Abstract
Retroviral recombination can occur between two viral RNA molecules (intermolecular) or between two sequences within the same RNA molecule (intramolecular). The rate of retroviral intramolecular recombination is high. Previous studies showed that, after a single round of replication, 50 to 60% of retroviral recombinations occur between two identical sequences within a Moloney murine leukemia virus-based vector. Recombination can occur at any polymerization step within the retroviral replication cycle. Although reverse transcriptase is assumed to contribute to the template switches, previous studies could not distinguish between changes introduced by host RNA polymerase II (Pol II) or by reverse transcriptase. A cell culture system has been established to detect the individual contribution of host RNA Pol II, host DNA polymerase or viral reverse transcriptase, as well as the recombination events taking place during minus-strand DNA synthesis and plus-strand DNA synthesis in a single round of viral intramolecular replication. Studies in this report demonstrate that intramolecular recombination between two identical sequences during transcription by host RNA Pol II is minimal and that most recombinations occur during minus-strand DNA synthesis catalyzed by viral reverse transcriptase.
Retroviral recombinations generate new variants that can survive various environmental challenges. Retroviral particles contain two copies of the single-stranded RNA genome. Recombination between these two RNA molecules (intermolecular) occurs at a high rate (7). Recombination can also occur between two identical sequences within the same RNA molecule (intramolecular) (23). In previous studies, the rate of such intramolecular recombination is determined as 50 to 60% during a single round of retroviral replication (14, 24).
After a retrovirus enters a cell, the reverse transcriptases within the virion copy the viral RNA genome into a double-stranded DNA, which is subsequently integrated into host chromosomal DNA and becomes a provirus. The provirus is then replicated by cellular DNA polymerases as an integral element of host chromosome. Transcription of the provirus is carried out by cellular RNA polymerase II (Pol II), which produces both viral genomic RNAs and mRNAs encoding viral proteins. Viral proteins and viral RNAs are assembled into virions, and progeny viruses are released from the infected cells. Therefore, during the retroviral life cycle, retroviral genetic information is transmitted by viral reverse transcriptase, host DNA polymerase, and host RNA Pol II. Reverse transcription by viral reverse transcriptase can be further divided into two steps: minus-strand DNA synthesis with the viral RNA molecule as a template and plus-strand DNA synthesis with minus-strand DNA as a template. Each of these polymerization steps has the potential to introduce variations or mutations into progeny viruses.
In vivo systems for studying retroviral recombination or mutation include viral vectors, helper cells, and target cells. Viral vectors carry all viral cis-acting sequences required for viral replication and packaging; the trans-acting sequences in the vectors are replaced with gene(s) of interest. The helper cells constitutively provide viral proteins in trans so that the viral vector introduced into them can be packaged into infectious virions (vector viruses). Vector viruses are used to infect target cells, which do not express any viral proteins, and therefore do not support the production of new viruses. This strategy ensures that the life cycle of the viral vectors is restricted to a single round of replication (26). The rates of retroviral recombination are calculated by ratios of the number of recombinants to the number of the total progeny viruses.
The high fidelity of DNA replication suggests that recombination at the DNA level should be much less frequent than that incurred by reverse transcriptase. The fidelity of mammalian cellular RNA Pol II in retroviral template transcription is not yet known. Since retroviral genomic RNA is first transcribed from DNA by RNA Pol II, any deletion or recombination occurring during this process will contribute to genomic variation in retroviral replication. The present demonstrates that deletion during retroviral replication by host RNA Pol II was minimal. A previous study indicated that the frequency of deletion by DNA polymerase was also negligible (14); therefore, reverse transcription was the step associated with the high rate of intramolecular recombination.
Retroviral reverse transcriptase copies both minus-strand DNA and plus-strand DNA. Although most retroviral intermolecular recombinations occur during minus-strand DNA synthesis (25), it is unclear whether intramolecular recombinations also occur during minus-strand synthesis. We demonstrate here that, like intermolecular recombination, intramolecular recombination of Moloney murine leukemia virus (MLV) also occurs during minus-strand DNA synthesis. However, Bowman et al. (3) reported that recombination between two identical sequences within the same spleen necrosis virus RNA occur at similar frequencies during minus- and plus-strand DNA synthesis. This conclusion was based on observations obtained with a pool of viruses harvested after transfection, which did not allow the unambiguous determination of the structure and amount of the precursors to recombination (17, 24). Transfection could also introduce mutations and recombination, which could have interfered with their results. In this report, MLV-based vectors were constructed. The data demonstrated that plus-strand deletion did not occur.
MATERIALS AND METHODS
Nomenclature.
Plasmids are designated as, for example, pJZ481; viruses made from these plasmids are designated as, for example, JZ481.
Vector construction.
The construction of the MLV-based retroviral vectors JZ481 and JZ419 were described previously. Briefly, from the 5′ to 3′ direction, JZ481 carried two mutated copies of the color reporter gene, gfp, an internal ribosome entry segment (IRES) sequence, and a drug resistance gene, neo (Fig. 1A) (14). JZ419 is identical to JZ481 except that the two mutated gfp genes were replaced with a functional gfp gene (21). JZ442 (Fig. 2A) and JZ442+3′Hyg (Fig. 2B) were described previously (24). JZ442 encoded a hyg gene and a gfp genes, whereas JZ442+3′Hyg encoded the same hyg and gfp genes and an additional 290-bp 3′ hyg segment downstream of the gfp gene. JZ442 PPT+3′Hyg (Fig. 2C) was identical to pJZ442+3′Hyg except that the 290-bp 3′ hyg sequence was located downstream of the ppt and attL sequence. PJZ442+3′Hyg was digested with ClaI and NheI, followed by repair with Klenow fragment and ligation by T4 ligase, to delete the 75-bp ClaI-NheI sequence (positions 4415 and 4490) encoding the ppt and attL regions. The resulting plasmid was designated pJZ589. pJZ508 was identical to pJZ442 (Fig. 2A) except that pJZ508 contained a ClaI site and a NotI sites at the 3′ end of the hyg gene. The AseI-NheI fragment of pJZ508 containing the 5′ long terminal repeat, the hyg gene, the IRES, the gfp gene, and the 75-bp of ppt and attL region was inserted into the AseI-XbaI sites of pJZ589, in which a XhaI was located at the 5′ end of the 290-bp hyg sequence. The resulting plasmid was designated pJZ442 PPT+3′Hyg (Fig. 2C). In addition to restriction mapping, the 3′ end of gfp gene, the MLV 3′-nontranslated region, ppt, attL, the 290-bp 3′ hyg sequence, and the 5′ U3 of pJZ442 PPT+3′Hyg were sequenced. The sequences coincided with the structure predicted in Fig. 2C.
