible host species I. setosa was grown from seeds obtained from the International Potato Center (Lima, Peru). Plants were inoculated by grafting with infected scions of sweet potato cv. Tanzania and grown in a 25 to 30°C greenhouse at the Swedish University of Agricultural Sciences (Uppsala, Sweden). Sodium halide lamps were used to extend day length to 16 h during the winter.
Cloning and sequencing of SPCSV-specific reverse transcription (RT)-PCR products.
SPCSV virions were purified from infected plants of I. setosa (10), and RNA was extracted from the virions (36). RNA concentration was determined by absorbance at 260 nm with a UV spectrophotometer. For amplification of the 3′ ends of viral genomic RNAs, 100 ng of purified viral RNA was polyadenylated with a poly(A)-polymerase (Amersham Biosciences AB, Uppsala, Sweden) according to the manufacturer's instructions. Purified viral RNA or purified polyadenylated viral RNA was then used to generate cDNA with 8-mer random primers or an anchored poly(dT) primer (5′ GCT GTC AAC GAT ACG CTA CGT AAC GGC ATG ACA GTG [T]18-3′), respectively, and RNase H-free reverse transcriptase (Superscript II; Invitrogen Ltd., Paisley, United Kingdom) according to the manufacturer's instructions.
Initially, the CP gene on RNA2 and a part of the RdRp gene on RNA1 were PCR amplified with primers CP1 and CP2 (4) and degenerate primers RdRpIV-F (5′ GAA/G ATA/T/C GAC/T TTC/T AAAA/TC/GN TTC/T GAC/T AAA/G-3′) and RdRpVI-R (5′ C/GGA A/GA/TA AAT NAA NC/GA/TA/GTC A/GTC NCC-3′), respectively. Based on the nucleotide sequences of these fragments, new specific primers were designed and used together with degenerate primers to amplify the remainder of the genome with a step-by-step walking strategy. The 3′ ends of the RNAs were amplified from the anchored poly(dT) cDNA with the anchor primer (5′ GCT GTC AAC GAT ACG CTA CGT AAC-3′) and upstream primers specific for each RNA molecule.
The 5′ ends of RNAs were amplified with the GeneRacer kit (Invitrogen Ltd.), which is designed to amplify only 5′ ends of m7GpppN-capped RNA molecules. Briefly, the procedure involves (i) dephosphorylation of the 5′ ends of noncapped RNAs with calf intestinal phosphatase, (ii) removal of the cap structure from the capped RNAs with tobacco acid pyrophosphatase, thereby exposing the 5′ end, and (iii) ligation of an RNA oligonucleotide to the 5′ phosphate groups of the decapped RNAs with T4 RNA ligase. The 5′ ends of putative sgRNAs were also amplified from total RNA extracted from leaves of SPCSV-infected I. setosa plants with the GeneRacer kit along with virus-specific primers designed to anneal 300 to 700 nt downstream from the expected 5′ end of the sgRNA. To verify that only RNAs that were originally capped were amplified by this method, a control reaction was performed in which the order of steps i and ii was reversed prior to ligation of the RNA oligonucleotide to the 5′ end.
The PCR-amplified fragments were ligated into the pCR4-TOPO vector (Invitrogen Ltd.) and cloned in Escherichia coli strain TOP10 (TOPO TA cloning kit; Invitrogen Ltd.). Sequences were determined from both directions with the DYEnamic ET terminator cycle sequencing kit (Amersham Biosciences AB) with M13 primers according to the manufacturer's recommendations. To sequence fragments larger than 500 bp, either additional sequence-specific primers were designed for the fragment or a set of nested deletions was generated with the Erase-a-Base system (Promega Corp, Madison, Wis.). The sequencing reactions were analyzed with an automated DNA sequencer (ABI Prism 377; Perkin Elmer Applied Biosystems, Foster, Calif.). The sequence for each genomic region analyzed was determined based on three clones obtained from at least two independent RT-PCRs. The 5′ ends of sgRNAs were determined from two independent GeneRacer reactions for each sgRNA, for which the RNA was obtained from two independent RNA extractions from different plants.
Nucleotide sequence analyses.
Sequence analyses and alignments and the phylogenetic analyses were performed with programs from the Wisconsin package (version 8, September 1994; Genetics Computer Group, Madison, Wis.). Nucleotide and protein sequences not determined in this study were obtained from the EMBL sequence database (http://www.ebi.ac.uk/embl/index.html) (Table 1). Database searches for protein similarities were performed with the Blast2 search tool at the EMBL site (http://dove.embl-heidelberg.de/Blast2/). Protein profiles and domains were analyzed at the Prosite database of protein families and domains (http://www.expasy.ch/prosite/) or at the Pfam protein families database (The Sanger Centre; http://www.sanger.ac.uk/cgi-bin/Pfam/). Transmembrane helices in proteins were predicted with the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM-2.0/) (40, 44).
TABLE 1.
Sequences of members of the family Closteroviridae used in this study
| Group | Virus (acronym) | Reference |
|---|---|---|
| Criniviruses | Cucurbit yellow stunting disorder virus (CYSDV) | 57 |
| Tomato chlorosis virus (ToCV) | 63 | |
| Lettuce infectious yellows virus (LIYV) | 37 | |
| Tomato infectious chlorosis virus (TICV) | 57 | |
| Beet psuedo yellows virus (BPYV) | 57 | |
| Potato yellow vein virus (PYVV) | 52 | |
| Closteroviruses | Apricot stem pitting-associated virus (ASPaV) | Unpublished (AJ305307) |
| Beet yellow virus (BYV) | 2 | |
| Beet yellow stunt virus (BYSV) | 34 | |
| Citrus tristeza virus (CTV) | 33 | |
| Grapevine leafroll-associated virus 1 (GLRaV-1) | 20 | |
| Grapevine leafroll-associated virus 2 (GLRaV-2) | 65 | |
| Grapevine leafroll-associated virus 3 (GLRaV-3) | 41 | |
| Grapevine leafroll-associated virus 4 (GLRaV-4) | 53 | |
| Grapevine leafroll-associated virus 5 (GLRaV-5) | 24 | |
| Grapevine leafroll-associated virus 7 (GLRaV-7) | 53 | |
| Little cherry virus (LChV) | 30 | |
| Little cherry virus 2 (LChV-2) | 51 | |
| Olive leaf yellowing-associated virus (OLYaV) | Unpublished (Y18128) | |
| Pineapple mealybug wilt-associated virus 2 (PMWaV-2) | 43 |
RNA secondary structures were predicted with the program RNAstructure, version 3.6 (http://rna.chem.rochester.edu/RNAstructure.html) (42).
RNA extraction and Northern analysis.
The second topmost leaf and the subsequent five lower leaves were studied in the SPCSV-infected I. setosa plants for the temporal appearance of sgRNA. Total RNA was extracted with the Trizol LS reagent (Invitrogen Ltd.) according to the manufacturer's recommendations. The quality of the extracted RNA was evaluated under UV light following electrophoresis in a standard formaldehyde gel (1%) and staining with ethidium bromide (54).
