Abstract
Vaccinia virus (VV), a member of the poxvirus family, is unique among most other DNA viruses in that both transcription and DNA replication occur in the cytoplasm of the host cell. It was recently shown by electron microscopy (EM) that soon after viral DNA synthesis is initiated in HeLa cells, the replication sites become enwrapped by the membrane of the endoplasmic reticulum (ER). In the same study, a novel VV membrane protein, the E8R gene product, that may play a role in the ER wrapping process was identified (N. Tolonen, L. Doglio, S. Schleich, and J. Krijnse Locker, Mol. Biol. Cell 12:2031-2046, 2001). In the present study, the gene product of E8R was characterized both biochemically and morphologically. We show that E8R is made predominantly early in infection but is packaged into the virion. On two-dimensional gel electrophoresis, the protein appeared as a single spot throughout the VV life cycle; however, in the assembled virion, the protein underwent several modifications which resulted in a change in its molecular weight and its isoelectric point. EM of labeled cryosections of infected HeLa cells showed that the protein localized to the ER and to membranes located on one side of the Golgi complex as early as 1 h postinfection. Late in infection, E8R was additionally associated with membranes of immature virions and with intracellular mature viruses. Although E8R is predominantly associated with membranes, we show that the protein is associated with viral cores; the protein is present in cores made with NP-40-dithiothreitol as well as in incoming cores, the result of the viral entry process, early in infection. Finally, we show that E8R can be phosphorylated in vitro by the viral kinase F10L. It is able to bind DNA in vitro, and this binding may be modulated by phosphorylation by F10L. A putative role of the E8R gene product throughout the VV life cycle is discussed.
Vaccinia virus (VV) is the best-characterized member of the Poxviridae. The 190-kb double-stranded DNA genome has been completely sequenced and codes for over 200 proteins, approximately 100 of which are associated with the virion (11). These proteins are divided into three temporal classes. The early proteins are synthesized prior to DNA replication, while the intermediate and late proteins are made after the onset of replication. The late proteins generally code for viral structural proteins required for virion assembly (28).
The first morphological evidence of viral assembly is the formation of crescent-shaped membranes, which we and others believe are derived from a membrane cisterna of the smooth endoplasmic reticulum (ER) and are composed of two tightly apposed membranes (32, 37, 38). From these crescents, immature virions (IVs) are formed; on electron microscopy (EM), IVs appear as perfect spheres delineated by a double membrane enclosing an electron-dense central portion. VV core proteins, such as p25 (L4R) and 4a (A10L), have been shown to localize to the central electron-dense structure (10, 44), while membrane proteins, such as p16 (A14L) and p21 (A17L), localize, as expected, to the membranes (23, 33). Initially, IVs do not contain the viral genome; upon DNA uptake, in a poorly understood process (13, 14), the particles mature into the first infectious form, intracellular mature viruses (IMVs). The transition from the spherical IVs to the IMVs results in the formation of a biochemical and morphological distinct core structure, a process that coincides with the cleavage of a number of core proteins (20, 43, 45; B. Moss and E. N. Rosenblum, Letter, J. Mol. Biol. 81:267-269, 1973). A fraction of the IMVs becomes enwrapped by a cisterna derived from the trans-Golgi network to form intracellular enveloped viruses (IEVs) (36). The IEVs utilize microtubules to facilitate their intracellular transport to the plasma membrane (16, 31, 46). Upon fusion of the outer IEV membrane with the plasma membrane, actin tails are made at the cell surface. These tails propel extracellular enveloped viruses away from the cell on the tip of a long filopodium, a process that may aid virus dissemination (16, 31, 46).
The first morphological evidence of VV infection is the appearance, at approximately 2 h after infection, of structures that are enriched in DNA and that are the sites of viral DNA replication (2, 4, 21). Early EM observations showed that the factory region appeared to lie free in the cytoplasm (7). However, in a recent study, it was shown that soon after initial replication, the sites of cytoplasmic DNA synthesis became enwrapped by membranes of the ER (39). It was shown that ER wrapping coincided with a peak in DNA synthesis, suggesting that the ER facilitates replication. By green fluorescent protein tagging of VV early genes, we identified a novel VV membrane protein, the E8R gene product, that may play an important role in the wrapping process. The E8R protein has two putative transmembrane domains; consistently, upon extraction of lysates of infected cells with Triton X-114 (TX-114), the protein partitioned in the detergent phase, as expected for a membrane protein. Using antibodies raised against the N terminus of E8R, we showed by immunofluorescence that the protein displayed a ring-like pattern around DNA factories. When the protein was localized by EM with either an anti-green fluorescent protein or an anti-E8R antibody, E8R was concentrated in the ER around the factory region and within this pool was predominantly found in the ER membrane, facing the replication site. Finally, our data suggested that the 120-amino-acid-long N-terminal portion was exposed in the cytoplasm and could thus interact with the DNA or DNA-binding proteins and mediate the binding of the ER to the factory region. The combined data thus suggested that the E8R gene product may play a role in the observed wrapping process (39).
Protein phosphorylation is a common mechanism for the regulation of cellular and viral processes (9, 17). The protein kinases and phosphatases activate or deactivate cellular or viral events in response to various stimuli. The phosporylation state of a protein can influence its ability to interact with other factors. Among the many facets of protein function affected by phosphorylation are subcellular localization, participation in multiprotein complexes, and interaction with nucleic acids.
Phosphorylation and dephosphorylation events may also regulate VV morphogenesis. The VV genome codes for one functional phosphatase and two kinases with essential roles in VV morphogenesis. The B1 gene codes for a serine/threonine protein kinase that is made early in infection. Accordingly, temperature-sensitive mutants with lesions in this gene have been shown to be deficient in the process of viral DNA replication (30). An additional protein kinase is coded for by the F10L gene (24) and is able to phosphorylate both serine/threonine and tyrosine residues (3, 8). F10L is expressed at late times of infection and is encapsidated in the virus (24). Temperature-sensitive mutants with lesions in this gene are blocked at the stage of virus assembly; at the restrictive temperature, late proteins are synthesized, but at the EM level, no crescent-shaped membranes or IVs are made (40). It was subsequently found that the kinase phosphorylates both serine/threonine and tyrosine residues of a number of viral proteins, such as p11 (F17R), p16 (A14L), p25 (L4R), and p21 (A17L) (3, 8, 19, 41, 47). The H1 gene codes for a dually specific protein phosphatase which is also encapsidated in the virion (15). The repression of H1 synthesis results in the production of particles that are deficient in early transcription (25). Consistent with its phosphatase activity, it was found that in such H1-deficient particles, a number of viral proteins are hyperphosphorylated (25). The combined data thus suggested that phosphorylation and dephosphorylation events may play a key role in theVV life cycle.
In the present study, we show that E8R is an early protein that is synthesized before DNA replication. Although made predominantly early, the protein is packaged into the virion, where it associates with the viral core and undergoes a number of modifications. Finally, we show that E8R is able to bind DNA in vitro and that its affinity for DNA is decreased upon phosphorylation by the F10L kinase.
MATERIALS AND METHODS
Cells, virus, drug treatment, and antibodies.
HeLa cell line C3, obtained from the American Type Culture Collection (Manassas, Va.), was grown in Dulbecco's modified Eagle's medium containing penicillin, streptomycin, and 5% heat-inactivated fetal calf serum. HeLa cells were grown overnight in 6-cm dishes before infection with semipurified virus diluted in serum-free Dulbecco's modified Eagle's medium. When needed, hydroxyurea (HU; 5 or 10 mM; Sigma) and rifampin (100 μg/ml; Sigma) were added to the cells immediately after infection. For all experiments, VV strain western reserve (WR) was used. The virus was propagated in HeLa cells, semipurified, and plaque titrated as described by Pedersen et al. (29). The generation of the antibodies to E8R, p21 (A17L), p16 (A14L), and p25 (L4R) has been described elsewhere (27, 29, 35, 39). The antibody to hemagglutinin (HA) was obtained from Babco (Richmond, Calif.). The antibody to glutathione S-transferase (GST) was a gift from Eric Karsenti (European Molecular Biology Laboratory, Heidelberg, Germany). Anti-rabbit antibody coupled to fluorescein isothiocyanate was obtained from Jackson Immunoresearch Laboratories (Dianova, Hamburg, Germany).
EM and immunofluorescence.
