Abstract
The assembly and maturation of the coat protein of a T=4, nonenveloped, single-stranded RNA virus, Nudaurelia capensis ω virus (NωV), was examined by using a recombinant baculovirus expression system. At pH 7.6, the coat protein assembles into a stable particle called the procapsid, which is 450 Å in diameter and porous. Lowering the pH to 5.0 leads to a concerted reorganization of the subunits into a 410-Å-diameter particle called the capsid, which has no obvious pores. This conformational change is rapid but reversible until slow, autoproteolytic cleavage occurs in at least 15% of the subunits at the lower pH. In this report, we show that expression of subunits with replacement of Asn-570, which is at the cleavage site, with Thr results in assembly of particles with expected morphology but that are cleavage defective. The conformational change from procapsid to capsid is reversible in N570T mutant virus-like particles, in contrast to wild-type particles, which are locked into the capsid conformation after cleavage of the coat protein. The reexpanded procapsids display slightly different properties than the original procapsid, suggesting hysteretic effects. Because of the stability of the procapsid under near-neutral conditions and the reversible properties of the cleavage-defective mutant, NωV provides an excellent model for the study of pH-induced conformational changes in macromolecular assemblies. Here, we identify the relationship between cleavage and the conformational change and propose a pH-dependent helix-coil transition that may be responsible for the structural rearrangement in NωV.
Conformational changes induced by low pH are critical in the life cycles of numerous viruses. Large-scale rearrangements occur in the fusion glycoproteins of enveloped viruses such as Semliki Forest virus (14), tick-borne encephalitis virus (19), and influenza virus (4, 7) during entry, and the transitions are required for fusion with the endosomic membrane. The mechanism for the transition is not understood in the alphaviruses and flaviviruses, while it is known to be the large-scale reorganization of a helix in influenza virus hemagglutinin. In many cases, cleavage of the precursor fusion polypeptide is known to be required for the transitions to occur (e.g., reference 8). Among nonenveloped, single-stranded RNA animal viruses, the detailed analysis of such a transition has only been reported for the tetravirus Nudaurelia capensis ω virus (NωV), where a large-scale, pH-induced conformational change provides the transition from a procapsid form to a capsid form during assembly. Previously, Canady and colleagues reported the characterization of NωV virus-like-particles (VLPs) at pH 7.5 and pH 5 with electron cryomicroscopy (cryoEM) (6) and authentic NωV virions by crystallography (15), as well as the kinetics of the transition for a rapid pH drop and the controlled titration of the transition (5).
NωV is a T=4, nonenveloped, icosahedral virus with a bipartite positive-sense RNA genome belonging to the tetravirus family (for reviews, see references 10 and 12). RNA1 codes for the RNA-dependent RNA polymerase, and RNA2 codes for the coat protein subunit. The capsid is composed of 240 copies of the 70-kDa (α) coat protein that, after assembly, undergoes an autocatalytic cleavage resulting in 62-kDa (β) and 8-kDa (γ) products (2). Because a cell culture capable of supporting authentic NωV has not been identified, we have studied the structure, assembly, and maturation of the coat protein via expression in a recombinant baculovirus system. Expression of the single, coat protein gene results in the spontaneous assembly of VLPs that encapsidate both the mRNA of its coat protein and cellular RNA (1). Analysis of particles purified at pH 5.0 by cryoEM and image reconstruction showed that the VLPs are morphologically identical to the authentic virions, with an icosahedral shape and an average diameter of 410 Å (6). These particles are referred to as the mature capsids. VLPs purified at pH 7.6 have the same protein composition as the mature capsids but are porous, round particles with a diameter of 450 Å and are referred to as procapsids (6). The coat proteins of the procapsid particles remain in the uncleaved, α state. Lowering the pH of these particles to 5.0 results in a structural transition to the mature capsid in less than 100 ms, but with a delayed autoproteolytic cleavage with a half-life of several hours (5). To date, this is the only RNA animal virus reported to undergo large structural rearrangements in the maturation process. Larger DNA viruses such as herpesvirus and several bacteriophages, however, display similar conformational changes during maturation, suggesting a similar stepwise strategy and mechanism for assembling a complex capsid.
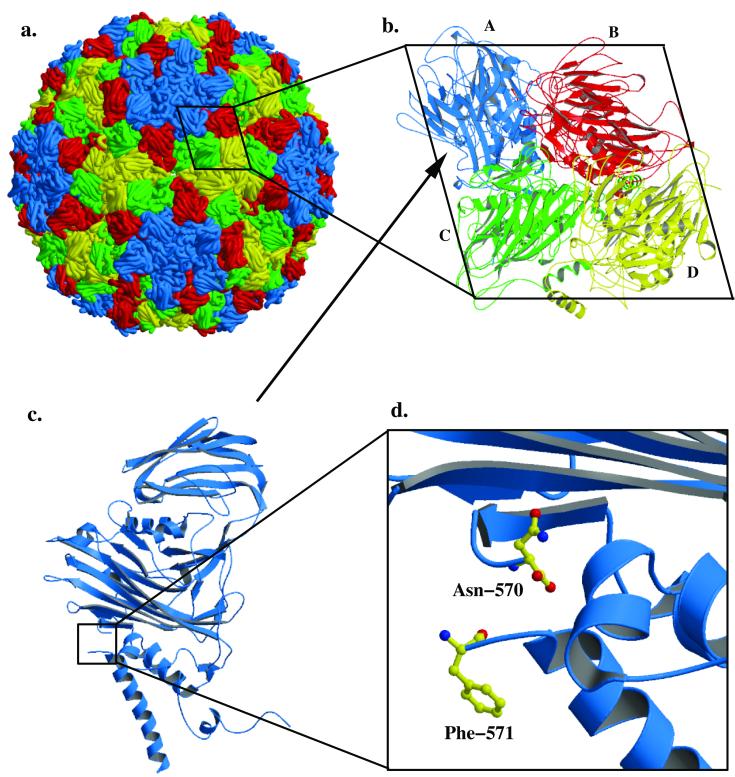
The 2.8-Å crystal structure of authentic NωV showed that the coat protein subunits assemble into a T=4 polyhedral capsid (Fig. 1a) (15). The icosahedral asymmetric unit of NωV consists of four quasiequivalent subunits (Fig. 1b). Each subunit undergoes a cleavage in the authentic virus that, by analogy with the VLPs, must occur only after the mature capsid conformation has been achieved. Figure 1c shows a ribbon drawing of the A subunit in the authentic virus after cleavage has occurred. A portion of the γ peptide is visible as a detached α helix. Initial studies demonstrated that after cleavage occurred in the majority of subunits, the NωV VLPs were “locked” in the capsid conformation (6) with no evidence of reversibility when the pH was raised to 7.5. Subsequent, time-resolved, small-angle X-ray scattering studies revealed that the conformational change of NωV VLPs was reversible until about 15% of the subunits were cleaved (5).
