Abstract
Proteolytic processing of the respiratory syncytial virus F (fusion) protein results in the generation of the disulfide-linked subunits F1 and F2 and in the release of pep27, a glycopeptide originally located between the two furin cleavage sites FCS-1 (RKRR136) and FCS-2 (RAR/KR109). We made use of reverse genetics to study the importance of FCS-2 and of pep27 for BRSV replication in cell culture. Replacement of FCS-2 in the F protein of recombinant viruses by either of the sequences NANR109, RANN109 or SANN109, respectively, abolished proteolytic processing at this position, whereas the cleavage of FCS-1 was not affected. All mutants replicated in calf kidney and Vero cells in the absence of exogenous trypsin, although somewhat higher titers of BRSV containing the NANR109 or the RANN109 motif were achieved in the presence of trypsin. The virus mutants showed a reduced cytopathic effect which was lowest in the case of the SANN109 mutant. These findings demonstrate that cleavage at FCS-2 is dispensable for replication of respiratory syncytial virus in cell culture. A deletion mutant containing FCS-1 but lacking FCS-2 and most of pep27 replicated in cell culture as efficiently as the parental virus, indicating that this domain of the F protein is not essential for virus maturation and infectivity.
Human respiratory syncytial virus (HRSV) and Bovine respiratory syncytial virus (BRSV) are closely related members of the genus Pneumovirus within the family Paramyxoviridae. HRSV is the most important viral agent of pediatric respiratory tract disease worldwide, causing bronchiolitis and pneumonia (7). A very similar disease is caused by BRSV in calves (1, 2, 20, 33).
The envelope of the respiratory syncytial viruses (RSV) contains three glycoproteins: the attachment protein G, the small hydrophobic protein SH, and the fusion protein F. Several studies indicate that both the G and the SH proteins are dispensable for virus replication in cell culture but may have some accessory function in the host (4, 18, 19, 35, 39). The F protein mediates fusion between the viral and cellular membrane and is therefore essential for virus replication. Since fusion does not require low pH, cells infected with RSV can fuse with adjacent cells resulting in multinucleated syncytia. Syncytium formation can also be observed with cells transfected with the F gene, although coexpression of F together with G and/or SH protein has been reported to enhance fusion activity (16, 29). Recent studies suggest that certain glycosaminoglycans of the cell surface are required for HRSV infection (13, 14, 23, 27). The G protein, as well as the F protein, have been demonstrated to bind to these carbohydrate structures (10, 11, 18, 23).
The primary sequence of the F protein from different serotypes of HRSV and BRSV is highly conserved but shows only little homology with other paramyxovirus fusion proteins. However, with respect to size, locations of hydrophobic domains, heptad repeats, and cysteine residues the RSV F protein shares many structural features with other paramyxovirus fusion proteins. A property that is even more common and also found with other virus families is the synthesis of the fusion protein as an inactive precursor F0 that has to be proteolytically cleaved to become fusion active (21, 22). This posttranslational modification results in the exposition of a hydrophobic fusion peptide at the N terminus of the membrane-anchored fragment. The fusion peptide is supposed to play a crucial role in the fusion process. The majority of viral fusion proteins, including the RSV F proteins, contain a multibasic cleavage motif of the consensus sequence RX(K/R)R immediately upstream of the fusion peptide. This sequence is recognized by the ubiquitous subtilisin-like endoprotease furin of the trans-Golgi network (21, 22). A few viruses are not activated by furin. Their fusion proteins usually contain a monobasic cleavage site that is cleaved by trypsin-like proteases. The type of the cleavage motif has been shown to be an important determinant for virus pathogenicity (21). A unique feature of the RSV F proteins is the cleavage of F0 at two conserved furin consensus sequences, RAR/KR109 (FCS-2) and KKRKRR136 (FCS-1), resulting in the generation of three proteolytic fragments, the large membrane-anchored subunit F1 with the hydrophobic fusion peptide at its N terminus, the small subunit F2 which is linked to F1 via a disulfide bridge, and a small peptide composed of 27 amino acids (pep27) originally located between the two cleavage sites (12, 41). All three products have been shown to contain N-linked oligosaccharide side chains (40, 41). Analysis of the two cleavage sites by site-directed mutagenesis revealed that efficient syncytium formation in transfected cells requires cleavage at both sites (12, 41). Moreover, complete cleavage at both sites was shown to be associated with a conformational change in the molecule (12). In the present study, we made use of reverse genetics to analyze the role of the second furin motif and the intervening peptide in proteolytic activation of BRSV.
MATERIALS AND METHODS
Cells.
BSR-T7/5 cells were grown in Eagle minimal essential medium with Earle salts (EMEM) supplemented with 5% fetal calf serum, nonessential amino acids, and 0.5 mg of G418 sulfate (Calbiochem-Novabiochem)/ml. Vero cells were maintained in Dulbecco modified Eagle medium with 5% fetal calf serum. Proximal tubule cells of calf kidney (PT-11 cells) were kindly provided by R. Riebe (Bundesanstalt für Viruserkrankungen der Tiere, Insel Riems, Germany). The cells were grown in EMEM containing 10% fetal calf serum.
Plasmid construction.