HeLa cells were grown overnight in 6-cm dishes to form 70% confluent monolayers. For EM, cells were infected at 37°C for 30 min at either a multiplicity of infection (MOI) of 10 and fixed 6 h postinfection or at an MOI of 200 and fixed 60 min postinfection. Fixed cells were prepared for cryosectioning as described previously (42). For indirect immunofluorescence, HeLa cells grown on coverslips were infected at an MOI of 40 with VV WR and fixed at 3 and 6 h postinfection with paraformaldehyde. The cells were rinsed with phosphate-buffered saline (PBS) and blocked with 5% fetal calf serum in PBS for 10 min at room temperature. To detect E8R, the peptide antibody was used diluted 1:100. The cells were also labeled with Hoechst stain (Sigma) at 0.5 mg/ml to visualize the DNA.
Metabolic labeling, SDS-PAGE, Western blotting, and isoelectric focusing.
For metabolic labeling, WR-infected HeLa cells were grown in 3.5-cm dishes and labeled for 30 min at 2.5 and 6 h postinfection with 50 μCi of 35S-methionine/dish. The cells were either lysed immediately or chased for 3 h. The cells were washed twice with ice-cold PBS, 100 μl of lysis buffer (1% NP-40 in 10 mM Tris-Cl [pH 9]) was added, and the cells were left on ice for 30 min. The cells were scraped from the dishes, and the nuclei were pelleted by centrifugation. For Western blotting, infected HeLa cells were grown in 6-cm dishes, and lysates were prepared as described above at 3 and 6 h postinfection. The radiolabeled and nonradiolabeled lysates were run on NEPHGE as described by Jensen et al. (18) followed by sodium dodecyl sulfate (SDS)-15% polyacrylamide gel electrophoresis (PAGE) in the second dimension.
Expression and purification of GSTE8R or GSTNE8R.
HeLa cells were grown overnight in 10-cm dishes to form 60% confluent monolayers. The cells were infected with VVT7 (a vaccinia virus recombinant expressing T7 polymerase) for 45 min at 37°C and then washed with PBS and Optimem (Gibco BRL, Life Technologies). The infected cells were transfected for 5 h with Lipofectin (Gibco BRL, Life Technologies) and 40 μg of GSTE8R, GSTNE8R, or GST. After 5 h of transfection, the transfection medium was replaced with 500 μl of buffer containing 25 mM HEPES-KOH (pH 7.4), 50 mM potassium acetate, and 0.1% NP-40, and the cells were left on ice for 30 min. The cells were scraped from the dishes and pelleted by centrifugation, and the supernatant was collected. Glutathione-Sepharose 4B beads (30 μl; Amersham Pharmacia Biotech, Uppsala, Sweden) were added to the supernatant, and the mixture was incubated for 1 h by head-over-head rotation. The beads were washed four times with the above-described buffer, and GSTE8R or GST was eluted in a volume of 30 μl from the beads with the same buffer containing 10 mM glutathione (Sigma).
The pGEX F10L vector was used for the transformation of bacterial strain BL21(DE3). Bacterial cells were grown in Luria-Bertani culture medium initially at 37°C and then were transferred to 20°C at an optical density at 600 nm of 0.4. Recombinant protein expression was induced by the addition of 0.1 mM isopropyl-β-d-galactopyranoside (IPTG) at an optical density at 600 nm of 0.8. After overnight growth, the bacterial culture was centrifuged, and the pellet was either collected or dissolved in fresh medium and cultured for an additional 2 h before being collected. The pellet corresponding to 1.5 ml of cell culture was sonicated in 400 μl of 20 mM potassium phosphate buffer (pH 7.3)-1 mM phenylmethylsulfonyl fluoride-1 mg of lysozyme/ml-10 μg of DNase/ml. After centrifugation at 13,000 rpm for 10 min with a bench centrifuge (Heraeus), the supernatant was mixed with 30 μl of glutathione-Sepharose 4 Fast Flow beads equilibrated in 20 mM potassium phosphate buffer-150 mM NaCl. Incubation was carried out for 30 min at room temperature with a sample mixer (Dynal). The suspension was briefly centrifuged, the supernatant was discarded, and the beads were resuspended in PBS, washed for 30 min, and finally pelleted. The supernatant was removed, and 30 μl of 20 mM Tris-Cl (pH 8) containing 10 mM glutathione was used to elute the proteins.
DNA mobility shift assays.
To prepare radiolabeled nucleic acids, the 35-mer oligonucleotide 5′TGCAGGTCGACTCTAGAGGATCCCCGGGTACCGAG3′ was incubated for 10 min at 95°C and labeled in a reaction mixture containing [γ-32P]ATP and T4 polynucleotide kinase (Roche Biochemicals). The reaction mixture was incubated at 37°C for 1 h, and oligonucleotides were separated from unincorporated nucleotides through a Nick column (Amersham Pharmacia Biotech). The percent incorporation of [γ-32P]ATP and the recovery of radiolabeled oligonucleotides were assessed by liquid scintillation counting. Electrophoretic mobility shift assays were performed with 20-μl reaction mixtures containing 20 mM Tris-Cl (pH 7.5), 5 mM dithiothreitol (DTT), 50 mM KCl, 2.5 mM MgCl2, 0.5 mM EDTA, and 10% glycerol. Nucleic acids and proteins were mixed at various concentrations. Reaction mixtures were incubated for 20 min at room temperature and then loaded directly onto a 6% native polyacrylamide gel. Radioactivity was visualized by autoradiography.
NP-40-DTT extraction, TX-114 treatment, membrane pelleting, and in vitro translation of E8R.
NP-40-DTT extraction of purified virus and TX-114 extraction of cell lysates were carried out essentially as described previously (18). The preparation of postnuclear supernatants (PNS) and the separation of membranes from soluble proteins with or without Na2CO3 treatment were performed as described by Jensen et al. (18). The proteins were detected by Western blotting and enhanced chemiluminescence (ECL) with the use of the antibodies indicated above or by autoradiography of metabolically labeled samples. In vitro translation of pGADHAE8R (see below) was done with a T7-coupled transcription and translation system (Promega, Madison, Wis.) as described previously (23)
Cloning of E8R, the N-terminal portion of E8R, and F10L.
E8R was amplified by PCR from the VV WR genome with the following primers: 5′CGGGGATCCGGTGGTATGGCTGCCACCGTTCCG3′ (upstream) and 5′CGGCTGCAGTTACAACAGTTGTACGTC3′ (downstream). The BamHI restriction site in the upstream primer and a PstI site downstream of the gene were digested and used to clone the gene into the BamHI/PstI-digested pTM1GST vector (a gift from Michael Merchlinsky). F10L was amplified by PCR with the following primers: 5′CGGGGATCCATGGGTGTTGCCAATGATTCATCC3′ (upstream; containing a BamHI site) and 5′GGCTCGAGTTAGTTTCCGCCATTTATC3′ (downstream; containing an XhoI site). The restriction sites were used to clone the gene into the pGEX-4T-1 vector (Amersham Pharmacia Biotech). The E8R N-terminal portion was amplified by PCR with the following primers: 5′CCGCTCGAGTTACTCGTTTGTTGGAAGAGCAGA3′ (downstream; containing an XhoI site) and the upstream primer used to insert E8R into the pTM1GST vector. The restriction sites were used for cloning into the pGEX-4T-1 vector. To create a second vector which allowed expression driven by the T7 polymerase, the E8R gene was amplified by PCR from the VV genome with the following primers: 5′CGGGGATCCAAATGGCTGCCACCGTT3′ (upstream) and 5′CGGCTCGAGTTACAACAGTTGTACGTC3′ (downstream). The BamHI and XhoI restriction sites were used to clone the gene into the BamHI/XhoI-digested pGADHAT7 vector (Clontech).
Protein kinase assays.
Protein kinase assays were performed with a 20-μl solution containing 50 mM Tris-HCl (pH 7.5), 1 mM DTT, 5 mM MgCl2, 100 μM [γ-32P]ATP, 250 ng of GSTF10L, and 1 μg of GSTE8R or GST. Reactions were carried out at 37°C for 20 min and were terminated by the addition of SDS-PAGE sample buffer. Proteins were separated by SDS-PAGE and visualized by autoradiography.
RESULTS
Generation of antibody and identification of the E8R protein on two-dimensional (2D) gels.