FIG. 1.
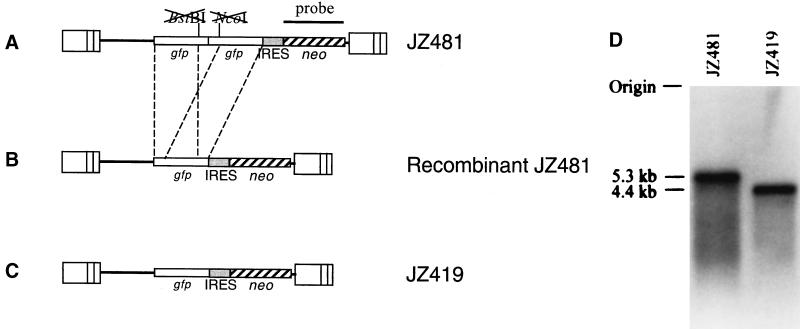
Structure of retrovirus vector used for determining of the recombination rate between two identical sequences within the same RNA molecule. (A) Structure of retrovirus vector containing two mutated gfp genes. JZ481 contains two nonfunctional gfp sequences in tandem positions and the neo gene. The 3′ end of the first gfp and the 5′ end of the second gfp are mutated. The neo gene is expressed from the encephalomyocarditis viral IRES. (B) Structure of a recombinant provirus. After one round of replication, the downstream gfp sequence recombines with the identical upstream gfp sequence, resulting in a functional gfp gene. The dashed lines between JZ481 and the recombinant provirus indicate identical sequences. (C) Structure of retrovirus vector encoding wild-type gfp gene. The structures of JZ419 and the expected recombinant JZ481 are identical. (D) Northern analysis of viral RNAs. Viral RNAs were extracted from viruses released from clear PG13 cells infected with JZ481 and PG13 cells infected with JZ419 and hybridized with a probe of the neo gene sequence. The approximate molecular sizes are shown on the left.
FIG. 2.
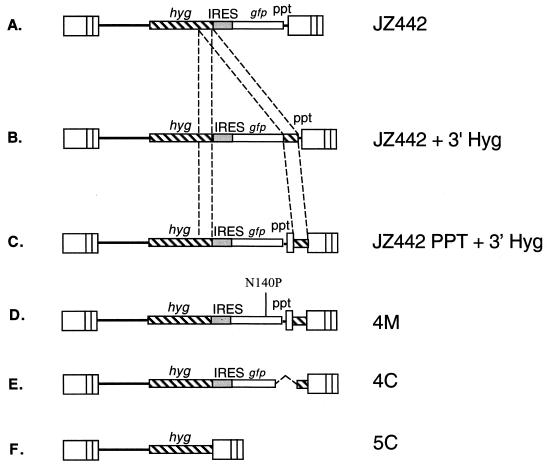
Structure of retrovirus vector used for intramolecular recombination. (A) JZ442 encodes a hyg gene and a gfp genes. (B) JZ442+3′Hyg encodes the same hyg and gfp genes and an additional 290-bp 3′ hyg segment downstream of the gfp gene. (C) JZ442 PPT+3′Hyg is identical to pJZ442+3′Hyg except that the 3′ hyg gene sequence is located downstream of the ppt and attL sequence. The dashed lines indicate identical sequences. (D, E, and F) Structure of step 3 proviruses.
Transfections, infections, and cells.
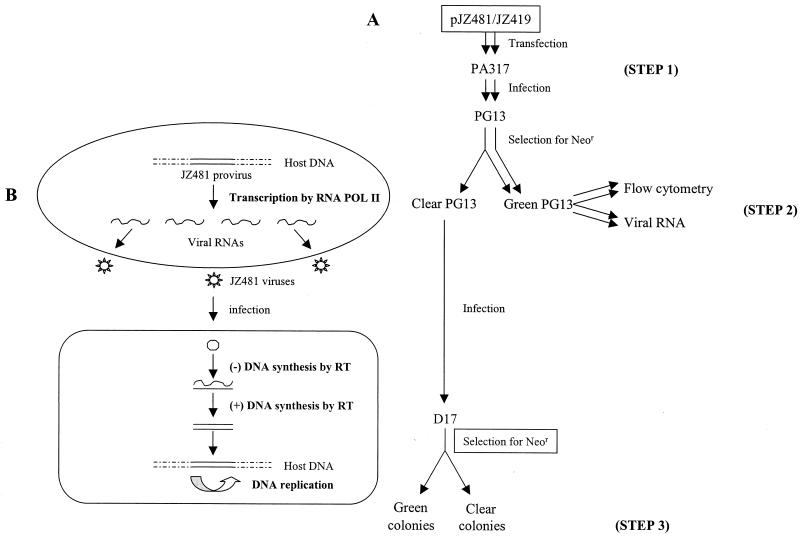
PA317 and PG13 are NIH 3T3-derived MLV-based helper cell lines that support the propagation of MLV-based vector virus (15, 16). D17 is a dog osteosarcoma cell line obtained from the American Type Culture Collection (CRL 8468). Transfections, infections, and the maintenance of the cells were previously described (26). Briefly, plasmid DNA of pJZ481 (or JZ442 PPT+3′Hyg), which carries a neo (or hyg) resistant gene, was transfected into the MLV amphotropic helper cell line PA317 (Fig. 3, step 1). The supernatant media containing the viruses were used to infect the MLV xenotropic helper cell line PG13 cells. After selection with an analog of neomycin, G418 (or hygromycin), well-separated colonies were individually cloned (Fig. 3, step 2). Viruses released from each clone were used to infect the D17 target cells. Visible D17 colonies appeared ca. 12 days after selection with G418 (or hygromycin) (Fig. 3, step 3).
FIG. 3.
(A) Outline of the experimental approach. Plasmid DNAs of pJZ481 and pJZ419 were transfected into an MLV amphotropic helper cell line, PA317, respectively (step 1). Supernatant medium containing the viruses was used to infect an MLV xenotropic helper cell line PG13. After selection with neomycin, clear and green neomycin-resistant (Neor) colonies were formed (step 2). PG13 cells were analyzed by flow cytometry. Viruses released from infected PG13 cells were analyzed by Northern blotting and were also used to infect D17 cells. The infected D17 cells were selected for neomycin resistance (step 3). Individual neomycin-resistant colonies were analyzed by fluorescence microscopy. (B) Schematic illustration of the experimental strategy for the study of a single round of JZ481 virus replication. The viral life cycle is represented in the form of DNA (provirus in PG13 cells), RNA (viruses released from PG13 cell), and DNA (provirus in target D17 cell), which depicts a single round of viral replication. Three polymerases are involved in four polymerization events (shown in boldface letters) during the transmission of retroviral genetic information.
