Abstract
Cauliflower mosaic virus (CaMV) open reading frame III (ORF III) codes for a virion-associated protein (Vap), which is one of two viral proteins essential for aphid transmission. However, unlike the aphid transmission factor encoded by CaMV ORF II, Vap is also essential for systemic infection, suggesting that it is a multifunctional protein. To elucidate the additional function or functions of Vap, we tested the replication of noninfectious ORF III-defective mutants in transfected turnip protoplasts. PCR and Western blot analyses revealed that CaMV replication had occurred with an efficiency similar to that of wild-type virus and without leading to reversions. Electron microscopic examination revealed that an ORF III frameshift mutant formed normally structured virions. These results demonstrate that Vap is dispensable for replication in single cells and is not essential for virion morphogenesis. Analysis of inoculated turnip leaves showed that the ORF III frameshift mutant does not cause any detectable local infection. These results are strongly indicative of a role for Vap in virus movement.
Plant viruses replicate within an infected cell, move from cell to cell, and are transported from plant to plant. Most of the seven open reading frames (ORFs) of cauliflower mosaic virus (CaMV), a plant pararetrovirus, have been assigned to one or other of these functions (27). ORFs IV, V, and VI are required for basic virus replication, encoding the precursor of the capsid proteins (CP), the precursor of protease and reverse transcriptase/RNase H (Pol), and the transactivator/viroplasmin (Tav), respectively (K.K. and T.H., submitted for publication). ORF I, encoding a factor allowing cell-to-cell movement (Mov) (30, 32), is required for movement within the plant, and ORF II, encoding aphid transmission factor (Atf) (1, 34), is needed for insect transmission. ORF VII is dispensable for systemic infection and is therefore difficult to assign. The most poorly understood function of CaMV is encoded by ORF III, although it is known to be essential for systemic infection (4, 6, 15). The protein product of ORF III is associated with the virions, giving it the name “virion-associated protein” (Vap) (5, 10). However, the association is relatively weak, and Vap is lost upon the urea treatment required to release virus particles from the virus inclusion body (viroplasm). Vap-related genes have been found in all caulimoviruses, but there is no clear corresponding protein in retroviruses. The Vap of CaMV and other caulimoviruses has a high α-helical content, part of which is required for tetramerization via an N-terminal coiled-coil structure (20, 29, 31). The C-terminal part of Vap includes domains for RNA and DNA binding (15) and for interaction with the CP (22). Finally, Vap interacts, via its N-terminal part, with Atf (23).
The molecular interactions of Vap have formed the basis of various interpretations of its functions. Thus, it was speculated that interactions with CP and with nucleic acids have a role in guiding reverse transcription, packaging of the genomic DNA, and/or virus assembly (10, 20, 21). On the other hand, it was shown that Vap is required for aphid transmission in addition to and via interaction with Atf (23). If purely involved in insect transmission, one would expect it to be dispensable for infection of a single plant upon mechanical inoculation, yet this is not the case (4, 6,15). Accordingly, one can assume that Vap has some additional function, either in basic virus replication or in movement. To distinguish between these possibilities, a protoplast transfection system (32) was employed. The CaMV genome and mutant derivatives were cloned with redundant termini, corresponding to the long terminal repeats (LTRs) of retroviruses, and introduced into turnip protoplasts. Progeny viral genomes were identified by PCR and distinguished from input DNA by their resistance to the restriction enzyme DpnI. The results showed that Vap is dispensable for basic replication, ruling out an essential function in aiding reverse transcription, genome packaging, or assembly. We infer that Vap has a role in virus movement within the infected plant, in addition to its role in transmission from plant to plant.
MATERIALS AND METHODS
Plasmids.
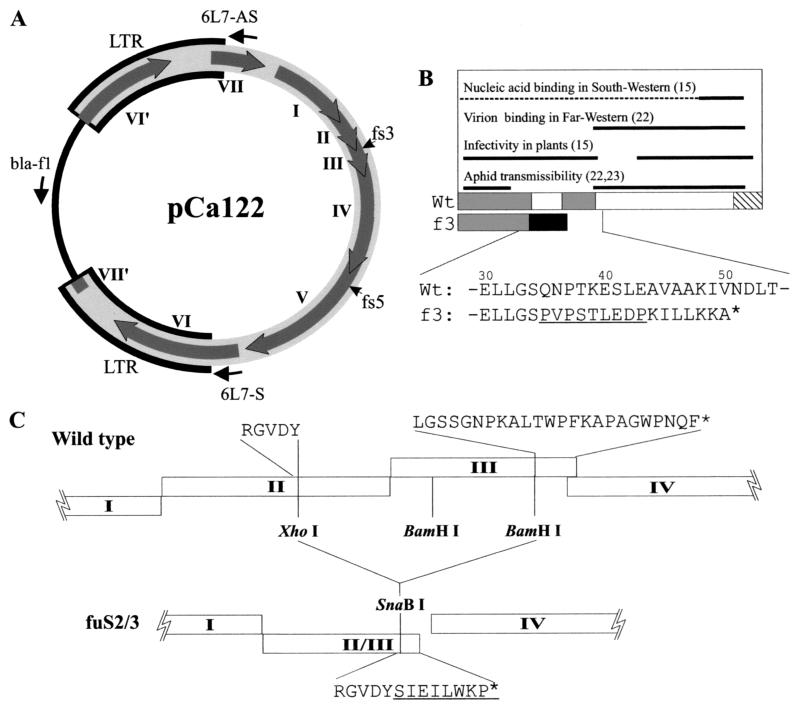