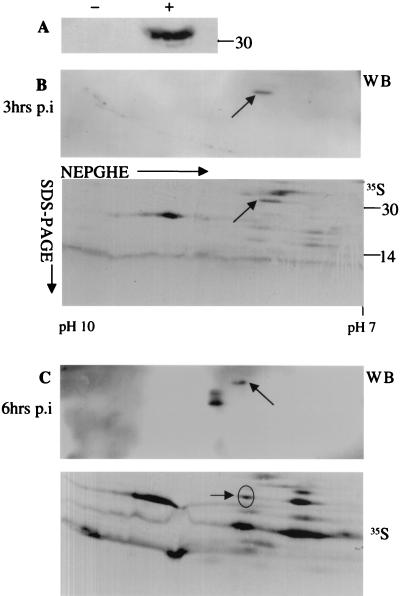
As mentioned above, the E8R gene product has been identified as a protein with a putative role in the process of ER wrapping around VV replication sites. In order to characterize the E8R protein, a peptide antibody to the extreme N terminus was raised, corresponding to amino acid residues 7 to 21. The specificity of the antibody was analyzed by Western blotting. As shown in Fig. 1A, the antibody recognized a single band that migrated at about 30 kDa in infected but not in uninfected cells.
FIG. 1.
Identification of 35S-labeled E8R in 2D gels. (A) Lysates of uninfected cells (−) and cells infected for 3 h (+) were separated by SDS-PAGE. The proteins were transferred to nitrocellulose and probed with an anti-E8R antibody. The E8R antibody detected one band in infected cells but not in uninfected cells. (B and C) Lysates of HeLa cells infected for 3 h (B) and 6 h (C) were prepared and mixed with lysates of infected cells metabolically labeled for 30 min at 2.5 and 6 h postinfection (p.i). The proteins were separated by NEPHGE followed by SDS-PAGE. The proteins were transferred to PVDF membranes, which were exposed for 1 week for autoradiography (35S). The same membranes were then subjected to Western blotting (WB) with the anti-E8R antibody. A comparison between the autoradiogram and the Western blot allowed the identification of E8R in the total 35S pattern (arrows). The pI range for NEPHGE is shown at the bottom of the gel. Numbers on the right indicate the positions of molecular mass marker proteins (in kilodaltons).
Because the antibody failed to immunoprecipitate the E8R protein, the radioactively labeled spot corresponding to the E8R gene product was first identified in total 35S-labeled cell lysates separated on 2D gels. Infected cells were metabolically labeled from 2.5 to 3 h and from 6 to 6.5 h postinfection, and cell lysates were separated on 2D gels, blotted onto polyvinylidene difluoride (PVDF) membranes, and exposed for autoradiography. After exposure for several days to obtain a satisfactory autoradiogram of the total 35S-labeled proteins, the same piece of membrane was incubated with the antibody against E8R, and the protein was detected by ECL. By Western blotting, a single spot of about 30 kDa and a pI of about 9, as predicted from the E8R sequence, was seen in cell lysates prepared at 3 h postinfection (Fig. 1B). A comparison of the total 35S pattern to the ECL results allowed us to unequivocally identify the E8R gene product in the total 35S-labeled cell lysates labeled from 2.5 to 3 h postinfection (Fig. 1B). At 6 h postinfection, however, Western blotting revealed three spots (Fig. 1C). In the corresponding total 35S-labeled cell lysates labeled from 6 to 6.5 h postinfection, only one of the three spots was barely visible (Fig. 1C), while the other two spots that were seen by Western blotting could not be unequivocally identified.
E8R is predominantly made early in infection, is packaged into virions, and is stable.
We next asked whether the protein was synthesized in a temporal fashion during viral infection. Lysates of infected cells were metabolically labeled at different times postinfection and under different conditions, and the proteins were separated on 2D gels. As explained above, a comparison of the Western blot and the 35S pattern enabled us to unequivocally identify the E8R gene product in total lysates separated on 2D gels.
To determine whether the protein was made early in infection, we first tested whether the protein was synthesized in the absence of viral DNA replication, a definition of VV early proteins. Lysates of infected cells were metabolically labeled from 2.5 to 3 h postinfection in the absence or in the presence of HU, separated on 2D gels, and prepared for autoradiography. In parallel, unlabeled cell lysates were prepared at 3 h postinfection, and the E8R protein was detected by Western blotting.
By Western blotting, a single spot was detected with or without HU, indicating that E8R is an early protein (Fig. 2A and B). Accordingly, in the corresponding autoradiogram, the metabolically labeled spot of the E8R gene product was seen with or without HU, although the protein appeared less intensely labeled in the presence of the drug. Since equal amounts of trichloroacetic acid-precipitable counts were loaded onto the first dimension, this result suggested that E8R is synthesized to a slightly lesser extent in the presence of HU than in the absence of the drug.
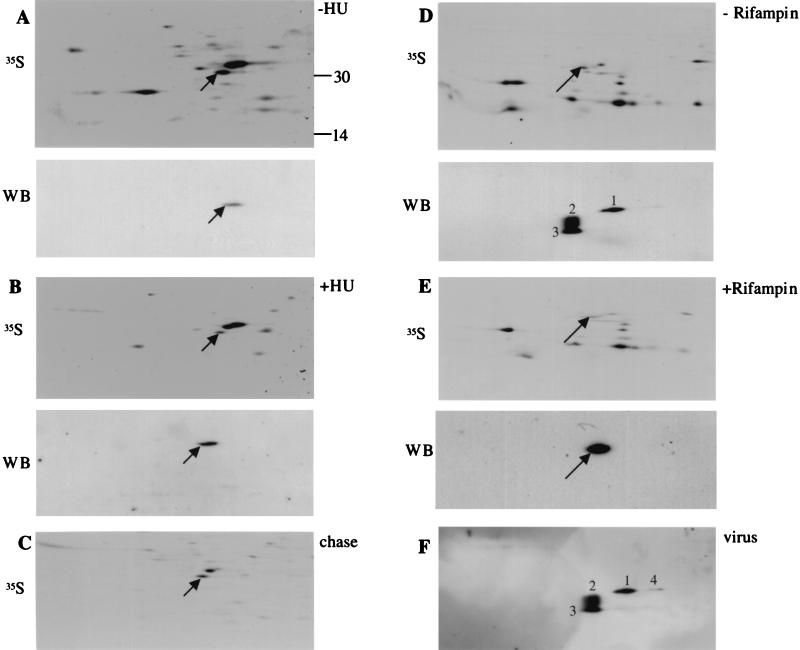
FIG. 2.
E8R is made early in infection and is packaged into virions. (A and B) Cells infected in the presence (B) or in the absence (A) of HU were lysed either at 3 h postinfection (WB) or after metabolic labeling from 2.5 to 3 h postinfection (35S). (C) Infected cells were labeled from 2.5 to 3 h postinfection, chased for 3 h in the presence of rifampin, and then lysed. Rifampin was used in the chase to avoid the incorporation of the E8R protein into virions. This was done because once packaged into virions, proteins are difficult to solubilize in first-dimension lysis buffer, resulting in a significant loss of proteins from infected cell lysates. (D and E) Infected cells were lysed at 6.5 h postinfection in the presence (E) or in the absence (D) of rifampin. 35S indicates lysates labeled from 6 to 6.5 h. (F) Purified virus. All samples were separated by NEPHGE and SDS-PAGE followed by autoradiography (35S; labeled samples) or Western blotting (WB; unlabeled samples) with an anti-E8R antibody. The arrows indicate E8R. The numbers in panels D and F indicate the different forms of E8R detected by Western blotting in the absence of rifampin or in the purified virus (see text for details).
We next tested in a similar manner whether E8R was also made late in infection. First, lysates of cells infected for 6 h in the presence or in the absence of rifampin were separated on 2D gels and blotted onto PVDF membranes. Since rifampin blocks the formation of IMVs, this drug was used to test whether the assembly of IMVs affected the migration of the protein in some way. A well-described modification that occurs during IMV formation is the cleavage of a number of core proteins, a process that is blocked by rifampin (20). Finally, to test whether E8R was packaged into virions, purified IMVs were also separated on 2D gels.
By Western blotting of cell lysates treated with rifampin, we could detect only one spot (Fig. 2E) whereas, in agreement with the above results, in the untreated samples three spots were readily detected (Fig. 2D). For convenience, we have numbered these different spots 1 to 3 in Fig. 2. Spot 1 was seen in both treated and untreated cell lysates and corresponded to the single spot seen at 3 h postinfection. Spot 2 was smaller and more basic than spot 1, while spot 3 was even smaller but had the same pI as spot 2. The protein also appeared to be packaged into virions; in purified IMV preparations, four spots could be resolved by Western blotting (Fig. 2F). Spots 1, 2, and 3 corresponded to those detected in the untreated cell lysates; in addition, we noted a fourth spot (4) which was more acidic than but had the same molecular weight as spot 1. Upon closer inspection of the Western blot of untreated cell lysates, spot 4 could also be detected but was generally much fainter than the corresponding spot detected in purified virions (compare Fig. 2D and F).