FIG. 1.
(a) Space filling model of NωV based on the 2.8-Å crystal structure of the authentic, mature virus, displaying A subunits in blue, B subunits in red, C subunits in green, and D subunits in yellow. (b) Ribbon diagram of the icosahedral, asymmetric unit of NωV. (c) Side view of the A subunit after the autoproteolytic cleavage event has occurred. Only the N terminus (residues 571 to approximately 590) of the γ-peptide is visible in the A and B subunits, and an additional helix is visible in the C and D subunits from residues 608 to 641. (d) The autoproteolytic cleavage occurs between Asn-570 and Phe-571. Mutation of Asn-570 to Thr results in cleavage-defective particles which undergo structural rearrangements reversibly as a function of pH.
The atomic structure of mature, infectious NωV particles provided an insight into the cleavage mechanism of the coat protein subunit. The structure showed that cleavage of the subunit occurs between Asn-570 and Phe-571 (Fig. 1d). An asparagine residue is located at the scissile bond in the nodaviruses and was demonstrated to be required for an autoproteolytic cleavage event in Flock House virus (FHV), a member of the nodavirus family (21). The cleavage event is critical in the life cycle of authentic FHV, because cleavage-defective FHV mutants were shown to be uninfectious (20). Comparisons of the cleavage sites of FHV and NωV revealed a striking similarity, although FHV is a T=3 virus. Based on these observations, we reasoned that replacement of Asn-570 with a glutamine (N570Q), threonine (N570T), or aspartic acid (N570D) would prevent cleavage and that the large-scale conformational change would be reversible. Here, we show that replacement of Asn-570 with Thr, Gln, or Asp results in normally assembled, cleavage-defective particles. The cleavage-defective mutants undergo the same conformational change as the wild-type particles, but the coat protein does not cleave. In contrast to wild-type VLPs, we demonstrate that the conformational change in the Asn-to-Thr cleavage-defective particles is bidirectional as a function of pH. These results demonstrate that the autoproteolytic cleavage in the wild-type particles disables the driving force responsible for the conformational change. We also demonstrate that the Asn at the cleavage site plays a critical role in the cleavage mechanism of NωV maturation, as it does in FHV.
MATERIALS AND METHODS
Site-directed mutagenesis of the coat protein cleavage site.
The plasmid pFastBAC-1, containing a cDNA copy of the coat protein gene of NωV, was used to generate the following point mutations at asparagine 570: N570D, N570Q, and N570T. Amplification primers matched the sequences at the 5′ and 3′ ends of the coat protein gene and contained 5′ BamHI and 3′ XbaI restriction sites for subcloning into the baculovirus transfer vector pBacPAK9 (see below). Mutations and restriction sites in the open reading frame of α coat protein were generated by overlap extension PCR (11). Briefly, oligonucleotide primers were designed in inverted tail-to-tail directions to amplify the entire gene with the target sequence and sticky 5′ BamHI and 3′ XbaI restriction sites. The sequences of primers used in this experiment will be supplied upon request. PCR conditions were as follows: 10 cycles at 95°C for 5 s, 60°C for 15 s, and 72°C for 2 min, followed by 10 cycles at 95°C for 5 s, 55°C for 15 s, and 72°C for 2 min, and completed with 10 cycles at 95°C for 5 s, 50°C for 15 s, and 72°C for 2 min. Products of the expected size were purified by agarose gel electrophoresis and a QIAEX II gel extraction kit (QIAGEN).
Construction of recombinant baculovirus.
Recombinant Autographa californica mononuclear polyhedrosis virus expressing NωV wild-type VLPs was as previously reported (6). NωV mutated coat protein constructs were ligated into the BamHI and XbaI multiple cloning site of the baculovirus transfer vector pBacPAK9 (Clontech) as previously described (9). Following purification, the plasmid DNA was transformed into DH5α competent cells and purified (Promega Wizard kit). Sequences were verified using the Sanger dideoxy sequencing method. The vector pBacPAK9, containing the NωV mutated coat protein gene, was transfected into Spodoptera frugiperda (Sf21) cells with Bsu36I-linearized BacPAK6 viral DNA, and baculovirus stocks were amplified and titers were determined as described previously (9). Individual plaque isolates confirming expression of the NωV coat protein gene were amplified. Infectivity titers were determined on monolayers of Sf21 cells by standard procedures (16).
Isolation of VLPs.
Sf21 cells were propagated at 27°C in TC100 medium supplemented with 10% (vol/vol) fetal bovine serum as previously described (21). A total of 8 × 106 Sf21 cells were plated on 100-mm-diameter tissue culture dishes and infected with recombinant baculovirus expressing the coat protein of NωV wild type, N570D, N570Q, or N570T. A multiplicity of 5 PFU/cell in 1 ml of TC100 medium was used for infections. After 1 h of rocking, an additional 5 ml of TC100 medium was added to each plate and the cells were incubated at 27°C without agitation.