The BRSV (strain ATue51908) antigenomic plasmid was described previously (3). The unique restriction sites for BstEII and NheI in this plasmid define a 2,700-bp fragment containing the M2 and almost the whole F gene lacking only the first 130 nucleotides of its open reading frame. This fragment was cloned into a modified pCR3.1 vector (Invitrogen) containing a new single BstEII restriction site. The single EcoRI restriction site of pCR3.1 was removed by the cloning step. The resulting plasmid was designated BRSV-F/M2-cassette. The BRSV fusion protein mutants bF:R106N/K108N and bF:K108N/R109N assembled in the pTM1 expression vector have been previously described (41). A BstEII/EcoRI-fragment of 540 bp containing the mutations was used to replace the corresponding region in the BRSV-F/M2 cassette. The mutant bF:R106S/K108N/R109N was generated in the same vector by using an overlapping PCR technique. Two PCR products were amplified from the bF:K108N/R109N template by using Pfu DNA polymerase (Promega). Nucleotides 1 to 327 of the F open reading frame were amplified by using forward primer bF-S(1-27) (5′-AATCCATGGCGACAACAACCATGAGGATGATC; start codon underlined) and a reverse mutagenesis primer (5′-CGTTGTTTGCTGAACTGAAGGAGG). Nucleotides 304 to 744 of the F gene were amplified by using a forward mutagenesis primer (5′-GCCTCCTTCAGTTCAGCAAACAACGGG) and reverse primer bF-AS(724-744) (5′-ACTGAGAGGTGTGGTAATACC). The two PCR products were separated by agarose gel electrophoresis and purified by gel extraction. Equimolar amounts of the purified fragments were combined, heated for 2 min at 95°C for denaturation, and annealed to each other at 60°C for 30 s. Oligonucleotide-primed DNA synthesis with Pfu DNA polymerase resulted in a completely double-stranded DNA fragment that was amplified by PCR after addition of the bF-S(1-27) and bF-AS(724-744) primers. The product was digested with BstEII and EcoRI and used to replace the corresponding fragment in the BRSV-F/M2 cassette. The whole region that was replaced was sequenced to confirm the nucleotide exchanges. The F protein deletion mutant bF:Δ106-130 lacking nucleotides 316 to 390 was assembled in a similar way by using the oligonucleotides 5′-GAACCGGCCTCCTTCAGTAAGAAGAGAAAAAGGAGATTTTTAGGATTC and 5′-TCTCCTTTTTCTCTTCTTACTGAAGGAGGCCGGTTCATTTTGCATAAG as forward and reverse mutagenesis primers, respectively. Finally, each of the BstEII/NheI fragments of the parental and mutated BRSV-F/M2 cassettes were used to replace the corresponding region in the BRSV antigenomic plasmid.
Recovery of recombinant BRSV.
Recombinant BRSVs were rescued from the supernatant of BSR-T7/5 cells transfected with the antigenomic plasmid, together with four plasmids directing the expression of the viral polymerase complex (3). The viruses were propagated on PT-11 cells. At 5 to 6 days after infection, when the cytophatic effect became obvious, the supernatants were taken; adjusted to 50 mM HEPES (pH 7.5), 0.1 MgSO4, and 10% fetal calf serum; and subjected to low-speed centrifugation to remove cell debris. The supernatants were divided into aliquots, flash-frozen in liquid nitrogen, and stored at −80°C. Low-passage stocks (maximum of four passages) were used throughout all experiments. The identity of the recombinant viruses was verified by reverse transcription-PCR (RT-PCR; see below) and DNA sequencing of the PCR products.
Growth kinetics and virus titration.
Multistep replication of the recombinant BRSVs was analyzed by using PT-11 and Vero cells. Confluent cell monolayers seeded the day before in six-well plates were inoculated at 37°C for 3 h with each virus at a multiplicity of infection (MOI) of 0.1. Four wells were infected in parallel with each virus. After adsorption, the inoculum was removed and the cells were washed three times with medium before the addition of 2.5 ml of medium (without fetal calf serum). Two wells of each infection received medium containing 0.5 μg of acetylated trypsin (Sigma)/ml. At the indicated times, aliquots of 250 μl were taken and replaced by the same volume of fresh medium. The aliquots were adjusted to 0.1 M MgSO4, 50 mM HEPES (pH 7.5), and 10% fetal calf serum; flash-frozen in liquid nitrogen; and stored at −80°C until titration.
The viruses were titrated in duplicate on Vero cells grown in 24-well dishes to 90% confluence. The cells were inoculated with 10-fold dilutions of each virus for 3 h at 37°C and overlaid with medium containing 2% fetal calf serum and 0.9% methylcellulose (Sigma). After incubation for 3 days, the medium was removed and the cells were washed twice with phosphate-buffered saline (PBS) and then fixed with 3% paraformaldehyde for 20 min at room temperature. Excess paraformaldehyde was quenched with 0.1 M glycine in PBS for 5 min. The cells were permeabilized with 0.2% Triton X-100 in PBS for 5 min at room temperature and then incubated for 90 min at room temperature with a monoclonal antibody directed to the RSV matrix protein (monoclonal antibody 18G6D1; diluted 1:40 in PBS). The cells were washed three times with PBS and incubated for 1 h at room temperature with a horseradish peroxidase-linked goat antiserum directed to mouse immunoglobulin (1:300 in PBS). After three wash steps, the cells were incubated for 10 min with AEC peroxidase substrate (1.7 mM 3-amino-9-ethylcarbazole and 0.1% H2O2 in 50 mM sodium acetate buffer [pH 5.0]). Immunostained cells were counted under an inverse light microscope.