When we compared the autoradiograms of total cell lysates labeled from 6 to 6.5 h postinfection to the Western blots, we could barely detect a tiny spot corresponding to spot 1, but spots 2, 3, and 4 were not seen with this method.
The data suggested that the E8R protein is made predominantly early in infection and acquires a number of unknown modifications that lead to changes in its pI and its molecular weight once it is packaged into the virion. Since E8R was barely synthesized late in infection, our data also implied that E8R molecules synthesized early were being packaged into virions late in infection. This possibility was addressed indirectly by monitoring the stability of E8R in a pulse-chase experiment. If E8R made early was being packaged late, then we expected the protein to be very stable. Cell lysates labeled from 2.5 to 3 h postinfection were chased for 3 h, separated on 2D gels, and prepared for autoradiography. As shown in Fig. 2C, the spot corresponding to E8R was readily detected as one of the most prominent spots in such samples (compare Fig. 2A and C). Since equal amounts of trichloroacetic acid-precipitable counts were analyzed, it appeared that E8R did not undergo substantial degradation over the 3-h chase period.
The E8R gene product is partially associated with membranes and is associated with the viral core.
The E8R sequence predicts two stretches of hydrophobic amino acids (amino acid residues127 to 145 and 236 to 258) that can adopt an α-helical configuration, a property of transmembrane domains, suggesting that the protein may span membranes twice.
As expected of a membrane protein, the E8R protein was localized by EM to the nuclear envelope, ER, and ER around the factory region. Moreover, when lysates of infected cells were extracted with TX-114, E8R was found completely in the detergent phase, a property of hydrophobic proteins such as integral membrane proteins (Fig. 3A). The extraction of cell lysates with TX-114 is not necessarily proof that a protein is membrane associated, however. Proteins may partition into the TX-114 detergent phase because they contain a sufficiently long stretch of hydrophobic residues and not necessarily because they are associated with membranes (see below).
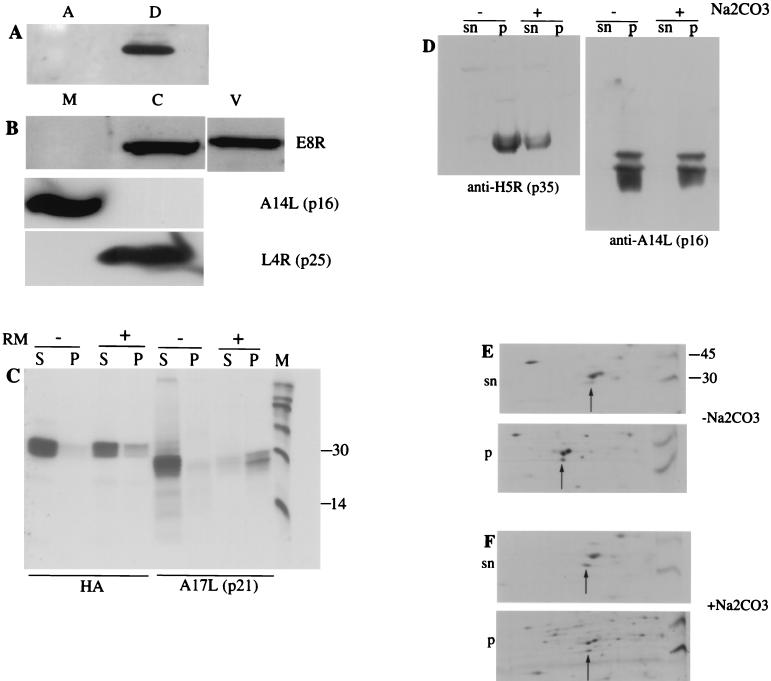
FIG. 3.
E8R is partially associated with membranes and associates with viral cores. (A) Cell lysates prepared at 6 h postinfection in the presence of rifampin (thisdrug was added to prevent virion formation; once packaged into virions, VV proteins may be pelleted irrespective of their biochemical properties) were extracted with 1% TX-114 to separate the aqueous (A) and detergent (D) phases. The fractions were transferred to PVDF membranes and probed with an anti-E8R antibody followed by ECL. (B) Purified IMVs were incubated with 1% NP-40 and 20 mM DTT for 30 min at 37°C, after which the samples were centrifuged through a 36% (wt/vol) sucrose cushion to pellet the cores. The membrane (M) and core (C) fractions were probed with antibodies to E8R, to A14L (p16), and to L4R (p25). V, untreated IMV. (C) HAE8R was translated in vitro in the presence (+) or absence (−) of rough microsomes. Soluble (S) proteins were separated from membrane proteins (P) by centrifugation. The proteins contained in the soluble and membrane fractions were immunoprecipitated with the corresponding antibody to A17L (p21) or to HAE8R. (D) A PNS sample of cells infected for 6 h in the presence of rifampin (see above) was prepared. Half of the PNS sample was treated for 20 min on ice with 0.1 M Na2CO3 (+), while the other half was left untreated (−). Soluble (sn) proteins were separated from membrane (p) proteins by centrifugation. The fractions were transferred to PVDF membranes and probed with anti-H5R (p35) and anti-A14L (p16) antibodies followed by ECL. (E and F) A PNS sample prepared from infected cells labeled with 35S from 2.5 to 3 h postinfection was treated (F) or not treated (E) with Na2CO3. Soluble (sn) proteins were separated from membrane (p) proteins, and the fractions were separated by NEPHGE and SDS-PAGE followed by autoradiography. Arrows indicate E8R. Numbers on the right indicate the positions of molecular mass marker proteins (in kilodaltons).
As shown above, the E8R protein is packaged into the virus. A common way to determine the localization of proteins in IMVs is the treatment of purified virions with NP-40 and DTT. Typical membrane proteins can be separated from the insoluble core and its associated proteins by centrifugation. Because the E8R protein displayed a number of characteristics typical of membrane proteins, we expected it to be in the membrane fraction under these conditions. Purified IMVs were treated with NP-40 and DTT. After separation of the membrane and core protein-containing fractions, the samples were loaded onto SDS-polyacrylamide gels and blotted onto PVDF membranes. The blots were probed with the antibody to E8R and with two antibodies to VV proteins known to be associated with viral cores and membranes. Core protein p25 (L4R) partitioned exclusively in the core fraction, whereas membrane protein p16 (A14L) was found in the membrane fraction. Unexpectedly, E8R was found exclusively in the core fraction (Fig. 3B).
Because of this unexpected result, we reinvestigated the membrane association of E8R in more detail. We first tested whether E8R was inserted into microsomal membranes in vitro. For this test, E8R was tagged at its N terminus with the HA epitope (HAE8R), enabling us to immunoprecipitate the protein with anti-HA antibodies. E8R then was translated in vitro in the presence or absence of rough microsomal membranes, and membrane proteins were separated from soluble proteins by centrifugation. As shown in Fig. 3C, membrane protein p21 (A17L), which we used as a control, clearly was inserted into microsomes, since the protein was pelleted when microsomes were present during its translation but not when microsomes were omitted, as shown before (23). Only a fraction (less than half of the total protein) of the E8R protein, however, was pelleted along with microsomes, suggesting that in vitro its insertion into membranes is inefficient (Fig. 3C).
We next tested whether E8R behaved as a membrane protein in infected cells. Infected cells were metabolically labeled from 2.5 to 3 h postinfection, and PNS were prepared. Half of the PNS were treated with sodium carbonate, a treatment that extracts peripherally associated (but not integral) proteins from membranes (12). It was previously observed that VV core proteins (which generally lack transmembrane domains) are pelleted in the absence of carbonate treatment but are solubilized after this treatment (6, 18). These observations may mean that VV core proteins are indeed (directly or indirectly) peripherally associated with membranes. An alternative explanation is that VV core proteins occur in large complexes that are pelleted independently of membranes but that these putative complexes are dissolved by high-pH (carbonate) treatment.
Treated or untreated PNS were layered onto a sucrose cushion, and soluble and membrane proteins were separated by centrifugation (18). As a control, a PNS from cells infected for 6 h was treated in an identical way, and viral control proteins were detected by Western blotting. These samples were probed with antibodies to p16 (A14L) and p35 (H5R), membrane and soluble proteins, respectively. As expected, p16 was pelleted along with membranes in treated and untreated PNS. Like most VV core proteins, p35 was pelleted in the absence of sodium carbonate treatment but was located entirely in the soluble fraction in its presence (Fig. 3D). When the E8R protein detected in metabolically labeled PNS was subjected to the same treatment, we found that without carbonate extraction, about 80% of the protein was pelleted (Fig. 3E). With carbonate treatment, the protein was almost equally distributed between the pellet and the soluble fraction (Fig. 3F). We therefore also examined whether the E8R protein associated with membranes in a posttranslational manner. Cells were pulse-labeled from 2.5 to 3 h postinfection and chased for 1 h, and PNS were subjected to the same treatment as that described above. After such a chase, however, E8R again was partially pelleted along with membranes, suggesting that it did not associate with membranes posttranslationally (data not shown).
