Abstract
Localization of RNA replication to intracellular membranes is a universal feature of positive-strand RNA viruses. Replication complexes of flock house virus (FHV), the best-studied alphanodavirus, are located on outer mitochondrial membranes in infected Drosophila melanogaster cells and are associated with the formation of membrane-bound spherules, similar to structures found for many other positive-strand RNA viruses. To further study FHV replication complex formation, we investigated the subcellular localization, membrane association, and membrane topology of protein A, the FHV RNA-dependent RNA polymerase, in the yeast Saccharomyces cerevisiae, a host able to support full FHV RNA replication and virion formation. Confocal immunofluorescence revealed that protein A localized to mitochondria in yeast, as in Drosophila cells, and that this mitochondrial localization was independent of viral RNA synthesis. Nycodenz gradient flotation and dissociation assays showed that protein A behaved as an integral membrane protein, a finding consistent with a predicted N-proximal transmembrane domain. Protease digestion and selective permeabilization after differential epitope tagging demonstrated that protein A was inserted into the outer mitochondrial membrane with the N terminus in the inner membrane space or matrix and that the C terminus was exposed to the cytoplasm. Flotation and immunofluorescence studies with deletion mutants indicated that the N-proximal region of protein A was important for both membrane association and mitochondrial localization. Gain-of-function studies with green fluorescent protein fusions demonstrated that the N-terminal 46 amino acids of protein A were sufficient for mitochondrial localization and membrane insertion. We conclude that protein A targets and anchors FHV RNA replication complexes to outer mitochondrial membranes, in part through an N-proximal mitochondrial localization signal and transmembrane domain.
The lack of broadly effective therapies for positive-strand RNA virus infections motivates the identification and characterization of common features of viral replication as potential targets for novel antiviral agents. A universal feature of positive-strand RNA virus replication is the essential involvement of host cell membranes. For different RNA viruses, RNA replication complexes form on membrane structures derived from diverse intracellular organelles, including the endoplasmic reticulum (30, 34, 39, 45, 52, 53), Golgi apparatus (52), lysosomes (13, 26, 33, 52), endosomes (13, 26), and mitochondria (11, 37). Viral proteins that target replication complexes to intracellular membranes have been identified, such as the poliovirus 3AB protein (21), the Semliki Forest virus nsP1 protein (40), the brome mosaic virus 1a protein (7), and the carnation Italian ringspot virus (CIRV) 36-kDa replicase protein (48). The identification of such viral targeting proteins and other features of intracellular membrane association have provided valuable insights into viral replication complex formation and function.
Flock house virus (FHV), the best-studied alphanodavirus in the Nodaviridae family, shares viral replication features with positive-strand RNA viruses from other families (2), and has been useful to investigate fundamental aspects of viral replication. FHV directs RNA replication and virion assembly in a wide variety of cells, including insect (14, 57), plant (56), mammalian (1, 23), and yeast (31, 43, 44) cells, implying that any host components required for FHV RNA replication are widely conserved. FHV contains a 4.5-kb bipartite genome in a nonenveloped icosahedral capsid (54, 55). The larger 3.1-kb RNA species (RNA1) encodes protein A, a 112-kDa protein with sequence motifs (41) and in vivo functions (1, 23, 31, 43) of a viral RNA-dependent RNA polymerase (RdRp). The smaller 1.4-kb RNA species (RNA2) encodes the capsid precursor protein (12, 54). During replication, FHV produces a subgenomic, 0.4-kb RNA3 that is colinear with the 3′ end of RNA1, and encodes a 10-kDa protein B of unknown function (2). FHV RNA replication occurs on outer mitochondrial membranes in infected Drosophila melanogaster cells (37). By confocal immunofluorescence microscopy, we demonstrated colocalization between protein A and both mitochondria and newly synthesized viral RNA. In addition, electron microscopy studies showed mitochondrial clustering and the formation of 40- to 60-nm membrane-bound spherules restricted to the mitochondrial intermembrane space and connected to the outer mitochondrial membrane, and immunogold electron microscopy localized protein A to the outer mitochondrial membrane (37). Similar mitochondrial membrane-associated structures are seen after infection with the related alphanodaviruses Nodamura and Boolarra virus (3, 15). Moreover, membrane-bound spherules are associated with RNA replication by many other viruses, including tombusviruses (11) and togaviruses (13, 17, 26, 33), suggesting that characterizing these structures and the mechanisms of their localization may identify general principles in positive-strand RNA virus replication.
FHV replication in Saccharomyces cerevisiae has been shown to duplicate many features of FHV replication in insect cells, including independent replication of RNA1 (31, 43), downregulation of subgenomic RNA3 production by RNA2 (44), and formation of infectious virions (44). One useful feature of FHV replication in yeast is the ability to separate the protein coding and replication template functions of RNA1. An RNA1 derivative with a protein A frameshift can serve as a nontranslatable template for RNA replication and subgenomic mRNA synthesis when protein A is provided in trans from a second, nonreplicatable FHV RNA1 derivative with modified 5′ and 3′ noncoding sequences. Such trans replication in yeast has been used to identify cis-acting signals, including the long-distance base pairing essential for RNA1 replication and subgenomic RNA3 synthesis (31) and for FHV-directed foreign gene expression (42, 43).
In this report, we used FHV replication in S. cerevisiae to identify the intracellular localization, membrane association characteristics, membrane topology, and organellar targeting signals of FHV protein A. The ability to separate the translation and replication template functions of FHV RNA1 allowed us to construct and functionally test protein A derivatives. We demonstrate by confocal immunofluorescence microscopy and membrane flotation assays that FHV protein A localized to mitochondrial membranes independently of viral RNA replication and that protein A was an integral membrane protein inserted into the outer mitochondrial membrane with the N terminus located in the intermembrane space or matrix and the C terminus exposed to the cytoplasm. Furthermore, deletion studies and gain-of-function assays with protein A-green fluorescent protein (GFP) fusions demonstrated that the N terminus of protein A contained sequences sufficient for mitochondrial localization and membrane insertion.
MATERIALS AND METHODS
Yeast strain, transformation, and culture conditions.
The haploid yeast strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) was used for all experiments. Yeast were transformed by using the E-Z Transformation Kit (Zymo Research, Orange, Calif.), plated onto selective minimal medium with glucose (2% [wt/vol]) as the carbon source, and incubated at 30°C for 2 to 3 days until distinct colonies developed. Individual clones were transferred to liquid media and grown at 30°C for 42 to 48 h until early saturation phase. Yeast were washed once with distilled water, resuspended at 107/ml in liquid media, and induced with galactose (2% [wt/vol]) to initiate DNA-directed FHV RNA expression as previously described (43), except that yeast were induced for 24 h at 26°C for all experiments. Two independently derived clones were grown and induced separately for each experimental group, and samples were combined from an equal number of cells for membrane flotation, protease digestion, and immunofluorescence analyses.
Antibodies.
Rabbit polyclonal antisera against FHV protein A was generated with gel-purified protein A expressed in Escherichia coli as previously described (37). Rabbit polyclonal antisera against the yeast mitochondrial proteins p32 and translocase of the outer membrane (Tom) 40 were the generous gifts of Debkumar Pain (University for Medicine and Dentistry of New Jersey). Rabbit polyclonal antisera against the yeast matrix heat shock protein Ssc1 was the generous gift of Elizabeth Craig (University of Wisconsin—Madison). Rabbit polyclonal antibodies against GFP and the influenza virus hemagglutinin (HA) epitope tag were from Santa Cruz Biotechnology (Santa Cruz, Calif.), and mouse monoclonal antibodies against HA (clone HA-7) were from Sigma (St. Louis, Mo.). Mouse monoclonal antibodies against the yeast proteins cytochrome oxidase subunit III (CoxIII), 3-phosphoglycerate kinase, and mitochondrial porin were from Molecular Probes (Eugene, Oreg.). All secondary antibodies for immunofluorescence and immunoblotting were from Jackson Immunoresearch (West Grove, Pa.).
Plasmids.