RT-PCR.
Total RNA was prepared from Vero cells 2 days after infection (RNeasy [Qiagen]) and reverse transcribed by using Expand reverse transcriptase (Roche Diagnostics) and random hexamers for priming. Nucleotides 1 to 744 of the F gene were amplified from the cDNA by PCR with oligonucleotides bF-S(1-27) and bF-AS(724-744) and according to the following protocol: initial denaturation at 94°C for 1.5 min, followed by 35 two-step cycles (each composed of denaturation at 94°C for 30 s and annealing-elongation at 60°C for 30 s), with a final elongation step at 72°C for 7 min. The PCR products were separated on a 2% agarose gel, stained with ethidium bromide, and analyzed on a UV transluminator.
Radioimmunoprecipitation.
Confluent monolayers of PT-11 cells grown on 35-mm dishes (ca. 106 cells per dish) were inoculated with 250 μl of serum-free EMEM containing rBRSV at an MOI of 0.1. After 2 h of adsorption, the inoculum was replaced by 2.5 ml of EMEM containing 5% fetal calf serum. At 40 h postinfection, the cells were washed twice with PBS, starved for 1 h in methionine-cysteine-deficient EMEM, and then incubated for 1 h with 250 μl of the same medium supplemented with 50 μCi of [35S]methionine-[35S]cysteine (Tran35S-label [ICN]). Immunoprecipitation of the F protein from cell lysates was performed as recently described (41).
Western blot.
Confluent monolayers of PT-11 cells grown in 25-cm2 flasks were inoculated in duplicate with the indicated recombinant BRSVs (MOI of 0.1) for 3 h at 37°C. After removal of the inoculum, the cells were maintained in medium with 10% fetal calf serum for 24 h at 37°C, washed three times with PBS, and then maintained in medium without fetal calf serum for a further 96 h. The duplicate cells received medium supplemented with 0.5 μg of acetylated trypsin/ml. At 5 days postinfection, the cell culture supernatants were harvested and subjected to low-speed centrifugation (2,000 × g, 15 min, 4°C) to remove detached cells. An aliquot of each supernatant was taken and titrated by plaque assay as described above. Viruses in the remaining supernatant (ca. 4.5 ml) were pelleted through a 25% sucrose cushion by ultracentrifugation (105,000 × g, 60 min, 4°C) and dissolved in sodium dodecyl sulfate (SDS) sample buffer. The volume was adjusted corresponding to the calculated virus titers. Aliquots (10 μl) of the viruses were run on an SDS-10% polyacrylamide gel under nonreducing conditions and transferred to nitrocellulose by the semidry blotting technique (24). The membrane was incubated overnight at 4°C with PBS containing 1% bovine serum albumin, washed three times with PBS containing 0.1% Tween 20, and incubated for 1 h at room temperature with a mixture of three different monoclonal antibodies directed to the RSV F protein (each diluted 1:1,000 in PBS). The antibodies used were RSV3216 (Serotec) and clones 7.901 and 47F kindly provided by Jose Antonio Melero (Madrid, Spain) and Claes Örvell (Stockholm, Sweden), respectively. The blots were washed as described above, and primary antibody was detected by incubation with a biotinylated goat anti-mouse immunoglobulin serum (1:1,000 in PBS; Amersham-Pharmacia), followed by three wash steps and incubation with a streptavidin-peroxidase complex (1:2,000 in PBS; Amersham-Pharmacia). Both incubations were performed for 60 min at 4°C. Finally, the nitrocellulose was washed as described above and incubated for 1 min with a chemiluminescent peroxidase substrate (BM chemoluminescence blotting substrate, Roche Diagnostics). The resulting light emission was visualized by short exposure of the membrane to an Biomax autoradiography film.
RESULTS
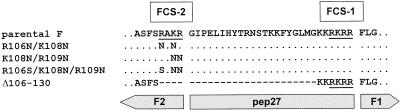
The RSV fusion protein contains two conserved furin consensus sequences: FCS-1 is located immediately upstream of the fusion peptide, whereas FCS-2 is separated from the fusion peptide by a stretch of 27 amino acids, designated pep27 (Fig. 1). Previously, we reported that changing FCS-2 of the HRSV (RARR109) or the BRSV (RAKR109) fusion protein to either NANR109 or NANR109 abolished cleavage by furin, whereas cleavage at FCS-1 was not affected (41). In the modified FCS-2 motifs, single arginine residues either at position 106 or position 109 were preserved making these sites susceptable to trypsin-like proteases. To evaluate the role of FCS-2 in the proteolytic activation of RSV, we generated two recombinant BRSV mutants, rBRSV-F(R106N/K108N) and rBRSV-F(K108N/R109N), differing from the parental virus in the same amino acid exchanges described above (Fig. 1). The mutants were rescued after transfection of BSR-T7/5 cells with the modified antigenomic plasmid together with four plasmids directing the expression of the polymerase complex (3). Since we first hypothesized that cleavage at FCS-2 might be required to activate fusion activity of the virus mutants, we added trypsin to the cell culture supernatant. However, the recovery of the mutants was also successful in the absence of exogenous trypsin. To exclude the possibility that endogenous trypsin-like proteases could cleave at the preserved arginine residues, we generated another recombinant BRSV, rBRSV-F(R106S/K108N/R109N), in which FCS-2 was replaced by the amino acid sequence SANN109 (Fig. 1). Like the other two mutants, this virus was efficiently recovered from transfected BSR-T7/5 cells. In addition to mutants containing amino acid exchanges, we constructed and rescued a recombinant virus, rBRSV-F(Δ106-130), with a deletion of 25 amino acids in the F protein. The deletion comprised FCS-2 and most of pep27, retaining only FCS-1 and the two basic amino acids at positions 131 and 132 (Fig. 1). Virus stocks were prepared by two passages on PT-11 (bovine calf kidney) cells. To verify the identity of the recombinant virus mutants, total RNA was extracted from infected cells and the region between nucleotides 5570 and 6313 of the RNA genome was amplified by RT-PCR. Although the PCR product derived from the parental virus genome was ca. 750 bp long, the deletion mutant was characterized by a PCR product of 670 bp, thus confirming the deletion within the F gene (data not shown). All changes introduced into the F gene were also verified by sequencing of the RT-PCR products.