At 4 days postinfection, cells were lysed with 0.5% (vol/vol) NP-40. After 15 min on ice, cell debris was pelleted in a Beckman JA17 rotor at 10,000 rpm for 10 min at 4°C. RNase A was added to the supernatant to a final concentration of 10 μg/ml and incubated at 27°C for 30 min. VLPs were pelleted through 4.0 ml of 30% (wt/wt) sucrose cushion prepared in pH 7.6 buffer (250 mM NaCl, 50 mM Tris HCl [pH 7.6]) at 25,000 rpm in a Beckman SW28 rotor for 5.5 h at 11°C. Pellets were resuspended overnight in 400 μl of pH 7.6 buffer. Particles converted to capsid were done so by overnight dialysis (room temperature) against pH 5.0 buffer (250 mM NaCl, 50 mM sodium acetate [pH 5.0]). Insoluble material was pelleted for 1 min in a tabletop centrifuge at maximum speed (16,000 × g), and the supernatant was loaded on a 10-to-40% (wt/wt) sucrose gradient prepared in pH 7.6 buffer for procapsids and pH 5.0 buffer for capsids. Particles were sedimented for 1.25 h at 40,000 rpm and 11°C in a Beckman SW41 rotor. Fractions were collected on an ISCO gradient fractionator at 0.75 ml/min and 0.5 min/fraction. Radiolabeled samples were individually mixed with 5 ml of scintillation fluid (Packard Ultima Gold), and radioactivity was counted using a Packard liquid scintillation analyzer.
Electrophoretic analysis of proteins and immunoblot analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gels were used according to the procedure described by Laemmli (13). Slab gels contained 10% (wt/vol) acrylamide in the resolving gel and 4% (wt/vol) acrylamide in the stacking gel. Protein samples were combined with an equal volume of 2× sample buffer (24 mM Tris-HCl [pH 6.8], 10% glycerol, 0.8% SDS, 5.8 mM 2-mercaptoethanol, 0.04% bromophenol blue) and heated to 99°C for 10 min. After electrophoresis for 75 min at 200 V, gels were fixed and stained with Coomassie brilliant blue according to standard procedures. Immunoblot analysis was performed to verify the expression of the NωV coat protein from the plaque-picked, recombinant baculovirus stocks and is described elsewhere (9).
Radiolabeling of VLPs.
For 35S-labeled VLPs, cells were rinsed with 2 ml of methionine-cysteine-deficient medium at 24 h postinfection. Each 100-mm dish was covered with 3 ml of fresh methionine-cysteine-deficient medium (JRH Biosciences) and rocked gently for 1 h at room temperature. Medium was then replaced with 2 ml of fresh medium containing 200 μCi of [35S]EasyTag Express protein labeling mix (NEN) per ml. Plates were rocked for 3 more h before the addition of 3.5 ml of fetal bovine serum-supplemented TC100 medium. The plates were incubated at 27°C until 4 days postinfection, when VLPs were isolated as described above.
For 3H-labeled VLPs, infections were carried out as described above. At 24 h after infection, 25 μCi of [5,6-3H]uridine (ICN Pharmaceuticals, Inc.) was added directly to each 100-mm dish. Plates were incubated at 27°C until 4 days postinfection, when VLPs were isolated as described above.
Electron microscopy.
Samples were negatively stained as described previously (9), except that pH 7.6 buffer and pH 5.0 buffer were used for procapsid and capsid particles, respectively. Prior to negative staining, samples were concentrated to approximately 0.5 mg/ml (Biomax-100, 100K nominal molecular weight limit; Millipore). Concentrations were estimated by absorbance, using a ɛ260 of 5.
RESULTS
Asparagine 570 is critical for cleavage, but not for the conformational change in NωV.
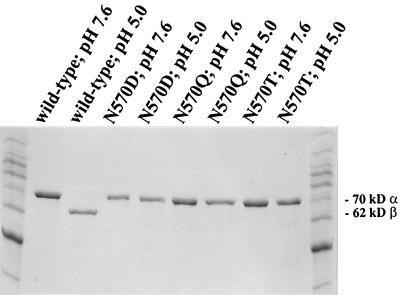
Gel electrophoresis of the wild-type and three Asn-570 mutant VLPs (Fig. 2) showed that all constructs possess coat proteins in the uncleaved α state at pH 7.6. At pH 5.0, however, the wild-type coat protein shows cleavage while all three of the Asn-570 mutant coat proteins remain in the uncleaved α form. Because of superior expression levels, the NωV N570T construct was used as a representative of the point mutation for all subsequent experiments.
FIG. 2.
An SDS-10% polyacrylamide gel illustrating cleavage-defective NωV particles. After the pH is decreased to 5.0, the majority of the α coat protein subunits in wild-type particles undergo an autocatalytic cleavage event into 62-kDa β and 8-kDa γ (which run off the gel) products. Cleavage products were undetectable by Coomassie stain in all three mutations, regardless of the pH.