A CaMV DNA clone with artificial LTRs (pCa122) (Fig. 1A) is infectious in plants and is able to give rise to CaMV replication in transfected turnip protoplasts (32). A frameshift mutant, pCafs3, which has a 26-bp insertion of GGA TCC CCG GTA CCG TCG ACT CTA GAG GAT CC in the BamHI site (underlined) at nucleotide (nt) 1927 (numbering according to Gardner et al. [8]), was constructed as described previously (31). In this mutant, ORF III terminates eight codons downstream of the insertion (Fig. 1B). A second ORF III-defective mutant, pCafuS2/3, was constructed by deleting an XhoI-BamHI fragment (positions1648 to 2148) from pCa122 (Fig. 1C). Since ligation between the blunted XhoI and BamHI sites resulted in in-phase fusion of ORFs II and III with restoration of the XhoI site, the latter was destroyed to cause a frameshift by end-filling and religation (confirmed by sequencing). An ORF V frameshift mutant, pCafs5, has a 22-bp inserted sequence of GAT ATC GAC TCT AGA GGA TCC CCG CGT CGA TAT C in the ClaI site (underlined) at position 3959. This insertion site is within the protease domain of Pol, and therefore the mutant should not exhibit any of the ORF V-encoded enzymatic activities. An ORF I frameshift mutant clone, pCafs1, was described previously as pCa822-3 (32). The Tav-expressing plasmid p35S-P6 was described by Kobayashi et al. (18). The CaMV clone pCa′S′ corresponds to a single-copy CaMV Strasbourg strain DNA cloned into pBR322 at the unique SalI site. All plasmids were propagated in a dam methylase-positive strain of Escherichia coli (DH5α).
FIG. 1.
(A) Map of infectious CaMV clone pCa122. Thick gray line, CaMV DNA; thick black outline, redundant regions (LTRs); arrows on the circle, CaMV ORFs I, II, III, IV, V, VI, and VII (and disrupted ORFs VI′ and VII′); thin black line, vector sequence; thin arrows outside the circle, positions of the PCR primers; fs3 and fs5, mutation sites in pCafs3 and pCafs5, respectively. (B) Schematic representation of CaMV Vap functional domains. Boxes show wild-type (Wt) and ORF III frameshift mutant (f3) coding regions. Gray, hatched, and black boxes show the coiled-coil domain(s), the proline-rich domain, and the altered sequence in the frameshift mutant, respectively. Essential regions for reported Vap functions are shown as thick black lines above the protein (references in parentheses). The dotted line shows the region possibly involved in nucleic acid binding, since the role of the N-terminal region in this function has not yet been clarified. Partial amino acid sequences of the wild-type and the frameshift mutant Vap (numbering from the N terminus) are shown, with the inserted sequence underlined. (C) Arrangement of coding sequences between ORFs I and IV in pCa122 (wild type) and pCafuS2/3. Open boxes, CaMV ORFs as indicated. Amino acid sequences flanking the deletion borders are shown for both the wild type and the mutant fuS2/3.
Preparation, transfection, and culture of protoplasts.
Turnip protoplasts were prepared, transfected, and cultured essentially as described by Tsuge et al. (32), with slight modification. Brassica rapa cv. Just Right protoplasts were also used in some experiments. Protoplasts were washed three to five times with Ca2+-free 0.5 M mannitol and twice with 0.5 M mannitol containing 1 mM CaCl2 and divided into aliquots of 1 × 106 to 2 × 106 cells/tube. The protoplasts were resuspended in 0.6 ml of solution T (0.5 M mannitol, 40 mM CaCl2, 10 mM morpholineethanesulfonic acid-KOH [pH 5.8]) containing a total of 100 μg of plasmid DNA, immediately followed by the addition of 0.9 ml of 40% (wt/vol) polyethylene glycol 3000 (PEG; Merck) in solution T. In all transfections with pCa122 or its derivatives, p35S-P6 was cotransfected to enhance virus replication (18). After treatment with PEG for 30 min and washing with high-Ca2+, high-pH buffer, the protoplasts were cultured at 25°C under conditions of 16 h of light followed by 8 h of darkness.
Preparation of virion fraction.
A virion-enriched fraction was prepared essentially as described by Gardner and Shepherd (9). The protoplast pellet was suspended in 0.7 ml of 200 mM Tris-HCl (pH 7.5), containing 20 mM EDTA, 1.5 M urea, and 2% Triton X-100, and well lysed by repeated pipetting on ice. The lysate was cleared by centrifugation at 10,000 × g for 10 min at 4°C, layered on a 0.3-ml sucrose cushion (15% sucrose, 50 mM Tris-HCl [pH 7.5], 5 mM EDTA) in a 1-ml ultracentrifuge tube, and centrifuged at 80,000 rpm for 1 h at 4°C in a Beckman TLA100.2 rotor. The supernatant was carefully removed to obtain the virion fraction as a pellet.
Isolation of DNA.