For the Northern blots, 10 to 20 μg of RNA was separated by formaldehyde gel electrophoresis, transferred to a nylon membrane (Hybond-N; Amersham Biosciences AB), cross-linked with UV light, prehybridized, hybridized, and washed in hybridization tubes as described by Sambrook et al. (54). After being washed, the blots were wrapped in polyethylene plastic and exposed to a phosphor exposure cassette (Molecular Dynamics, Kensing, United Kingdom) for 1 to 3 days. The cassette was then scanned with a PhosphorImager (Molecular Dynamics), and the results were analyzed with ImageQuant software (Molecular Dynamics). All membranes were also exposed to standard X-Ray film (Boehringer Mannheim Corp., Indianapolis, Ind.).
ssRNA probes radiolabeled with [α-32P]UTP (Amersham Biosciences AB) were made by transcription from the appropriate linearized plasmid with T3 or T7 RNA polymerase (USB Corporation, Cleveland, Ohio) according to the manufacturer's recommendations.
Nucleotide sequence accession numbers.
The sequences have been deposited in the EMBL nucleotide sequence database under accession numbers AJ428554 (SPCSV RNA1) and AJ428555 (SPCSV RNA2).
RESULTS AND DISCUSSION
In this study, the complete genomic sequence of SPCSV was determined to provide the second example of a viral genome in the genus Crinivirus. The computer-assisted analyses of the sequences obtained showed that the SPCSV genome consists of two genomic RNA molecules of 9,407 nt and 8,223 nt, containing five and seven predicted ORFs, respectively (Fig. 1A). Viral RNA was obtained from purified virions, and at least three independently RT-PCR-amplified products were sequenced for each segment of the genome to exclude possible PCR-based errors, and the terminal sequences of the genomic RNAs were determined from several independent cDNA samples and PCR clones to validate the data. The sequence data indicated that SPCSV possesses the second largest positive-strand ssRNA genome (after CTV) known for a plant virus. Also, SPCSV and LIYV, the only two criniviruses characterized to date at the molecular level, have several significant differences in their genome structures, and, furthermore, SPCSV contains an ORF for a putative RNase III protein that is unique among plant and animal viruses.
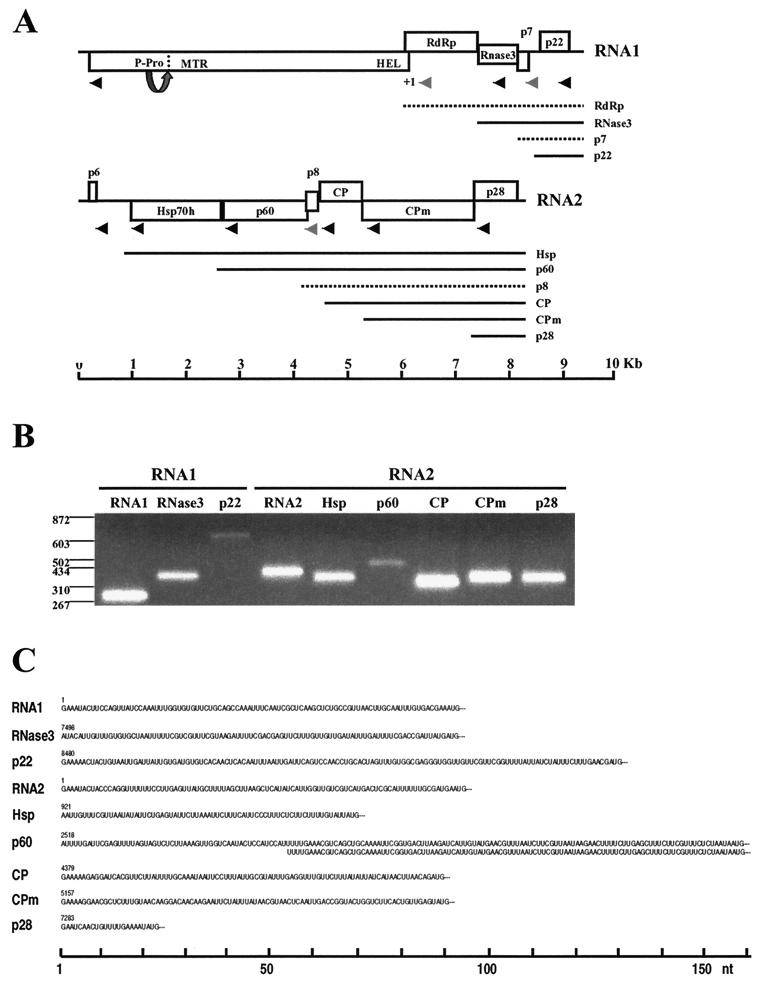
FIG. 1.
Genomic structure of SPCSV and expression of sgRNAs. (A) The genomic RNA1 and RNA2 are represented by a line, with the ORFs indicated by boxes. ORFs are shown above, below, or in the middle of the line, indicating that they are found in different reading frames. On RNA1, +1 indicates a putative +1 ribosomal frameshift site. The functional domains predicted from the deduced amino acid sequence are indicated inside the boxes, or, if no function could be predicted, the approximate molecular weight is indicated. P-Pro, putative papain-like leader proteinase, for which the arrow and dotted line indicate the predicted autocatalytic cleavage site; MTR, methyltransferase domain; HEL, helicase domain; RdRp, RNA-dependant RNA polymerase domain; RNase3, RNase III-like domain; Hsp70h, heat shock protein 70 family homologue; CP, coat protein; CPm, minor coat protein. The lines below the genomic RNA indicate sgRNAs, each of which was named according to the first ORF at the 5′ end. Dashed lines indicate hypothetical sgRNAs that were not detected with the GeneRacer method. Arrowheads indicate the positions of primers used for amplification of the 5′ ends from the genomic and sgRNAs with GeneRacer, the products of which are shown in panel B (2% agarose gel; positions of molecular size markers [in nucleotides] are shown to the left). Grey arrowheads in panel A indicate that no product was obtained in GeneRacer with these primers. (C) The 5′ untranslated sequences of the sgRNAs are shown, as determined from PCR products obtained with GeneRacer. Amplification of the p60 sgRNA 5′ end resulted in two fragments, of which one was 53 nt shorter but otherwise identical.
We extended the genome analysis of SPCSV to assay sgRNAs produced from SPCSV in infected plants. With an approach designed to amplify only the 5′ ends of m7GpppN-capped RNAs, the sequences of the 5′ ends of seven sgRNAs were determined. These data simultaneously provided compelling evidence that SPCSV sgRNAs are capped, a result previously unreported for viruses of the family Closteroviridae. Furthermore, Northern analyses suggested differences in temporal and quantitative accumulation of the different genomic and subgenomic RNAs in infected leaves of test plants.
Since the sgRNAs detected are probably used for expression of various viral proteins, they are discussed in the context of the genome structure and ORFs of SPCSV. Therefore, the data on detection of the sgRNAs and determination of their 5′ termini are described first, followed by a detailed discussion of the SPCSV genome structure.
Genome expression strategy.
Since viruses replicate in cellular environments that are dedicated to translation of monocistronic mRNAs, all ORFs except the most 5′ proximal ORF in a viral positive-strand ssRNA will be disadvantaged for translation initiation. Therefore, several additional mechanisms, such as production of 3′ coterminal sgRNAs, frameshifting, and polyprotein processing, are probably used by clostero- and criniviruses to regulate their gene expression (2, 20, 30, 33, 34, 37, 41, 43, 65). In this study, our aim was to detect at least a few of the expected sgRNAs of SPCSV and to map their 5′ termini in relation to the genomic RNAs. It was unknown whether the sgRNAs of Closteroviridae viruses are capped, but since the genomic RNA of Beet yellows virus (BYV) is m7GpppN-capped (32), this possibility was considered, and therefore the GeneRacer protocol designed to amplify only the 5′ ends of m7GpppN-capped RNAs was used.