Standard molecular biology procedures were used for all cloning steps unless otherwise indicated, and all products generated by PCR or with synthesized oligonucleotides were verified by automated sequencing. FHV RNA1 expression plasmids pF1, pF1fs, and pFA have been previously described (31, 43). pF1 and pF1fs are HIS3-selectable yeast 2μ plasmids that contain full-length cDNA for FHV RNA1 under control of the galactose-inducible-glucose-repressible GAL1 promoter and are terminated by a self-cleaving hepatitis δ ribozyme to generate authentic viral 3′ termini (Fig. 1A). RNA1 from pF1 and pF1fs can serve as a template for protein A-directed RNA1 replication and subgenomic RNA3 synthesis. RNA1 from pF1 can also serve as mRNA for protein A translation, whereas RNA1 from pF1fs contains a four-nucleotide frameshifting insertion in the 5′ coding region (43). pFA is a LEU2-selectable yeast centromeric plasmid that contains full-length cDNA for FHV RNA1 under control of the GAL1 promoter but is flanked upstream by the GAL1 leader sequence and downstream by the CYC1 polyadenylation signal sequence (Fig. 1A). RNA1 from pFA cannot serve as a template for replication but can be translated to provide protein A for RNA replication in the presence of a template from pF1fs.
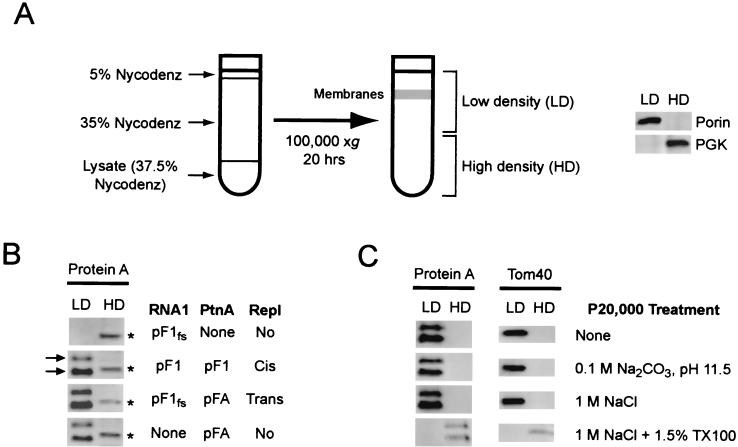
FIG. 1.
FHV protein A localizes to mitochondria. (A) Schematic of plasmids used for RNA1 and protein A expression. Authentic viral 3′ termini are generated in pF1 and pF1fs by a hepatitis δ ribozyme (Rz). A frameshift in pF1fs at the indicated location disrupts translation, whereas the GAL1 leader (L) and CYC1 polyadenylation signal (An) in pFA disrupt its activity as a template for replication but not its translation. (B) Yeast transformed with pF1fs (top row), pF1 (second row), pF1fs plus pFA (third row), or pFA alone (bottom row) were immunostained with rabbit anti-protein A and mouse anti-CoxIII, followed by Texas red-labeled goat anti-rabbit and fluorescein isothiocyanate-labeled goat anti-mouse secondary antibodies. Representative confocal images for protein A (left images, red), CoxIII (middle images, green), and merged signals (right images) are shown. The merged images represent a digital superimposition of red and green signals, where areas of fluorescence colocalization are yellow. Northern blot analysis showed viral RNA replication, determined by the presence of negative-strand RNA1 and positive-strand subgenomic RNA3, in yeast transformed with pF1 and pF1fs plus pFA, but not in yeast transformed with pFA or pF1fs alone (31, 43; data not shown). (C) Yeast were immunostained with rabbit anti-protein A and mouse anti-vacuolar ATPase. Representative confocal images for protein A (left image, red), vacuolar ATPase (middle image, green), and merged signals (right image) are shown.
The protein A expression plasmids pFA-N/HA and pFA-C/HA were derived from pFA and were designed to express protein A with an N- or C-terminal HA epitope tag and an eight-amino-acid spacer (GGSGGSGG). We constructed pFA-C/HA by using mutually primed extension of overlapping oligonucleotides as previously described for the bacterial expression plasmid pET-FHVPA (37), substituting HA for the hexahistidine epitope tag. To construct pFA-N/HA, we used mutually primed extension of overlapping oligonucleotides that contained 68 nucleotides from the 5′ noncoding region of pFA, 78 nucleotides that encoded the HA epitope tag, the eight-amino acid-spacer, and the first nine amino acids from the protein A N terminus. The annealed and extended fragment was inserted into the PstI/AvrII region of pFA to generate pFA-N/HA.
Plasmids with protein A deletion mutants were derived from pFA-C/HA. We generated a series of 10 deletions that covered all but amino acids 1 to 7 of protein A by using convenient restriction enzyme sites and blunt-end closure where appropriate. The deletion mutant designations and corresponding restriction enzyme sites were Δ8-230 (AvrII/NheI), Δ99-230 (BsiWI/NheI), Δ99-686 (BsiWI/RsrII), Δ231-988 (NheI/BlpI), Δ336-686 (BsaBI/RsrII), Δ399-826 (MscI/MscI), Δ399-921 (MscI/NcoI), Δ473-686 (BglII/RsrII), Δ609-921 (NcoI/NcoI), and Δ687-1004 (RsrII/BspEI). We generated a series of five overlapping N-proximal protein A deletion mutants by PCR to delete amino acids 9 to 45, 89, 135, 209, or 245. Primer pairs corresponding the SexAI site within RNA1 and the desired deletion location with an engineered AvrII site were used to amplify fragments that were inserted into AvrII/SexAI region of pFA-C/HA to generate the deletion mutants Δ9-45, Δ9-89, Δ9-135, Δ9-209, and Δ9-245.
The GFP fusion protein expression plasmids were derived from pGFP, a LEU2-selectable yeast 2μ plasmid that contains a yeast-optimized cDNA for GFP (8) under control of the GAL1 promoter. We constructed pT70TM/GFP by using mutually primed extension of overlapping oligonucleotides that encoded the first 30 amino acids of Tom70 (35) with flanking NsiI sites. The annealed and extended fragment was digested and inserted into the PstI site at the N terminus of GFP. We generated a series of protein A-GFP fusions by PCR to attach protein A amino acids 1 to 46, 81, and 121, 42 to 81, and 75 to 121 to the N terminus of GFP. Primers pairs with engineered flanking PstI sites were used to amplify fragments from pFA that were inserted into the PstI site of pGFP to generate pPA1-46/GFP, pPA1-81/GFP, pPA1-121/GFP, pPA42-81/GFP, and pPA75-121/GFP. The initial construction procedure for GFP fusion protein expression plasmids added the amino acid sequence MQM to the GFP junction site. To prevent potential downstream translation initiation, we changed the junction sequence to the amino acids SG for all GFP fusion plasmids by oligonucleotide-directed mutagenesis.
Immunofluorescence microscopy.
We performed confocal immunofluorescence microscopy as previously described for FHV-infected Drosophila cells (37), with modifications for yeast preparation (46). For selective permeabilization experiments, fixed spheroplasts were permeabilized with either 0.2% (vol/vol) Triton X-100 plus 0.002% (wt/vol) saponin or 0.002% saponin alone for 10 min prior to immunofluorescence staining.
Northern blot analysis.
Total RNA was isolated from intact yeast by using hot acidic phenol (43), quantitated spectrophotometrically, and stored at −80°C until analysis. RNA was separated on 1.4% agarose-formaldehyde gels, blotted onto nylon membranes, and probed for positive- and negative-strand FHV RNA1 and RNA3 as previously described (37). We quantitated RNA levels with ImageQuant software (Molecular Dynamics, Piscataway, N.J.).
Immunoblot analysis.
For total protein isolation, yeast were washed with distilled water and resuspended in 20 mM Tris (pH 8.0), 0.2 M NaCl, 10 mM MgCl2, 1 mM EDTA, and 5% (wt/vol) glycerol. Yeast cell walls were disrupted by mechanical shearing with glass beads, sodium dodecyl sulfate (SDS) was added to 1% (wt/vol), and samples were heated to 100°C for 10 min. Glass beads and insoluble cellular debris were removed by centrifugation at 10,000 × g for 5 min, and lysates were stored at −20°C until analysis. Cell lysates were separated on reducing SDS-polyacrylamide gels, transferred to polyvinylidene difluoride membranes, and immunoblotted as previously described (37). We quantitated protein levels with Lumi-Imager software (Boehringer Mannheim, Indianapolis, Ind.).