Total DNA from the transfected protoplasts was isolated by standard procedures. Protoplasts were lysed in 0.4 ml of DNA extraction buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 500 mM NaCl, 1% sodium dodecyl sulfate [SDS]) containing 0.5 mg of proteinase K per ml. This was incubated for 4 to 16 h at 37°C. DNA was then extracted twice with Tris-buffered phenol and twice with chloroform-isoamyl alcohol (24:1) before being precipitated in 1 ml of ethanol. The DNA was purified with a QIAquick PCR purification kit (QIAGEN) according to the manufacturer's specifications. DNA from the virion fraction was isolated as described above, except that the fraction was treated with DNase prior to DNA extraction as follows. The virion fraction pellet was suspended in 0.2 ml of 100 μg of DNase I per ml in DNase buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 10 mM MgCl2, 1 mM dithiothreitol) and incubated for 1 h at 37°C before addition of 200 μl of 2× concentrated DNA extraction buffer containing 1 mg of proteinase K per ml and processing as described above.
PCR analysis of viral and plasmid DNA.
Total DNA or virion fraction DNA from about 5 × 104 transfected protoplasts was analyzed for both the progeny viral DNA and the input plasmid DNA. To detect viral progeny DNA and plasmid DNA, PCR primer sets 6L7-S (CTG GAG ACT GAG AAA ATC AGA C; nt 5744 to 5765) and 6L7-AS (TGA AAC CTT ACG GTG GTA AAC; nt 158 to 138, complementary) and 6L7-S and bla-f1 (CAG CAA TAA ACC AGC CAG CC; nt 3463 to 3482 of pBR322) (numbering according to Watson [33]), respectively, and Taq DNA polymerase (Gibco-BRL) were used. The PCR conditions used with these primers were 94°C for 5 min and 27 or 35 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 2 min, followed by 72°C for 10 min. For specific detection of newly replicated progeny viral DNA, total DNA was treated with DpnI to digest Dam-methylated plasmid DNA prior to PCR analysis (see Results). The ORF III region was amplified with the primers Vap-PN (GCA TCG ATT GAA ATG GCT AAT CTT AAT CAA) and Vap-PC (GCG TCG ACC CTA AAA TTG ATT CGG CCA TCC) under the conditions described above, but with Pfu DNA polymerase (Promega). The amplified DNA fragments were sequenced with the same primers as for PCR by the dye terminator method on an ABI PRISM 377 DNA sequencer. pCa′S′ was used as a positive control for PCR (with primers 6L7-S and 6L7-AS), because it lacks interruptions within the target region.
Western blot analysis of virion proteins.
Virion fractions were examined for CaMV CP by Western blotting. Virion fraction pellets were dissolved in 10 mM Tris-HCl-1 mM EDTA (pH 8.0). The protein was denatured and fractionated by SDS-polyacrylamide gel electrophoresis as described by Laemmli (19). The fractionated protein was transferred to polyvinylidene difluoride (PVDF) membrane (Immobilon P; Millipore) with 10 mM CAPS (Na 3-[cyclohexylamino]-1-propane sulfonate) buffer (pH 10.5) containing 5% methanol. CaMV CP was detected with a rabbit antibody raised against recombinant CaMV CP, goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase (HRP) conjugate (Bio-Rad), and Supersignal West Pico chemiluminescent substrate (Pierce).
ISEM.
The virion fractions from six transfections each with pCa122 or pCafs3 were pooled and used for immunosorbent electron microscopy (ISEM). After coating with anti-CaMV CP antiserum, EM grids were extensively washed and absorbed with the virus preparations resuspended in 50 mM Tris-HCl (pH 7.5). They were then washed again and covered with anti-CaMV CP antiserum and finally negatively stained with 1% uranyl acetate. Samples were analyzed with a Zeiss EM910 transmission EM operated at 80 kV.
Assays to detect CaMV infection in inoculated leaves.
Turnip plants were inoculated with 10 μg of pCa122, pCafs1, or pCafs3 per leaf at the three-leaf stage as described previously (32). Local infection with CaMV was examined by hammer blotting (28) at 5, 8, and 10 days postinfection (dpi) and 3 weeks postinfection by immunofluorescence analysis of protoplasts. After removal of the midrib, leaves inoculated with one of the plasmids described above were placed between two sheets of Whatman 3MM filter paper, wrapped with plastic film, and gently hit with a hammer so that sap from broken leaf tissue could be uniformly adsorbed by the filter paper. After removal of leaf debris with tweezers, the blots were fixed with methanol at room temperature for 10 min and kept in 70% ethanol at 4°C until analysis. The blots were washed extensively with Tris-buffered saline (TBS) containing 1% Triton X-100 until the green color was almost completely removed and then blocked in TBS containing 2% skim milk and 0.2% NaN3 overnight at room temperature. CaMV CP was detected with a rabbit antibody raised against recombinant CaMV CP and goat anti-rabbit IgG-HRP conjugate (Bio-Rad), as in Western blotting, and visualized with diaminobenzidine as a substrate. Protoplasts were isolated from strips (approximately 1 mm) of inoculated leaves as described previously (7). After being washed twice with 0.5 M mannitol, the protoplasts were fixed as described previously (7) and stained for CaMV CP with rabbit anti-recombinant CaMV CP antibody and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG antibody.
RESULTS
DNA replication of a CaMV ORF III frameshift mutant in transfected protoplasts.
Plasmid pCa122 (Fig. 1) contains the wild-type CaMV genome provided with artificial equivalents to retroviral LTRs, including promoter and polyadenylator sequences. This allows the escape by transcription of CaMV pregenomic RNA, which then serves as a template for reverse transcriptase to produce viral DNA. Upon inoculation of turnip plants, pCa122 leads to systemic infection, while pCafs5 and pCafs3 (Fig. 1B), which carry frameshift mutations in the protease region of ORF V and in ORF III, respectively, do not (data not shown).