These combined data suggested that the E8R protein may exist in two pools in infected cells, one pool that is stably associated with membranes and another pool that may be less stably integrated into membranes (see Discussion).
Localization of E8R by immunofluorescence microscopy.
As mentioned above, it was previously shown by immunofluorescence that at 3 h postinfection, E8R displays a ring-like pattern around the DNA synthesis site (39).
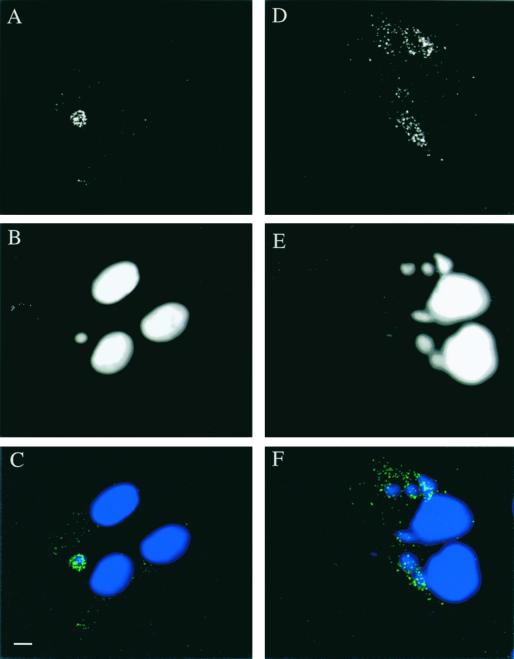
Since E8R appeared to be packaged into virions, in the present study the localization of E8R was also investigated at a late stage of infection. Infected HeLa cells were fixed at 3 h to confirm previous data and at 6 h postinfection. Fixed cells were doubly labeled with anti-E8R antibody and Hoechst stain to visualize the sites of viral DNA synthesis. As shown in Fig. 4A to C, at 3 h postinfection, clear labeling around the factory region could be detected, consistent with previous results. At 6 h postinfection, E8R was found predominantly in granular structures which did not obviously colocalize with the factory region (Fig. 4D to F).
FIG. 4.
Immunofluorescence localization of E8R in infected HeLa cells at 3 h postinfection (A, B, and C) and at 6 h postinfection (D, E, and F). (A and D) Labeling with antibodies to E8R (green channel in C and F). (B and E) Labeling with Hoechst stain (blue channel in C and F). (C and F) Merges. The labeling shows a fluorescence-labeled ring around the viral factories (C). Late in infection, however, the labeling shows a granular pattern that does not colocalize with the viral factories (F). Bar, 2 μm.
Localization of the E8R gene product by EM.
Next, the protein was localized at the ultrastructural level at different stages of the VV life cycle. The localization by EM of E8R at 3 h postinfection was recently described in detail, and the protein was shown to localize to the ER and to concentrate in the inner of the two ER membranes surrounding the DNA replication site (39). The results described above suggested that E8R is predominantly made early, is packaged into virions late in infection and, surprisingly, is associated with the NP-40- and DTT-insoluble core fraction. We decided therefore to localize the protein early in infection before DNA replication had started, as well as late in infection during virion assembly.
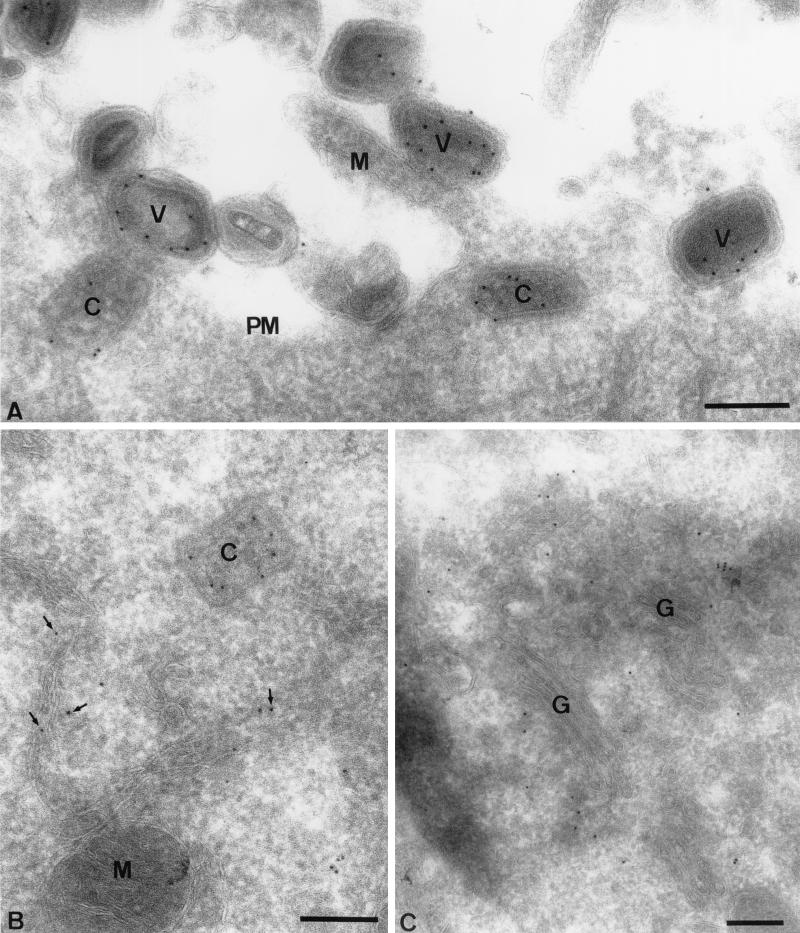
To monitor the fate of the E8R gene product during virus entry as well as its intracellular location early in infection, cells were infected at a high MOI (200) and fixed at 1 h postinfection. Cryosections were prepared and labeled with anti-E8R antibody. At the plasma membrane of these sections, many virions could be detected that were clearly labeled with anti-E8R antibody, consistent with the fact that the protein is packaged into virus particles (Fig. 5A). The labeling seemed to be associated with the core membrane rather than the virion surface. Intracellular cores located underneath the plasma membrane, which had apparently recently penetrated into the cytoplasm, could also be seen. These cores were clearly labeled with anti-E8R antibody, and the labeling was predominantly associated with the outer aspect of the core in a manner similar to, for instance, the labeling seen with anti-A4L or anti-A10L antibody (Fig. 5A and B) (29). A previous study on VV entry showed that IMV membrane proteins, like p21 (A17L), are lost at the plasma membrane and localize to membrane remnants attached to the cell surface (22). No labeling of E8R, other than in intact particles, was detected at the plasma membrane, strongly suggesting that upon virion entry, the protein was entirely associated with the incoming cores.
FIG. 5.
Localization of E8R by EM at 1 h postinfection. HeLa cells were infected at an MOI of 200 and fixed at 1 h postinfection, and cryosections were labeled with an anti-E8R antibody. (A) Plasma membrane (PM). Attached on its extracellular side are several IMVs (V) that are labeled with the antibody. One virion is attached to a microvillus (M), the formation of which is induced by the virion entry process (22). Underneath the plasma membrane are two intracellular cores (C) that are also labeled. (B) Labeled intracellular core (C) close to ER membranes that are also labeled (arrows). M, mitochondrion. (C) Golgi region (G). While the Golgi complex is generally not labeled, the tubular-vesicular membranes associated with this organelle are. Bars, 200 nm.
Already at this early time of infection (1 h postinfection), abundant E8R labeling could be detected on intracellular membranes; E8R was found on the ER as well as on tubular-vesicular membranes associated with one side of the Golgi complex (Fig. 5B and C). Although the origin of these membranes was not further investigated, we expected them to be part of the ER intermediate compartment. This labeling on intracellular membranes was not detected when cells were infected in the presence of cycloheximide, suggesting that it represented newly synthesized E8R protein (data not shown).