Membrane flotation.
Yeast were spheroplasted in 1 M sorbitol-0.1 M potassium phosphate (pH 7.6) for 15 min at 30°C, washed once, resuspended in flotation buffer (50 mM HEPES [pH 7.2], 10 mM KCl, 5 mM MgCl2, 1 mM EDTA) with a yeast protease inhibitor cocktail containing 1 mM 4-(2-aminoethyl)benzenesulfonyl fluoride, 5 mM 1,10-phenanthroline, 22 μM pepstatin A, and 14 μM trans-epoxysuccinyl-l-leucylamido(4-guanidino)butane and then stored on ice for 10 min. Spheroplasts were disrupted with 10 strokes of a Dounce homogenizer, and lysates were centrifuged at 500 × g for 5 min to pellet nuclei, unlysed cells, and large debris. Nycodenz was added to postnuclear lysates to a final concentration of 37.5% (wt/vol), loaded under a 5% to 35% Nycodenz discontinuous gradient, and centrifuged to equilibrium in a Beckman TLS-55 swing-bucket rotor at 100,000 × g for 20 h. After centrifugation, the upper half of the gradient was removed manually and designated the low-density (LD) fraction, and the lower half was designated the high-density (HD) fraction. Equal volume samples from each fraction were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotted as described above. For membrane dissociation assays, postnuclear lysates were centrifuged at 20,000 × g for 10 min to pellet membranes and their associated proteins. Pellets were resuspended in flotation buffer, 0.1 M Na2CO3 [pH 11.5], 1 M NaCl, or 1 M NaCl with 1.5% Triton X-100, incubated on ice for 30 min, and fractionated in Nycodenz gradients as described above.
Protease digestion.
We used membrane fractions prepared by the method of Glick and Pon (16) for protease digestion assays. Spheroplasts were lysed by Dounce homogenization in lysis buffer (20 mM HEPES [pH 7.2], 0.5 M sorbitol, 50 mM potassium phosphate) with the protease inhibitor cocktail described above and then centrifuged at 1,000 × g for 5 min to pellet the nuclei, unlysed cells, and large debris. Postnuclear lysates were centrifuged at 12,000 × g for 10 min, pellets were resuspended in lysis buffer and centrifuged at 1,000 × g for 5 min, and the supernatant was recovered and used as the membrane fraction. Proteinase K (0 to 10 μg/ml) was added in the presence or absence of 0.1% Triton X-100, samples were incubated on ice for 30 min, and protease activity was terminated by the addition of SDS-PAGE sample buffer and heating to 100°C for 10 min. Samples were separated by SDS-PAGE and analyzed by immunoblotting as described above. Preliminary experiments showed that exogenous proteinase K at the concentrations used in protease digestion experiments was not inhibited by residual protease inhibitors present in the membrane fraction (data not shown).
Sequence analysis and transmembrane domain (TMD) prediction.
We performed sequence analysis with the following programs: DAS (9) (www.sbc.su.se/∼miklos/DAS/), Topred2 (62) (www.sbc.su.se/∼erikw/toppred2/), Tmpred (www.ch.embnet.org/software/TMPRED/), HMMTOP (59) (www.enzim.hu/hmmtop/), and TMHMM v.2 (25) (www.cbs.dtu.dk/services/TMHMM-2.0/).
RESULTS
Protein A localizes to mitochondria in yeast.
We previously found that protein A localizes with viral RNA replication to outer mitochondrial membranes in FHV-infected Drosophila DL-1 cells (37). To determine whether yeast duplicated the localization of protein A in insect cells, we investigated protein A localization in yeast with confocal immunofluorescence microscopy (Fig. 1). We used the inner mitochondrial membrane protein CoxIII (6) as a mitochondrial marker. All experiments were repeated multiple times, and representative results are shown.
In wild-type yeast or yeast transformed with pF1fs, a plasmid that expresses an RNA1 template but no protein A due to an early frameshift (Fig. 1A) (31, 43), yeast mitochondria appeared as a branched, peripherally distributed network (Fig. 1B, top row), a finding consistent with previously reported yeast mitochondrial morphology (18). In yeast transformed with pF1, a plasmid that expresses wild-type RNA1 and protein A (Fig. 1A) and hence initiates RNA replication (31, 43), CoxIII distribution was altered and displayed a more localized, clumped pattern that colocalized with protein A (Fig. 1B, second row), similar to the mitochondrial clustering that occurs in FHV-infected Drosophila cells (37). To address the possibility that FHV RNA replication or protein A expression altered the distribution of CoxIII to include nonmitochondrial membranes, we also used antibodies against Ssc1, a mitochondrial matrix-specific heat shock protein (32) in confocal immunofluorescence experiments, and found that protein A also colocalized with Ssc1 (data not shown). As controls for other intracellular membrane compartments, immunofluorescence microscopy showed no significant localization of protein A to either vacuoles (Fig. 1C) or endoplasmic reticulum (data not shown). Thus, based on the colocalization of protein A with both a mitochondrial inner membrane and a mitochondrial matrix protein, and the absence of colocalization with nonmitochondrial markers, we concluded that yeast reproduced the mitochondrial localization of protein A in FHV-infected Drosophila cells (37).
Protein A localization to mitochondria is independent of other viral proteins and RNA replication.
In infected Drosophila cells, the presence of all FHV RNA and protein components precluded determining whether other viral factors contributed to protein A localization (37). Figure 1B showed that mitochondrial localization of protein A did not require FHV RNA2 or its encoded capsid proteins. Moreover, when RNA2 was coexpressed and coreplicated with RNA1, leading to capsid protein expression and encapsidation, the colocalization and globular patterns of protein A and CoxIII immunofluorescence were indistinguishable from those with RNA1 replication alone (data not shown). These patterns also remained unchanged when protein A was expressed in trans to replicate an RNA1 frameshift derivative (Fig. 1B, third row) or when protein A was expressed alone, without a replicatable RNA1 template or protein B (Fig. 1B, bottom row). Thus, protein A localized to mitochondria and induced a clumped, globular mitochondrial distribution in the absence of any other viral proteins, subgenomic RNA3 synthesis, or RNA replication.
Protein A is membrane-associated in yeast.
We previously found that protein A from FHV-infected Drosophila cells fractionated as a membrane-associated protein (37). To investigate the intracellular membrane association of protein A in yeast, we initially performed differential centrifugation experiments with postnuclear yeast lysates. Protein A was recovered almost exclusively in the 20,000 × g pellet fraction (data not shown), suggesting that protein A was membrane associated. However, protein A expression in yeast may lead to formation of sedimentable aggregates, similar to FHV protein A expression in E. coli (37) and expression of some brome mosaic virus 1a derivatives in yeast (10). To discriminate between protein A sedimentation due to membrane association or aggregation, we used equilibrium density gradient centrifugation to examine the flotation behavior of protein A from yeast lysates (Fig. 2). Postnuclear lysates were adjusted to 37.5% Nycodenz, loaded under a 5 to 35% Nycodenz gradient, and centrifuged to equilibrium (Fig. 2A). In initial experiments, we divided gradients into six equal volume fractions and found that membrane-associated proteins reproducibly floated into the upper half of Nycodenz gradients, whereas cytosolic proteins remained in the bottom fractions (data not shown). Thereafter, we divided gradients into two equal fractions, and examined the upper LD and lower HD fractions individually by immunoblot assay. The LD fraction contained a visible membrane layer and the majority of the yeast outer mitochondrial integral membrane porin protein, whereas the majority of the cytosolic protein 3-phosphoglycerate kinase (PGK) partitioned into the HD fraction (Fig. 2A).
FIG. 2.