FIG. 1.
Amino acid changes introduced into the F protein of recombinant BRSVs. A section of the BRSV fusion protein comprising amino acids 102 to 139 is shown. The furin consensus sequences are underlined, and the positions of the amino acids located N terminally of the furin cleavage site are indicated. Identical amino acids are represented by dots, and dashes indicate deleted amino acids. The three proteolytic cleavage products F1, F2, and pep27 are indicated by bars.
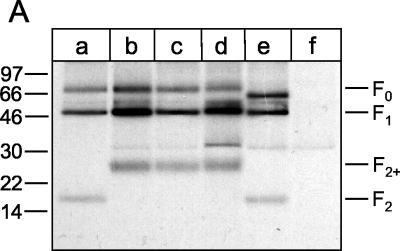
We have previously shown that FCS-2 cleavage mutants are characterized by a large size F2 subunit designated F2+ (41). The difference in molecular weight between F2 and F2+ is due to the glycosylated pep27 that remains attached to F2. This phenotypical marker allowed us to distinguish between parental and mutant rBRSVs. The F proteins were immunoprecipitated from metabolically labeled PT-11 cells 2 days after infection and analyzed by Tricine-SDS-PAGE under reducing conditions (Fig. 2A). The parental F protein (lane a) appeared as three distinct bands: the precursor F0 (72 kDa), the large subunit F1 (50 kDa), and the small subunit F2 (17 kDa). Instead of F2, all three FCS-2 mutants showed the characteristic F2+ band of 26 kDa (lanes b to d), indicating that the modified motifs were resistant to furin cleavage. Immunoprecipitation of the F deletion mutant revealed a different pattern. Due to the absence of the glycosylated pep27, F(Δ106-130) differed from the parental F protein in the smaller size of its precursor F0 (lane e).
FIG. 2.
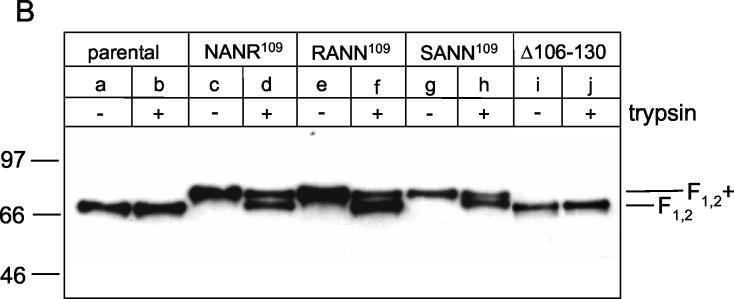
Proteolytic processing of parental and mutant F proteins of recombinant BRSVs. (A) PT-11 cells were infected with recombinant BRSVs at an MOI of 0.1. At 40 h after infection, the cells were metabolically labeled with [35S]methionine-[35S]cysteine for 1 h, and F protein was immunoprecipitated from the cell lysates. The immunoprecipitates were separated by Tricine-SDS-10% polyacrylamide gel electrophoresis under reducing conditions and detected by autoradiography [lane a, rBRSV-F(parental); lane b, rBRSV-F(R106N/K108N); lane c, rBRSV-F(K108N/R109N); lane d, rBRSV-F(R106S/K108N/R109N); lane e, rBRSV-F(Δ106-130); lane f, noninfected cells]. (B) PT-11 cells were infected with recombinant BRSVs at an MOI of 0.1 and maintained in medium either in the absence or presence of trypsin (as indicated at the top of the gel). After 4 days, the viruses were harvested from the cell culture supernatant and then pelleted by ultracentrifugation. The viruses were solubilized by SDS sample buffer and run on an SDS-8% polyacrylamide gel under nonreducing conditions [lanes a and b, rBRSV-F(parental); lanes c and d, rBRSV-F(R106N/K108N); lanes e and f, rBRSV-F(K108N/R109N); lanes g and h, rBRSV-F(R106S/K108N/R109N); lanes i and j, rBRSV-F(Δ106-130)]. F protein was detected by conventional Western blot technique with a mixture of three monoclonal antibodies directed to this protein. The relative positions of standard proteins (with the molecular masses indicated in kilodaltons) are shown on the left.