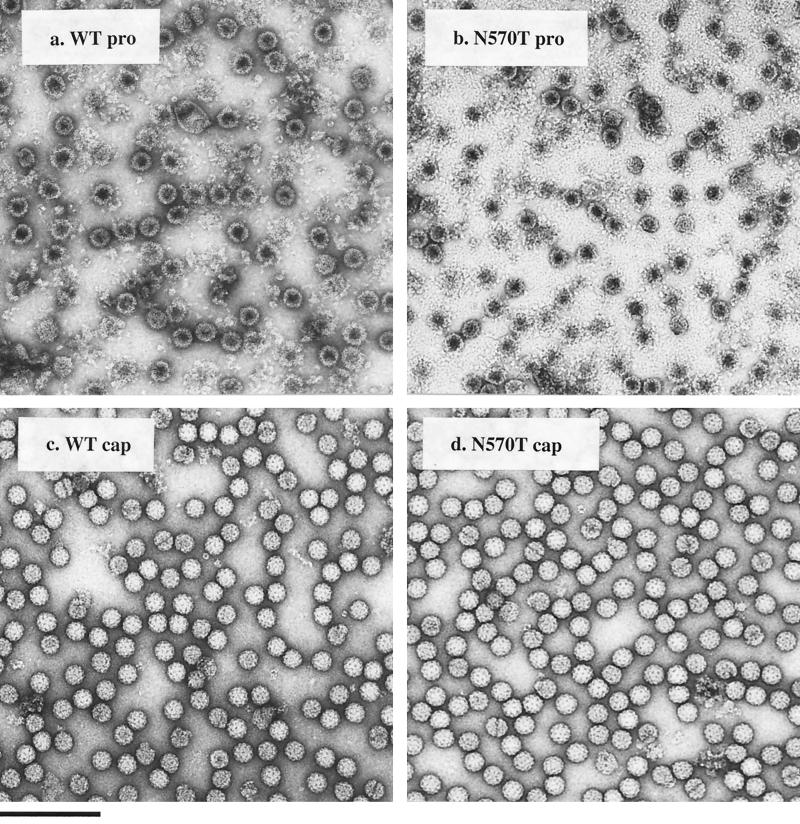
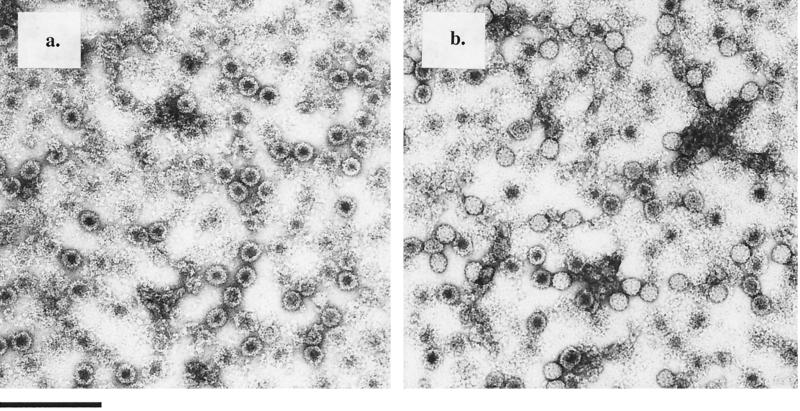
Electron microscopy of purified, negatively stained wild-type procapsid particles (Fig. 3a) established two morphological phenotypes: (i) procapsid particles with a dark center, indicating that they are porous and penetrated by the stain; and (ii) several procapsid particles that are unstable, generating a background of broken particles in the negatively stained electron micrographs. Electron microscopy analysis of negatively stained N570T procapsids (Fig. 3b) showed that they were morphologically indistinguishable from the wild-type procapsids.
FIG. 3.
Electron micrographs of negatively stained, gradient-purified NωV particles. (a) Wild-type procapsid; (b) N570T procapsid; (c) wild-type capsid; (d) N570T capsid. Wild-type and N570T procapsids are porous, displaying dark centers from stain penetration. In contrast to the capsid, both wild-type and N570T procapsids are unstable, with several broken particles in the micrograph. Although uranyl acetate is a fixative, it is possible that the low pH of the stain contributes to the rupturing of some of the procapsid particles. N570T procapsids and capsids are morphologically indistinguishable from the wild-type procapsids and capsids, respectively. Bar, 200 nm.
The micrographs of the mature capsid conformation (Fig. 3c) showed that the particles are stable and impenetrable to the stain. Like wild-type capsid particles, the N570T capsid particles were impenetrable to the stain, with fewer broken particles than the procapsid population (Fig. 3d).
NωV wild-type procapsid and capsid sediment differently on a 10-to-40% sucrose gradient.
Sucrose gradient-purified [3H]uridine-labeled wild-type particles were purified as procapsid (pH 7.6) and converted to the capsid conformation by dialysis against 1 liter of pH 5.0 buffer at 25°C for approximately 24 h. The particles were then left at room temperature overnight to ensure cleavage of at least 90% of the coat protein subunits. The capsid VLPs were returned to pH 7.6 by dialysis and sedimented on a 10-to-40% sucrose gradient also prepared in pH 7.6 buffer. As expected, mature capsids prepared in this way displayed a single homogenous peak of radioactivity in the sucrose gradient. The fractions containing the majority of mature, [3H]uridine-labeled wild-type particles were pooled and stored at 4°C. Cleavage of the coat protein subunit was verified by gel electrophoresis (data not shown.) These particles were used as a sedimentation marker for subsequent experiments.
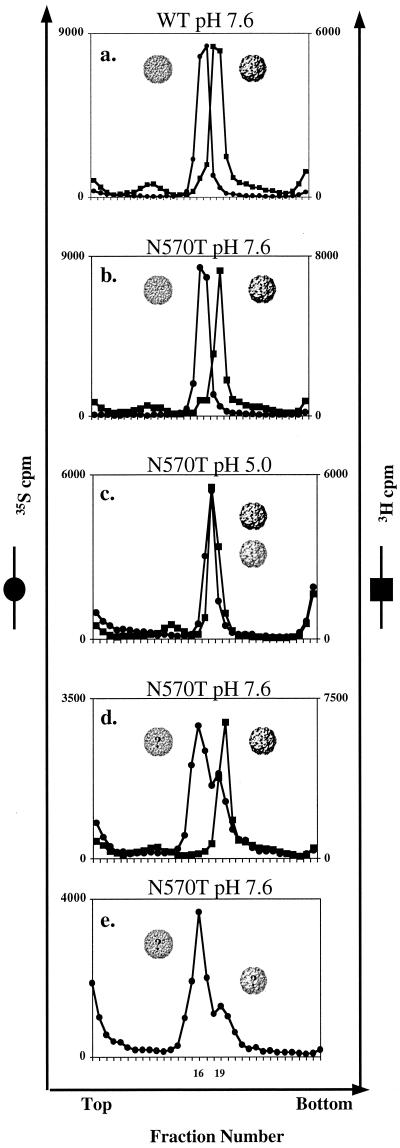
[35S]Met-Cys-labeled wild-type particles were purified in pH 7.6 buffer as procapsids and stored at 4°C. After overnight dialysis against 1 liter of pH 7.6 buffer at room temperature to remove remaining sucrose, the procapsid sample was mixed with [3H]uridine-labeled NωV wild-type capsid, as a sedimentation marker, and centrifuged on a 10-to-40% sucrose gradient in pH 7.6 buffer. Figure 4a shows that by differential labeling the two populations of wild-type particles can be resolved after sedimentation on the same sucrose gradient. The procapsid particles consistently sedimented two or three fractions slower than the capsid population of particles under these conditions.