FHV protein A is tightly membrane associated. (A) Schematic of equilibrium density Nycodenz gradient fractionation procedure. Yeast lysates were loaded under 5 to 35% discontinuous Nycodenz gradients and centrifuged to equilibrium, and equal volume fractions from the upper LD and lower HD regions were recovered and analyzed by immunoblotting. (B) Equilibrium density Nycodenz gradient fractions of lysates from yeast transformed with pF1fs, pF1, pF1fs plus pFA, or pFA alone were immunoblotted with rabbit anti-protein A. Protein A appeared as a doublet (arrows) in the LD fractions, whereas a host protein band that cross-reacted with the protein A antisera was present in the HD fractions (asterisks). (C) Postnuclear lysates from yeast transformed with pF1 were centrifuged at 20,000 × g for 10 min, pellets were resuspended in the indicated buffer and subjected to Nycodenz gradient fractionation, and fractions were immunoblotted with rabbit antisera against protein A or the outer mitochondrial integral membrane protein Tom40 (20). The yeast protein detected by the protein A antisera in Fig. 2B (asterisks) was not recovered in the 20,000 × g pellet, and therefore was not present in the HD fraction after Nycodenz gradient fractionation.
To investigate the membrane association of protein A in the presence or absence of viral RNA replication, yeast transformed with pF1fs, pF1, pF1fs plus pFA, or pFA alone were lysed, and the lysates analyzed by Nycodenz gradient flotation and immunobloting with antisera against protein A (Fig. 2B). In the HD fractions, protein A antisera revealed only a cross-reacting host protein band, which was present in all lysates (Fig. 2B, asterisks), including lysates from yeast transformed with only the protein A frameshift plasmid, pF1fs. For lysates from yeast expressing pF1, pF1fs plus pFA, or pFA alone, protein A partitioned into the LD fraction. Thus, protein A was membrane associated in yeast independently of FHV RNA replication. Protein A appeared as a doublet in yeast LD fractions (arrows in Fig. 2B), whereas the same protein A antisera reacts with a single band in infected Drosophila cell lysates (37). The upper band in yeast LD fractions likely represented full-length protein A, and the lower band likely represented a C-terminal degradation product, since flotation analysis with terminally epitope-tagged protein A derivatives demonstrated that the upper band contained intact N and C termini, whereas the lower band contained only an intact N terminus (data not shown).
Protein A fractionates as an integral membrane protein.
We further examined the flotation behavior of protein A in yeast lysates under conditions designed to dislodge peripherally associated membrane proteins (4). Postnuclear lysates from yeast transformed with pF1 were centrifuged at 20,000 × g, and pelleted membranes and their associated proteins were resuspended in lysis buffer, 0.1 M Na2CO3 [pH 11.5], 1 M NaCl, or 1 M NaCl with 1.5% Triton X-100 and incubated for 30 min prior to Nycodenz gradient fractionation (Fig. 2C). Protein A remained in the LD fraction even at pH 11.5 or in the presence of 1 M NaCl but was recovered in the HD fraction in the presence of 1.5% Triton X-100. The mitochondrial integral membrane protein Tom40 (20) behaved equivalently under these conditions. Thus, protein A was tightly membrane associated in yeast and had biochemical characteristics of an integral membrane protein. For both protein A and Tom40, full-length protein recovery was reduced when membrane pellets were resuspended in buffer containing Triton X-100 (Fig. 2C). A similar reduced recovery was seen with other membrane proteins in the presence of detergent (see below, Fig. 8C, and data not shown), suggesting that endogenous proteases were released upon detergent treatment and were active despite the presence of protease inhibitors (see Materials and Methods).
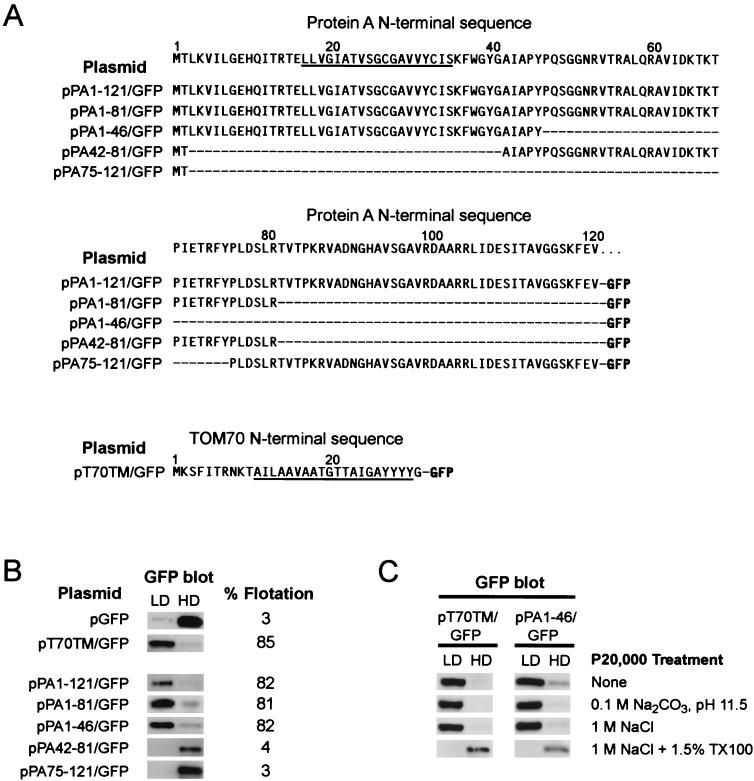
FIG. 8.
FHV protein A amino acids 1 to 46 impart integral membrane protein characteristics to GFP. (A) Sequences of GFP N-terminal fusions. The protein A N-terminal sequence is shown at the top for reference. The underlined amino acids are the core predicted TMD for protein A (see Fig. 3). Dashes indicate no amino acid and are included for alignment purposes. The Tom70 N-terminal sequence is also shown, with the TMD underlined. (B) Yeast transformed with plasmids containing the GFP fusion constructs shown in panel A were processed for Nycodenz gradient fractionation as described in the Fig. 2 legend. Flotation efficiencies represent the percentage of the total protein recovered in the LD fraction and are averages of three independent experiments. Representative rabbit anti-GFP immunoblots are shown. (C) Yeast lysates were prepared for Nycodenz gradient fractionation as described in the Fig. 2C legend and immunoblotted with rabbit anti-GFP.
Protein A contains a predicted TMD.
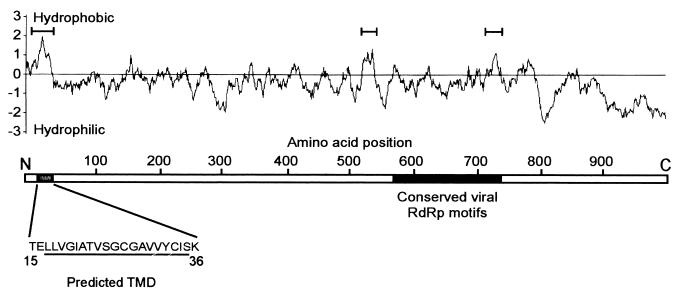
The behavior of protein A in Drosophila cells (37) and yeast lysates (Fig. 2) suggested that it was an integral membrane protein. We analyzed the protein A amino acid sequence by using several programs to identify potential hydrophobic regions and TMDs (Fig. 3). For reference, the conserved viral RdRp palm motifs (41) of protein A are located between amino acids 575 and 740. We identified three hydrophobic regions within protein A: the first around amino acids 3 to 50, the second around amino acids 520 to 550, and the third around amino acids 720 to 750 (Fig. 3, bracketed regions). Four sequence analysis programs (DAS, Topred2, Tmpred, and HMMTOP) predicted with high probability that residues 15 to 36 contained a stretch of amino acids with sufficient hydrophobicity and length to span a lipid bilayer. One program (TMHMM v.2) predicted the same region but with only moderate probability, whereas none of the programs predicted that the most hydrophobic internal regions (amino acids 520 to 550 and 720 to 750) had sufficient hydrophobicity to span a lipid bilayer.
FIG. 3.
Schematic representation of FHV protein A with hydrophobicity plot. The primary sequence of the predicted TMD in the N-proximal region between amino acids 15 and 36 is shown. The underlined amino acids represent the core predicted TMD identified by five different structural prediction programs (see the text). The protein A region with conserved viral RdRp palm motifs is indicated in black; the highly conserved GDD motif is located at amino acids 690 to 692. Hydrophobicity was calculated by using the method of Kyte and Doolittle (28) with a window size of 17 amino acids. Regions with a maximum hydrophobicity of >1 are indicated with brackets.