Northern analyses with probes prepared from the sequences of the most 3′ proximal ORFs of SPCSV RNA1 and RNA2 revealed several sgRNAs (Fig. 2). Their 5′ ends were subsequently RT-PCR amplified from total RNA of SPCSV-infected I. setosa leaves with the GeneRacer protocol along with primers designed to anneal 300 to 700 nt downstream from the expected 5′ terminus of each sgRNA (Fig. 1A). Distinct bands were obtained with all primers (Fig. 1B) except p7, RdRp (RNA1), and p8 (RNA2). No bands were obtained with total RNA extracted from healthy plants. Also, results were negative if the RNA was first decapped and subsequently dephosphorylated prior to ligation of the RNA oligonucleotide. These control experiments proved that the amplified fragments were SPCSV specific and obtained from capped RNA.
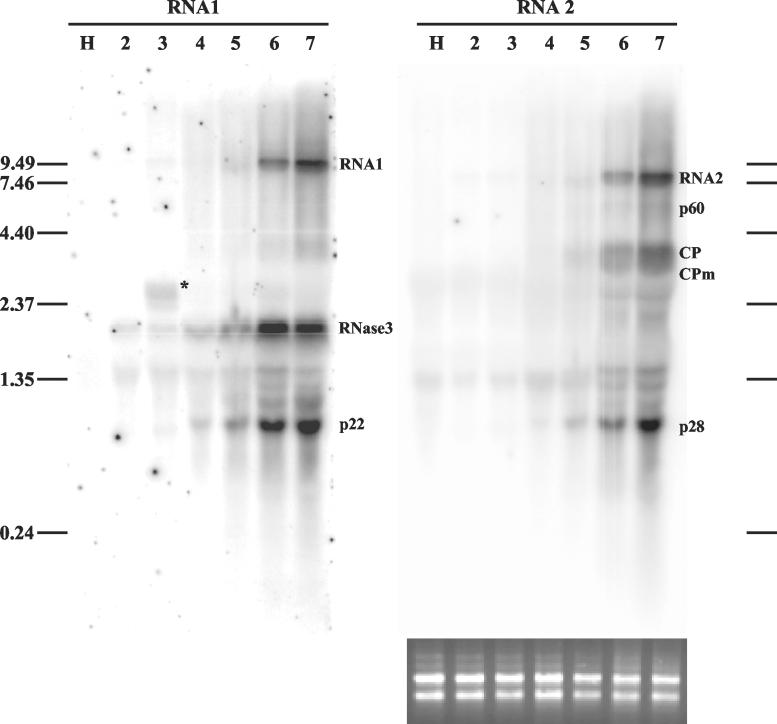
FIG. 2.
Northern analysis of genomic RNAs and sgRNAs of SPCSV in infected leaves of I. setosa. Total RNA extracted from SPCSV-infected leaves (lanes 2 to 7) and leaves of healthy plants (lane H) was first hybridized with an ssRNA probe corresponding to the negative strand of p28, the most 3′-proximal gene of RNA2. The same blot was subsequently used for hybridization with an ssRNA probe corresponding to the negative strand of p22, the most 3′-proximal gene of RNA1. Lanes 2 to 7 each contain total RNA extracted from six top leaves of the plant. The second leaf from the top (lane 2) was very young, was still folded, and had only very recently been invaded by SPCSV, in contrast to the lower leaves, which had been SPCSV infected for at least 2 weeks. The youngest leaf was too tiny for sampling. The positions of genomic RNA1 and RNA2 and the sgRNAs, identified by size, are indicated. The sgRNA corresponding to Hsp70h was not detected. The asterisk in lane 3 indicates a possible defective RNA (see text). The positions of the RNA molecular size markers (in kilobases) are indicated to the left and to the right. Equal loading of RNA was checked by ethidium bromide staining (shown below the RNA2 panel, the blot that was hybridized first).
The exact nucleotide position of the 5′ end of each sgRNA was determined by sequencing of the cloned GeneRacer PCR products obtained from two independent RNA extractions, each from a different plant. For all sgRNAs, the independent clones that were sequenced had identical 5′ ends and sequences, except for the p60 fragment, which consisted of two products of different lengths (Fig. 1C). The same two products were found when the p60 products from two separate RNA extractions were amplified and sequenced, indicating that the products were genuine and that the p60 sgRNA has two alternative 5′ ends.
No fragments corresponding to the hypothetical sgRNAs of RdRp, p7 (RNA1), or p8 (RNA2) were obtained by the GeneRacer approach, even after two cycles with nested amplification, and Northern analyses revealed only faint bands corresponding to the expected sizes for these putative sgRNAs. These data may imply that SPCSV uses other mechanisms for the translation of p7 and p8 or that these proteins are not expressed. Alternatively, the p7 and p8 sgRNAs of SPCSV may not be capped, and/or they may accumulate only at low levels. For example, Tobacco mosaic virus expresses CP and p30 from sgRNAs, but only the CP sgRNA is capped (reviewed in reference 12). The putative expression strategy of RdRp is dealt with later.
The lengths of the 5′ untranslated regions (UTRs) in the sgRNAs ranged from 20 nt (p28 sgRNA) to 159 nt (the larger p60 sgRNA) (Fig. 1C). The sgRNAs of p22, CP, CPm, and p28 start with a guanine, followed by two to five adenines, similar to the genomic RNAs. The remaining sgRNAs, however, do not seem to have shared 5′ sequence motifs except that the first three nucleotides are adenine or uracil. Disregarding the first nucleotide of the longer p60 sgRNA, the 5′ nucleotides of both p60 sgRNAs are UUUUGA, suggesting a role for this sequence in the initiation of the p60 sgRNA.
Except for the most 5′-proximal nucleotides, the remainder of the 5′ regions of the sgRNAs revealed no significant sequence similarity or predictable RNA secondary structures (both strands analyzed; data not shown). In an attempt to reveal possible conservation in the putative control elements for sgRNA production, genomic sequences immediately upstream from the sgRNA 5′ termini were compared, but no obvious resemblance was detected (both strands analyzed; data not shown). These data are consistent with those of Gowda et al. (25), who found little similarity between the sequences or predicted structures of the controller elements of CTV sgRNAs. Moreover, mutational analyses of the CP sgRNA controller element in CTV indicated no functional significance for the predicted secondary structures (25).
Temporal and quantitative expression of sgRNAs.
Expression of sgRNAs in closteroviruses (26, 46) and LIYV (64) is temporally and quantitatively regulated. Also, our previous studies showed only low or nondetectable levels of SPCSV in young leaves, in contrast to the high virus titers detected in mature leaves (35). We hypothesized that these results mean that the phloem of the youngest leaves provided only limited support for SPCSV replication and/or movement, in contrast to the mature phloem in the veins of the more developed leaves. Consequently, viral RNA accumulation at earlier and later phases of infection could be compared by assaying the extracts of leaves harvested at different developmental stages.
RNA was extracted from the second topmost leaf and the subsequent five lower leaves of vigorously growing vines of I. setosa plants infected with SPCSV (the topmost leaf was too tiny for analysis). Northern analyses were performed with ssRNA probes complementary to the 3′-proximal region of RNA1 (p22) or RNA2 (p28) (Fig. 2). The experiment was carried out four times on three different plants, and the signal strengths of the different bands were quantified with a PhosphorImager. The results of expression kinetics consistently showed that the bands specific to the RNA1 sgRNAs were readily detectable prior to those of the RNA2 sgRNAs (Fig. 2, compare lanes 2 and 3 in the panels for RNA1 and RNA2). Genomic RNA1 and RNA2 were detected in the youngest leaves (faint bands on lane 2), but their concentration increased much more slowly than the concentration of RNase 3 and p22 sgRNAs produced from RNA1 (lanes 2 to 5). A significant increase in the concentration of the genomic RNAs was observed only from leaf 5 (lane 5) to leaf 6 (lane 6), suggesting that efficient genome replication requires mature phloem cells present in the leaf veins of the well-developed leaves.