Since the FCS-2 mutants might be activated by cellular proteases secreted into the medium, we also analyzed the F proteins incorporated into mature virus particles. At 4 days postinfection, the virions were pelleted from the clarified supernatants through a 25% sucrose cushion, separated by SDS-polyacrylamide gel electrophoresis under nonreducing conditions, and analyzed by the Western blot technique (Fig. 2B). The F protein of the parental rBRSV appeared as a 72-kDa disulfide-linked complex (F1,2) composed of F1 and F2 (lane a). Addition of trypsin to the cell culture supernatant did not change this pattern (lane b). The presence of F2+ in the FCS-2 cleavage mutants caused a molecular weight shift of the disulfide-linked complex to ca. 82 kDa (F1,2+) (lanes c, e, and g). A 72-kDa band that would indicate cleavage by endogenous proteases was not detected with any of the mutants. However, the addition of trypsin to the cell culture medium led to a partial cleavage of rBRSV-F(R106N/K108N) (lane d) and rBRSV-F(K108N/R109N) (lane f) and also of rBRSV-F(R106S/K108N/R109N) (lane h). However, cleavage of the latter mutant did not result in the F1,2 complex of 72 kDa, indicating that cleavage has occurred at another basic amino acid within pep27. The deletion mutant showed the same pattern as the parental virus (compare lanes a and b with lanes i and j). After proteolytic release of pep27 from the parental F protein, there is no major difference between the two F proteins that could be detected by the Western blot.
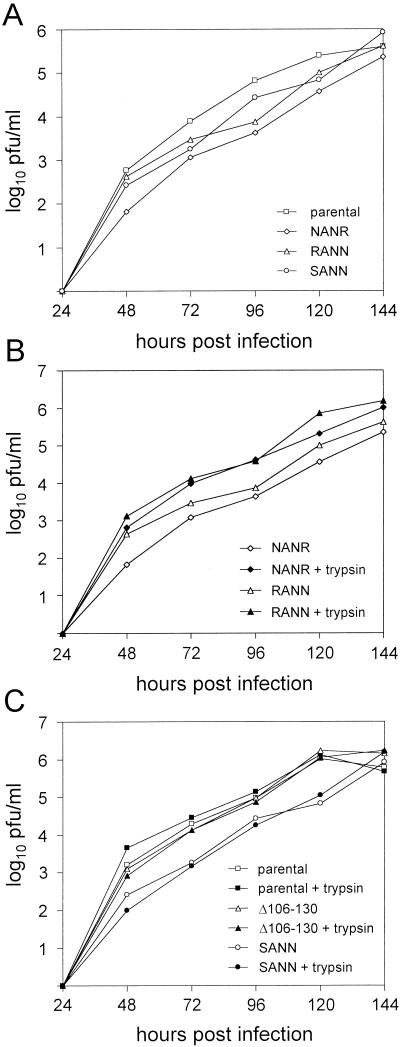
The growth characteristics of the parental and mutant rBRSVs were analyzed by using the bovine kidney PT-11 cell line, as well as African green monkey kidney (Vero) cells. The cells were infected in duplicate with the viruses at an MOI of 0.1, and supernatants were collected over a 6-day period at 24-h intervals. The virus titers were quantitated in duplicate by a plaque assay facilitated by immunological staining of the matrix protein. In the absence of trypsin (Fig. 3A), the FCS-2 cleavage mutants showed a somewhat reduced virus release in the beginning, but at day 6 postinfection they reached the titers of the parental virus. In the case of the mutants rBRSV-F(R106N/K108N) and rBRSV-F(K108N/R109N), this growth retardation was compensated for by the addition of acetylated trypsin (0.5 μg/ml) to the cell culture supernatant (Fig. 3B). In contrast, trypsin did not affect replication of either the parental virus or the mutant rBRSV-F(R106S/K108N/R109N) (Fig. 3C). The deletion mutant rBRSV-F(Δ106-130) replicated in PT-11 cells with a kinetics comparable to that of the parental virus. Likewise, the presence of trypsin had no effect on replication of this virus. Very similar growth kinetics were observed with Vero cells, although BRSV generally grew to lower titers in this cell line (not shown). On the other hand, BRSV caused a much more pronounced cytophatic effect in Vero cells than in PT-11 cells. At day 3 postinfection, we observed giant multinucleated cells in the Vero cell monolayer infected with the parental rBRSV (Fig. 4). Syncytium formation was also induced by the FCS-2 cleavage mutants; however, the syncytia were of smaller size and contained much fewer nuclei, indicating that the mutations introduced into the FCS-2 cleavage site affect cell-to-cell fusion. The syncytia formed by the mutants rBRSV-F(R106N/K108N) and rBRSV-F(K108N/R109N) grew to almost the parental virus level during the following 24 h. In striking contrast, the size of syncytia formed by the mutant rBRSV-F(R106S/K108N/R109N) did not change with time (data not shown). The deletion mutant showed a phenotype similar to that of the triple mutant, e.g., formation of very small syncytia that did not increase in size after longer incubation. Taken together, these results indicate that both FCS-2 and the intervening peptide pep27 are dispensable for virus replication in cell culture.
FIG. 3.