The majority of the protein A sequence is exposed to the cytoplasm.
The prediction that protein A contained a potential N-proximal TMD (Fig. 3) and the consistent flotation and membrane dissociation results (Fig. 2) prompted us to investigate the membrane topology of protein A by protease digestion (Fig. 4). Pelleted membrane fractions were digested with increasing concentrations of proteinase K in the absence or presence of nonionic detergent to disrupt membranes and analyzed by immunoblotting with polyclonal antisera against protein A. Proteinase K-digested protein A even at low protease concentrations, and the addition of Triton X-100 did not significantly alter the degradation pattern (Fig. 4A). In contrast, the mitochondrial matrix protein p32 showed detergent-dependent proteinase K susceptibility (Fig. 4B), a finding consistent with its membrane-protected matrix localization (38). The protein A antisera used in these experiments was raised against full-length protein (37) and reacted with every protein A deletion mutant described below (see Fig. 6 for deletion map), suggesting that the antisera reacted with multiple, distributed protein A epitopes. These results implied that a significant portion of the membrane-bound protein A sequence was exposed to the cytoplasm.
FIG. 4.
FHV protein A is susceptible to protease digestion in the absence of detergent. Membrane fractions from yeast expressing protein A were incubated with increasing concentrations of proteinase K in the absence or presence of 0.1% Triton X-100 and analyzed by immunoblotting with rabbit anti-protein A (A) or anti-p32 (B). The background yeast band detected by the protein A antisera (asterisks, Fig. 2B) was not recovered in the membrane fraction used in protease digestion experiments. Molecular size (MW) markers in kilodaltons are shown on the left in panel A.
FIG. 6.
Membrane association of FHV protein A deletion mutants. Flotation analysis was done as described in the Fig. 2 legend with C-terminally HA epitope-tagged protein A deletion mutants covering all but amino acids 1 to 7 of protein A (A) or with a nested series of N-proximal deletions (B). Protein A schematic with amino acid positions and the locations of the predicted TMD (amino acids 15 to 36; see Fig. 3), with viral RdRp motifs shown at the top. Deleted regions are represented by solid lines. Deletion designations are shown on the left and represent the amino acids included within the deleted region. Flotation efficiencies represent the percentage of the total protein recovered in the LD fraction after Nycodenz gradient fractionation and are averages of at least three independent experiments. Representative mouse anti-HA immunoblots are shown.
Protein A is a transmembrane protein.
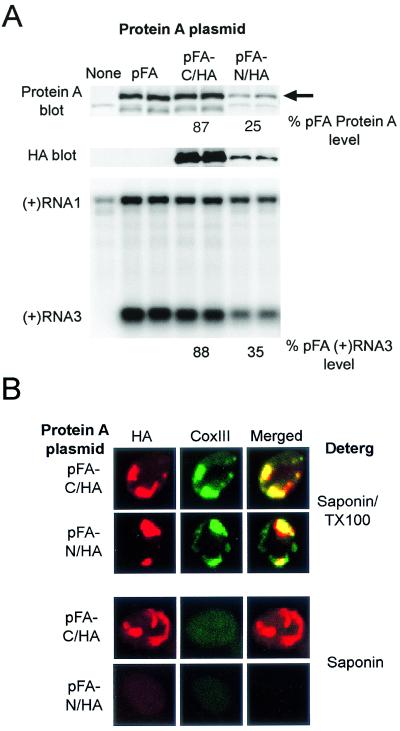
To further investigate the membrane topology of protein A, we constructed protein A derivatives with the HA epitope tag attached to either the N or C terminus. We first examined whether the epitope tags influenced protein A accumulation, function, or membrane-association (Fig. 5A). The C-terminal HA tag reduced accumulation of full-length protein A by 23%, whereas the N-terminal HA tag reduced accumulation by 75% (Fig. 5A, top blot). For both protein A HA-tagged derivatives, the level of RNA replication, measured by positive-strand subgenomic RNA3 accumulation (Fig. 5A, bottom blot), was reduced similarly to that of protein A accumulation, implying that the specific activities of the HA-tagged protein A derivatives were similar to those of wild-type protein A. Moreover, the N- and C-terminal HA tags did not alter protein A flotation behavior, since both protein A derivatives partitioned into the LD fraction of Nycodenz gradients (data not shown) in parallel to wild-type protein A (Fig. 2B). Thus, the epitope-tagged derivatives of protein A reproduced the biochemical behavior and in vivo function of wild-type protein A.
FIG. 5.
FHV protein A is a transmembrane protein. (A) Protein A expression and viral RNA replication in yeast transformed with pF1fs alone or pF1fs plus pFA, pFA-C/HA, or pFA-N/HA. Total protein samples from an equivalent number of yeast per sample were separated by SDS-PAGE and immunoblotted with rabbit anti-protein A (upper blot) or mouse anti-HA (middle blot). Total protein staining showed equivalent protein loading in all lanes (data not shown). The full-length protein A band (arrow) was quantitated by densitometry, and the values shown are averages of two independent experiments and represent the relative protein A accumulation compared to that in yeast transformed with pF1fs plus pFA. We separated 2 μg of total RNA per sample on denaturing formaldehyde-agarose gels and blotted this with a 32P-labeled complementary riboprobe that detected positive-strand RNA1 and RNA3 (lower blot). The positions of RNA1 and RNA3 are indicated on the left. Positive-strand RNA3 was quantitiated by densitometry, and the values shown are averages of two independent experiments and represent the relative positive-strand RNA3 accumulation compared to that in yeast transformed with pF1fs plus pFA. The positive-strand RNA1 band in the far left control lane represents DNA-directed transcription from pF1fs and not viral RdRp-directed replication (43). (B) Selective membrane permeabilization of yeast expressing HA-tagged protein A derivatives. Yeast transformed with pF1fs plus pFA-C/HA or pFA-N/HA were permeabilized with the indicated detergent and immunostained with mouse anti-CoxIII and rabbit anti-HA as described in the Fig. 1 legend. Representative confocal immunofluorescence images for HA (left images, red), CoxIII (middle images, green), and merged signals (right images) are shown.
To confirm the protease digestion results (Fig. 4) and directly investigate the membrane orientation of protein A, we used a selective membrane permeabilization strategy coupled with confocal immunofluorescence microscopy (Fig. 5B). Low concentrations of saponin selectively permeabilize the cell plasma membrane, while leaving mitochondrial membranes intact (50). Yeast transformed with pF1fs and pFA-C/HA or pFA-N/HA were fixed with formaldehyde, spheroplasted, permeabilized with either 0.002% saponin plus 0.2% Triton X-100 or saponin alone, and then immunostained with antibodies against HA and CoxIII. CoxIII, an oxidative phosphorylation chain enzyme located in the inner mitochondrial membrane (6), was monitored as a control to assay selective mitochondrial permeabilization to antibodies. When we permeabilized yeast containing pFA-C/HA (Fig. 5B, top row) or pFA-N/HA (Fig. 5B, second row) with saponin and Triton X-100, we observed a clumped distribution of HA-tagged protein A that colocalized with CoxIII immunofluorescence, similar to the pattern observed with wild-type protein A (Fig. 1B). Thus, the HA epitope tags did not alter the colocalization of protein A and CoxIII, and both the N and the C termini of protein A were accessible when the mitochondrial membranes were permeabilized.
When we permeabilized yeast with saponin alone, CoxIII immunofluorescence was lost (Fig. 5B, lower two rows), indicating that the outer mitochondrial membrane remained impermeable to antibodies. Although HA-specific antibodies still detected the protein A C terminus in yeast permeabilized with saponin alone (Fig. 5B, third row), the N terminus was not detected (Fig. 5B, bottom row). Polyclonal antisera against full-length protein A detected both HA-tagged versions of protein A in yeast permeabilized with saponin alone (data not shown). Thus, protein A was a transmembrane protein with the C terminus exposed to the cytoplasm.
Membrane association of protein A deletion mutants.