Our data suggest that the sgRNAs from RNA1 are expressed prior to those of RNA2 during SPCSV infection. This finding is similar to that for LIYV, in which the sgRNAs formed from RNA1 as well as the genomic RNA1 accumulate up to 3 days earlier than RNA2 genomic and sgRNAs in infected protoplasts (64).
An additional fragment corresponding to RNA1 was detected in some samples (asterisk in lane 3, Fig. 2). This RNA accumulated to high titers in some plants and probably represents a defective RNA molecule because it hybridized with probes from the 3′ end as well as the 5′ end of RNA1 but not with a probe corresponding to the RdRp region (data not shown).
Nucleotide sequence analyses of RNA1.
RNA1 of SPCSV encodes the putative replication module, consisting of ORF1a/b, which encodes conserved domains of the viral methyltransferase, helicase, and RNA-dependent RNA polymerase (RdRp). In closteroviruses (49) and in the crinivirus LIYV (38), this module is sufficient for replication of viral RNA in protoplasts. Two of the three additional ORFs present in SPCSV RNA1 are unique to SPCSV (Fig. 1A).
RNA1 ORF1a and ORF1b: the replication module.
The first AUG codon found at position 90 in RNA1 is at an optimal context for translation initiation (39) and is therefore likely to be the start codon for ORF1, which continues until the first stop codon at position 6053. The putative protein of 227 kDa contains the conserved domains of methyltransferase (amino acids 549 to 906) and helicase (amino acids 1690 to 1954), as identified with Pfam (Protein Families Database, The Sanger Centre). The region between the methyltransferase and helicase domains contains no significant similarity to any known proteins. However, the TMHMM program (40, 44) predicts two transmembrane domains at amino acid positions 1245 to 1267 and 1363 to 1387, suggesting that this protein may be localized to membranes, analogous to the replicase of BYV (17, 18).
Alignment of the amino acid sequences of the region upstream from the methyltransferase domain in SPCSV and LIYV revealed a putative papain-like cysteine proteinase (P-Pro) domain, similar to other viruses of the family Closteroviridae (2, 30, 33, 37, 38, 65). Autoproteolysis at the predicted cleavage site between Gly-492 and Ala-493 by the catalytic residue His-477 would yield a putative leader proteinase of 56 kDa and a larger protein of 171 kDa in SPCSV. Besides being required for proteolytic cleavage of the ORF1a/b-encoded proteins, the 54 N-proximal amino acids of the leader proteinase of BYV function as a replication enhancer (47, 49). The leader proteinases of closteroviruses also exhibit specialized functions during viral movement (48).
The predicted ORF1b starts at nt position 6004 of RNA1, overlapping ORF1a by 49 nt, and continues until the stop codon (UGA) at nt position 7569. Analysis by Pfam indicated that ORF1b encodes a conserved RdRp domain that may be expressed by a +1 ribosomal frameshift in several viruses of the Closteroviridae (2, 20, 30, 33, 34, 37, 41, 43, 65). Many frameshifting models have been proposed (reviewed in reference 1), but none seem applicable to all clostero- and criniviruses.
ORFs 1a and 1b of SPCSV are oriented in a 0/+1 configuration, which is similar to the replication-associated genes of other clostero- and criniviruses. However, the nucleotide sequences surrounding the putative frameshifting site (nt 6051) in SPCSV do not contain “shifty” heptamers, and no stable secondary structures or overlapping codons are detectable, i.e., elements implicated in frameshifting mechanisms cannot be found (2, 37, 43). However, alignment of sequences that are upstream from the putative frameshift region for clostero- and criniviruses revealed some level of similarity to the well-studied +1 frameshifting site in the prfB gene of E. coli (reviewed in reference 19) in all viruses except CTV and the closteroviruses belonging to the mealybug-transmitted lineage (31). The frameshifting in SPCSV RNA1 would yield a fusion protein of 286 kDa.
RNA1 ORF2: RNase III-like protein.
The predicted ORF2 (nt 7471 to 8272) overlaps ORF1b by 89 nt. However, the first AUG is not in an optimal context for translation initiation. Accordingly, the 5′ end of an sgRNA was mapped to nt position 7498 (Fig. 1C) downstream from the putative start codon. Additional AUG codons were found downstream at nt positions 7583 and 7586, the latter of which is in an optimal context for translation initiation. Four independent clones covering ORF2 were sequenced, three of which also overlapped the flanking ORFs. Also, Northern analyses with the sequence of SPCSV ORF2 as a probe revealed signals exclusively in SPCSV-infected plants and no signals in healthy plants. During infection (compare lanes 2 and 3 in the panel RNA1 in Fig. 2), the sgRNA corresponding to ORF2 (RNase3) was detected earlier than the sgRNA (p22) corresponding to the last ORF on RNA1 (Fig. 1).
Translation from the AUG at position 7586 would result in a protein of 27 kDa. Screening of this deduced protein for conserved domains with the Prosite database revealed a conserved N-proximal RNase III domain (112 amino acids) and a C-proximal double-stranded RNA-binding motif, i.e., a typical structure for RNase III proteins (11). Blast analysis revealed that the greatest similarity was to putative Arabidopsis thaliana protein BAB02825 (46% similarity), which also contains RNase III and double-stranded RNA-binding motifs (Fig. 3). Protein BAB02825, in turn, is similar to the Arabidopsis CAF protein (29); both of these are related to Dicer of Drosophila melanogaster, an RNase implicated in the initiation of RNA silencing (7). According to the database searches, the double-stranded DNA virus Paramecium bursaria Chlorella virus 1 (accession number AAC96831), which infects green algae, is the only other virus that encodes a predicted RNase III-like protein.
FIG. 3.
Amino acid sequence alignment of the RNase III-like protein encoded by SPCSV (SPCSV1-ORF2) and homologous proteins encoded by eukaryotic organisms including Arabidopsis thaliana (CAF-like protein BAB02825 and CAF) (29) and Drosophila melanogaster (Dicer) (7). The numbers shown at the beginning and end of each sequence indicate the number of residues that precede or follow in each case. Gaps between sequences (marked with dots) indicate adjoining sequences and are due to the alignment program. Amino acids common to the putative SPCSV RNase III and at least one other protein are indicated above the alignment, and the residues conserved in all sequences are in bold. Shading indicates the position of the RNase III signature box (11).
RNA1 ORF3: a small hydrophobic protein.
The third ORF (nt 8277 to 8450) in SPCSV RNA1 putatively encodes a small hydrophobic protein (SHP) of 7 kDa that contains a predicted transmembrane helix between amino acids 26 and 48. Hydrophobic proteins of similar size but with little sequence similarity are encoded by all closteroviruses and LIYV and are considered one of the hallmarks of the Closteroviridae. The 6-kDa SHP of BYV is essential for viral cell-to-cell movement (5). However, the genomic location of the SPCSV SHP gene is unique because SHP in the closteroviruses and LIYV is found immediately upstream of Hsp70h. The unique position of SHP in SPCSV is even more striking considering that all other movement-associated proteins are encoded by RNA2. Since the sgRNAs of SPCSV produced from RNA1 accumulate earlier in infection than those produced from of RNA2 (Fig. 2), the SPCSV SHP may have an as yet unknown function early in infection. However, it should be noted that we did not detect any sgRNA corresponding to SHP, in contrast to the sgRNAs corresponding to the putative RNase III- and p22-encoding sequences flanking SHP on RNA1 (Fig. 1 and 2). Thus, expression of the putative SHP protein in SPCSV requires further study.