Isolation of viral RNA.
Fresh media were added to virus-producing PG13 cells 24 h prior to harvesting. Cell debris was removed from the supernatant media by low-speed centrifugation. Viruses were concentrated by centrifugation with an SW41 rotor at 25,000 rpm for 1.5 h. Viral RNAs were isolated from the concentrated viruses by using NuceoSpin kit (Clontech, Palo Alto, Calif.) as described by the manufacturer.
Northern analysis.
Northern blotting was performed by standard methods (27). Briefly, 1 μg of viral RNAs was electrophoresd on a 1% agarose gel. The RNAs were then transferred onto a nitrocellulose membrane, which was probed with a 32P-labeled DNA fragment encompassing the neo gene (Fig. 1).
Preparation of Hirt fractionation.
A total of 2 × 10 5 cells were infected with JZ442 PPT+3′Hyg of step 2 clone 5 with the highest titer (multiplicity of infection = 1:1). Cells were lysed for 20 min. by 1 ml of 0.6% sodium dodecyl sulfate (SDS) and 0.01 M EDTA at 36 h after infection (9). After being incubated at 4°C for overnight, the sample was centrifuged at 17,000 × g for 30 min at 4°C to remove the major proteins and the SDS. Then, 5 M NaCl was added to a final concentration of 1 M, and the DNA in a Hirt fractionation was precipitated with isopropanol. The remaining EDTA was removed by Wizard PCR DNA purification kit (Promega, Madison, Wis.).
Amplification of proviruses and sequencing analysis.
The proviruses of step 2 were amplified with primer GFP 2312 (CTGGAGTTCGTGACCG), which annealed within the 3′ end of gfp gene, and primer U3 3469 (GCTGGACCGCATCTGG), which annealed within the 3′ U3. Hirt DNA and proviral DNA in step 3 cells were amplified by primer Hygro 1685 (GCTTGTATGGAGCAGCAGACG), which annealed with the 3′ end of the hyg upstream the 290-bp sequence, and primer U3 3469. The sequencing primers were GFP 2312 (CTGGAGTTCGTGACCG), MLV U3 (ATGCCTTGCAAAATGG), GFP 1627 (TGAGCAAGGGCGAGGAGCTG), and GFP 2011 (GAACCGCATCGAGCTGAA).
Fluorescence microscopy.
A fluorescence inverted microscope (Zeiss Axiovert 25) with a mercury arc lamp (100 W) and a fluorescent filter set (CZ909) consisting of a 470- to 40-nm exciter, a 515-nm emitter, and a 500-nm beamsplitter were used to detect green fluorescent protein (GFP) in living cells.
Flow cytometric analysis.
Flow cytometric analysis was conducted by using a MoFlo Cytomation (Becton Dickinson) with a 488-nm argon ion laser operating at a constant power output at a flow rate of 500 to 600 cells per s with a total accumulation of 20,000 cells/run. The FL-1 emission channel was used to monitor the intensity of the GFP.
RESULTS
Experimental protocol to distinguish between deletions made by RNA Pol II and by reverse transcriptase.
Virus JZ481 carried, from the 5′ to 3′ direction, two mutated gfp color reporter genes (4), an IRES, and a drug resistance gene, neo (Fig. 1A). The 3′ end of the first gfp and the 5′ end of the second gfp gene were mutated (25), so that neither of the two gfp genes was functional. The rates of reverse mutation of the two frameshifts had been shown to be lower than 10−5 (25). Intramolecular recombination between the identical sequences within the two defective gfp genes resulted in a recombinant virus that encoded a functional gfp gene (Fig. 1B) (14). Cells infected with parental-type viruses encoding two nonfunctional gfp sequences formed clear (or colorless) colonies under a fluorescence microscope (14), whereas cells transduced with recombinant viruses with a functional gfp gene appeared green. The IRES sequence of encephalomyocarditis virus origin allowed ribosomes to bind to the internal AUG and independently initiate the translation of the neo gene (1). The drug resistance gene, neo, was expressed by the IRES, and the gfp gene was expressed from the 5′ end of the RNA transcript by a traditional ribosome scanning model. Cells infected with viral vector JZ481 can survive neomycin (G418) selection and form green or clear colonies depending on whether or not intramolecular recombination occurs during vector virus replication. The number of the green neomycin-resistant colonies represented the number of recombinant viruses, whereas the number of clear colonies represented the number of the parental-type viruses. Using this approach, the rates of recombinations are determined by the ratio of the number of the green colonies to the number of the total colonies (green colonies plus clear colonies).
To determine the overall rates of recombination during a single replication cycle (Fig. 3), the vector viruses of JZ481 were introduced into PG13 cells by infection as described in Materials and Methods to eliminate deletion caused by transfection (24). PG13 cells infected with JZ481 were designated step 2 cells. Viruses released from step 2 cells were unable to infect cells of murine origin, including PG13 itself (16). Furthermore, the target cells (D17) do not express any viral structural proteins; therefore, the infection of D17 cells with JZ481 viruses released from step 2 PG13 cells represented a single round of viral replication (Fig. 3) (26). A previous study demonstrated that, due to recombination between the two gfp genes, 51% ± 2% of the viruses underwent deletion after a single round of replication (14). The rate was redetermined here to be 46.2% ± 5.9% (Table 1).
TABLE 1.
Microscopic analysis of D17 cells infected with JZ481a
| JZ481 clone | No. of colonies
|
Overall rate | Plus-strand rate (%) | |||
|---|---|---|---|---|---|---|
| Green | Mix colored | Clear | Total | |||
| 1-1 | 50 | 1 | 73 | 124 | 41.1 (%) | <0.8 |
| 1-3 | 52 | 0 | 52 | 104 | 50.0 | 0 |
| 3-1 | 68 | 1 | 58 | 127 | 54.3 | <0.8 |
| 3-2 | 37 | 0 | 45 | 82 | 45.1 | 0 |
| 3-3 | 35 | 0 | 51 | 86 | 40.7 | 0 |
| Total | 242 | 2 | 279 | 523 | 46.2 ± 5.9 | <0.3 ± 0.4 |
Each clone was an individual clear colony of PG13 cells containing a parent-type JZ481 provirus. The viruses released from each clone were used to infect D17 cells. Infected cells were selected for neomycin resistance. Neomycin-resistant colonies were analyzed by using a fluorescence microscope. The overall rate of recombination after one round of replication was determined by the ratio of the sum of the green and mix-colored colonies to the number of total neomycin-resistant colonies. The rate of recombination during plus-strand DNA synthesis is less than the ratio of the number of mix-colored colonies to that of the total number of colonies.