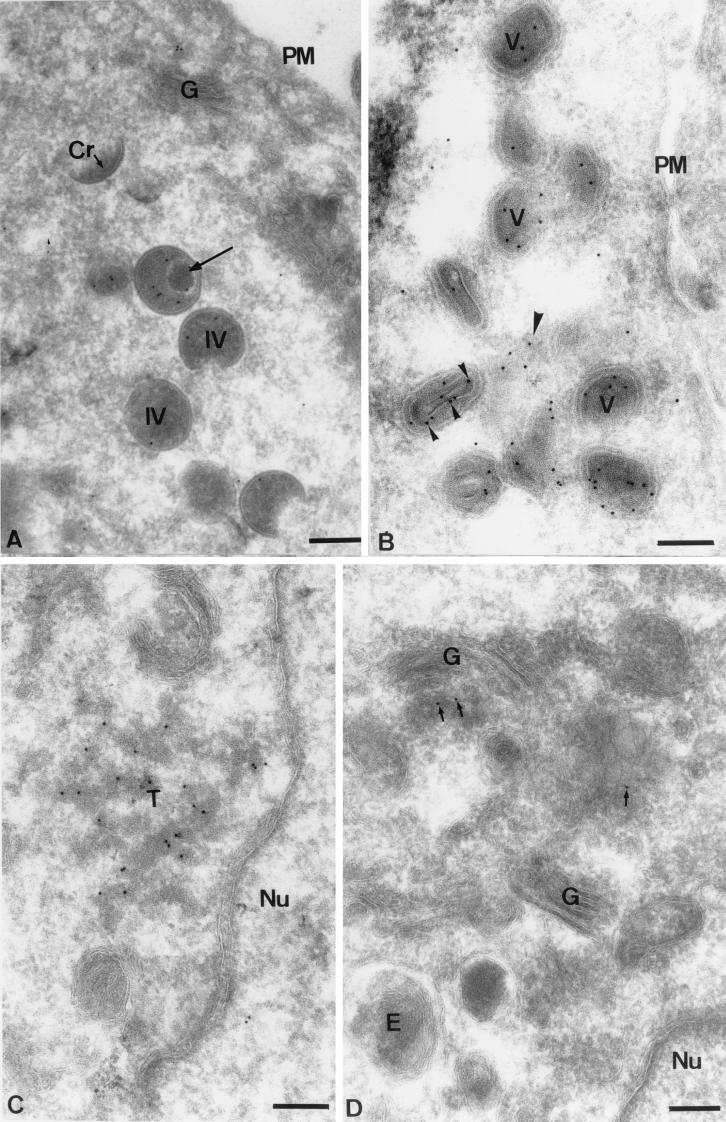
To localize the E8R protein late in infection, sections of cells fixed at 6 h postinfection were prepared and labeled with anti-E8R antibody. At this time of infection, the bulk of the labeling appeared to be associated with IVs and IMVs (Fig. 6A and B). The labeling was found to be associated with the inner aspect of the IV membranes, similar to that seen with the abundant IMV membrane protein p16 (A14L) (Fig. 6A) (35). Most importantly and in apparent contrast to the fact that E8R is associated with the viral core, the labeling on the IVs did not resemble the typical pattern seen for core proteins like A4L, A10L, and L4R. The latter generally show uniform labeling of the central, electron-dense part of the IVs rather than the IV membranes (6, 10, 44). Interestingly, in profiles in which IVs that had apparently taken up the viral genome were seen, the E8R labeling appeared to be redistributed more toward the inside of the IVs, away from the inner membrane (Fig. 6A). In the IMVs, we found that the labeling was mostly associated with the inside of the particle, the expected site of the IMV core, consistent with the results described above (Fig. 6B). At this late time of infection, labeling could also be detected on intracellular membranes. Tubular-vesicular membranes were found to be labeled in regions close to where the IMVs accumulated (Fig. 6B). A small amount of labeling could also be detected on one side of the Golgi complex (Fig. 6D). Occasionally, electron-dense tubular membranes could be seen to be abundantly labeled (Fig. 6C). These membrane structures did not seem to bear any obvious relationship to the viral assembly sites and appeared to be randomly located in the cell. Although the origin of these tubular membranes was not further investigated, they resembled very much tubular membranes of the ER-IC network. We believe that the latter labeling seen by EM corresponds to the rather prominent, vesicular labeling that we observed by immunofluorescence microscopy and that did not colocalize with the Hoechst stain-positive viral DNA regions.
FIG. 6.
Localization of E8R by EM at 6 h postinfection. HeLa cells were infected at an MOI of 10. (A) Several IVs (IV) and one crescent (Cr). The E8R labeling on the IVs appears to be associated with the inner membrane. One IV contains a nucleoid (long arrow); the labeling seems to be concentrated more toward the inside of the IV, away from the inner membrane, closer to the viral DNA. PM, plasma membrane. (B) Section through a cluster of IMVs (V), showing that these are all labeled on their inner aspects. In one particle, a cross-section through the viral core, with which the labeling is associated (small arrowheads), can be seen. Labeling also can be observed on membranes (large arrowhead) close to the IMVs. (C) E8R also abundantly labels tubular membranes (T) that, in this example, are close to the nucleus (Nu). (D) The Golgi complex (G) is not labeled, while a small amount of labeling is seen on one side of this organelle (arrows). E, endosome. Bars, 200 nm.
In conclusion, the EM data demonstrate that, apart from the E8R labeling that was associated with the viral core, the protein was associated with viral and cellular membranes, both early and late in infection (see Discussion).
The E8R protein and its N-terminal portion are phosphorylated by the VV F10L kinase.
As described above, once the protein is packaged into the virus, it undergoes a series of modifications that lead to the production of three new forms, two of which have decreased molecular weight and one of which appears to be more acidic. The latter modification could be expected to result from phosphorylation, perhaps by the F10L gene product. The latter kinase has been shown to be a late protein that is packaged into the virion.
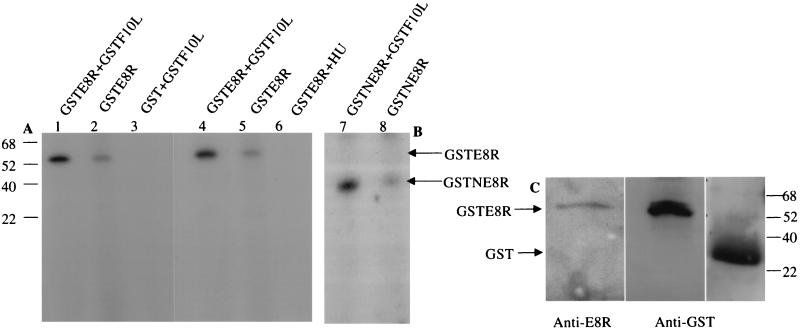
To determine whether the F10L kinase was able to phosphorylate the E8R protein, the GSTF10L protein expressed in Escherichia coli was incubated with GSTE8R or GST alone in the presence of [γ-32P]ATP. Initially, we tried to express and purify GSTE8R from E. coli, but the yields of the protein were always very poor (data not shown). We therefore expressed GSTE8R in VV-T7-infected cells; the fusion protein was isolated from infected cells and incubated with GSTF10L purified from bacteria. The reaction of GSTF10L with GST alone did not result in the phosphorylation of GST, whereas upon coincubation of GSTF10L and GSTE8R, a phosphorylated 55-kDa protein could be detected (Fig. 7A). Western blots of the same samples confirmed that the 55-kDa protein corresponded to GSTE8R (Fig. 7C). Identical results were obtained when the 120-amino-acid N-terminal portion of E8R (GSTNE8R; amino acids 1 to 120) was expressed in cells and incubated with the F10L kinase (Fig. 7B). Unexpectedly, when isolated GSTE8R (or GSTNE8R) was incubated in the presence of [γ-32P]ATP but without F10L, the same phosphorylated, albeit much fainter, band was detected (Fig. 7A and B). Since GSTE8R was expressed in a viral background, we considered the possibility that the phosphorylation of the protein was due to F10L that came from infected cells and that apparently was copurified with GSTE8R. To test this hypothesis, GSTE8R samples were prepared in the presence of 10 mM HU to block DNA replication and the synthesis of late proteins, such as the F10L kinase. When GSTE8R isolated from HU-treated cells was incubated in the presence of [γ-32P]ATP but without GSTF10L, the phosphorylated 55-kDa band could not be detected (Fig. 7A). The combined data thus show that E8R can be phosphorylated by the late viral kinase F10L, that phosphorylation may occur at the 120-amino-acid N terminus, and that the kinase may be copurified with E8R.
FIG. 7.