RNA1 ORF4: p22 protein.
The most 3′-proximal ORF (ORF4; nt 8606 to 9181) in RNA1 encodes a putative protein of 22 kDa (p22). p22 shows no significant sequence similarity to any known proteins, but the proteins encoded from the most 3′-proximal gene of the BYV genome and RNA1 of LIYV are similar in size and are known to enhance RNA replication (49) and accumulation of RNA2 (64), respectively. The ORF4 start codon is found 154 nt downstream from the ORF3 stop codon. The 5′ end of an sgRNA corresponding to the putative p22 gene was mapped to nt position 8480, 129 nt upstream from the p22 gene start codon (Fig. 1C).
Nucleotide sequence analyses of RNA2.
RNA2 of SPCSV contains seven predicted ORFs: p6, Hsp70h, p60, p8, CP, CPm, and p28 (Fig. 1). The genomic structure of SPCSV RNA2 is colinear with that of LIYV RNA2, with the exception that the first ORF of LIYV RNA2 encodes SHP, whereas in SPCSV, SHP is encoded by RNA1 (see above).
RNA2 ORF1: a putative p6 protein.
The first AUG codon of RNA2 is found at nt position 91. While it is not in an optimal context for translation initiation, if it is in fact operational, then it would start ORF1 (nt 91 to 240), which encodes a putative protein (p6) of 5.8 kDa which has no significant similarity to any known proteins and no apparent stretches of hydrophobic amino acid residues. The region between the stop codon of the putative p6 gene and the start codon of the next gene, Hsp70h, contains a multitude of AUG codons. However, none of the ORFs initiated by these putative start codons encodes a peptide larger than 3 kDa, and therefore we did not consider them in our present study. Alignment of the SPCSV and LIYV RNA2 sequences upstream from the start codon of the putative Hsp70h gene shows considerable sequence conservation, with several identical stretches of 8 to 12 nt, regardless of the difference in size of this region in the two viruses (986 nt in SPCSV and 692 nt in LIYV). While the partly conserved sequences may suggest that the 5′ region of RNA2 has an as yet unknown functional significance in criniviruses, no similar RNA secondary structures were found in SPCSV and LIYV.
RNA2 ORF2: a heat shock protein homologue, Hsp70h.
The predicted ORF2 (nt 987 to 2651) may be translated from the sgRNA, of which the 5′ end was mapped to nt position 921 (Fig. 1C). The translation start codon of ORF2 is in an optimal context, and the predicted protein of 61 kDa contains the typical signatures of the Hsp70 protein family as identified by Pfam. The Hsp70 homologues (Hsp70h) of clostero- and criniviruses are highly conserved and physically associated with virus particles (45, 58). They are essential for proper virion assembly as well as virus movement (6, 50, 55).
RNA2 ORF3: a p60 protein.
The predicted ORF3 (nt 2673 to 4229) encodes a putative protein of 60 kDa, which is homologous to proteins of other crini- and closteroviruses encoded at the corresponding genomic position. Two sgRNAs that have their 5′ ends 159 nt and 105 nt upstream from the ORF3 start codon were identified (Fig. 1C). The deduced p60 amino acid sequence contains no motifs with recognized functional significance according to Prosite and Pfam analyses. Among the sequences available in protein databases, the corresponding protein of LIYV is the most similar (29.8% amino acid identity) to SPCSV p60. The CTV p61 protein, which corresponds to SPCSV p60, is essential for viral cell-to-cell movement (5) and virion assembly (55).
RNA2 ORF4: a p8 protein.
The fourth predicted RNA2 ORF (nt 4211 to 4432) overlaps ORF3 by 19 nt and encodes a putative protein of 8.4 kDa. No significant similarity was found with any known proteins with Blast, but pairwise alignment with the putative protein encoded by the ORF present at the corresponding position in LIYV revealed moderate amino acid sequence identity (29.3%). The putative p8 protein seems to be unique to the genus Crinivirus, since no similar protein has been detected in closteroviruses. However, no sgRNA corresponding to p8 was detected (Fig. 1 and 2), and therefore p8 expression requires further study.
RNA2 ORF5 and ORF6: coat proteins.
The predicted ORF5 (nt 4465 to 5241) starts 32 nt downstream from the p8 stop codon and encodes the putative CP of 29 kDa. The 5′ end of the putative sgRNA for ORF5 was mapped to nt position 4379 (Fig. 1C). Pairwise alignment with the corresponding protein of LIYV indicated an amino acid sequence identity of 28.8%.
The predicted ORF6 (nt 5244 to 7298) starts just 2 nt downstream from the CP stop codon and encodes the putative CPm of 79 kDa. The 5′ end of the putative sgRNA for ORF6 was mapped to nt position 5157 (Fig. 1C). Alignment of the C-proximal 170 amino acids of SPCSV and LIYV CPm showed 37.5% identity (Fig. 4). However, the SPCSV CPm was 1.5-fold larger than that of LIYV (52 kDa), due primarily to differences within the N-proximal portions of the proteins. Studies of clostero- and crinivirus particles have revealed that CPm assembles within a 75- to 85-nm stretch at one end of the virus particle (3, 21, 58, 66) and is essential for viral cell-to-cell movement (5, 6). LIYV CPm has been implicated in virus transmissibility with the whitefly vector (58).
FIG. 4.
Alignment of amino acid sequences for the coat protein (CP), N-proximal minor coat protein (nCPm), and C-proximal minor coat protein (cCPm) of the criniviruses LIYV and SPCSV. The numbers of the first and the last amino acids shown in the alignment are indicated, and an asterisk indicates the end of the protein. Gaps between sequences (marked with dots) indicate adjoining sequences and are due to the alignment program. Conserved residues in each pairwise alignment are indicated in bold. Residues conserved in all sequences are highlighted by shading, and the S, R, and D residues conserved in all filamentous plant virus CPs are also indicated by an exclamation mark. Consensus (Cons.) indicates the similar or identical amino acid residues for the CP sequences (above the alignment) or the cCPm sequences (below the alignment). The amino acids that were similar or identical in the CP and cCPm sequences are indicated between the alignments. In the consensus: a, an acidic residue (D or E); b, a basic residue (H, K, or R); f, a hydrophobic residue (A, F, I, L, M, P, V, or W); o, an aromatic residue (F, W, or Y); p, a polar residue (C, G, N, Q, S, T, or Y).
Pairwise comparison of the deduced amino acid sequences of SPCSV CP and CPm revealed two regions of conservation, one within the N-proximal portion of CPm (nCPm) and another one within the C-proximal part (cCPm). cCPm shows higher conservation (22.2% identity) with CP than does nCPm, but both regions of CPm contain the conserved residues serine, arginine, and aspartic acid (S, R, and D), as in all filamentous virus coat proteins (14) (Fig. 4), including LIYV CPm (37).
RNA2 ORF7: a p28 protein.