Determination of the frequency of deletion during transcription by host RNA Pol II.
After the replication cycle of JZ481 began with a provirus in PG13 cells (Fig. 3, step 2 cells), as many as 46% of the viruses recombined during one round of retroviral replication. The deletion between the two mutated gfp genes could occur during transcription by the host RNA Pol II and/or during reverse transcription by the viral reverse transcriptase (Fig. 3). If a sizable portion of the deletions are caused by RNA Pol II, the synthesis of functional GFP proteins would generate notable fluorescence that could be monitored by fluorescence intensities in a flow cytometric analysis. The fluorescence intensity should delineate the concentration of the functional GFP proteins or the amount of functional gfp transcripts in the cells.
In PG13 lines containing recombinant JZ481 proviruses, all of the viral RNA transcripts encode functional gfp; thus, the fluorescence intensity of these cells represents the amount of total viral RNA transcripts in the cells. In PG13 lines encoding parental-type JZ481 proviruses, most of the viral transcripts are of the parental type (encoding nonfunctional gfp), hence contributing no fluorescence to the host, but the amount of total viral RNA transcripts should be equal to that in cells with recombinant JZ481. If a portion of the viral RNAs have undergone deletion by recombination, thus encoding functional gfp, the amount of these deletion transcripts would be commensurate to the fluorescence they produced. Therefore, the ratio of the fluorescence intensity of PG13/parent-type JZ481 to that of PG13/recombinant JZ481 represents the percentage of recombinant transcripts present in the parent-type cells, or in other words, the frequency of deletion during transcription by host RNA Pol II.
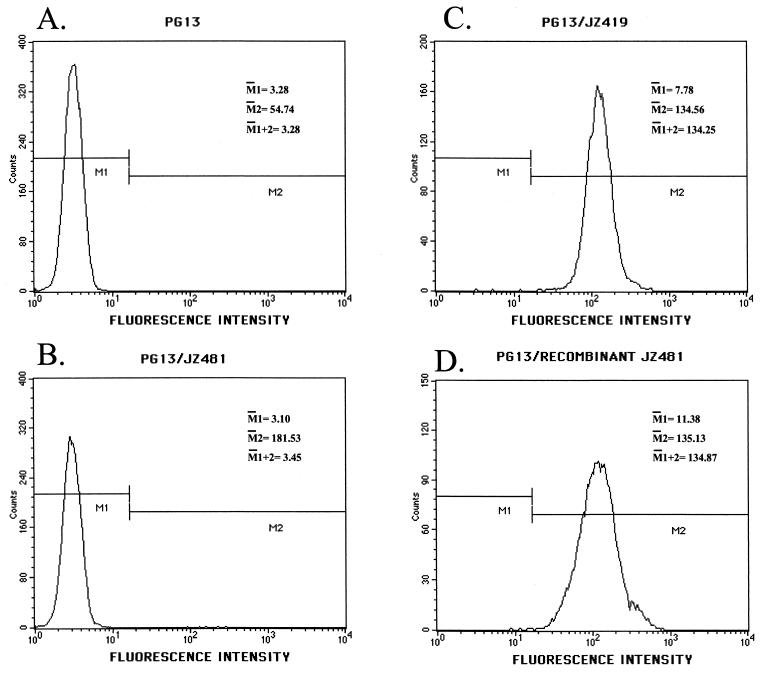
Clear (parent-type) and green (recombinant) neomycin-resistant PG13 colonies (Fig. 3, step 2) were individually subcloned and then analyzed by flow cytometry. To compare the recombinant GFP with wild-type GFP, a PG13 cell line carrying JZ419 provirus was also analyzed (21). JZ419 encodes the same sequences as a recombinant JZ481 (Fig. 1B and C). The fluorescence intensity of PG13 carrying the recombinant JZ481 was the same as that of PG13/JZ419, which encoded a wild-type gfp gene (Fig. 4C and D). The fluorescence intensity of PG13 cells infected with parent-type JZ481 was similar to that of uninfected PG13 cells. (Fig. 4A and B).
FIG. 4.
Flow cytometric analysis of virus-infected PG13 cells. PG13 cells were infected with viruses JZ481 or JZ419, respectively. Infected PG13 cells were selected for neomycin resistance. Individual neomycin-resistant clones were isolated and analyzed by flow cytometry.
Uninfected PG13 cells exhibited a mean fluorescence intensity of 3.28 (Fig. 4A), which denoted a background. After subtraction of the background from both PG13/parent-type JZ481 and PG13/recombinant JZ481, the ratio of these two differences (3.45 − 3.28)/(134.87 − 3.28) represented the percentage of GFP proteins in PG13/parent-type JZ481 cells. Therefore, the frequency of the functional JZ481 transcript in the PG13/JZ481 was only 0.1%, indicating that no significant amount of recombinant RNAs were used as messenger RNAs. Compared to the overall rate of retroviral recombination between the two mutated gfp genes (46%), the cellular contribution by RNA Pol II to the total retroviral recombination was minimal.
In addition to analysis of the mRNA in the helper cells, the viral RNAs packaged into virions were also analyzed. A deleted JZ481 transcript contained the same sequence as the JZ419 transcript. Viruses released from PG13/JZ481 and PG13/JZ419 were concentrated, and viral RNAs were isolated for analysis by Northern blotting by using a radiolabeled probe that hybridized to the neo gene sequence, which is encoded by both the parent-type and the recombinant viral RNAs (Fig. 1). Northern analysis showed that parent-type virion contained a 5.3-kb viral RNA, whereas the virions released from PG13/JZ419 cells contained a viral RNA of 4.4 kb. Had a significant amount of recombinant RNAs been transcribed by the RNA Pol II and subsequently packaged into virions, 4.4-kb RNAs should be produced by PG13/JZ481. Analysis by a density photometer showed that the amount of the deleted fragments of JZ481 was <6% of the undeleted JZ481. The Northern analysis revealed that very little, if any, amount of deleted viral RNAs were transcribed in the helper cells (Fig. 1D) and that most deletions must have occurred during reverse transcription.
Determination of the relative rates of intramolecular recombination during minus-strand and plus-strand DNA synthesis.