E8R is phosphorylated in vitro by the F10L kinase. GSTE8R was expressed in HeLa cells by infection with VV-T7 followed by transfection of pTMGSTE8R. GST or GSTF10L was expressed in and purified from E. coli. (A) Purified GSTE8R plus GSTF10L (lane 1), GSTE8R alone (lane 2), and GST plus GSTF10L (lane 3) were incubated for 20 min at 37°C in the presence of [γ-32P]ATP and then subjected to SDS-PAGE. GSTE8R is phosphorylated by the F10L kinase (lane 1), whereas there is no band corresponding to GST (lane 3). Note that in the sample with GSTE8R alone (lane 2), a faint 55-kDa band can also be detected. Purified GSTE8R plus GSTF10L (lane 4), GSTE8R alone (lane 5), and GSTE8R isolated from cells infected and transfected in the presence of HU (lane 6) were incubated as described above. In the sample treated with HU, the band corresponding to GSTE8R cannot be detected. (B) GSTNE8R plus GSTF10L (lane 7) and GSTNE8R (lane 8) were incubated as described for panel A. Whereas a phosphorylated band corresponding to GSTNE8R can readily be detected in lane 7, only a faint band can be seen in the absence of the kinase in lane 8. (C) Corresponding Western blots of GSTE8R or GST isolated from infected and transfected cells or bacteria, respectively, with anti-GST and anti-E8R antibodies to show the expected sizes of the proteins. The numbers to the left of panel A and to the right of panel C indicate the positions of the molecular mass markers (in kilodaltons).
The E8R protein binds DNA in vitro.
Our EM data suggested that E8R might mediate the binding of the ER to the newly synthesized viral DNA. Moreover, the labeling for E8R was found on the cytosolic side of the membranes and, since the antibody was raised against the extreme N terminus of the protein, these data suggested that the 120-amino-acid N terminus preceding the first transmembrane domain of the protein was exposed in the cytoplasm and might mediate this binding.
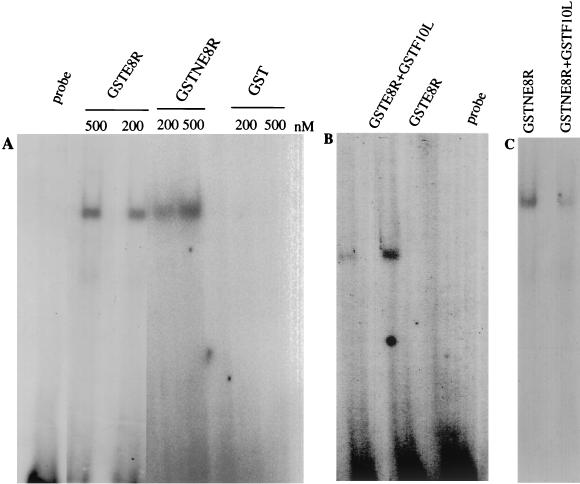
To characterize the affinity of E8R for double-stranded DNA, an electrophoretic gel mobility shift assay was performed. Two different concentrations of purified GSTE8R, GSTNE8R, or GST alone were incubated with a radiolabeled 35-mer oligonucleotide, and the complexes were resolved from free DNA by gel electrophoresis. At the two concentrations tested for both GSTE8R and GSTNE8R, but not for GST alone, a labeled band corresponding to the formation of a complex between the protein and the DNA was resolved (Fig. 8A).
FIG. 8.
GSTE8R binds DNA in vitro. GSTE8R was expressed as described in the legend to Fig. 7. GSTNE8R, GST, and GSTF10L were expressed and purified from bacteria. (A) Purified GSTE8R, GSTNE8R, and GST at the indicated concentrations were incubated for 20 min at room temperature in the presence of a radiolabeled oligonucleotide. The complexes were resolved by electrophoresis on a native gel. GSTE8R and GSTNE8R bind DNA, whereas no band can be detected in the samples incubated with GST. (B) Purified GSTE8R plus GSTF10L and GSTE8R alone were first incubated for 20 min in the presence of 1 mM ATP and then incubated for 20 min in the presence of a radiolabeled oligonucleotide. GSTE8R alone is able to bind DNA, whereas in the presence of the kinase, the binding is significantly reduced. The lane on the extreme left in panel A or the extreme right in panel B shows the probe without added protein. (C) The same experiment as in panel B was done with GSTNE8R and shows that coincubation with F10L also significantly reduces the ability of the N terminus to bind DNA.
We next tested whether phosphorylation by F10L in vitro affected the DNA-binding properties of E8R. Purified GSTE8R or GSTNE8R was incubated with or without GSTF10L in the presence of 1 mM ATP. A radiolabeled 35-mer oligonucleotide was then added to the mixture, and the complexes were resolved from free DNA by gel electrophoresis. Reaction of GSTE8R or GSTNE8R alone resulted in a strongly labeled band corresponding to the formation of a complex between the protein and the DNA, whereas upon coincubation of GSTF10L and GSTE8R or GSTNE8R, the signal corresponding to the labeled band was significantly reduced (Fig. 8B and C). These results suggested that the E8R protein as well as its 120-amino-acid N terminus binds DNA in vitro and that this binding may be modulated by phosphorylation.
DISCUSSION
E8R is made predominantly early in infection but is packaged into the virus.
The E8R gene product was previously identified as a membrane protein with a putative role in the process of ER wrapping around DNA replication sites.
In the present study, the E8R protein was characterized biochemically and morphologically. We show that the E8R gene product was synthesized predominantly early in infection; the protein was made in the absence of VV replication and could readily be detected in cell lysates metabolically labeled early in infection but was barely detectable at late times. Furthermore, the protein was detected on intracellular membranes by EM as early as 1 h postinfection. Both Western blotting and EM also demonstrated that the protein was packaged into virions late in infection, and we consistently found that the protein was very stable. The fact that the protein was made early but packaged into virions is not without precedent, as the B1R gene product has been shown to behave in an identical manner (1). Upon incorporation into virions, the protein apparently associated with the viral core, as shown both biochemically and by EM.
E8R is partially membrane associated.
As predicted from two protein prediction programs (the dense alignment surface method [5] and the PHDhtm program [34]), the E8R sequence may have two transmembrane domains. Both programs predicted two stretches of hydrophobic amino acids in the E8R sequence that are long enough to span a membrane and that can adopt an α-helical configuration (39). Consistently, upon extraction of cell lysates with TX-114, the protein partitioned in the detergent phase. The protein appears to be predominantly associated with cellular (as well as viral) membranes throughout the viral life cycle (39; this study), as shown by EM. We were therefore initially surprised to discover that E8R was associated with the viral core rather than with the IMV membranes.
In the present study, we therefore tested biochemically whether the protein behaved like a true membrane protein. Upon in vitro translation in the presence of rough microsomes, the bulk of the protein, surprisingly, remained in the soluble fraction. The latter result can perhaps be attributed to inefficient membrane insertion of E8R in vitro. The first transmembrane domain of the protein is preceded by 120 amino acids, and this rather long N-terminal portion may inhibit efficient membrane insertion in vitro. Our data show, however, that in infected cells as well, about half of the E8R protein may not be stably integrated into membranes. In a number of studies, sodium carbonate extraction of PNS of cell lysates has been used to determine the subcellular localization of VV proteins. Membrane proteins such as p21 (A17L) and p16 (A14L) are pelleted with membrane fractions irrespective of carbonate extraction. Core proteins, however, are pelleted in the absence of carbonate extraction but are completely solubilized in its presence (see Results) (6, 18, 23, 35). The E8R protein behaved in an unexpected manner in that both in the presence and in the absence of sodium carbonate, it was partially membrane associated and partially soluble. Given the fact that all of the protein localized to membranes at all times of infection (apart from the labeling on the viral core), as determined by EM, we suggest that a pool of E8R is not stably membrane anchored and is solubilized upon preparation of PNS.
E8R binds DNA in vitro.
It was previously speculated that E8R has a putative role in mediating the binding of the ER to viral DNA. Here we show that the protein was indeed able to bind DNA in vitro. Our data show that the protein does bind DNA and that the N-terminal portion is sufficient for binding. Our 2D gel analyses show that, once encapsidated in the virion, the protein undergoes a number of modifications that result in a decreased molecular weight as well as a shift toward a more acid pI. The latter shift is typical of phosphorylation. Attempts to prove that the protein becomes phosphorylated once packaged into the virus were, however, unsuccessful. We were unable to detect a 32Pi-labeled spot in 2D gels corresponding to (the modified) E8R. This result is most likely due to the fact that the protein is predominantly made early in infection and at this stage is not phosphorylated. A subset of these molecules synthesized hours before assembly may then become phosphorylated once packaged into the virus late in infection. This condition most likely explains our failure to detect phosphorylated E8R in assembled virions by 32Pi labeling. Our in vitro data nevertheless show that E8R (but not B1R; data not shown) is a good substrate for the late kinase F10L. We speculate therefore that the observed shift in pI may be due to phosphorylation by this kinase, which is known to be encapsidated in the particle upon IMV formation (24). Moreover, we show that phosphorylation by F10L reduces the ability of E8R to bind DNA in vitro.