The predicted ORF7 (nt 7303 to 8031) encodes a putative protein of 27.8 kDa that is similar in size and genomic position to LIYV p26 (37). No significant similarity to any protein sequence available in the databases was found with Blast, but pairwise comparison of the putative SPCSV p28 and LIYV p26 showed 20.3% identity over a 73-amino-acid stretch in the C-proximal region. No obvious counterpart to the putative p28 is encoded by closteroviruses, suggesting that ORF7 is specific to the genus Crinivirus. The 5′ end of the putative sgRNA for ORF7 was mapped to nt position 7283 (Fig. 1C).
5′ and 3′ UTRs.
The putative 5′ UTRs of SPCSV RNA1 and RNA2 are 89 and 90 nt in length, respectively. The first eight nucleotides of the 5′ UTRs in these RNAs are identical. Prediction of secondary structure within the first 200 nt of RNA1 and RNA2 (including the 5′ UTRs) suggests three stable RNA stem-loops at positions 20 to 47, 57 to 104, and 113 to 168.
The 3′ UTRs of RNA1 (226 nt) and RNA2 (192 nt), while different in length, are identical over the last 208 nt with the exception of two nucleotides. These 3′ UTRs are predicted to form identical stable secondary structures. It is likely that these regions have a regulatory role, e.g., in the initiation of viral RNA replication or particle assembly (15). Unlike the SPCSV RNAs, the corresponding 3′ UTRs of LIYV RNA1 and RNA2 are not identical (<31%), suggesting functional differences for the 3′ UTRs in these two criniviruses.
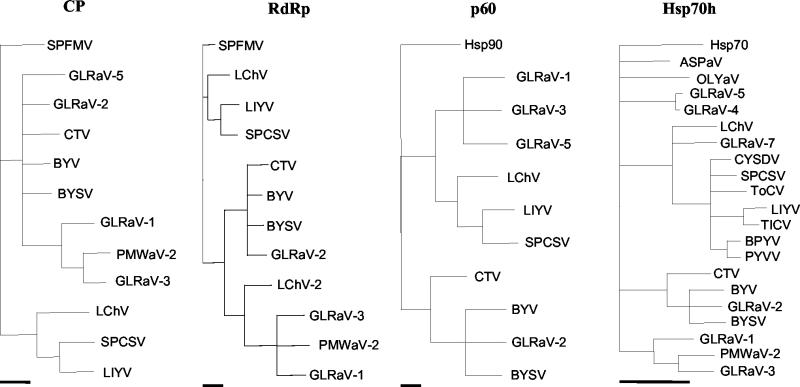
Phylogenetic analyses.
Phylogenetic analyses were carried out on the amino acid sequences of the conserved proteins CP, RdRp, p60, and Hsp70h of the clostero- and criniviruses. The data revealed that SPCSV sequences consistently clustered together with those of the crinivirus LIYV, and the sequences of these two viruses clustered with the closterovirus Little cherry virus (Fig. 5), in agreement with the findings of Karasev (31). Analysis of the conserved N-proximal ATPase domain of Hsp70h placed all criniviruses in a cluster that excluded all closteroviruses (Fig. 5). The SPCSV Hsp70h was most similar to that of Tomato chlorosis virus (63) and Cucurbit yellow stunting disorder virus (57).
FIG. 5.
Phylogenetic analysis of the amino acid sequences deduced for the coat protein (CP), RNA-dependent RNA polymerase (RdRp), p60, and the N-proximal ATPase domain of the Hsp70h of clostero- and criniviruses. Alignments were made with the Clustal algorithm (27), and the trees were generated with the maximum parsimony algorithm (16, 22). Clades supported by less than 70% of the bootstrap replicates were collapsed. In the analysis of the CP and RdRp sequences, the sequence of the corresponding protein of Sweet potato feathery mottle virus (genus Potyvirus, family Potyviridae; BAA07546) was used as an outgroup, whereas in the analysis of the p60 and Hsp70h sequences, the Hsp90 of Arabidopsis thaliana (NP_194150) and Hsp70 of tomato (Lycopersicon esculentum; P34935) were used as outgroups, respectively. The bars below each tree indicate a distance of 10 parsimonious steps. See Table 1 for virus acronyms and references.
Phylogenetic comparisons of our SPCSV Hsp70h sequence with those of other SPCSV isolates reported in databases showed that the isolate used in this study grouped together with other isolates originating from East Africa, particularly with those in the same serological subdivision (results not shown).
Conclusions.
Determination of the complete genomic sequence of SPCSV, the second crinivirus genome studied to date, revealed a number of new features of crinivirus genome composition. Prior to this study, only partial sequences of the SPCSV Hsp70h and CP genes had been described, providing only limited grounds for phylogenetic and evolutionary studies in the genus Crinivirus. The phylogenetic analyses of this study support the close relatedness of the two criniviruses SPCSV and LIYV compared to the closteroviruses. On the other hand, the differences between the SPCSV and LIYV genomes show that the genus Crinivirus, similar to the genus Closterovirus, contains considerable genetic variation. The most striking novel features of SPCSV compared to LIYV are the identical 3′ regions of SPCSV RNA1 and RNA2, the placement of SHP within RNA1 instead of RNA2, and the predicted ORF for an RNase III-like protein on RNA1. Consequently, the genome of SPCSV is considerably larger than LIYV and appears to be the second largest among the positive-strand ssRNA genomes of plant viruses characterized to date, after CTV.
Our study revealed seven SPCSV sgRNAs produced from RNA1 and RNA2, and the 5′ ends of these sgRNAs were determined relative to the genomic RNAs. The method used for amplification of the 5′ ends of the sgRNAs was specific to capped RNAs, which strongly suggests that the seven SPCSV sgRNAs characterized in this study are capped. These data constitute a comprehensive basis for a more detailed analysis of SPCSV gene functions in the future.
Two SPCSV genes, those for RNase3 and Hsp70h, will be of particular interest. The Closteroviridae are unique among viruses in that they appear to have recruited an Hsp70 gene into their genome, most likely from host cells in which they replicate. The RNase3 gene, encoding a putative RNase III-like protein in SPCSV, is novel and seems to provide another example of the curious possibility that the Closteroviridae have incorporated host mRNA sequences into their genomes during evolution. The only other virus encoding a similar protein seems to be Paramecium bursaria Chlorella virus 1. Cellular RNase III enzymes are involved in the maturation of almost every class of prokaryotic and eukaryotic RNA (reviewed in reference 11). Also, they play an important role in RNA silencing (7), a common defense mechanism against viruses (reviewed in reference 60). Having a novel gene conceivably provides SPCSV with novel or alternative means for carrying out essential functions during the infection cycle. Therefore, delineating the putative enzymatic activities of the RNase III-like protein and its function(s) during the virus infection cycle will be objectives of our future studies.
Acknowledgments
We are grateful to Kristiina Mäkinen for helpful discussions on ribosomal frameshifting and R. W. Gibson for critical reading of the manuscript.
Financial support from Sida/SAREC (grant SWE-1997-141) and the European Union (grant ICA4-CT-2000-30007) is gratefully acknowledged.