To identify regions within protein A that mediated membrane association, we constructed a series of deletion mutants covering all but amino acids 1 to 7 of protein A (Fig. 6). To equalize detection sensitivity for immunoblotting and immunofluorescence analyses, all protein A deletion mutants contained a C-terminal HA tag. The total accumulation of protein A deletion mutants varied from 69% (Δ99-230) to 202% (Δ399-921) compared to full-length protein A. When the deletion mutants were expressed with pF1fs to provide an RNA1 template, none of the mutants were able to support RNA replication, including mutants with deletions outside the conserved viral RdRp motifs (data not shown).
We analyzed the membrane association of protein A deletions mutants by Nycodenz gradient flotation (Fig. 6A). Flotation efficiencies of deletion mutants varied from 48% (Δ8-230) to 87% (Δ399-921), and there was no correlation between accumulation and flotation efficiency (R = 0.07, P > 0.8). None of the deletions reduced protein A flotation into the membrane-enriched LD fraction by more than twofold compared to the 91% flotation efficiency for full-length protein A (Fig. 6A). Although the deletion analysis did not identify any region within protein A that was solely responsible for membrane association, deletion mutant Δ8-230 showed the largest reduction in flotation efficiency. Thus, protein A membrane association was more sensitive to N-proximal deletions than to internal or C-proximal deletions, but more than one region was likely involved in the overall association of protein A with membranes.
Sequence analysis identified two internal regions of protein A with moderate predicted hydrophobicity (Fig. 3) that might contain additional membrane association domains (see also Discussion). However, the membrane association deletion studies described above and the immunofluorescence localization studies with deletion mutants described below indicated that the N-proximal region of protein A was involved in both membrane association and mitochondrial localization. Thus, we further investigated sequences within the N terminus of protein A, the single largest contributor to membrane association, by using a series of nested N-proximal deletions (Fig. 6B). To provide a common N terminus, we started the nested deletion series after amino acid 8. Similar to the deletion mutants in Fig. 6A, the accumulation of the nested N-proximal deletion mutants varied from 71% (Δ9-89) to 162% (Δ9-45) of full-length protein A, and none of the N-proximal deletions supported RNA replication when expressed with an RNA1 template (data not shown).
Protein A N-proximal deletion mutants with the largest (Δ9-245 and Δ9-209) and smallest (Δ9-45) deletions all had similar flotation efficiencies of 41 to 42% (Fig. 6B), which were comparable to the 48% flotation efficiency seen with Δ8-230 (Fig. 6A). This suggested that the primary contribution of the N-proximal region to membrane association contained amino acids 9 to 45. However, the intermediate deletions in Fig. 6B showed changes in flotation efficiency that did not correlate with primary sequence changes. For example, Δ9-135 showed only 4% flotation, whereas larger deletions encompassing the same region had 10-fold greater flotation efficiencies. Conversely, Δ9-89 had a calculated flotation efficiency of 64%, greater than both larger and smaller deletions. These results suggest that changes in secondary or tertiary structure influenced protein A membrane association, potentially through protein misfolding in the setting of large sequence deletions and altered accessibility of flanking sequences.
Amino-proximal deletions alter the intracellular distribution of protein A.
To determine whether the membrane association of protein A deletion mutants, as measured by flotation, correlated with CoxIII colocalization, we analyzed all deletion mutants shown in Fig. 6 by confocal immunofluorescence microscopy. A representative set of deletion mutants is shown in Fig. 7. All protein A mutants with deletions outside the N-terminal 98 amino acids showed a clustered protein A distribution and colocalization with CoxIII (e.g., Fig. 7, Δ99-230, Δ99-686, Δ399-921, and Δ687-1004), similar to full-length protein A (Fig. 1B; also Fig. 7, top row). In contrast, protein A mutants with deletions in the N-terminal 98 amino acids displayed a more diffuse protein A intracellular distribution (illustrated in Fig. 7 for Δ9-89 and Δ9-135) that was distinct from the clustered protein A distribution and CoxIII colocalization seen with full-length protein A (Fig. 1B; see also Fig. 7, top row). Thus, the colocalization of protein A with CoxIII was disrupted by N-proximal but not internal or C-proximal deletions. However, because of the diffuse distribution of N-proximal protein A deletion mutants, we cannot rule out that a fraction of these proteins localized to mitochondria or other intracellular membrane compartments, which may explain their 41 to 64% flotation efficiency (Fig. 6A and B; see also Discussion).
FIG. 7.
Subcellular localization of FHV protein A deletion mutants. Deletions are described in Fig. 6. Yeast were immunostained with mouse anti-CoxIII and rabbit anti-HA as described in the Fig. 1 legend. Representative confocal immunofluorescence images for HA (left images, red), CoxIII (middle images, green), and merged signals (right images) are shown.
Protein A amino acids 1 to 46 impart integral membrane protein characteristics to GFP.
To determine whether the protein A N terminus was sufficient to target a heterologous protein to membranes, we performed gain-of-function studies with fusion proteins composed of protein A N-terminal sequences attached to the N terminus of GFP (Fig. 8A). As a positive control, we attached the first 30 amino acids of the yeast outer mitochondrial membrane import receptor protein Tom70 to GFP. The Tom70 N-terminal 30 amino acids contain a TMD and signal-anchor sequence that targets and anchors Tom70 into the outer mitochondrial membrane (35). The total accumulation of full-length GFP fusion proteins relative to wild-type GFP varied from 11% for PA1-121/GFP and PA42-81/GFP to 43% for PA75-121/GFP.
We analyzed the membrane association of the GFP fusion proteins by Nycodenz gradient flotation (Fig. 8B). Wild-type GFP showed only 3% flotation efficiency, whereas addition of the Tom70 N-terminal 30 amino acids or protein A N-terminal 121 amino acids increased flotation efficiencies to 85 or 82%, respectively. Addition of shorter protein A N-terminal regions, including amino acids 1 to 81 and 1 to 46, also increased GFP flotation efficiencies to 81 and 82%, respectively, similar to the flotation efficiency with PA1-121/GFP. Consistent with this observation, GFP fusion proteins that contained protein A amino acids 42 to 81 or 75 to 121 did not show higher flotation efficiencies than GFP alone. Thus, protein A amino acids 1 to 46 were sufficient to mediate GFP membrane association.
We further investigated the membrane affinity of T70TM/GFP and PA1-46/GFP under conditions designed to dislodge peripherally associated membrane proteins (Fig. 8C), similar to the experiments described above for full-length protein A (Fig. 2C). Both T70TM/GFP and PA1-46/GFP fusion proteins remained in membrane-associated LD fractions at pH 11.5 or in the presence of 1 M NaCl but were recovered in the HD fraction in the presence of 1.5% Triton X-100, albeit at reduced levels. Thus, protein A amino acids 1 to 46 were sufficient to impart integral membrane protein characteristics to GFP.
Protein A amino acids 1 to 46 target GFP to mitochondria.
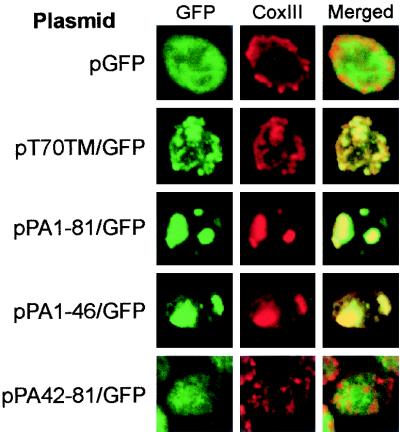
To investigate the subcellular localization of the GFP fusion proteins, we used confocal immunofluorescence microscopy (Fig. 9). Yeast expressing GFP showed a diffuse cytoplasmic GFP distribution that was distinct from the mitochondrial network identified by CoxIII immunofluorescence (Fig. 9, top row). In contrast, yeast expressing T70TM/GFP showed colocalization of GFP and CoxIII immunofluorescence (Fig. 9, second row), confirming the mitochondrial targeting characteristics of the Tom70 N-terminal 30 amino acids (35). Yeast expressing PA1-121/GFP (data not shown), PA1-81/GFP (Fig. 9, third row), and PA1-46/GFP (Fig. 9, fourth row) also showed colocalization of GFP and CoxIII immunofluorescence, although the GFP and CoxIII immunofluorescence patterns were more localized and clustered, similar to the pattern observed with full-length protein A (Fig. 1B). Yeast expressing PA42-81/GFP (Fig. 9, bottom row) and PA75-121/GFP (data not shown) showed a more diffuse GFP distribution that was distinct from CoxIII immunofluoresence. Thus, protein A amino acids 1 to 46 were sufficient to target GFP to mitochondria.