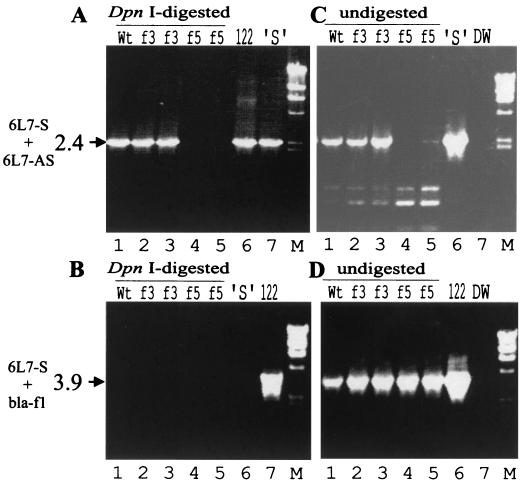
In plant protoplast transfections with the same plasmids, replication of the viral genome can be assessed by PCR of total DNA after digestion with DpnI, which cleaves the original Dam-methylated plasmid, but not progeny viral DNA. Primers 6L7-S and 6L7-AS were designed to yield a 6.5-kb fragment from undigested plasmid DNA and a 2.4-kb fragment from newly replicated viral DNA (Fig. 1A). Another primer pair (virus-based 6L7-S and plasmid-based bla-f1) would yield a 3.9-kb fragment from the undigested plasmid. In protoplasts transfected with pCa122, a 2.4-kb DNA fragment was detected as expected (Fig. 2A). The 2.4-kb fragment was also observed with pCafs3, showing that this viral genome is replication competent, but not with the ORF V mutant pCafs5. Neither the 6.5-kb fragment nor the 3.9-kb fragment could be visualized (Fig. 2A and B), showing that the input plasmid DNA was removed. However, if DpnI digestion was omitted, the 2.4- and 3.9-kb fragments could be detected from all three constructs by using the viral and plasmid primers, respectively (Fig. 2C and D). This shows that the input plasmids survive the handling, but can be effectively removed by DpnI treatment. Interestingly, only very little 6.5-kb fragment could be seen (Fig. 2A, lane 6). We interpret this as being due to a template switch of the Taq DNA polymerase between the redundant CaMV sequences causing recombination and leading to the 2.4-kb fragment from plasmid DNA. It is likely that PCR of the 2.4-kb fragment, once created, competes successfully with amplification of the much larger 6.5-kb fragment.
FIG. 2.
PCR analysis of CaMV DNA replication in transfected protoplasts. Turnip protoplasts were transfected with 20 μg of p35S-P6 and 30 μg of pBluescript plus 50 μg of pCa122 (wild type [Wt]; lane 1), pCafs3 (f3; lanes 2 and 3), or pCafs5 (f5; lanes 4 and 5). Total DNA from the transfected protoplasts was analyzed by PCR with the primer sets 6L7-S and 6L7-AS (A and C) and 6L7-S and bla-f1 (B and D), before (C and D) or after (A and B) DpnI digestion. 122, pCa122; ′S′, pCa′S′; DW, DNA-free control for PCR; M, molecular weight standard (HindIII-digested lambda phage DNA). The sizes (in kilobase pairs) and positions of PCR products are shown on the left together with the names of the primers used.
In summary, these results show that the conditions used enable detection of CaMV genome replication via reverse transcription in transfected plant protoplasts and that full-length ORF III product (i.e., Vap) is not required for this process.
Encapsidation of CaMV ORF III frameshift mutant progeny DNA.
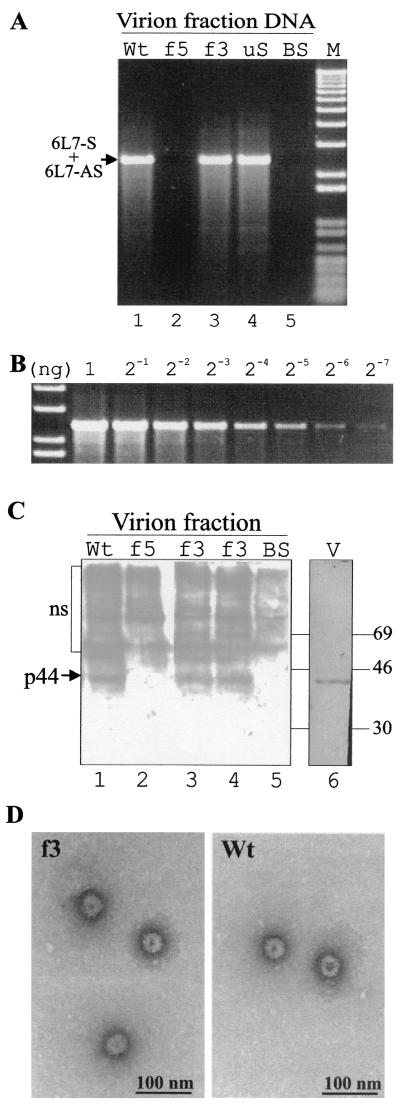
To test whether the progeny viral DNA is encapsidated, DNA was extracted from the virion fraction obtained from protoplasts at 72 h posttransfection. The DNA isolation protocol included the isolation of a virion-enriched fraction by using a urea-Triton procedure and DNase pretreatment. The virion-enriched fraction did not yield the plasmid-derived 3.9-kb DNA fragment by PCR with the primer pair designed to detect the plasmid (data not shown), showing that the input DNA was efficiently eliminated by the DNase pretreatment. In contrast, PCR with the viral primer pair still gave rise to amplification of 2.4-kb DNA fragments from protoplasts transfected with pCa122 and pCafs3, but not pCafs5 (Fig. 3A). These experiments show that DNA produced from the Vap mutant had accumulated in a urea- and DNase-protected form, much like the wild-type viral genome, again suggesting that Vap is dispensable for CaMV replication in single cells.