REFERENCES
- 1.Agranovsky, A. A. 1996. Principles of molecular organization, expression and evolution of Closteroviruses: over the barriers. Adv. Virus Res. 47:119-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agranovsky, A. A., E. V. Koonin, V. P. Boyko, E. Maiss, R. Frötschl, N. A. Lunina, and J. G. Atabekov. 1994. Beet yellows closterovirus: complete genome structure and identification of a leader papain-like thiol protease. Virology 198:311-324. [DOI] [PubMed] [Google Scholar]
- 3.Agranovsky, A. A., D. E. Lesemann, E. M. Maiss, R. Hull, and J. G. Atabekov. 1995. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc. Natl. Acad. Sci. USA 92:2470-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alicai, T., N. S. Fenby, R. W. Gibson, E. Adipala, H. J. Vetten, G. D. Foster, and S. E. Seal. 1999. Occurrence of two serotypes of sweet potato chlorotic stunt virus in East Africa and their associated differences in coat protein and HSP70 homologue gene sequences. Plant Pathol. 48:718-726. [Google Scholar]
- 5.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 268:192-200. [DOI] [PubMed] [Google Scholar]
- 6.Alzhanova, V. V., A. J. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70 homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 8.Carey, E. E., R. W. Gibson, S. Fuentes, M. Machmud, R. O. M. Mwanga, G. Turyamureeba, L. Zhang, D. Ma, F. Abo El-Abbas, R. El-Bedewy, and L. F. Salazar. 1999. The causes and control of virus diseases of sweetpotato in developing countries: is sweetpotato virus disease the main problem? CIP Program Report 1997-1998, p. 241-248. International Potato Center, Lima, Peru.
- 9.Cohen, J., and G. Loebenstein. 1991. Role of a whitefly-transmitted agent in infection of sweet potato by cucumber mosaic virus. Plant Dis. 75:291-292. [Google Scholar]
- 10.Cohen, J., A. Frank, H. J. Vetten, D. E. Leseman, and G. Loebenstein. 1992. Purification and properties of closterovirus-like particles associated with a whitefly transmitted disease of sweet potato. Ann. Appl. Biol. 121:257-268. [Google Scholar]
- 11.Conrad, C., and R. Rauhut. 2002. Ribonuclease III: new sense from nuisance. Int. J. Biochem. Cell. Biol. 34:116-129. [DOI] [PubMed] [Google Scholar]
- 12.Dawson, W. O., and K. M. Lehto. 1990. Regulation of tobamovirus gene expression. Adv. Virus. Res. 38:307-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Feo, L., S. F. Nome, E. Biderbost, S. Fuentes, and L. F. Salazar. 2000. Etiology of sweet potato chlorotic dwarf disease in Argentina. Plant Dis. 84:35-39. [DOI] [PubMed] [Google Scholar]
- 14.Dolja, V. V., V. P. Boyko, A. A. Agranovsky, and E. V. Koonin. 1991. Phylogeny of capsid proteins of rod-shaped and filamentous RNA plant viruses: two families with distinct patterns of sequence and probably structure conservation. Virology 184:79-86. [DOI] [PubMed] [Google Scholar]
- 15.Dreher, T. W. 1999. Functions of the 3′ untranslated regions of positive strand RNA viral genomes. Annu. Rev. Phytopathol. 37:151-174. [DOI] [PubMed] [Google Scholar]
- 16.Eck, R. V., and M. O. Dayhoff. 1966. Atlas of protein sequence and structure. National Biomedical Research Foundation, Silver Spring, Md.
- 17.Erokhina, T. N., R. A. Zinovkin, M. V. Vitushkina, W. Jelkmann, and A. A. Agranovsky. 2000. Detection of beet yellows closterovirus methyltransferase-like and helicase-like proteins in vivo with monoclonal antibodies. J. Gen. Virol. 81:597-603. [DOI] [PubMed] [Google Scholar]
- 18.Erokhina, T. N., M. V. Vitushkina, R. A. Zinovkin, D. E. Lesemann, W. Jelkmann, E. V. Koonin, and A. A. Agranovsky. 2001. Ultrastructural localization and epitope mapping of the methyltransferase-like and helicase-like proteins of Beet yellows virus. J. Gen. Virol. 82:1983-1994. [DOI] [PubMed] [Google Scholar]
- 19.Farabaugh, F. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fazeli, C. F., and M. A. Rezaian. 2000. Nucleotide sequence and organization of ten open reading frames in the genome of grapevine leafroll-associated virus 1 and identification of three subgenomic RNAs. J. Gen. Virol. 81:605-615. [DOI] [PubMed] [Google Scholar]
- 21.Febres, V. J., L. Ahoulin, M. Mawassi, A. Frank, M. Bar-Joseph, K. L. Manjunath, R. F. Lee, and C. L. Niblett. 1996. The p27 protein is present at one end of citrus tristeza virus particles. Phytopathology 86:1331-1335. [Google Scholar]
- 22.Fitch, W. M. 1977. On the problem of discovering the most parsimonious tree. Am. Nat. 111:223-257. [Google Scholar]
- 23.Gibson, R. W., I. Mpembe, T. Alicai, E. E. Carey, R. O. M. Mwanga, S. E. Seal, and H. F. Vetten. 1998. Symptoms, aetiology and serological analysis of sweet potato virus disease in Uganda. Plant Pathol. 47:95-102. [Google Scholar]
- 24.Good, X., and J. Monis. 2001. Partial genome organization, identification of the coat protein gene, and detection of Grapevine leafroll-associated virus-5. Phytopathology 91:274-281. [DOI] [PubMed] [Google Scholar]
- 25.Gowda, S., T. Satyanarayana, M. A. Ayllón, M. R. Albiach-Martí, M. Mawassi, S. Rabindran, S. M. Garnsey, and W. O. Dawson. 2001. Characterization of the cis-acting elements controlling subgenomic mRNAs of Citrus tristeza virus: production of positive- and negative-stranded 3′ terminal and positive-stranded 5′ terminal RNAs. Virology 286:134-151. [DOI] [PubMed] [Google Scholar]
- 26.Hagiwara, Y., V. V. Peremyslov, and V. V. Dolja. 1999. Regulation of closterovirus gene expression examined by insertion of a self-processing reporter and by Northern hybridization. J. Virol. 73:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higgins, D. G., and P. M. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 28.Hoyer, U., E. Maiss, W. Jelkmann, D. E. Lesemann, and H. J. Vetten. 1996. Identification of the coat protein gene of a sweet potato sunken vein closterovirus isolate from Kenya and evidence for a serological relationship among geographically diverse closteroviruses isolates from sweet potato. Phytopathology 86:744-750. [Google Scholar]
- 29.Jacobsen, S. E., M. P. Running, and E. M. Meyerowitz. 1999. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126:5231-5243. [DOI] [PubMed] [Google Scholar]
- 30.Jelkmann, W., B. Rechtner, and A. A. Agranovsky. 1997. Complete genome structure and phylogenetic analysis of little cherry virus, a mealybug-transmissible closterovirus. J. Gen. Virol. 78:2067-2071. [DOI] [PubMed] [Google Scholar]
- 31.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 32.Karasev, A. V., A. A. Agranovsky, V. V. Rogov, N. A. Miroshnichenko, V. V. Dolja, and J. G. Atabekov. 1989. Virion RNA of beet yellows closterovirus: cell-free translation and some properties. J. Gen. Virol. 70:241-245. [Google Scholar]
- 33.Karasev, A. V., V. P. Boyko, S. Gowda, O. V. Nikolaeva, M. E. Hilf, E. V. Koonin, C. L. Niblett, K. Cline, D. J. Gumpf, R. F. Lee, S. M. Garnsey, D. J. Lewandowski, and W. O. Dawson. 1995. Complete sequence of the citrus tristeza virus RNA genome. Virology 208:511-520. [DOI] [PubMed] [Google Scholar]