The frequency of recombination at the proviral level (DNA) is only 10−5 events/cell division (Table 2) (14). Flow cytometry and Northern analysis indicated that 99.9% of recombinations took place during reverse transcription. Retroviral reverse transcription includes the synthesis of minus-strand and plus-strand DNAs. Therefore, reverse transcriptase-promoted template switching may occur during minus-strand DNA synthesis and/or during plus-strand synthesis. A previous study showed that most, if not all, retroviral intermolecular recombinations between two different viral vectors occurred during minus-strand DNA synthesis (25). To study whether most intramolecular recombinations also occur during the minus-strand DNA synthesis, individual step 3 colonies of the JZ481-infected DNA cells were analyzed (Fig. 3, step 3). If recombination takes place during minus-strand DNA synthesis, since plus-strand DNA is made by using the minus-strand as a template, the resulting double-stranded viral DNAs will encode recombinant JZ481 on both strands. After integration into the host cell chromosome, the recombinant virus-infected target cell will grow to a green colony. If an intramolecular recombination takes place during plus-strand DNA synthesis, since the minus-strand DNA still carries the parental-type gfp, the resulting double-stranded viral DNA will be a heteroduplex with a portion of the minus strand looping out. When such a duplex integrates into host DNA and the cellular repair system cannot correct the mismatched sequences within the viral double-stranded DNA, subsequent cell division would result in two genotypically different daughter cells; one receives the parent-type provirus, and the other receives the recombinant provirus. While their progeny would form a single colony, it will have a mixed phenotype, with half clear cells and half green cells. The D17 cell line was chosen as target cell in the present study because it is unable to repair mismatches within the retroviral double-stranded DNA (22, 25). When neomycin-resistant colonies of JZ481-infected D17 cells were analyzed under a fluorescence microscope, only 2 of 244 green colonies examined were mixtures of green and clear cells (Table 1). In addition to recombination during plus-strand DNA synthesis, mix-colored colonies could be also resulted either from a mutation of the gfp gene during plus-strand DNA synthesis or from two separate infection events (25). Therefore, at least 99% of the retroviral intramolecular recombinations had occurred during minus-strand DNA synthesis by reverse transcriptase (Table 2).
TABLE 2.
Relative recombination rates during retroviral life cycle for JZ481
| Stage in cyclea | Rate (%) | Distributionf (%) |
|---|---|---|
| Minus-strand DNA synthesis | 45.8b | >99.15 |
| Plus-strand DNA synthesis | <0.3c | <0.65 |
| Viral RNA transcription | <0.1d | <0.2 |
| Provirus replication | 0.85 × 10−5e | Negligible |
| Total | 46.2 | 100 |
Four stages comprise one single round of a retroviral life cycle.
The rate was determined by subtracting the portions of plus-strand DNA synthesis (0.3%) and of viral RNA transcription (0.1%) from the overall rate (46.2%).
The rate of recombination during plus-strand DNA synthesis was determined as described in Table 1.
The rate of recombination during viral RNA transcription was estimated on the basis of flow cytometry analysis.
The frequency of recombination at the DNA level was determined as the number of green cells per cell division. The mean value is 0.85 × 10−5 (14).
The relative distribution of the recombination events during transmission of the viral genetic information through the four types of macromolecules was determined by the ratio of the rate at each stage over the overall rate (46.2%).
Intramolecular recombinations did not occur during plus-strand DNA synthesis.
JZ442+3′Hyg carried the hyg gene, and a 290-bp repeat sequence of the 3′ hyg gene was inserted into the 3′ untranslated region of the gfp gene (Fig. 2B). The rate of deletion between the two identical sequences was found to be 62% (24). Our previous study demonstrated that a deletion occurring between the two identical sequences within the same viral RNA molecule was the result of intramolecular recombinations (23). Furthermore, we demonstrated that intermolecular recombination occurred during minus-strand DNA synthesis (25). In the present, we demonstrated that intramolecular recombinations occur also during minus-strand DNA synthesis. However, this result conflicts with a study done by Bowman et al. (3), who reported that in a spleen necrosis virus-based system the recombination between two direct repeats occurred at the same frequency during minus- and plus-strand DNA syntheses. To further investigate intramolecular recombinations, an MLV-based vector was constructed under the same principle as that reported by Bowman et al. JZ442 PPT+3′Hyg (Fig. 2C) was identical to JZ442 +3′Hyg (Fig. 2B), except that the MLV ppt and attL sequences were moved to a position upstream of the second 3′ hyg sequence. During reverse transcription, ppt is the sequence for initiation of plus-strand DNA synthesis exactly before the attL, which is essential for retroviral integration into the host chromosomal DNA (8). If recombination between the two identical hyg sequences occurred during minus-strand DNA synthesis, in addition to deletion of the gfp sequence, the ppt and attL would also be deleted. As a consequence of the absence of ppt, the plus-strand DNA synthesis could not initiate and the reverse transcription could not be completed. If a recombination occurred during plus-strand DNA synthesis, the minus-strand DNA would still contain the ppt and attL, and it was proposed that a plus-strand DNA could initiate (3). In this case the deleted plus-strand DNA and undeleted minus-strand DNA would form a mismatched viral double-stranded DNA, which would integrate into the cell chromosome. Provirus derived from the deleted plus-strand DNA would lack the gfp gene. Cells with the provirus without the gfp gene were clear under a fluorescence microscope. Observations by Bowman et al. were based on viruses collected from transfected helper cells (3). However, previous studies have indicated that during transfection deletion between two identical sequences occurred at a very high frequency (24); furthermore, transfection could not allow the unambiguous determination of the DNA structure or the quantitation of the precursors to recombination (17).