FIG. 3.
Detection of progeny viral DNA, coat protein, and virions in transfected protoplasts. (A) Turnip protoplasts were transfected with 20 μg of p35S-P6 and 30 μg of pBluescript plus 50 μg of pCa122 (wild type [Wt]; lane 1), pCafs5 (f5; lane 2), pCafs3 (f3; lane 3), pCafuS2/3 (uS; lane 4), or 100 μg of pBluescript (BS; lane 5). Virion fraction DNA from the transfected protoplasts was analyzed by PCR with primers 6L7-S and 6L7-AS. M, molecular weight standard (1-kb-plus ladder; GIBCO-BRL). (B) PCR with primers 6L7-S and 6L7-AS was performed with serially diluted pCa′S′ in parallel with those shown in panel A to serve as a standard for approximate PCR quantification. (C) Turnip protoplasts were transfected with 20 μg of p35S-P6 and 30 μg of pBluescript plus 50 μg of pCa122 (Wt; lane 1), pCafs5 (f5; lane 2), pCafs3 (f3; lanes 3 and 4), or 100 μg of pBluescript (BS; lane 5). Virion fractions from the transfected protoplasts were analyzed for CaMV capsid protein by Western blotting. Purified CaMV (2 μg) transferred onto PVDF membrane was stained with Coomassie brilliant blue (V; lane 6). The position of the major (p44) capsid protein is indicated by an arrow. ns, nonspecific signals in lanes 1 to 5. The positions of molecular mass standards (in kilodaltons) are shown on the right. (D) Virion fraction was prepared from protoplasts transfected with 20 μg of p35S-P6 and 30 μg of pBluescript plus 50 μg of pCa122 (Wt) or pCafs3 (f3). The virions were captured on anti-CaMV CP antibody-coated grids, labeled with the same antibody, and examined under an electron microscope.
Next, we examined whether CaMV CP cosediments with the progeny viral DNA in virion fractions from transfected protoplasts. Western blotting of virion fractions from pCa122- and pCafs3-transfected protoplasts detected similar amounts of 44-kDa CP (Fig. 3C), but this protein was not detected in pCafs5-transfected protoplasts (Fig. 3C). These results indicate that CaMV can form virions in the absence of Vap. The lack of virus-like particles in pCafs5-transfected protoplasts is probably due to the absence of enzymatic functions required for virus replication.
It is unlikely that CaMV CP and the viral DNA cosediment in the same fraction without interacting, because CaMV pregenomic RNA has been shown to interact with viral CP in vitro (11), and CaMV DNA replication depends on the presence of the CP precursor (K.K. and T.H., submitted). It is, however, still possible that only abnormal capsid protein-DNA complexes are formed, providing protection of the DNA in the absence of Vap. ISEM was employed to clarify this question. Since the virus extraction involves urea, thus removing the majority of Vap from wild-type virions, it is not surprising that the progeny from the ORF III frameshift mutant was indistinguishable from those of the wild type (Fig. 3D). This shows that normal CaMV virions can be formed in the absence of Vap and confirms the notion of Leh et al. (22) that Vap binds to the surface of virions and is not an integral part of the capsid.
Stability of the ORF III frameshift mutant.
Replication via reverse transcription leads to high reversion rates of CaMV mutants (e.g., reference 26), and the possibility that a reversion of the ORF III (Vap) frameshift mutation occurred in our experiments had to be seriously considered. Therefore, the entire ORF III was amplified by PCR from the DNA obtained from the virion fractions of pCa122- and pCafs3-transfected protoplasts. The nucleotide sequence of the PCR product was directly determined with the same primers. No alteration of the nucleotide sequence in the ORF III region was detected in any of the progeny DNAs, confirming the stability of the mutation throughout the duration of the experiment.
Replication of a CaMV ORF II/III deletion mutant in transfected protoplasts.
Although the results presented above clearly indicated that the ORF III frameshift mutant could replicate in transfected protoplasts, this mutant can express the first 34 amino acid residues of ORF III, which may potentially have a role in viral DNA replication and/or encapsidation. To examine if CaMV Vap is totally dispensable for viral replication in transfected protoplasts, we tested the replication in protoplasts of a second CaMV mutant clone. In CaMV ORF II/III deletion mutant pCafuS2/3, the C-terminal region of ORF III was fused to the N-terminal half of ORF II with a +1 frameshift, and thus this mutant cannot express any portion of CaMV ORF III (Fig. 1C). This mutant produced DNase-protected progeny viral DNA in transfected protoplasts at levels similar to those of the wild type and pCafs3 (Fig. 3A), confirming that CaMV Vap is totally dispensable for CaMV replication in single cells. PCR analysis with 27 reaction cycles—i.e., within the range of linear amplification of the target DNA (Fig. 3B)—revealed that progeny viral DNA levels from both mutants tested (pCafs3 and pCafuS2/3) were comparable to that from the wild type (Fig. 3A).
Replication of the ORF III frameshift mutant in inoculated turnip leaves.