Multistep replication of recombinant BRSV mutants in PT-11 cells. Duplicate cell monolayers in six-well dishes were infected with the indicated viruses at an MOI of 0.1 and incubated at 37°C with medium either in the absence (open symbols) or presence (solid symbols) of 0.5 μg of acetylated trypsin/ml. Aliquots were taken at the indicated times, stored at −80°C, and titrated in parallel by plaque assay on Vero cells. Each point shown is the mean titer from two wells of infected cells. For reasons of presentation, the figure has been divided into three parts. (A) Comparison of parental rBRSV (parental) with rBRSV-F(R106N/K108N) (NANR), rBRSV-F(K108N/R109N) (RANN), and rBRSV-F(R106S/K108N/R109N) (SANN); (B) effect of trypsin on replication of the mutants rBRSV-F(R106N/K108N) (NANR) and rBRSV-F(K108N/R109N) (RANN); (C) effect of trypsin on replication of parental virus (parental), rBRSV-F(Δ106-130) (Δ106-130), and rBRSV-F(R106S/K108N/R109N) (SANN).
FIG. 4.
Cytophatic effect induced in Vero cells by parental and mutant BRSVs. The cells were photographed at 72 h postinfection with rBRSVs containing the indicated mutant F proteins (MOI = 0.1).
DISCUSSION
The fusion protein of RSV resembles many other viral fusion proteins in the location of a furin recognition site immediately upstream of the fusion peptide. Cleavage at this site by furin or a related cellular protease results in the location of the fusion peptide at the N terminus of the membrane-anchored subunit and is associated with a conformational change as shown for influenza virus hemagglutinin and the fusion proteins of simian virus 5 and RSV (6, 9, 12). Many viral fusion proteins require this posttranslational modification in order to become fusion active (21, 22). For example, blocking this step by specific furin inhibitors has been shown to reduce the infectivity of human immunodeficiency virus type 1 (15). In addition, recombinant measles was demonstrated to require on exogenous trypsin for activation of infectivity if the furin motif of the viral fusion protein was changed into a trypsin-like motif (26). However, reverse genetics showed that the conserved furin motifs found in the Ebola virus glycoprotein and in the human cytomegalovirus glycoprotein B are dispensable for virus growth in cell culture (28, 34). A major difference between these and the former viruses is the location of the furin cleavage site distantly from the postulated hydrophobic fusion domains.
A unique feature of RSV is the additional cleavage of the F protein at a second furin consensus sequence, FCS-2, separated from the fusion peptide by 27 amino acids. Previous studies by using a plasmid-driven or vaccinia virus-based expression system revealed that F-mediated syncytium formation was significantly affected when either FCS-1 or FCS-2 were changed into a furin-resistant motif by site-directed mutagenesis, indicating that cleavage at both sites might be important for activation of the RSV fusion protein (12, 41). In the present study, we learned from reverse genetics that cleavage at FCS-2 is not essential for virus infectivity though the FCS-2 cleavage mutants did not grow as efficiently as the parental virus during the first replication cycles. If a single arginine was left with the modified motif (NANR109 or RANN109), the addition of trypsin to the cell culture supernatant compensated for this growth retardation, whereas it had no supporting effect on rBRSV-F(R106S/K108N/R109N) that did not contain any arginine or lysine residues in the modified FCS-2. Accordingly, trypsin treatment caused a partial cleavage of the mutants rBRSV-F(R106N/K108N) and rBRSV-F(K108N/R109N), whereas there was no evidence for cleavage by endogenous trypsin-like proteases. However, the mutant F protein of rBRSV-F(R106S/K108N/R109N) was also cleaved by trypsin but probably at a different site. Potential cleavage sites within pep27 are Arg119 and Lys-Lys124. The amino acid changes made with the FCS-2 also led to a reduced syncytium formation by the recombinant BRSV mutants. These findings suggest that the fusion activity of the FCS-2 mutants is not abolished but impaired. Probably, the glycosylated pep27 that remains attached to the F2 subunit of the FCS-2 cleavage mutants interferes with conformational rearrangements necessary for optimal fusion activity (12). In accordance with this view, the deletion mutant lacking pep27 did not show any growth retardation and addition of trypsin had no supporting effect on this virus. Nevertheless, the deletion mutant also showed a drastically reduced syncytium formation activity in Vero cells. It should be noted that the mature parental F protein differs from the deletion mutant with respect to the C terminus of its F2 subunit. Although the former ends with the sequence RAKR109, the latter has two additional amino acids and terminates with the sequence KKRKRR111. This C terminus did not impair the infectivity (virus-to-cell fusion) of rBRSV-F(Δ106-130) as indicated by the growth kinetics. However, it might interfere with cell-to-cell fusion, suggesting that the structural requirements for these two processes differ from each other. Differences between virus-to-cell fusion and syncytium formation have also been reported for the fusion protein of other enveloped viruses (8, 32, 37).