FIG. 4.
(a) Sedimentation profile of wild-type, [35S]Met-Cys-labeled procapsid and mature, [3H]uridine-labeled capsid. Both popu-lations of particles are in the same buffer (pH 7.6) and were mixed before sedimentation on a 10-to-40% sucrose gradient. The [3H]uridine-labeled capsid particles are used as a sedimentation marker. The two differentially labeled populations were sedimented on the same 10-to-40% sucrose gradient at pH 7.6. The procapsid population of particles consistently sedimented two or three fractions slower than the capsid population of particles. (b) [35S]Cys-Met-labeled NωV N570T was purified at pH 7.6, mixed with [3H]uridine-labeled wild-type capsid, and sedimented on the same sucrose gradient at pH 7.6. Analogous to wild-type procapsid particles, N570T particles sediment two or three fractions slower than [3H]uridine-labeled wild-type capsid at pH 7.6, suggesting that the N570T particles are in the procapsid conformation at that pH. (c) [35S]Cys-Met-labeled NωV N570T was dialyzed to pH 5.0 overnight and mixed with [3H]uridine-labeled wild-type capsid. The two differentially labeled samples were sedimented on the same 10-to-40% sucrose gradient at pH 5.0. The cosedimentation of the two populations suggests that the N570T particles are in the capsid conformation at this pH. (d) The [35S]Cys-Met-labeled NωV N570T particles were dialyzed back to pH 7.6 overnight, mixed with [3H]uridine-labeled wild-type capsid, and sedimented on the same 10-to-40% sucrose gradient at pH 7.6. The majority of N570T particles sedimented approximately four fractions slower than capsids and approximately one fraction slower than the original procapsid particles. A small percentage of the particles continued to sediment with the [3H]uridine-labeled wild-type capsid. (e) Even after 4 days of dialysis back to pH 7.6, a small percentage of the N570T particles continued to sediment faster than the majority of the particles.
Conformational change in NωV N570T particles is reversible.
[35S]Met-Cys-labeled NωV N570T procapsid particles were purified and dialyzed in pH 7.6 buffer and mixed with [3H]uridine-labeled NωV wild-type capsid. The mixture was sedimented on a 10-to-40% sucrose gradient prepared in pH 7.6 buffer (Fig. 4b). Analogous to wild-type procapsids, the [35S]Met-Cys-labeled NωV N570T procapsid particles consistently sedimented two or three fractions slower than the [3H]uridine-labeled NωV wild-type capsid particles under identical conditions.
The transition from the [35S]Met-Cys-labeled NωV N570T procapsid to capsid conformation was induced by dialysis against 1 liter of pH 5.0 buffer for approximately 24 h at room temperature. The dialyzed sample was mixed with [3H]uridine-labeled NωV wild-type capsid, and the mixture was run on a 10-to-40% sucrose gradient prepared in pH 5.0 buffer. Figure 4c shows that the N570T mutant [35S]Met-Cys-labeled particles cosedimented with the [3H]uridine-labeled wild-type capsid marker, indicating that the N570T mutant particles are now in the capsid conformation.
The reverse transition from capsid to procapsid of the NωV N570T mutants was induced by overnight dialysis against 1 liter of pH 7.6 buffer at room temperature. The mutant particles were mixed with [3H]uridine-labeled NωV wild-type capsid and run on a 10-to-40% sucrose gradient prepared in pH 7.6 buffer. In contrast to the wild-type particles in the capsid conformation, Fig. 4d shows that the majority of the NωV N570T particles sedimented differently than the [3H]uridine-labeled NωV wild-type capsid particles. The majority of the particles, however, consistently sedimented about a fraction slower than the original procapsid particles. The peak representing the reexpanded particles is also broader, suggesting some heterogeneity of the particles. A small population of the particles continued to cosediment with the [3H]uridine-labeled NωV wild-type capsid. The percentage of NωV N570T particles that cosedimented with NωV wild-type capsid particles varied from preparation to preparation but was typically less than half.
The reexpanded N570T particles were dialyzed for an additional 3 days (4 days total) against 1 liter of pH 7.6 buffer at room temperature. This sample was centrifuged on a 10-to-40% sucrose gradient prepared in pH 7.6 buffer with no [3H]uridine-labeled wild-type capsid marker. The peak representing the majority of particles was sharper than it was after only 24 h of dialysis, but it continued to sediment a fraction slower than the original procapsid particles (Fig. 4e). The small population of particles that initially cosedimented with the wild-type capsid continued to sediment at the same position.
NωV N570T particles sediment at two rates after reexpansion treatment.
We analyzed the two populations of unlabeled, reexpanded N570T particles by negative-stain electron microscopy (Fig. 5). Fractions 16 (peak) and 19 (shoulder) (Fig. 4e) were concentrated to approximately 0.5 mg/ml and analyzed by electron microscopy.
FIG. 5.