FIG. 9.
FHV protein A amino acids 1 to 46 target GFP to mitochondria. Yeast transformed with the indicated plasmids were immunostained with rabbit anti-GFP and mouse anti-CoxIII as described in the Fig. 1 legend, except that the secondary antibodies were fluorescein isothiocyanate-labeled donkey anti-rabbit and rhodamine-labeled donkey anti-mouse antibodies. Representative confocal immunofluorescence images for GFP (left images, green), CoxIII (middle images, red), and merged signals (right images) are shown.
DISCUSSION
In this report, we investigated the intracellular localization, membrane association, membrane topology, and organellar targeting signals of FHV protein A in S. cerevisiae. We drew four main conclusions. First, FHV replication in yeast recapitulated cell biology features of FHV replication in Drosophila cells (37), such as mitochondrial localization of protein A and the induction of mitochondrial clustering. Second, protein A alone was sufficient for mitochondrial localization in the absence of RNA replication, nonstructural protein B, or capsid proteins. Third, protein A was an outer mitochondrial transmembrane protein with the N terminus located in the inter- membrane space or matrix and the C terminus exposed to the cytoplasm. Fourth, the N-terminal 46 amino acids of protein A contain sequences sufficient for mitochondrial localization and membrane insertion. These observations address several important aspects of FHV replication complex formation and provide a framework for further studies on the mechanisms of positive-strand RNA virus replication.
The protein A N terminus contains an outer mitochondrial membrane targeting signal.
The N-proximal region of protein A, identified in the deletion studies as a primary contributor to protein A-membrane association (Fig. 6) and a crucial determinant for mitochondrial localization (Fig. 7), was validated as a membrane association domain through gain-of-function analysis. GFP fusion studies demonstrated that the first 46 amino acids of protein A were sufficient for efficient membrane association with integral membrane protein characteristics (Fig. 8), a finding consistent with the presence of a predicted TMD between amino acids 15 to 36 (Fig. 3). The GFP fusion studies also demonstrated that the first 46 amino acids of protein A were sufficient for mitochondrial localization of GFP (Fig. 9). Most mitochondrial proteins are nuclear encoded and posttranslationally imported into mitochondria and therefore must contain signals for proper localization (19). For example, proteins destined for the mitochondrial matrix contain a cleavable, N-terminal matrix targeting signal, consisting of 20 to 60 amino acids that can form an amphipathic α-helix with one hydrophobic and one positively charged face (61). This matrix targeting signal interacts with mitochondrial import receptors to mediate translocation through the mitochondrial membranes (19). The mechanisms of targeting and membrane insertion of outer mitochondrial membrane proteins are less well defined. Mitochondrial outer membrane proteins are synthesized in the cytosol without a cleavable presequence and are targeted to the outer mitochondrial membrane via either N- or C-proximal sequences (36). The targeting sequences include a hydrophobic TMD with flanking sequences enriched in basic amino acids. Although most mitochondrial membrane proteins characterized to date are anchored via C-proximal sequences, the import receptor components Tom20 (24) and Tom70 (35) are targeted and anchored to the mitochondrial outer membrane via N-proximal sequences, similar to the topology of FHV protein A.
A BLAST search using protein A amino acids 1 to 46 failed to reveal significant similarity with known outer mitochondrial membrane proteins, a finding consistent with the absence of identified primary sequence motifs for proteins targeted to outer mitochondrial membranes (19). However, the protein A N terminus does contain similarities to features previously identified as important for mitochondrial membrane localization of some other proteins. Localization of Tom20 to the outer mitochondrial membrane requires a TMD with moderate hydrophobicity in the range 1.97 to 2.15, followed within the next five amino acids by a net positive charge (24). Similarly, a basic amino acid immediately following the TMD was essential for outer mitochondrial membrane localization of a cytochrome b5 isoform (27). Consistent with these findings, the predicted core TMD of protein A (underlined in Fig. 3) has a hydrophobicity of 1.96 and is immediately followed by a lysine.
The N-terminal 46 amino acids were the primary determinants of protein A association with membranes and were crucial and sufficient for mitochondrial localization, although flotation analysis of protein A deletion mutants implied that one or more other regions C-terminal to amino acid 230 also contributed to membrane association (Fig. 6). While the protein A sequence after amino acid 230 lacks any additional predicted TMDs (Fig. 3), these sequences might contribute to membrane association in any of several ways. They might form an amphipathic helix with a hydrophobic face; be sites for posttranslational modifications that increase membrane affinity, such as the palmitoylation of Semliki Forest virus nsP1 (29); or interact with charged lipid head groups or other membrane proteins. Studies are in progress to identify additional protein A sequences that contribute to membrane association.
Protein A shares membrane association features with other viral replicase proteins.
Viral replicase proteins with integral membrane characteristics have been identified for hepatitis C virus (HCV) (22, 53), poliovirus (58), equine arteritis virus (60), CIRV (48), and tobacco etch virus (51), although the only other viral RdRp with an experimentally determined transmembrane topology is the HCV NS5B protein (53). HCV NS5B is associated with the endoplasmic reticulum membrane via a C-terminal TMD, whereas FHV protein A is associated with the outer mitochondrial membrane in part via an N-terminal TMD. The tombusvirus CIRV also encodes a transmembrane replicase protein (48), which is interesting in light of the similar mitochondrial changes induced by FHV and CIRV. Both viruses induce the formation of viral replication-associated membrane-bound spherules in the inner mitochondrial membrane space (37, 49), similar to structures seen on endosomal and lysosomal membranes with togavirus infections (13, 17, 26, 33). The CIRV 36-kDa replicase protein is responsible for mitochondrial membrane targeting (5) and is inserted into the outer mitochondrial membrane via two N-proximal TMDs (48). In addition, Rubino et al. demonstrated mitochondrial aggregation by electron microscopy when the CIRV 36-kDa protein was expressed in yeast as a GFP fusion protein (47). Preliminary electron microscopy studies in yeast expressing FHV protein A have shown similar ultrastructural changes (D. Miller, M. Schwartz, and P. Ahlquist, unpublished data), further paralleling FHV-induced mitochondrial clustering in Drosophila cells (37). Further ultrastructural studies will help definitively identify the intracellular structures induced by FHV replication in yeast, and additional studies on the cell biology of replication complex formation by viruses such as HCV, CIRV, and FHV may reveal parallels and common principles in positive-strand RNA virus replication.
Acknowledgments
We thank Kathleen Wessels, Dara Parrish, and Priscilla McDowell for assistance and Johan den Boon for helpful comments on the manuscript. We performed confocal immunofluorescence microscopy at the Keck Neural Imaging Laboratory at the University of Wisconsin—Madison.
This work was supported by National Institutes of Health grants K08 AI01770-01 and GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ball, L. A. 1995. Requirements for the self-directed replication of flock house virus RNA 1. J. Virol. 69:720-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A., and K. L. Johnson. 1998. Nodaviruses of insects, p. 225-267. In L. K. Miller and L. A. Ball (ed.), The insect viruses. Plenum Publishing, New York, N.Y.
- 3.Bashiruddin, J. B., and G. F. Cross. 1987. Boolarra virus: ultrastructure of intracytoplasmic virus formation in cultured Drosophila cells. J. Invertebr. Pathol. 49:303-315. [Google Scholar]
- 4.Bonifacino, J. S. 2000. Characterization of cellular proteins, p. 5.0.1-5.5.11. In K. S. Morgan (ed.), Current protocols in cell biology. John Wiley & Sons, Inc., New York, N.Y.