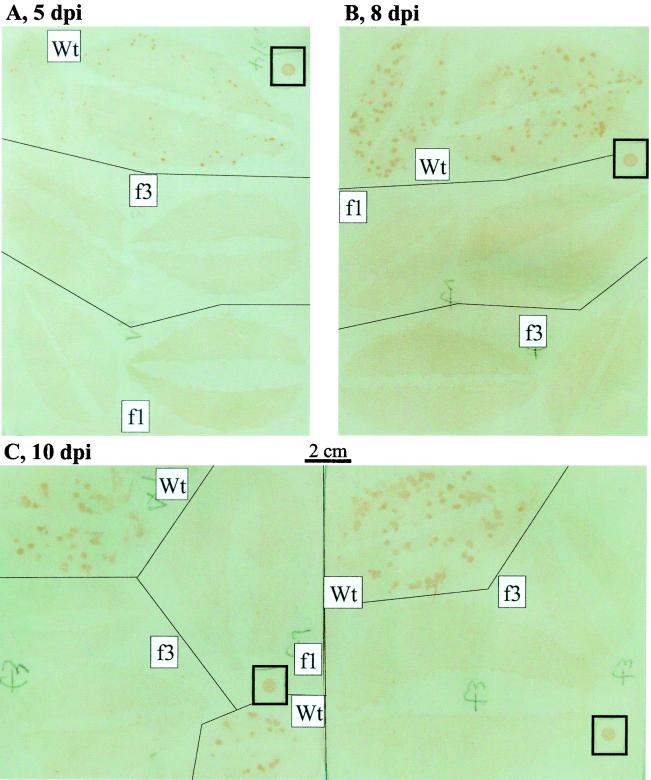
The inability of CaMV ORF III mutants to cause systemic infection was confirmed by PCR with primers 6L7-S and AS (data not shown). This result, together with the results presented above, suggests that ORF III-defective mutants are unable to support either cell-to-cell or long-distance movement. To discriminate between these possibilities, we inoculated turnip leaves with pCa122 or pCafs3 and tested for viral replication. An ORF I frameshift mutant, which is defective in the viral cell-to-cell movement function, was also tested in parallel. Hammer blotting detected replication and spread (i.e., the establishment of local infection) of wild-type virus, but failed to detect propagation of the mutants (Fig. 4). Immunofluorescence analysis of protoplasts prepared from the inoculated leaves demonstrated that, in pCa122-inoculated leaves, 16.9% (mean of three independent experiments) of the cells harbored the viral antigen. In contrast, no protoplasts with CaMV antigen were observed in pCafs3- or pCafs1-inoculated leaves. These results indicate that the ORF III frameshift mutant is unable to cause any detectable local infection in inoculated leaves.
FIG. 4.
Hammer blot analysis for local infection with CaMV in inoculated leaves. Turnip leaves inoculated with pCa122 (wild type [Wt]), pCafs3 (f3), or pCafs1 (f1) were harvested 5 (A), 8 (B), and 10 (C) dpi and analyzed by hammer blotting. Local infection with CaMV was detected as brown spots, which grew larger over time. The brown spots in small squares represent signals from 300 ng (in 3 μl) of purified CaMV. The faint bluish background is an artifact caused by the image scanner used, but was left uncorrected because it highlights background brown staining that represents leaf images.
DISCUSSION
Recently, Leh et al. used a reconstituted virus acquisition system (2) to show that Vap is indispensable for aphid transmission of CaMV and that it interacts with the CaMV insect transmission factor Atf (23). However, this cannot be its only function, since Vap is required for systemic infection upon artificial inoculation of host plants.
The molecular properties of Vap reported so far include nucleic acid (10), virion or CP (5, 21, 22, 31), and Atf (23) binding, as well as tetramer formation (20, 31). Based on these properties, a role for Vap in virion assembly, viral genome replication, and/or genome packaging of CaMV had been suspected. Unexpectedly, however, as shown in this study, CaMV Vap is dispensable for normal viral replication in single transfected protoplasts, ruling out an essential function in virus morphogenesis and genome packaging. Use of two different mutants demonstrated that Vap is totally dispensable for CaMV replication in single cells.
We have also shown that an ORF III frameshift mutant is unable to establish detectable local infection in turnip leaves. This was supported by negative results from hammer blotting (Fig. 4) and immunofluorescence analysis, although by the same methods, we have detected local infection with other CaMV mutants that cannot establish systemic infection (K.K. and T.H., unpublished data), supporting the conclusion that the ORF III frameshift mutant is indeed unable to cause local infection. Since the replication levels of the ORF III frameshift mutant were comparable to those of wild-type virus (Fig. 3A), the mutant progeny would have been detected if they had spread to adjacent cells. Therefore, the results from this study collectively suggest that Vap has a role in virus movement. Another viral ORF, ORF I, had already been shown to be dispensable for viral replication in primarily infected cells (30, 32), but to be necessary for cell-to-cell movement. Features of the product of ORF I, Mov, that identified it as a movement protein (MP) include amino acid sequence homology with tobamoviral MP (14), subcellular localization (24), and the ability to induce tubular structures in protoplasts and insect cells (16, 25). CaMV Vap may be an additional component of the CaMV cell-to-cell movement machinery, although it has not been reported to localize to any movement-related subcellular compartment, such as plasmodesmata or the cytoskeleton, in infected cells. Alternatively, Vap may be involved in intracellular movement, which would be necessary to deliver virions to the Mov-modified plasmodesmata. Unlike tobamoviral MP (12), Mov has not been shown to interact with the cytoskeleton (13), and the potential role of Vap in this aspect of the viral life cycle remains to be investigated.