Negative-stained electron micrographs of the two populations of NωV N570T particles after changing the pH from 7.6 to 5.0 and back to 7.6 revealed two populations. (a) The particles in the slower-sedimenting peak (fraction 16) have the same phenotypes as procapsids, displaying dark centers from stain penetration along with broken particles in the micrographs. (b) The faster-sedimenting shoulder population (fraction 19) revealed a heterogenous population of particles, with some resembling procapsids with stain-penetrated centers and others resembling the stain-impenetrable capsids. Bar, 200 nm.
The particles in fraction 16 resembled procapsid, displaying dark centers from stain penetration and some broken particles. The particles in fraction 19 appeared to be a heterogenous mixture of particles, each resembling either capsid or procapsid. The A260/A280 ratio of procapsid, capsid, and fraction 19 was approximately 1.20, while the ratio of fraction 16 was 1.08.
DISCUSSION
The procapsid-to-capsid transition in NωV has only been observed with particles produced in a baculovirus expression-assembly system. Procapsids are only observed in preparations isolated at pH 7.6; capsids are observed when particles are isolated at pH 5. It is likely that the procapsid is a transient intermediate in the formation of authentic virus particles if the assembly occurs in a low pH environment within the cell. A working model for assembly, consistent with this hypothesis, requires that virion formation occur in a low-pH cellular vacuole that depends on viral infection. Expression of only the coat protein in the Sf21 cells apparently does not generate a vacuole, thereby eliminating pH regulation from the assembly. Particles are still able to assemble at the neutral pH of the cytoplasm but are nonnative and are inhibited from maturing into the form of particle observed in the authentic virion. The maturation process occurs in vitro when the pH is reduced to 5.0.
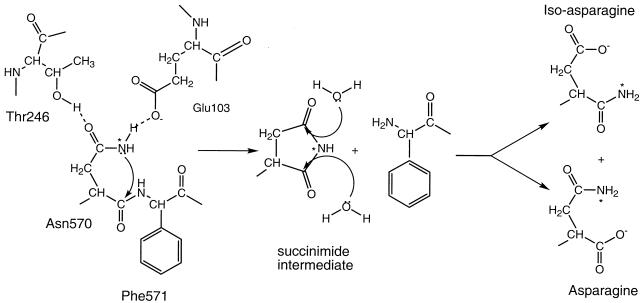
Authentic NωV undergoes an autocatalytic maturation cleavage, and this proteolysis occurs in the VLP capsids at pH 5.0. Like the maturation in nodaviruses (21, 25), results from the present study show that cleavage does not occur if Asn-570 is replaced by threonine, glutamine, or aspartic acid. The arrangement of residues at the cleavage site in nodaviruses and NωV was shown to be strikingly similar by crystallography (15), suggesting similar mechanisms for the cleavage in the two virus groups. Although not exhaustively tested, we believe that Asn is absolutely required at the cleavage site. The Asn residue is well documented as contributing to nonprogrammed protein “aging” through deamidation or cleavage on the C-terminal side of the Asn residue (e.g., references 3 and 17). An important example is the formation of cataracts; when these reactions occur in the crystalline protein it forms opaque aggregates in the eye lens (23). Deamidation at asparagine residues in proteins results from the nucleophilic attack of the peptide nitrogen of the adjacent residue, C-terminal to the Asn, on the side chain carbonyl carbon of the Asn residue, which causes the formation of the succinimide intermediate (Fig. 6). Hydrolysis of the succinimide intermediate gives rise to iso- (β) Asn and Asn in 3:1 proportions (24). While the deamidation is the major reaction that occurs at the susceptible Asn residues, cleavage of the peptide bond following the Asn is also known to occur as a reaction that proceeds via the formation of the succinimide intermediate (23). In the latter reaction, the side chain amide nitrogen of the Asn residue is responsible for the nucleophilic attack on its own main chain carbonyl carbon atom, causing the cyclic imide intermediate to form and releasing the C-terminal peptide. Hydrolysis of the imide ring produces Asn and β-Asn (aspartyl α-amide) (22). Such a cleavage reaction at Asn residues is also known to occur during protein splicing and is responsible for the C-terminal cleavage (18). We propose that this reaction is responsible for the cleavage in both noda- and tetraviruses and is kinetically enhanced by the presence of a catalytic acidic residue in the immediate vicinity of the Asn side chain. In FHV it was shown that altering either Asn-363 or Asp-75 reduced the rate of the reaction below measurability (25), while the present study showed that replacing Asn-570 has the same effect in NωV. The analogous residue to Asp-75 in NωV is Glu-103. Replacement of this residue in NωV affected assembly, and its role in cleavage could not be independently established. The work is ongoing and will be reported in a separate communication.
FIG. 6.
The proposed mechanism of the autoproteolytic cleavage of the NωV coat protein. Glu-103 and Thr-246 are believed to form hydrogen bonds with Asn-570, orienting the asparagine in the ideal position for cleavage to occur. Nucleophilic attack by the side chain of the asparagine on the carbonyl group of the peptide chain results in a succinimide intermediate of the cleaved protein. Hydrolysis opens the ring, yielding either asparagine or isoasparagine at the C terminus of the 62-kDa β-peptide.
Inhibition of the maturation cleavage allowed an investigation of the reversibility of the pH-dependent, large-scale quaternary structure of the VLPs. The noncleaving VLP used for the investigation was the N570T mutation. This mutation expressed and assembled with larger yields of particles than the wild-type sequence, as well as the other noncleaving mutations. The reason for the efficiency of production of this particular mutant was not obvious. N570T mutants underwent the transition to the capsid form at pH 5.0, and their sedimentation properties and negative-stain electron microscopy images were indistinguishable from those of the wild-type VLPs; however, cleavage was not detectable by SDS-gel electrophoresis in any of the mutations. As shown in previous studies, the transition from procapsid to capsid in the wild-type particles was not reversible after the subunits cleaved (5). By labeling the RNA packaged in the wild-type particles with tritiated uridine during expression, these particles were used as a convenient marker for capsid formation, as they sedimented at the same position after maturation, regardless of pH. As anticipated from the previous time-resolved study of the quaternary structure transition (5), the noncleaved capsid particles expanded to what appeared to be procapsid when the pH was raised to 7.6.