To demonstrate JZ442 PPT+3′Hyg (Fig. 2C) also encoded two identical sequences, the 3′ end of the plasmid of pJZ442 PPT+3′Hyg was sequenced. The sequences coincided with the structure predicted in Fig. 2C. The plasmid DNA of pJZ442 PPT+3′Hyg was used to transfect an amphotropic helper cell line, PA317 (15) (step 1). Virions from the cells can infect mouse cells. To eliminate the influence of deletion during transfection, viruses released from the transfected cells were used to infect a xenotropic helper cell line, PG13 derived from the mouse cell line NIH 3T3 (16). Viruses released from this xenotropic helper cell lines can infect the dog cell line D17 but not mouse cells such as PG13. This procedure ensures that there is only a single round of replication. After selection for hygromycin resistance, individual hygromycin-resistant colonies were examined under a fluorescence microscope. Seven well-separated green clones were isolated and used for further studies (step 2). The GFP-positive phenotype of these clones indicated that the cells encoded the gfp gene and that the two identical sequences had not yet recombined within the proviruses found in the green step 2 cells, since the gfp gene is located between the two identical sequences of the 290-bp 3′ hyg sequences. Furthermore, the 3′ end of proviruses from three step 2 clones were amplified and sequences, which coincided with the plasmid of pJZ442 PPT+3′Hyg. Therefore, in contrast to the report by Bowman et al., there could not be any deleted viral clones used in the present study. Virus released from step 2 cells were used to infect D17 cells, and the infected D17 cells were selected for hygromycin resistance. Individual hygromycin-resistant colonies were examined by fluorescence microscopy. The ratio of the number of clear plus mixed colonies to the number of total colonies was determined to be 0.9% ± 0.4% (Table 3), which is much lower than the deletion rate for JZ442+3′Hyg (62% ± 9%) but is similar to the rate of mutation for JZ442 (1.3% ± 0.4%), which did not contain any repeat sequences (Fig. 2A) (24). Therefore, although JZ442 PPT+3′Hyg viruses carried two 290 base repeats of identical sequence similar in structure to JZ442+3′Hyg, the rate of recombination between the two sequences was only 1% (0.9%/62%) of the rate of recombination for JZ442+3′Hyg. The only difference between the two vectors lies in the localization of the ppt and attR, in which JZ442+3′Hyg allows the generation of double-stranded viral DNA after the deletion of its minus-strand DNA but JZ442 PPT+3′Hyg does not. Therefore, this result is consistent with our conclusion that intramolecular recombinations occur during minus-strand DNA synthesis.
TABLE 3.
Analyses of JZ442 PPT + 3′ Hyg on D17 cellsa
| Clone | No. colonies
|
Rate of mutation (%) | Titer | ||
|---|---|---|---|---|---|
| Green | Clear | Mix colored | |||
| Clone 1 | 431 | 1 | 2 | 0.7 | 0.18 × 105 |
| Clone 2 | 487 | 1 | 1 | 0.4 | 1 × 105 |
| Clone 3 | 761 | 8 | 1 | 1.2 | 0.25 × 105 |
| Clone 4 | 782 | 8 | 5 | 1.6 | 0.27 × 105 |
| Clone 5 | 564 | 3 | 1 | 0.7 | 1.2 × 105 |
| Clone 6 | 172 | 2 | 0 | 1.2 | 0.34 × 105 |
| Clone 7 | 289 | 1 | 0 | 0.5 | 0.57 × 105 |
| Total | 3,486 | 24 | 10 | 0.9 ± 0.4b | (0.54 × 105) ± (0.40 × 105)c |
Viruses from step 2 PG13 cells, which contain a gfp gene, were used to infect D17 cells. Infected D17 cells were selected for hygromycin resistance. Each hygromycin-resistant colony was analyzed under a fluorescence microscope.
The rate of deletion of JZ442+3′ Hyg was 62%, whereas the rate mutation of JZ442 was 1% (24).
The titer of viruses released from JZ442+3′Hyg was (0.86 × 105) ± (0.38 × 105) (24).
To determine the nature of the clear step 3 D17 cells, three clones, one mixed colony (4M) and two clear colonies (4C and 5C) were amplified by PCR and sequenced. Provirus in clone 4 M (Fig. 2D) contained the same sequences as the plasmid DNA, which carried the complete gfp, the ppt, the 290-bp hyg sequences and the 3′ U3. The complete gfp sequence in provirus 5M indicated that its colorless phenotype did not result from a deletion between the two repeated hyg sequences. A mutation (N140P) was found in the 718-bp open reading frame of the gfp gene, as shown by sequences from both directions. The 4C provirus (Fig. 2E) carried a 95-nucleotide deletion at the 3′ end of the gfp gene, which was followed the 3′ 209-bp of the hyg gene and the U3, which was probably resulted from a nonhomologous recombination during minus-strand DNA synthesis, followed by an intermolecular transfer of the plus strong stop primer. The provirus of 5C (Fig. 2F) only contained the hyg gene followed by the U3, which should have resulted from a homologous intramolecular recombination between the two identical 290-bp 3′ hyg sequences during minus-strand DNA synthesis, followed by an intermolecular transfer of the plus strong stop DNA.
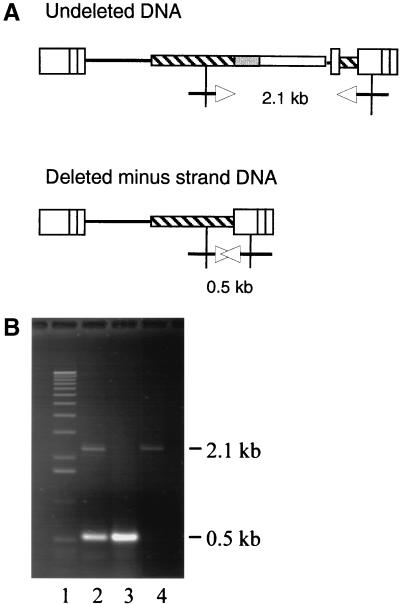
Due to the presence of the identical 290-bp sequences in the JZ442 PPT+3′Hyg, during minus-strand DNA synthesis deletion between the two repeats should occur at a high frequency. Although the deleted minus-strand DNA could neither be used to synthesize the plus-strand DNA nor be integrated into the cellular chromosomal DNA, the deleted and unintegrated minus-strand DNA should be found in the Hirt section (9). To demonstrate that deletions within JZ442 PPT+3′Hyg occurred during minus-strand DNA synthesis, DNAs in the Hirt portion were isolated as described in Methods and Materials. Unintegrated viral DNAs were amplified by PCR with two primers. The first primer annealed to the hyg seqeunce upstream the 3′ 290-bp end, whereas the second one annealed to the U3 region (Fig. 5A). The PCR product was resolved by electrophoresis, and two fragments were separated on an agarose gel (Fig. 5B). The 2.1-kb fragment represented the amplified sequence within the parent-type JZ442 PPT+3′Hyg, whereas the 0.5-kb fragment represented the sequence with the deletion between the two identical 290-bp within the JZ442 PPT+3′Hyg. A large portion of the unintegrated viral DNAs contained a deletion (Fig. 5B); however,<1% proviruses contained the deletion, indicating that most deleted DNA were not integrated into host chromosome.
FIG. 5.