- 34.Karasev, A. V., O. V. Nikolaeva, A. R. Mushegia, R. F. Lee, and W. O. Dawson. 1996. Organization of the 3′ terminal half of beet yellow stunt virus genome and implications for the evolution of closteroviruses. Virology 221:199-207. [DOI] [PubMed] [Google Scholar]
- 35.Karyeija, R. F., J. F. Kreuze, R. W. Gibson, and J. P. T. Valkonen. 2000. Synergistic interactions of a potyvirus and a phloem-limited crinivirus in sweet potato plants. Virology 269:26-36. [DOI] [PubMed] [Google Scholar]
- 36.Klaassen, V. A., M. L. Boeshore, V. V. Dolja, and B. W. Falk. 1994. Partial characterization of lettuce infectious yellows virus genomic RNAs, identification of the coat protein gene and comparison of its amino acid sequence with those of other filamentous RNA plant viruses. J. Gen. Virol. 75:1525-1533. [DOI] [PubMed] [Google Scholar]
- 37.Klaassen, V. A., M. L. Boeshore, E. V. Koonin, T. Tian, and B. W. Falk. 1995. Genome structure and phylogenetic analysis of lettuce infectious yellows virus, a whitefly-transmitted, bipartite closterovirus. Virology 208:99-110. [DOI] [PubMed] [Google Scholar]
- 38.Klaassen, V. A., D. Mayhew, D. Fisher, and B. W. Falk. 1996. In vitro transcripts from cloned cDNA of the lettuce infectious yellows closterovirus bipartite genomic RNAs are competent for replication in Nicotiana benthamiana protoplasts. Virology 222:169-175. [DOI] [PubMed] [Google Scholar]
- 39.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 40.Krogh, A., B. Larsson, G. Heijne, and L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 41.Ling, K. S., H. Y. Zhu, R. F. Drong, J. L. Slightom, J. R. McFerson, and D. Gonsalves. 1998. Nucleotide sequence of the 3′ terminal two-thirds of the grapevine leafroll-associated virus-3 genome reveals a typical monopartite closterovirus. J. Gen. Virol. 79:1299-1307. [DOI] [PubMed] [Google Scholar]
- 42.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 43.Melzer, M. J., A. V. Karasev, D. M. Sether, and J. S. Hu. 2001. Nucleotide sequence, genome organization and phylogenetic analysis of pineapple mealybug wilt-associated virus-2. J. Gen. Virol. 82:1-7. [DOI] [PubMed] [Google Scholar]
- 44.Möller, S., M. D. R. Croning, and R. Apweiler. 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17:646-653. [DOI] [PubMed] [Google Scholar]
- 45.Napuli, A. J., B. W. Falk, and V. V. Dolja. 2000. Interaction between HSP70-homolog and filamentous virions of the beet yellows virus. Virology 274:232-239. [DOI] [PubMed] [Google Scholar]
- 46.Navas-Castillo, J., M. R. Albiach-Martí, S. Gowda, M. E. Hilf, S. M. Garnsey, and W. O. Dawson. 1997. Kinetics of accumulation of Citrus tristeza virus RNAs. Virology 228:92-97. [DOI] [PubMed] [Google Scholar]
- 47.Peng, C.-W., and V. V. Dolja. 2000. Leader proteinase of the beet yellows closterovirus: mutation analysis of the function in genome amplification. J. Virol. 72:9766-9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng, C.-W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1998. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J. Gen. Virol. 72:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1999. HSP70 homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 96:14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rott, M. E., and W. Jelkmann. 2001. Detection and partial characterization of a second closterovirus associated with little cherry disease, little cherry virus-2. Phytopathology 91:261-267. [DOI] [PubMed] [Google Scholar]
- 52.Salazar, L. F., G. Müller, M. Querci, J. L. Zapata, and R. A. Owens. 2000. Potato yellow vein virus: its host range, distribution in South America and identification as a crinivirus transmitted by Trialeurodes vaporariorum. Ann. Appl. Biol. 137:7-19. [Google Scholar]
- 53.Saldarelli, P., A. Rowhani, G. Routh, A. Minafra, and M. Digiaro. 1998. Use of degenerate primers in a RT-PCR assay for the identification and analysis of some filamentous viruses, with special reference to clostero- and vitiviruses of the grapevine. Eur. J. Plant Pathol. 104:945-950. [Google Scholar]
- 54.Sambrook, J., E. E. Fritch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 55.Satyanarayana, T., S. Gowda, M. Mawassi, M. R. Albiach-Martí, M. A. Ayllón, C. Robertson, S. M. Garnsey, and W. O. Dawson. 2000. Closterovirus encoded HSP70 homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 278:253-265. [DOI] [PubMed] [Google Scholar]
- 56.Sim, J., R. A. Valverde, and C. A. Clark. 2000. Whitefly transmission of Sweetpotato chlorotic stunt virus. Plant Dis. 84:1250.. [DOI] [PubMed] [Google Scholar]
- 57.Tian, T., V. A. Klaasen, J. Soong, G. Wisler, J. E. Duffus, and B. W. Falk. 1996. Generation of cDNAs specific to lettuce infectious yellows closterovirus and other whitefly-transmitted viruses by RT-PCR and degenerate oligonucleotide primers corresponding to the closterovirus gene encoding the heat shock protein 70 homolog. Phytopathology 86:1167-1173. [Google Scholar]
- 58.Tian, T., L. Rubio, H.-H. Yeh, B. Crawford, and B. W. Falk. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis with partially purified virions and the whitefly Bemisia tabaci. J. Gen. Virol. 80:1111-1117. [DOI] [PubMed] [Google Scholar]
- 59.van Regenmortel, M. H., C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner. 2000. Virus taxonomy: the classification and nomenclature of viruses: the seventh report of the International Committee on Taxonomy of Viruses, p. 51. Academic Press, San Diego, Calif.
- 60.Voinnet, O. 2001. RNA silencing as a plant immune system against viruses. Trends Genet. 17:449-459. [DOI] [PubMed] [Google Scholar]
- 61.Winter, S., A. Purac, F. Leggett, E. A. Frison, H. W. Rossel, and R. I. Hamilton. 1992. Partial characterization and molecular cloning of a closterovirus from sweet potato infected with sweet potato virus disease complex from Nigeria. Phytopathology 82:869-875. [Google Scholar]
- 62.Wisler, G. C., J. E. Duffus, H.-Y. Liu, and R. H. Li. 1998. Ecology and epidemiology of whitefly-transmitted closteroviruses. Plant Dis. 82:270-280. [DOI] [PubMed] [Google Scholar]
- 63.Wisler, G. C., R. H. Li, H.-Y. Liu, D. S. Lowry, and J. E. Duffus. 1998. Tomato chlorosis virus: a new whitefly-transmitted, phloem-limited, bipartite closterovirus of tomato. Phytopathology 88:402-409. [DOI] [PubMed] [Google Scholar]
- 64.Yeh, H.-H., T. Tian, L. Rubio, B. Crawford, and B. W. Falk. 2000. Asynchronous accumulation of Lettuce Infectious Yellows Virus RNAs 1 and 2 and identification of an RNA1 trans enhancer of RNA 2 accumulation. J. Virol. 74:5762-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu, H.-Y., K.-S. Ling, D. E. Goszczynski, J. R. McFerson, and D. Gonsalves. 1998. Nucleotide sequence and genome organization of grapevine leafroll-associated virus-2 are similar to beet yellows virus, the closterovirus type member. J. Gen. Virol. 79:1289-1298. [DOI] [PubMed] [Google Scholar]
- 66.Zinovkin, R. A., W. Jelkmann, and A. A. Agranovsky. 1999. The minor coat protein of beet yellows closterovirus encapsidates the 5′ terminus of RNA in virions. J. Gen. Virol. 80:269-272. [DOI] [PubMed] [Google Scholar]