In the present study, we also show that, once packaged into the virus, a subset of E8R molecules undergo a decrease in molecular weight, suggesting that the protein may also undergo cleavage. Such cleavage is a common event in VV core proteins and coincides with the formation of IMVs (see above). Inspection of the E8R sequence, however, did not reveal the typical consensus sequence (Ala-Gly-X) that has been shown to be the cleavage site in VV core proteins (and at least one membrane protein) (3, 43, 45). Our data raise the question as to why E8R undergoes these modifications (putative cleavage and phosphorylation) in the assembled particle but not before virion formation. Although quite speculative at present, we propose that these modifications abolish the binding of E8R to DNA, as explained below.
Model for the role of E8R during the VV life cycle.
Based on the current results as well as on previous data (13, 14, 26), we propose that the E8R gene product can mediate the binding of viral DNA to ER membranes throughout the VV life cycle.
In a recent study, it was shown that upon core uncoating, early in infection, parental DNA is released into the cytoplasm, where it may associate with the cytosolic side of the ER (26). Since E8R is an early protein, it is synthesized before the process of core uncoating. We could consistently detect by EM as early as 1 h postinfection the protein on intracellular membranes, including the ER. We therefore suggest that newly synthesized E8R may mediate the binding of parental DNA, once released from the core, to ER membranes. Subsequently, later in infection, when DNA replication is initiated, E8R may mediate the binding of more ER cisternae to the newly synthesized DNA. This process eventually leads to the almost complete enclosure of the replication site by ER membranes, as shown before (39).
A model describing how the viral DNA could be incorporated into the viral particle late in infection was recently proposed. In that model, it was postulated that the membranes of the IVs are continuous with the ER, the latter membranes having attached to the cytosolic side the viral genome (13, 14, 38). DNA uptake by an IV occurs when the DNA-containing ER membrane is pushed inside the IV. In that model, the resulting IMV consists of a continuous S-shaped cisternal membrane. The genome is surrounded by one part of this cisterna, the latter being continuous with the outer IMV cisternal membranes that serve to seal the particle off. We propose that at the late stage of infection, the E8R protein might mediate the binding of the viral genome to the ER. We envision that the protein might mediate the binding of the genome to the cytosolic side of the ER that is in continuity with the viral crescents. Upon IMV assembly, it might initially mediate the association of the viral DNA with the cisternal membranes of the core. We consistently found that although the protein behaves (at least partially) as a membrane protein, it is associated with the viral core. Finally, we hypothesize that the observed modifications acquired by E8R once assembled in the particle subsequently abolish its binding to DNA (see above). This process would ensure that upon infection and core uncoating, parental DNA is able to leave the core and bind to newly synthesized E8R molecules in the ER. We are now in the process of testing some of these assumptions.
Acknowledgments
We thank Gareth Griffiths for critical reading of the manuscript and Concetta Ambrosino for help in setting up the DNA shift experiment.
Part of this work was supported by an EU TMR grant.
REFERENCES
- 1.Banham, A. H., and G. L. Smith. 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191:803-812. [DOI] [PubMed] [Google Scholar]
- 2.Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. [DOI] [PubMed] [Google Scholar]
- 3.Betakova, T., E. J. Wolffe, and B. Moss. 1999. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J. Virol. 73:3534-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cairns, H. J. F. 1960. The initiation of vaccinia infection. Virology 11:603-623. [DOI] [PubMed] [Google Scholar]
- 5.Cserzo, M., J. M. Bernassau, I. Simon, and B. Maigret. 1994. New alignment strategy for transmembrane proteins. J. Mol. Biol. 243:388-396. [DOI] [PubMed] [Google Scholar]
- 6.Cudmore, S., R. Blasco, R. Vincentelli, M. Esteban, B. Sodeik, G. Griffiths, and J. Krijnse Locker. 1996. A vaccinia virus core protein, p39, is membrane associated. J. Virol. 70:6909-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dales, S., and R. Kajioka. 1964. The cycle of multiplication of vaccinia virus in Earle's strain L cells. 1. Uptake and penetration. Virology 24:278-294. [DOI] [PubMed] [Google Scholar]
- 8.Derrien, M., A. Punjabi, M. Khanna, O. Grubisha, and P. Traktman. 1999. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J. Virol. 73:7287-7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edelman, A. M., D. K. Blumenthal, and E. G. Krebs. 1987. Protein serine/threonine kinases. Annu. Rev. Biochem. 56:567-613. [DOI] [PubMed] [Google Scholar]
- 10.Ericsson, M., S. Cudmore, S. Shuman, R. C. Condit, G. Griffiths, and J. K. Locker. 1995. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J. Virol. 69:7072-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essani, K., and S. Dales. 1979. Biogenesis of vaccinia: evidence for more than 100 polypeptides in the virion. Virology 95:385-394. [DOI] [PubMed] [Google Scholar]
- 12.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths, G., N. Roos, S. Schleich, and J. Krijnse Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analysis. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths, G., R. Wepf, T. Wendt, J. Krijnse Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guan, K. L., S. S. Broyles, and J. E. Dixon. 1991. A Tyr/Ser protein phosphatase encoded by vaccinia virus. Nature 350:359-362. [DOI] [PubMed] [Google Scholar]
- 16.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter, T. 1987. A thousand and one protein kinases. Cell 50:823-829. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao, S. Y., and W. R. Bauer. 1987. Biosynthesis and phosphorylation of vaccinia virus structural protein VP11. Virology 159:399-407. [DOI] [PubMed] [Google Scholar]
- 20.Katz, E., and B. Moss. 1970. Vaccinia virus structural polypeptide derived from a high-molecular-weight precursor: formation and integration into virus particles. J. Virol. 6:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kit, S., D. R. Dubbs, and T. C. Hsu. 1963. Biochemistry of vaccinia-infected mouse fibroblasts (strain L-M). III. Radioautographic and biochemical studies of thymidine-3H uptake into DNA of L-M cells and rabbit cells in primary culture. Virology 19:13-22. [DOI] [PubMed] [Google Scholar]
- 22.Krijnse Locker, J., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 11:2497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krijnse Locker, J., S. Schleich, D. Rodriguez, B. Goud, E. J. Snijder, and G. Griffiths. 1996. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J. Biol. Chem. 271:14950-14958. [DOI] [PubMed] [Google Scholar]
- 24.Lin, S., and S. S. Broyles. 1994. Vaccinia protein kinase 2: a second essential serine/threonine protein kinase encoded by vaccinia virus. Proc. Natl. Acad. Sci. USA 91:7653-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, K., B. Lemon, and P. Traktman. 1995. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J. Virol. 69:7823-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mallardo, M., E. Leithe, S. Schleich, N. Roos, L. Doglio, and J. Krijnse Locker. 2002. Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites. J. Virol. 76:5167-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallardo, M., S. Schleich, and J. Krijnse Locker. 2001. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs into distinct cytoplasmic structures. Mol. Biol. Cell 12:3875-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 29.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. Krijnse Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rempel, R. E., and P. Traktman. 1992. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J. Virol. 66:4413-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 32.Risco, C., J. R. Rodriguez, C. Lopez-Iglesias, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 2002. Endoplasmic reticulum-Golgi intermediate compartment membranes and vimentin filaments participate in vaccinia virus assembly. J. Virol. 76:1839-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rost, B., R. Casadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. Krijnse Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans-Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 39.Tolonen, N., L. Doglio, S. Schleich, and J. Krijnse Locker. 2001. Vaccinia virus DNA-replication occurs in ER-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12:2031-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Traktman, P., A. Caligiuri, S. A. Jesty, K. Liu, and U. Sankar. 1995. Temperature-sensitve mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J. Virol. 69:6581-6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. Krijnse Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 44.Vanslyke, J. K., and D. E. Hruby. 1994. Immunolocalization of vaccinia virus structural proteins during virion formation. Virology 198:624-635. [DOI] [PubMed] [Google Scholar]
- 45.Vanslyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 46.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, W. P, and W. R. Bauer. 1988. Purification and characterization of vaccinia virus structural protein VP8. Virology 167:578-584. [PubMed] [Google Scholar]