PCR amplification of Hirt fractionation. (A) Structure of undeleted DNA and deleted minus-strand DNA of JZ442 PPT+3′Hyg. The arrows represent the primers used to amplify the Hirt fractionation DNA: Hygro 1685 (GCTTGTATGGAGCAGCAGACG) and U3 3469 (GCTGGACCGCATCTGG). (B) Electrophoresis of amplified fragments: line 1, 1-kb DNA ladder (Invitrogen, Carlsbad, Calif.); line 2, PCR fragment of Hirt fractionation DNA; line 3, PCR fragments of the step 3 clone 5C; and line 4, PCR fragment of pJZ442 PPT+3′Hyg.
DISCUSSION
The first goal of the present study was to determine the contribution of RNA Pol II to retroviral recombination during a single round of retroviral replication. The design of the experimental study was different from previous studies in that (i) the rate of intramolecular recombination between two identical sequences was very high (46%), (ii) the vector used in this report encoded a gfp gene that allowed recombination to be monitored by flow cytometry, and (iii) recombination resulted in deletion that was detectable by Northern blot analysis. After a single round of replication using the retroviral vector with this tandem repeat sequence, recombination took place at a high rate (46.2% per cycle). The contributions of RNA Pol II and DNA polymerase to such recombinations were minimal. Moreover, most recombinations took place during minus-strand DNA synthesis and were catalyzed by reverse transcriptase.
Although RNA Pol II can switch between RNA templates (5), the present study is the first to explore the potential effect of eucaryotic RNA Pol II on retroviral recombination. RNA recombination has been observed mostly in studies of RNA viruses that use RNA as their sole genetic material and utilize RNA-dependent RNA polymerases (6). In a prokaryotic system, end-to-end template switching by RNA polymerase has also been reported (18, 19); such template switching generates hybrid RNA molecules in an in vitro system. In an in vivo study, Birge and Low postulated a template-switching model for the generation of a recombinant RNA by a prokaryotic RNA polymerase (2). The present study demonstrates that, in our cell culture system, RNA Pol II does not promote any detectable RNA recombination events in a mammalian cell line (NIH 3T3).
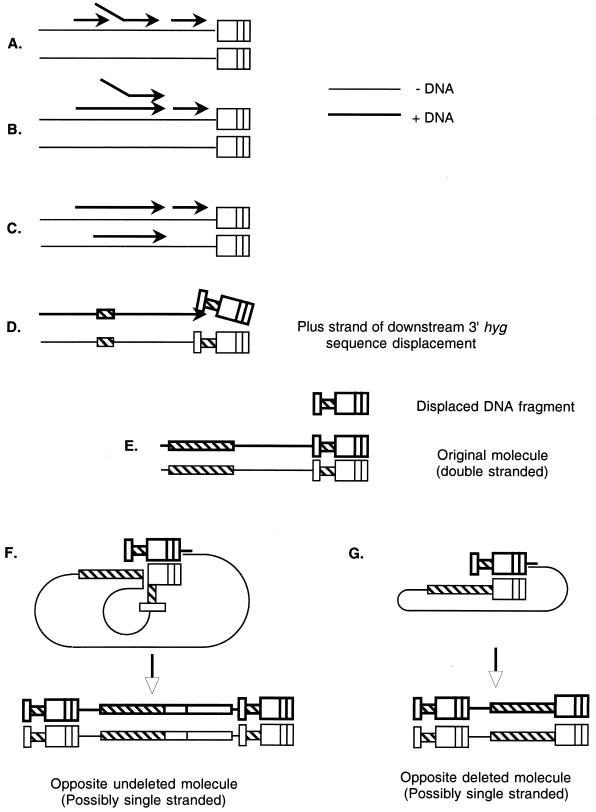
A previous study shows that the frequency of recombination at the proviral level is exceedingly low (ca. 10−5 per cell division) (14). Hence, it must be the reverse transcriptase that causes the high incidence of recombination. In addition, we demonstrated that at least 99% intramolecular recombination occurred during minus-strand DNA synthesis. This conclusion contrasts with the report by Bowman et al. (3). The discrepancy might be attributed to the difference between MLV and spleen necrosis virus systems. However, this is very unlikely considering the universal mechanism of reverse transcription. The current model for plus-strand replacement (12, 13) proposed that plus-strand DNA synthesis is initially discontinuous (Fig. 6A), and a fragment of product DNA might be displaced by a continuous DNA synthesis (Fig. 6B). The displaced DNA fragment may then hybridize to the minus-strand DNA synthesized from the other molecule of viral RNA used as a template (Fig. 6C).
FIG. 6.
Current recombination models do not support any intramolecular recombination occurring during plus-strand DNA synthesis. (A) Model of plus-strand replacement recombination. Plus-strand DNA synthesis is initially discontinuous. (B) A fragment of product DNA might be displaced by continuous DNA synthesis. (C) The displaced DNA fragment may then hybridize to the minus-strand DNA synthesized from the other molecule of viral RNA used as the template. (D) The plus-strand primer DNA is displaced by subsequent upstream DNA synthesis. (E) The displaced fragment could not hybridize to the homologous regions intramolecular, because its original template has been occupied by already-synthesized plus-strand DNA. (F) The opposite minus-strand DNA is possibly single stranded. An undeleted minus-strand DNA and an intermolecular transfer of plus strong stop primer resulted in an undeleted provirus. (G) A deleted minus-strand DNA and an intermolecular transfer of plus strong stop primer resulted in a deleted provirus.
We have demonstrated that recombinations between two identical sequences occur intramolecularly (23). During plus-strand DNA synthesis, recombination between two sequences on the same molecule (intramolecular) would be more difficult than recombination between two homologous molecules (intermolecular). It is noted that a replaced plus-strand DNA fragment could hardly hybridize intramolecularly. In vector JZ442 PPT+3′Hyg (Fig. 2C), the second hyg sequence was located downstream of the ppt. Since retroviral plus-strand DNA synthesis initiated at the ppt region, the downstream sequence should be copied first (Fig. 6D). Suppose plus-strand synthesis of MLV was discontinous; if this newly synthesized plus-strand DNA was displaced by the upstream DNA synthesis, it was theoretically able to hybridize to the homologous regions on both minus-strand DNAs. However, in reality, both hyg sequences on the original template would have been occupied by already-synthesized plus-strand DNA: the 5′ hyg sequence would be double stranded due to the subsequent plus-strand DNA synthesis; the downstream 3′ hyg sequence and the long terminal repeat would have base paired before and after the plus strong stop DNA was displaced (Fig. 6E). Therefore, the displaced DNA fragment of the plus strong stop primer could not hybridize to the original template, which means an intramolecular recombination would be impossible. Furthermore, the original double-stranded viral DNA was nonfunctional since the attL would not be available for integration (Fig. 6E). Although a deletion could be resulted from an intermolecular recombination event, it requires the displaced plus-strand fragment to hybridize to the other minus-strand template, which might be single stranded (Fig. 6F and G). However, the rate of intermolecular recombination (1%) (23) is much lower than the intramolecular rate (62%) (24). Taken together, these results suggest that fewer than 2% (1% of 62%) of the deletions could have resulted from intermolecular transfer of the plus strong stop DNA.