- 5.Burgyan, J., L. Rubino, and M. Russo. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967-1974. [DOI] [PubMed] [Google Scholar]
- 6.Capaldi, R. A. 1990. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem. 59:569-596. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack, B. P., G. Bertram, M. Egerton, N. A. Gow, S. Falkow, and A. J. Brown. 1997. Yeast-enhanced green fluorescent protein (yEGFP) a reporter of gene expression in Candida albicans. Microbiology 143:303-311. [DOI] [PubMed] [Google Scholar]
- 9.Cserzo, M., E. Wallin, I. Simon, G. von Heijne, and A. Elofsson. 1997. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 10:673-676. [DOI] [PubMed] [Google Scholar]
- 10.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Franco, A., M. Russo, and G. P. Martelli. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233-1237. [Google Scholar]
- 12.Friesen, P. D., and R. R. Rueckert. 1981. Synthesis of black beetle virus proteins in cultured Drosophila cells: differential expression of RNAs 1 and 2. J. Virol. 37:876-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallagher, T. M., and R. R. Rueckert. 1988. Assembly-dependent maturation cleavage in provirions of a small icosahedral insect ribovirus. J. Virol. 62:3399-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garzon, S., H. Strykowski, and G. Charpentier. 1990. Implication of mitochondria in the replication of Nodamura virus in larvae of the Lepidoptera, Galleria mellanolla (L.), and in suckling mice. Arch. Virol. 113:165-176. [DOI] [PubMed] [Google Scholar]
- 16.Glick, B. S., and L. A. Pon. 1995. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 260:213-223. [DOI] [PubMed] [Google Scholar]
- 17.Grimley, P. M., I. K. Berezesky, and R. M. Friedman. 1968. Cytoplasmic structures associated with an arbovirus infection: loci of viral ribonucleic acid synthesis. J. Virol. 2:1326-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hermann, G. J., and J. M. Shaw. 1998. Mitochondrial dynamics in yeast. Annu. Rev. Cell Dev. Biol. 14:265-303. [DOI] [PubMed] [Google Scholar]
- 19.Herrmann, J. M., and W. Neupert. 2000. Protein transport into mitochondria. Curr. Opin. Microbiol. 3:210-214. [DOI] [PubMed] [Google Scholar]
- 20.Hill, K., K. Model, M. T. Ryan, K. Dietmeier, F. Martin, R. Wagner, and N. Pfanner. 1998. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins. Nature 395:516-521. [DOI] [PubMed] [Google Scholar]
- 21.Hope, D. A., S. E. Diamond, and K. Kirkegaard. 1997. Genetic dissection of interaction between poliovirus RNA-dependent RNA polymerase and viral protein 3AB. J. Virol. 71:9490-9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hügle, T., F. Fehrmann, E. Bieck, M. Kohara, H. Kräusslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 23.Johnson, K. L., and L. A. Ball. 1997. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 71:3323-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanaji, S., J. Iwahashi, Y. Kida, M. Sakaguchi, and K. Mihara. 2000. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. J. Cell Biol. 151:277-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 26.Kujala, P., A. Ikäheimonen, N. Ehsani, H. Vihinen, P. Auvinen, and L. Kääriäinen. 2001. Biogenesis of the Semliki Forest virus RNA replication complex. J. Virol. 75:3873-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda, R., T. Ikenoue, M. Honsho, S. Tsujimoto, J. Mitoma, and A. Ito. 1998. Charged amino acids at the carboxy-terminal portions determine the intracellular locations of two isoforms of cytochrome b5. J. Biol. Chem. 273:31097-31102. [DOI] [PubMed] [Google Scholar]
- 28.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 29.Laakkonen, P., T. Ahola, and L. Kääriäinen. 1996. The effects of palmitoylation on membrane association of Semliki Forest virus RNA capping enzyme. J. Biol. Chem. 271:28567-28571. [DOI] [PubMed] [Google Scholar]
- 30.Lee, W., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host Δ9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindenbach, B. D., J. Sgro, and P. Ahlquist. 2002. Long-distance base pairing in Flock House virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 76:3905-3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, Q., J. Krzewska, K. Liberek, and E. A. Craig. 2001. Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 276:6112-6118. [DOI] [PubMed] [Google Scholar]
- 33.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meagner, and J. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 34.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein on intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McBride, H. M., D. G. Millar, J. Li, and G. C. Shore. 1992. A signal-anchor sequence selective for the mitochondrial outer membrane. J. Cell Biol. 119:1451-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mihara, K. 2000. Targeting and insertion of nuclear-encoded preproteins into the mitochondrial outer membrane. Bioessays 22:364-371. [DOI] [PubMed] [Google Scholar]
- 37.Miller, D. J., M. D. Schwartz, and P. Ahlquist. 2001. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J. Virol. 75:11664-11676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muta, T., D. Kang, S. Kitajima, T. Fujiwara, and N. Hamasaki. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363-24370. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, K. W., Y. van der Meer, N. Roos, and E. J. Snijder. 1999. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J. Virol. 73:2016-2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peränen, J., P. Laakkonen, M. Hynönen, and L. Kääriäinen. 1995. The alphavirus replicase protein nsP1 is membrane-associated and has affinity to endocytic organelles. Virology 208:610-620. [DOI] [PubMed] [Google Scholar]
- 41.Poch, O., I. Sauvaget, M. Delarue, and N. Tordo. 1989. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J. 8:3867-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price, D. B., P. Ahlquist, and L. A. Ball. 2002. DNA-directed expression of an animal virus RNA for replication-dependent colony formation in Saccharomyces cerevisiae. J. Virol. 76:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Price, D. B., M. Roeder, and P. Ahlquist. 2000. DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J. Virol. 74:11724-11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price, D. B., R. R. Rueckert, and P. Ahlquist. 1996. Complete replication of an animal virus and maintenance of expression vectors derived from it in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 93:9465-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize in the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Restrepo-Hartwig, M., and P. Ahlquist. 1999. Brome mosaic virus RNA replication proteins 1a and 2a colocalize and 1a independently localizes on the yeast endoplasmic reticulum. J. Virol. 73:10303-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rubino, L., A. Di Franco, and M. Russo. 2000. Expression of a plant virus non-structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J. Gen. Virol. 81:279-286. [DOI] [PubMed] [Google Scholar]
- 48.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 49.Russo, M., A. Di Franco, and G. P. Martelli. 1987. Cytopathology in the identification and classification of tombusviruses. Intervirology 28:134-143. [DOI] [PubMed] [Google Scholar]
- 50.Saks, V. A., V. I. Veksler, A. V. Kuznetsov, L. Kay, P. Sikk, T. Tiivel, L. Tranqui, J. Olivares, K. Winkler, F. Wiedemann, and W. S. Kunz. 1998. Permeabilized cell and skinned fiber techniques in studies of mitochondrial function in vivo. Mol. Cell. Biochem. 184:81-100. [PubMed] [Google Scholar]
- 51.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schlegel, A., T. H. Giddings, Jr., M. S. Ladinsky, and K. Kirkegaard. 1996. Cellular origin and ultrastructure of membranes induced during poliovirus infection. J. Virol. 70:6576-6588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schmidt-Mende, J., E. Bieck, T. Hügle, F. Penin, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. Determinants for membrane association of the hepatitis C virus RNA-dependent RNA polymerase. J. Biol. Chem. 276:44052-44063. [DOI] [PubMed] [Google Scholar]
- 54.Schneemann, A., V. Reddy, and J. E. Johnson. 1998. The structure and function of nodavirus particles: a paradigm for understanding chemical biology. Adv. Virus Res. 50:381-446. [DOI] [PubMed] [Google Scholar]
- 55.Scotti, P. D., S. Dearing, and D. W. Mossop. 1983. Flock house virus: a nodavirus isolated from Costelytra zealandica (White) (Coleoptera: Scarabaeidae). Arch. Virol. 75:181-189. [DOI] [PubMed] [Google Scholar]
- 56.Selling, B. H., R. F. Allison, and P. Kaesberg. 1990. Genomic RNA of an insect virus directs synthesis of infectious virions in plant. Proc. Natl. Acad. Sci. USA 87:434-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selling, B. H., and R. R. Rueckert. 1984. Plaque assay for black beetle virus. J. Virol. 51:251-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Towner, J. S., T. V. Ho, and B. L. Semler. 1996. Determinants of membrane association for poliovirus protein 3AB. J. Biol. Chem. 271:26810-26818. [DOI] [PubMed] [Google Scholar]
- 59.Tusnady, G., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 60.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.von Heijne, G. 1986. Mitochondrial targeting sequences may form amphiphilic helices. EMBO J. 5:1335-1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.von Heijne, G. 1992. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]