Two features of the particles obtained after raising the pH distinguished them from the native procapsid prior to exposure to pH 5.0. First, the majority of the particles sedimented one fraction slower than the original procapsid, and the major peak was always accompanied by a faster-sedimenting shoulder. Second, the negative-stained electron microscopy images revealed the stain-penetrable phenotype as the native procapsids in the slower-sedimenting peak and a heterogenous mixture of stain-penetrable procapsids and stain-impenetrable capsids in the faster-sedimenting shoulder. The fact that the A260/A280 ratio for all the particles in the peak of the reexpanded particles, where it falls from 1.20 to 1.08, would suggest a loss of encapsidated, cellular RNA during reexpansion. This would explain why these particles sediment a fraction slower than original procapsid particles.
The conversion of procapsids to capsids was previously shown to occur between pH 5.3 and 6.5, with a sharp transition in the titration curve (5). If the pH was lowered rapidly from 7.5 to 5 in a stop-flow apparatus, the transition occurred in less than 100 ms, the dead time of the solution X-ray scattering apparatus used to measure the transition (5). The reverse reaction, in which the particles reexpand, is slow, with some never making the transition after 4 days of dialysis against pH 7.6 buffer. The capsid-to-procapsid transition is probably slow because there are extensive protein-protein, and possibly protein-RNA, interactions in the capsid that must be broken during the expansion process. Titration curves were not developed for the N570T VLP transitions; however, qualitatively the procapsid-to-capsid transition occurs in the same pH range and occurs rapidly. The reverse reaction, however, is slow, requiring hours of dialysis against pH 7.6 buffer to convert the majority of capsids to the procapsid-like particles. This observation suggests that the transition can be viewed as having two components: the pH-dependent driving force and a “cargo” that is coupled to the driving force. Previous structural studies of the procapsid and capsid of wild-type particles demonstrated that almost all of the NωV subunit behaves as a rigid body during the transition and that the procapsid cryoEM density can be modeled with high fidelity by the X-ray coordinates determined from the capsid structure (6). The only region of density that showed an obvious quaternary and possibly secondary and tertiary structure change in the two particles occurred on the inside of the subunit in the region occupied by the helical domain of the proteins. It is known that these helices have different configurations in the different subunits observed in the T=4 particle and that they function as the switches that determine the angle of contact between twofold-related subunits in the authentic virus (15). Residues 608 to 641 are visible as helices in the C and D subunits of the X-ray structure of the capsid but are invisible in the A and B subunits. As previously discussed (6), the quasi- and icosahedral twofold axes of the procapsid are nearly indistinguishable from each other in the procapsid, while they differentiate into distinct, flat (dihedral angle, 180°) and bent (dihedral angle, 144°) twofold contacts in the capsid. A working hypothesis, currently being tested, proposes a pH-induced helix-coil transition that occurs in the interior, helical region of the subunit. It is likely that these regions exist as helices in the procapsids and that, after protonation at lower pH, they transform into coils initiating the conformational transition to capsids. The different environments established by flat dihedral angles between C and D subunits and bent dihedral angles between A and B subunits alter the pH of the helix-coil transition in those two environments. This becomes apparent in the crystal structure of NωV, where the C terminus is visible as a helix in the C and D subunits but is invisible in the A and B subunits. Protonation of specific residues at low pH initiates the helix-coil transitions of these polypeptides. The transition occurs at a pH near 6, suggesting histidine as a possible candidate for this protonation, based on pKa, but the influence of tertiary and quaternary structures may create environments that alter the pKa of various residues. The protonation would be proceeded by electrostatic repulsions causing the helices to unfold into random coils, thus initiating the driving force for the conformational change. This region of the subunit can be viewed as a pH-driven engine, while the remainder of the subunit is the cargo that is pushed or pulled as a rigid body. When cleavage occurs, the cargo is uncoupled from the engine and the particle remains in the capsid state regardless of pH. When the cleavage does not occur, the cargo remains coupled and the whole process is reversible (Fig. 7). The great number of protein-protein interactions created when the capsid forms undoubtedly causes the hysteresis and the slow reversible transition back to the procapsid.
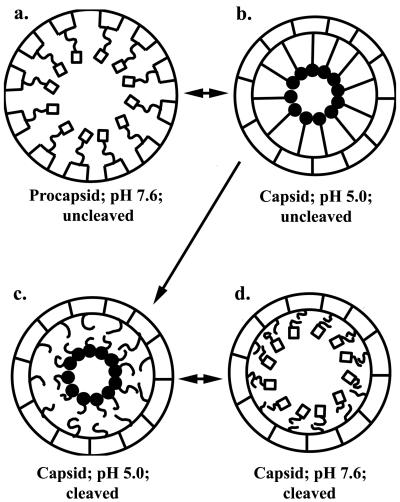
FIG. 7.
Schematic representation of the relationship of coat protein cleavage and the pH-induced conformational change in NωV. The region of the helical domain of each coat protein subunit believed to be responsible for the helix-coil transition represents residues 1 to 44 and 571 to 644 of the 644-residue protein. The diagram depicts this region as either a helix (□) or a random coil (•). (a) The particles were purified as procapsid at pH 7.6, with no cleavage occurring in the subunits. (b) Upon lowering the pH to 5.0, protonation of specific residues causes the helix to transform into a coil, initiating the quaternary structural rearrangement. If no cleavage occurs in the capsid at pH 5.0, the process is reversible. (c) Cleavage of wild-type particles locks the particles in the capsid conformation. (d) Altering the pH of the particles allows the helix-coil to operate reversibly, but cleavage uncouples this pH-driven engine from the rest of the cargo subunit, causing the surface of the particles to resemble capsids.