Caulimoviruses are thought to move from cell to cell as virions (17), but additional movement as (deoxy)ribonucleoprotein complexes (3) has not been excluded. Since Vap is known to bind both to CaMV particles (22) and to naked nucleic acids (10), either form of CaMV movement could provide a role for Vap in intercellular and/or intracellular transport functions.
We do not think that Vap has a role in initiating CaMV replication (such as uncoating, nuclear transport, etc.) in newly invaded cells after cell-to-cell movement, because both naked CaMV DNA and CaMV particles purified by the urea-Triton procedure—which are devoid of Vap—are able to infect host plants efficiently (2). One may claim that CaMV particles formed in the absence of Vap could not uncoat viral genome after invading the adjacent cells. Our results do not exclude this possibility, but it is not very likely that CaMV CP could be assembled into two functionally distinct forms of indistinguishable shape and size (Fig. 3D). Thus, the inability of Vap mutant CaMV to cause local infection in inoculated host plants is strongly indicative of a block in a local transport function early in infection.
We envisage a role for Vap in cell-to-cell transport, perhaps by guiding the viral particles to the cell membrane or by participating in their transfer across the cell wall. How might this function of Vap be linked to its established function in aphid transmission? Stavolone et al. (29) proposed that Vap, anchored via its interaction with CP, could act as the multifunctional “arm” of the virus particle, interacting with different viral and host factors to target different pathways throughout the infection cycle. One such factor is Atf, thereby allowing virus transport from plant-to-plant; another could involve a factor (possibly Mov) directing cell-to-cell movement. We are currently testing this model and searching for additional Vap-interacting partners to further our understanding of Vap function and the mechanism of CaMV movement in plants.
Acknowledgments
K.K. and S.T. contributed equally to this work.
We are very grateful to I. Furusawa (Kyoto University), in whose laboratory S.T. initiated this study, for continuous encouragement. We thank I. Furusawa and H. Rothnie for critical reading of the manuscript and K. Mise and K. Fujisaki for the hammer blotting protocol. Sincere thanks are due to all members of the Hohn laboratory, as well as members of the former laboratory of I. Furusawa for fruitful discussions and kind cooperation.
This work was supported by the Novartis Research Foundation (K.K., L.S., and T.H.) and by the Kyoto Prefecture (S.T.).
REFERENCES
- 1.Armour, S. L., U. Melcher, T. P. Pirone, D. J. Lyttle, and R. C. Essenberg. 1983. Helper component for aphid transmission encoded by region II of cauliflower mosaic virus DNA. Virology 129:25-30. [DOI] [PubMed] [Google Scholar]
- 2.Blanc, S., M. Cerutti, M. Usmany, J. M. Vlak, and R. Hull. 1993. Biological activity of cauliflower mosaic aphid virus transmission factor expressed in a heterologous system. Virology 192:643-650. [DOI] [PubMed] [Google Scholar]
- 3.Citovski, V., D. Knorr, and P. Zambryski. 1991. Gene I, a potential cell-to-cell movement locus of cauliflower mosaic virus, encodes an RNA-binding protein. Proc. Natl. Acad. Sci. USA 88:2476-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daubert, S. D., R. J. Shepherd, and R. C. Gardner. 1983. Insertional mutagenesis of the cauliflower mosaic virus genome. Gene 25:201-208. [DOI] [PubMed] [Google Scholar]
- 5.Dautel, S., T. Guidasci, M. Pique, J. L. Mougeot, G. Lebeurier, P. Yot, and J. M. Mesnard. 1994. The full-length product of cauliflower mosaic virus open reading frame III is associated with the viral particle. Virology 202:1043-1045. [DOI] [PubMed] [Google Scholar]
- 6.Dixon, L. K., I. Koenig, and T. Hohn. 1983. Mutagenesis of cauliflower mosaic virus. Gene 25:189-199. [DOI] [PubMed] [Google Scholar]
- 7.Furusawa, I., N. Yamaoka, T. Okuno, M. Yamamoto, M. Kohno, and H. Kunoh. 1980. Infection of turnip protoplasts with cauliflower mosaic virus. J. Gen. Virol. 48:431-435. [DOI] [PubMed] [Google Scholar]
- 8.Gardner, R. C., A. J. Howarth, P. Hahn, M. Brown Leudi, R. J. Shepherd, and J. Messing. 1981. The complete nucleotide sequence of an infectious clone of cauliflower mosaic virus by m13mp7 shotgun sequencing. Nucleic Acids Res. 9:2871-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, R. C., and R. J. Shepherd. 1980. A procedure for rapid isolation and analysis of cauliflower mosaic virus DNA. Virology 106:159-161. [DOI] [PubMed] [Google Scholar]
- 10.Giband, M., J.-M. Mesnard, and G. Lebeurier. 1986. The gene III product (P15) of cauliflower mosaic virus is a DNA-binding protein while an immunologically related P11 polypeptide is associated with the virion. EMBO J. 5:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guerra-Peraza, O., M. de Tapia, T. Hohn, and M. Hemmings-Mieszczak. 2000. Interaction of the cauliflower mosaic virus coat protein with the pregenomic RNA leader. J. Virol. 74:2067-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinlein, M., B. L. Epel, H. S. Padgett, and R. N. Beachy. 1996. Interaction of tobamovirus movement proteins with the plant cytoskeleton. Science 270:1983-1985. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Z., Y. Han, and S. H. Howell. 2000. Formation of surface tubules and fluorescent foci in Arabidopsis thaliana protoplasts expressing a fusion between the green fluorescent protein and the cauliflower mosaic virus movement protein. Virology 271:58-64. [DOI] [PubMed] [Google Scholar]