The original plus-strand displacement model for intermolecular homologous recombination proposed that the displaced fragments are able to hybridize with the opposite minus-strand DNA (Fig. 6B and C). If the plus-strand primer somehow hybridized with an undeleted opposite minus-strand DNA, the resulting double-stranded DNA would be undeleted (Fig. 6F). If the primer hybridized to an opposite minus-strand DNA that had undergone deletion during minus-strand DNA synthesis (Fig. 6G), the double-stranded DNA would lack the gfp gene and would be able to integrate. Since this process required an intermolecular transfer of the plus strong stop DNA, the frequency should be very low (10, 11, 20). Since the deleted proviruses did not result from recombination during plus-strand DNA synthesis, colonies of the infected cells should exhibit a clear phenotype instead of a mixed one. The proviruses from two clear colonies were analyzed and found to contain a deletion of the ppt region, which should have resulted from a minus-strand deletion, followed by an intermolecular transfer of the plus strong DNA primer. One step 3 provirus from a mixed colony did not contained any deletion, but a mutation was detected in the open reading frame of the gfp gene, indicating that the mixed colony had not resulted from a plus-strand recombination either. This mutation probably occurred during plus-strand DNA synthesis. Therefore, structure of the step 3 proviruses did not support plus-strand intramolecular recombination.
In summary, neither our experimental observations nor the current plus-strand replacement model (12, 13) support any intramolecular recombination during plus-strand DNA synthesis.
Acknowledgments
We thank W. Bargmann and A. Simmons for helpful comments on the manuscript. We thank J. Strange for flow cytometry analysis and M. Russ for sequencing analysis.
This research was supported by Public Health Service research grant CA70407.
REFERENCES
- 1.Adam, M. A., N. Ramesh, A. D. Miller, and W. R. Osborne. 1991. Internal initiation of translation in retroviral vectors carrying picornavirus 5′ nontranslated regions. J. Virol. 65:4985-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birge, E. A., and K. B. Low. 1974. Detection of transcribable recombination products following conjugation in rec+, recB−, and recC− strains of Escherichia coli K-12. J. Mol. Biol. 83:447-457. [DOI] [PubMed] [Google Scholar]
- 3.Bowman, R. R., W. S. Hu, and V. K. Pathak. 1998. Relative rates of retroviral reverse transcriptase template switching during RNA- and DNA-dependent DNA synthesis. J. Virol. 72:5198-5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chalfie, M., Y. Tu, G. Euskirchen, W. W. Ward, and D. C. Prasher. 1994. Green fluorescent protein as a marker for gene expression. Science 263:802-805. [DOI] [PubMed] [Google Scholar]
- 5.Chang, J., and J. Taylor. 2002. In vivo RNA-directed transcription, with template switching, by a mammalian RNA polymerase. EMBO J. 21:157-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chetverin, A. B. 1999. The puzzle of RNA recombination. FEBS Lett. 460:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin, J. M., S. H. Hughes, and H. Varmus. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 8.Gilboa, E., S. W. Mitra, S. Goff, and D. Baltimore. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93-100. [DOI] [PubMed] [Google Scholar]
- 9.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 10.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 11.Jones, J. S., R. W. Allan, and H. M. Temin. 1994. One retroviral RNA is sufficient for synthesis of viral DNA. J. Virol. 68:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junghans, R. P., L. R. Boone, and A. M. Skalka. 1982. Products of reverse transcription in avian retrovirus analyzed by electron microscopy. J. Virol. 43:544-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Junghans, R. P., L. R. Boone, and A. M. Skalka. 1982. Retroviral DNA H structures: displacement-assimilation model of recombination. Cell 30:53-62. [DOI] [PubMed] [Google Scholar]
- 14.Li, T., and J. Zhang. 2000. Determination of the frequency of retroviral recombination between two identical sequences within a provirus. J. Virol. 74:7646-7650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller, A. D., and C. Buttimore. 1986. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol. Cell. Biol. 6:2895-2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller, A. D., J. V. Garcia, N. von Suhr, C. M. Lynch, C. Wilson, and M. V. Eiden. 1991. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J. Virol. 65:2220-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murnane, J. P., M. J. Yezzi, and B. R. Young. 1990. Recombination events during integration of transfected DNA into normal human cells. Nucleic Acids Res. 18:2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nudler, E., E. Avetissova, V. Markovtsov, and A. Goldfarb. 1996. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science 273:211-217. [DOI] [PubMed] [Google Scholar]
- 19.Oostra, B. A., A. C. Arnberg, G. Ab, and M. Gruber. 1981. Terminal strand-switching of E. coli RNA polymerase transcribing a truncated DNA fragment. Biochim. Biophys. Acta 655:446-448. [DOI] [PubMed] [Google Scholar]
- 20.Panganiban, A. T., and D. Fiore. 1988. Ordered interstrand and intrastrand DNA transfer during reverse transcription. Science 241:1064-1069. [DOI] [PubMed] [Google Scholar]
- 21.Sapp, C. M., T. Li, and J. Zhang. 1999. Systematic comparison of a color reporter gene and drug resistance genes for the determination of retroviral titers. J. Biomed. Sci. 6:342-348. [DOI] [PubMed] [Google Scholar]
- 22.Tang, L. Y., and J. Zhang. 2000. The cellular mismatch repair system is able to repair mismatches within MLV retroviral double-stranded DNA at a low frequency. Nucleic Acids Res. 28:2302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang, J., and Y. Ma. 2001. Evidence for retroviral intramolecular recombinations. J. Virol. 75:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang, J., and C. M. Sapp. 1999. Recombination between two identical sequences within the same retroviral RNA molecule. J. Virol. 73:5912-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, J., L. Y. Tang, T. Li, Y. Ma, and C. M. Sapp. 2000. Most retroviral recombinations occur during minus-strand DNA synthesis. J. Virol. 74:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, J., and H. M. Temin. 1993. Rate and mechanism of nonhomologous recombination during a single cycle of retroviral replication. Science 259:234-238. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, J., and H. R. Bose, Jr. 1989. Acquisition of new proviral copies in avian lymphoid cells transformed by reticuloendotheliosis virus. J. Virol. 63:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]