The NωV particle displays the properties of a molecular machine as it undergoes the transitions required to achieve the formation of a complex quaternary structure. Unlike comparable transitions in bacteriophages, and fusion glycoproteins in alphaviruses, flaviviruses, and influenza viruses that depend on the generation of meta-stable particles and states that irreversibly mature, the NωV particle maturation is pH dependent and reversible as long as cleavage of the coat protein is inhibited, allowing a detailed analysis of the machinery by mutagenesis and biophysics.
Acknowledgments
We thank Vijay Reddy for critical revision of the manuscript and for assistance in generating Fig. 1.
This work was supported by NIH grant GM54076.
REFERENCES
- 1.Agrawal, D., and J. Johnson. 1995. Assembly of the T=4 Nudaurelia capensis omega virus capsid protein, posttranslational cleavage, and specific encapsidation of its mRNA in a baculovirus expression system. Virology 207:89-97. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal, D., and J. Johnson. 1992. Sequence and analysis of the capsid protein of Nudaurelia capensis ω virus, an insect virus with T=4 icosahedral symmetry. Virology 190:806-814. [DOI] [PubMed] [Google Scholar]
- 3.Artigues, A., A. Birkett, and V. Schirch. 1990. Evidence for the in vivo deamidation and isomerization of an asparaginyl residue in cytosolic serine hydroxymethyltransferase. J. Biol. Chem. 265:4853-4858. [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Canady, M., H. Tsuruta, and J. Johnson. 2001. Analysis of rapid, large-scale protein quaternary structural changes: time-resolved X-ray solution scattering of Nudaurelia capensis ω virus (NωV) maturation. J. Mol. Biol. 311:803-814. [DOI] [PubMed] [Google Scholar]
- 6.Canady, M. A., M. Tihova, T. N. Hanzlik, J. E. Johnson, and M. Yeager. 2000. Large conformational changes in the maturation of a simple RNA virus, Nudaurelia capensis ω virus (NωV). J. Mol. Biol. 299:573-584. [DOI] [PubMed] [Google Scholar]
- 7.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 8.Chen, J., K. H. Lee, D. A. Steinhauer, D. J. Stevens, J. J. Skehel, and D. C. Wiley. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95:409-417. [DOI] [PubMed] [Google Scholar]
- 9.Dong, X. F., P. Natarajan, M. Tihova, J. E. Johnson, and A. Schneemann. 1998. Particle polymorphism caused by deletion of a peptide molecular switch in a quasiequivalent icosahedral virus. J. Virol. 72:6024-6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hanzlik, T. N., and K. H. Gordon. 1997. The Tetraviridae. Adv. Virus Res. 48:101-168. [PubMed] [Google Scholar]
- 11.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson, P. Quirke, and G. R. Taylor (ed.), PCR, a practical approach. Oxford University Press, Oxford, United Kingdom.
- 12.Johnson, J., and V. Reddy. 1998. Structural studies of noda and tetraviruses, p. 171-223. In L. Miller and L. Ball (ed.), The insect viruses. Plenum, New York, N.Y.
- 13.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 14.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 15.Munshi, S., L. Liljas, W. Cavarelli, W. Bomu, B. McKinney, V. Reddy, and J. Johnson. 1996. The 2.8 Å structure of T=4 animal virus and its implications for membrane translocation of RNA. J. Mol. Biol. 261:1-10. [DOI] [PubMed] [Google Scholar]
- 16.O'Reilly, D. R., L. K. Miller, and V. A. Luckow. 1992. Baculovirus expression vectors. A laboratory manual. W. H. Freeman and Company, New York, N.Y.
- 17.Patel, K., and R. T. Borchardt. 1990. Chemical pathways of peptide degradation. II. Kinetics of deamidation of an asparaginyl residue in a model hexapeptide. Pharm. Res. 7:703-711. [DOI] [PubMed] [Google Scholar]
- 18.Perler, F. B., M. Q. Xu, and H. Paulus. 1997. Protein splicing and autoproteolysis mechanisms. Curr. Opin. Chem. Biol. 1:292-299. [DOI] [PubMed] [Google Scholar]
- 19.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 20.Schneemann, A., R. Dasgupta, J. E. Johnson, and R. R. Rueckert. 1992. Maturation cleavage required for infectivity of a nodavirus. J. Virol. 66:6728-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schneemann, A., R. Dasgupta, J. Johnson, and R. Rueckert. 1993. Use of recombinant baculovirus in synthesis of morphologically distinct virus-like particles of flock house virus, a nodavirus. J. Virol. 67:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephenson, R. C., and S. Clarke. 1989. Succinimide formation from aspartyl and asparaginyl peptides as a model for the spontaneous degradation of proteins. J. Biol. Chem. 264:6164-6170. [PubMed] [Google Scholar]
- 23.Voorter, C. E., W. A. de Haard-Hoekman, P. J. van den Oetelaar, H. Bloemendal, and W. W. de Jong. 1988. Spontaneous peptide bond cleavage in aging alpha-crystallin through a succinimide intermediate. J. Biol. Chem. 263:19020-19023. [PubMed] [Google Scholar]
- 24.Wright, H. T. 1991. Sequence and structure determinants of the nonenzymatic deamidation of asparagine and glutamine residues in proteins. Protein Eng. 4:283-294. [DOI] [PubMed] [Google Scholar]
- 25.Zlotnick, A., V. Reddy, R. Dasgupta, A. Schneemann, W. Ray, R. Rueckert, and J. Johnson. 1994. Capsid assembly in a family of animal viruses primes an autoproteolytic maturation that depends on a single aspartic acid residue. J. Biol. Chem. 269:13680-13684. [PubMed] [Google Scholar]