The second furin consensus sequence RAK/RR109 in the RSV fusion protein is highly conserved in all HRSV and BRSV strains isolated so far, suggesting that this cleavage site has a role in the viral life cycle. Our results indicate that cleavage at FCS-2 is not critical for virus replication in cell culture. However, it might be advantageous for RSV replication in vivo. One possible function of pep27 and the dual cleavage might be related to the host immune response. The RSV fusion protein has recently been shown to inhibit proliferation of T cells by cell-to-cell contact (31). In analogy to measles virus (38), proteolytic activation of RSV F might be necessary for this function. It will be interesting to determine, by using the mutants described here, how the proteolytic processing of the fusion protein may influence its inhibitory activity. Another possible function might be related to pep27 itself, the intervening peptide released upon furin cleavage at FCS-2 and FCS-1. Like FCS-2, this peptide is dispensable for virus replication in cell culture. However, the motif FYGLM129 in pep27 of BRSV suggests a possible role in the host. FYGLM129 matches the signature sequence FXGLM characteristic for the tachykinin family of bioactive peptides (36). Substance P and other members of this family exhibit multiple activities, including the induction of bronchoconstriction, mucus secretion, histamine release, vasodilation, and others (5, 17, 25, 30). It remains to be experimentally shown whether pep27 of BRSV or a further processed form of it exhibits a similar tachykinin-like activity. Although the pep27 peptides of all known BRSV isolates are highly homologous to one another, they show only little similarity with the pep27 of HRSV. In particular, the latter lacks the tachykinin motif, indicating that the pep27 peptides of BRSV and HRSV might exhibit different activities. Whatever the function of the two different peptides exactly is, we should take into account that both peptides might contribute to the pathogenicity of RSV. This idea is especially important for the development of live attenuated RSV vaccines or other virus vectors expressing the RSV F protein, as well as for DNA vaccines which are based on the F gene. In this regard, the deletion mutant rBRSV-F(Δ106-130) is of particular interest since it is expected to lack the proposed activity of pep27. Another feature of this mutant that makes it an interesting vaccine candidate is the reduced cytopathic effect in infected cells. Finally, this virus will help us to study the role of pep27 in infection of the host.
Acknowledgments
We thank R. Riebe for providing the PT-11 cell line. We acknowledge the help of Jose Antonio Melero and Claes Örvell, who made monoclonal antibodies available to us.
This work was supported by grants from the European Community (QLK2-CT-1999-00443) and the Deutsche Forschungsgemeinschaft (HE 1168/11-1/2) to G.H.
REFERENCES
- 1.Baker, J. C. 1991. Human and bovine respiratory syncytial virus: immunopathologic mechanisms. Vet. Q. 13:47-59. [DOI] [PubMed] [Google Scholar]
- 2.Baker, J. C., T. R. Ames, and R. J. F. Markham. 1986. Seroepizootiologic study of bovine respiratory syncytial virus in a dairy herd. Am. J. Vet. Res. 47:240-245. [PubMed] [Google Scholar]
- 3.Buchholz, U. J., S. Finke, and K.-K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukreyev, A., S. S. Whitehead, B. R. Murphy, and P. L. Collins. 1997. Recombinant respiratory syncytial virus from which the entire SH gene has been deleted grows efficiently in cell culture and exhibits site-specific attenuation in the respiratory tract of the mouse. J. Virol. 71:8973-8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos, M. M., and J. B. Calixto. 2000. Neurokinin mediation of edema and inflammation. Neuropeptides 34:314-322. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 7.Collins, P. L., K. McIntosh, and R. M. Chanock. 1996. Respiratory syncytial virus, p. 1313-1351. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed. Raven Press, New York, N.Y.
- 8.Dedera, D., and L. Ratner. 1991. Demonstration of two distinct cytopathic effects with syncytium formation-defective human immunodeficiency virus type 1 mutants. J. Virol. 65:6129-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutch, R. E., R. N. Hagglund, M. A. Nagel, R. G. Paterson, and R. A. Lamb. 2001. Paramyxovirus fusion (F) protein: a conformational change on cleavage activation. Virology 281:138-150. [DOI] [PubMed] [Google Scholar]
- 10.Feldman, S. A., R. M. Hendry, and J. A. Beeler. 1999. Identification of a linear heparin binding domain for human respiratory syncytial virus attachment glycoprotein G. J. Virol. 73:6610-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feldman, S. A., S. Audet, and J. A. Beeler. 2000. The fusion glycoprotein of human respiratory syncytial virus facilitates virus attachment and infectivity via an interaction with cellular heparan sulfate. J. Virol. 74:6442-6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Reyes, L., M. B. Ruiz-Argüello, B. Garcia-Barreno, L. Calder, J. A. Lopez, J. P. Albar, J. J. Skehel, D. C. Wiley, and J. A. Melero. 2001. Cleavage of the human respiratory syncytial virus fusion protein at two distinct sites is required for activation of membrane fusion. Proc. Natl. Acad. Sci. USA 98:9859-9864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallak, L. K., D. Spillmann, P. L. Collins, and M. E. Peeples. 2000. Glycosaminoglycan sulfation requirements for respiratory syncytial virus infection. J. Virol. 74:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hallak, L. K., P. L. Collins, W. Knudson, and M. E. Peeples. 2000. Iduronic acid-containing glycosaminoglycans on target cells are required for efficient respiratory syncytial virus infection. Virology 271:264-275. [DOI] [PubMed] [Google Scholar]