- 14.Hull, R., J. Sadler, and M. Longstaff. 1986. The sequence of carnation edge ring virus DNA: comparison with CaMV and retroviruses. EMBO J. 12:3083-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquot, E., A. Geldreich, M. Keller, and P. Yot. 1998. Mapping regions of the cauliflower mosaic virus ORF III product required for infectivity. Virology 242:395-402. [DOI] [PubMed] [Google Scholar]
- 16.Kasteel, D. T., M. C. Perbal, J. C. Boyer, J. Wellink, R. W. Goldbach, A. J. Maule, and J. W. van Lent. 1996. The movement proteins of cowpea mosaic virus and cauliflower mosaic virus induce tubular structures in plant and insect cells. J. Gen. Virol. 77:2857-2864. [DOI] [PubMed] [Google Scholar]
- 17.Kitajima, E. W., and J. A. Lauritis. 1969. Plant virions in plasmodesmata. Virology 37:681-684. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi, K., S. Tsuge, H. Nakayashiki, K. Mise, and I. Furusawa. 1998. Requirement of cauliflower mosaic virus open reading frame VI product for viral gene expression and multiplication in turnip protoplasts. Microbiol. Immunol. 42:377-386. [DOI] [PubMed] [Google Scholar]
- 19.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 20.Leclerc, D., L. Burri, A. V. Kajava, J. L. Mougeot, D. Hess, A. Lustig, G. Kleemann, and T. Hohn. 1998. The open reading frame III product of cauliflower mosaic virus forms a tetramer through a N-terminal coiled-coil. J. Biol. Chem. 273:29015-29021. [DOI] [PubMed] [Google Scholar]
- 21.Leclerc, D., L. Stavolone, E. Meier, O. Guerra-Peraza, E. Herzog, and T. Hohn. 2001. The product of ORF III in cauliflower mosaic virus interacts with the viral coat protein through its C-terminal proline rich domain. Virus Genes 22:159-165. [DOI] [PubMed] [Google Scholar]
- 22.Leh, V., E. Jacquot, A. Geldreich, M. Haas, S. Blanc, M. Keller, and P. Yot. 2001. Interaction between the open reading frame III product and the coat protein is required for transmission of cauliflower mosaic virus by aphids. J. Virol. 75:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leh, V., E. Jacquot, A. Geldreich, T. Hermann, D. Leclerc, M. Cerutti, P. Yot, M. Keller, and S. Blanc. 1999. Aphid transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO J. 18:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linstead, P. J., G. J. Hills, K. A. Plaskitt, I. G. Wilson, C. L. Harker, and A. J. Maule. 1988. The subcellular location of the gene I product of cauliflower mosaic virus is consistent with a function associated with virus spread. J. Gen. Virol. 69:1809-1818. [Google Scholar]
- 25.Perbal, M. C., C. L. Thomas, and A. J. Maule. 1993. CaMV gene I product (pI) forms tubular structures which extend from the surface of infected protoplasts. Virology 195:281-285. [DOI] [PubMed] [Google Scholar]
- 26.Pooggin, M. M., T. Hohn, and J. Fütterer. 1998. Forced evolution reveals the importance of short open reading frame A and secondary structure in the cauliflower mosaic virus 35S RNA leader. J. Virol. 72:4157-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rothnie, H. M., Y. Chapdelaine, and T. Hohn. 1994. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv. Virus Res. 44:1-67. [DOI] [PubMed] [Google Scholar]
- 28.Srinivasan, I., and S. A. Tolin. 1992. Detection of three viruses of clovers by direct tissue immunoblotting. Phytopathology 82:721. [Google Scholar]
- 29.Stavolone, L., E. Herzog, D. Leclerc, and T. Hohn. 2001. Tetramerization is a conserved feature of the virion-associated protein in plant pararetroviruses. J. Virol. 75:7739-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas, C. L., M. C. Perbal, and A. J. Maule. 1993. A mutation of cauliflower mosaic virus gene I interferes with virus movement but not virus replication. Virology 192:415-421. [DOI] [PubMed] [Google Scholar]
- 31.Tsuge, S., K. Kobayashi, H. Nakayashiki, K. Mise, and I. Furusawa. 1999. Cauliflower mosaic virus ORF III product forms a tetramer in planta: its implication in viral DNA folding during encapsidation. Microbiol. Immunol. 43:773-780. [DOI] [PubMed] [Google Scholar]
- 32.Tsuge, S., K. Kobayashi, H. Nakayashiki, T. Okuno, and I. Furusawa. 1994. Replication of cauliflower mosaic virus ORF 1 mutants in turnip protoplasts. Ann. Phytopathol. Soc. Jpn. 60:27-35. [Google Scholar]
- 33.Watson, N. 1988. A new revision of the sequence of plasmid pBR322. Gene 70:399-403. [DOI] [PubMed] [Google Scholar]
- 34.Woolston, C. J., S. N. Covey, J. R. Penswick, and J. W. Davies. 1983. Aphid transmission and a polypeptide are specified by a defined region of the cauliflower mosaic virus genome. Gene 23:15-23. [DOI] [PubMed] [Google Scholar]