- 15.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H.-D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 16.Heminway, B. R., Y. Yu, Y. Tanaka., K. G. Perrine, E. Gustafson, M. Bernstein, and M. S. Galinski. 1994. Analysis of respiratory syncytial virus F, G, and SH proteins in cell fusion. Virology 200:801-805. [DOI] [PubMed] [Google Scholar]
- 17.Joos, G. F., K. O. Swert, and R. A. Pauwels. 2001. Airway inflammation and tachykinins: prospects for the development of tachykinin receptor antagonists. Eur. J. Pharmacol. 429:239-250. [DOI] [PubMed] [Google Scholar]
- 18.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 19.Karron, R. A., D. A. Buonagurio, A. F. Georgiu, S. S. Whitehead, J. E. Adamus, M. L. Clements-Mann, D. O. Harris, V. B. Randolph, S. A. Udem, B. R. Murphy, and M. S. Sidhu. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc. Natl. Acad. Sci. USA 94:13961-13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimman, T. G., and F. Westenbrink. 1990. Immunity to human and bovine respiratory syncytial virus. Arch. Virol. 112:1-25. [DOI] [PubMed] [Google Scholar]
- 21.Klenk, H.-D., and W. Garten, W. 1994. Host cell proteases controlling viral pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 22.Klenk, H.-D., and W. Garten. 1994. Activation of viral spike proteins by host proteases, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Monograph 28. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Krusat, T., and H.-J. Streckert. 1997. Heparin-dependent attachment of respiratory syncytial virus (RSV) to host cells. Arch. Virol. 142:1247-1254. [DOI] [PubMed] [Google Scholar]
- 24.Kyhse-Anderson, J. 1984. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J. Biochem. Biophys. Methods 10:203-209. [DOI] [PubMed] [Google Scholar]
- 25.Lecci, A., S. Giuliani, M. Tramontana, F. Carini, and C. A. Maggi. 2000. Peripheral actions of tachykinins. Neuropeptides 34:303-313. [DOI] [PubMed] [Google Scholar]
- 26.Maisner, A., B. Mrkic, G. Herrler, M. Moll, M. A. Billeter, R. Cattaneo, and H.-D. Klenk. 2000. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 81:441-449. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, I., and J. A. Melero. 2000. Binding of human respiratory syncytial virus to cells: implication of sulfated cell surface proteoglycans. J. Gen. Virol. 81:2715-2722. [DOI] [PubMed] [Google Scholar]
- 28.Neumann, G., H. Feldmann, S. Watanabe, I. Lukashevich, and Y. Kawaoka. 2002. Reverse genetics demonstrates that proteolytic processing of the ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 76:406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pastey, M. K., and S. K. Samal. 1997. Analysis of bovine respiratory syncytial virus envelope glycoproteins in cell fusion. J. Gen. Virol. 78:1885-1889. [DOI] [PubMed] [Google Scholar]
- 30.Rogers, D. F. 2001. Motor control of airway goblet cells and glands. Respir. Physiol. 125:129-144. [DOI] [PubMed] [Google Scholar]
- 31.Schlender, J., G. Walliser, J. Fricke, and K.-K. Conzelmann. 2002. Respiratory syncytial virus fusion protein mediates inhibition of mitogen-induced T-cell proliferation by contact. J. Virol. 76:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid, E., Zurbriggen, A., Gassen, U., Rima, B., Ter Meulen, V., and J. Schneider-Schaulies. 2000. Antibodies to CD9, a tetraspan transmembrane protein, inhibit canine distemper virus-induced cell-cell fusion but not virus-cell fusion. J. Virol. 74:7554-7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stott, E. J., and G. Taylor. 1985. Respiratory syncytial virus. Brief review. Arch. Virol. 84:1-52. [DOI] [PubMed] [Google Scholar]
- 34.Strive, T., E. Borst, M. Messerle, and K. Radsak. 2002. Proteolytic processing of human cytomegalovirus glycoprotein B is dispensable for viral growth in culture. J. Virol. 76:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Techaarpornkul, S., N. Barretto, and P. E. Peeples. 2001. Functional analysis of recombinant respiratory syncytial virus deletion mutants lacking the small hydrophobic and/or attachment glycoprotein gene. J. Virol. 75:6825-6834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vanden Broeck, J., H. Torfs, J. Poels, W. Van Poyer, E. Swinnen, K. Ferket, and A. De Loof. 1999. Tachykinin-like peptides and their receptors: a review. Ann. N. Y. Acad. Sci. 897:374-387. [DOI] [PubMed] [Google Scholar]
- 37.von Messling, V., G. Zimmer, G. Herrler, L. Haas, and R. Cattaneo. 2001. The hemagglutinin of canine distemper virus determines tropism and cytopathogenicity. J. Virol. 75:6418-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weidmann, A., A. Maisner, W. Garten, M. Seufert, V. ter Meulen, and S. Schneider-Schaulies. 2000. Proteolytic cleavage of the fusion protein but not membrane fusion is required for measles virus-induced immunosuppression in vitro. J. Virol. 74:1985-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitehead, S. S., A. Bukreyev, M. N. Teng, C. Y. Firestone, M. St Claire, W. R. Elkins, P. L. Collins, and B. R. Murphy. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438-3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmer, G., I. Trotz, and G. Herrler. 2001. N-glycans of F protein differentially affect fusion activity of human respiratory syncytial virus. J. Virol. 75:4744-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimmer, G., L. Budz, and G. Herrler. 2001. Proteolytic activation of respiratory syncytial virus fusion protein: cleavage at two furin consensus sequences. J. Biol. Chem. 276:31642-31650. [DOI] [PubMed] [Google Scholar]