Abstract
We examined the influence of dose and method of antigen delivery on the dynamics and durability of T-cell responses to candidate human immunodeficiency virus (HIV) vaccines. Codon-optimized sequences from the HIV gag gene were inserted into alternative DNA vaccine vectors to express the coding sequence with or without the tissue plasminogen activator leader sequence. We delivered the vaccines by intramuscular injection as plasmid DNA without adjuvant or as plasmid DNA formulated with a novel block copolymer adjuvant (CRL8623) and then monitored the ensuing T-cell responses by using a gamma interferon enzyme-linked immunospot assay. We demonstrated persistence of the cell-mediated immune (CMI) response in rhesus macaques for at least 18 months following a four-dose vaccination regimen. The plasmid vaccine, with or without CRL8623, was immunogenic in macaques; however, the form coadministered with adjuvant exhibited improved T-cell responses, with a bias toward more antigen-specific CD8+ T cells. Finally, we examined the fine specificity of the T-cell response to the gag vaccines by testing the response of 23 vaccinated macaques to individual Gag 20-mer peptides. Collectively, the monkeys responded to 25 epitopes, and, on average, each monkey recognized a minimum of 2.7 epitopes. The results indicate that a broad and durable CMI response to HIV DNA vaccines can be induced in a relevant nonhuman primate model.
Evidence that vaccines capable of inducing a cell-mediated immune (CMI) response (1, 5, 11, 17, 18, 20) may be effective in mitigating infection by human immunodeficiency virus (HIV) is accumulating. Several approaches to the design of such vaccines have been proposed, and some have been implemented. Plasmid DNAs expressing relatively conserved (i.e., nonenvelope) proteins have been investigated (4), and a variety of viral vectors, including canarypox virus (6), modified vaccinia virus Ankara (22), and adenovirus (15), have been used to deliver HIV proteins intracellularly for induction of CMI responses. More recently, mixed-modality vaccine regimens have been used in an effort to overcome or circumvent preexisting immunity to viral vectors. Typically, these regimens include priming with a plasmid DNA vaccine followed by boosting with a protein-based vaccine (14) or with a viral vector (9) expressing the same HIV gene products as are encoded by the DNA vaccine. Characterization and monitoring of the resulting CMI responses by classical methods are both cumbersome and labor-intensive because of the major histocompatibility complex (MHC) diversity of the human population. Cytolytic T-lymphocyte (CTL) responses have been induced by a variety of vaccines, but CTL assays are difficult to perform and are not very quantitative. Lymphoproliferation assays have also been used, but again, these assays are not very quantitative, and they do not adequately measure CD8 T-cell responses.
Previously, Larsson et al. (13) described a recombinant vaccinia virus enzyme-linked immunospot (ELISPOT) assay for detection of HIV polymerase responses in HIV-positive subjects. This assay has the advantage of delivering target protein antigen into the cells' cytosol, where the expressed protein can be processed into peptides capable of restimulating T-cell immune responses. A disadvantage is that a live, albeit attenuated, virus must be used for the assay, and construction of the recombinant vaccinia virus vector is not trivial. An alternative is the use of peptides known to bind to MHC class I or class II molecules. Tetramer assays have proven useful in monitoring responses to dominant epitopes (25), but this approach appears to overestimate the number of responding T cells since it measures binding but not function (8). The tetramer assay is also MHC restricted and is limited to detecting responses to known epitopes. When studying an outbred population, assays reliant on measuring responses to a few dominant epitopes will underestimate the response to vaccines or natural infection. To circumvent these issues, we have developed a strategy for using the gamma interferon (IFN-γ) ELISPOT assay to monitor T-cell responses to HIV vaccines. We determined that pools containing as many as 50 peptides spanning the HIV Gag protein could elicit a response equivalent to that induced by infecting peripheral blood mononuclear cells (PBMCs) with a vaccinia virus vector expressing Gag. We further determined the fine specificity of the response to Gag by measuring the ELISPOT response to each individual peptide in the pool. This assay was used to show that immunization of rhesus monkeys with an HIV gag DNA vaccine induced long-lasting IFN-γ memory responses involving CD4 and CD8 T cells.
Equivalent IFN-γ ELISPOT responses to naturally processed Gag delivered with a vaccinia virus vector versus peptide pools.
We compared the use of peptide pools with the use of a recombinant vaccinia virus gag vector (vac-gag) for elicitation of IFN-γ production by individual T cells. Our first goal was to determine whether stimulation with naturally processed Gag would elicit a more robust memory T-cell response than that elicited to peptides. We compared the recall response to a vaccinia virus vector expressing Gag with that elicited by a pool of overlapping 20-mer peptides spanning the Gag protein. PBMCs from gag DNA-vaccinated monkeys were seeded in 96-well plates at a density of 4 × 105 cells per well and stimulated with a peptide pool consisting of 50 overlapping 20-mer peptides (offset by 10 amino acids) encompassing the Gag sequence encoded by the vaccine. In parallel, cells were stimulated with vac-gag by infecting 10% of the PBMCs with that vector at a multiplicity of infection of 5:1 for 2 h at 37°C and then adding these cells to the remaining PBMCs in the wells of the assay plate. Controls included cells cultured in medium alone or infected with a control vaccinia virus vector (vac-sc). PBMCs from immunized animals were assayed for the ability to secrete IFN-γ during in vitro restimulation with antigenic peptides, using a modification of published methods (7, 12, 24). Briefly, 96-well polyvinylidine difluoride-backed plates (MAIP NOB 10; Millipore, Bedford, Mass.) were coated with a mouse monoclonal antibody to IFN-γ (catalog no. 1598-00; R&D Systems), washed three times with phosphate-buffered saline (PBS), and then blocked with RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum. Cells were cultured at a density of 4 × 105 per well in 0.1 ml of medium for restimulation with an HIV Gag peptide pool. (Stock pools were stored in 100% dimethyl sulfoxide at 2-mg/ml concentrations of each peptide and were used at a final concentration of approximately 4 μg/ml, attained by dilution in culture medium.) After 18 to 24 h of incubation at 37°C, the plates were washed six times with PBS containing 0.005% Tween 20. Biotinylated goat anti-human recombinant IFN-γ polyclonal antiserum (catalog no. BAF285; R&D Systems) was then added, and the plates were incubated overnight at 4°C. The plates were then washed six times with PBS prior to the addition of streptavidin-alkaline phosphatase conjugate (catalog no. 7100-05; Southern Biotechnology Associates, Birmingham, Ala.). After incubation for 2 h at room temperature, the plates were washed three times with PBS-0.005% Tween 20 and then three times with PBS. Finally, a one-step nitroblue tetrazoleum-5-bromo-4-chloro-3-indolylphosphate substrate (catalog no. 34042; Pierce) was added for 5 to 15 min for development of spots. The reaction was stopped by rinsing the plates in water. After the plates had air dried, spots were counted by use of a stereomicroscope. Select wells were imaged by using an image analysis system from Resolution Technology (Columbus, Ohio). The results (Table 1) indicate that ELISPOT responses detected using the peptide pool were approximately equivalent to those elicited with vac-gag. HIV gag DNA-vaccinated monkeys had responses ranging from 78 to 666 spot-forming cells (SFC) per 106 PBMCs upon restimulation with the Gag peptide pool, and they had responses of 59 to 569 SFC per 106 PBMCs after restimulation with vac-gag. Background responses in the control wells in this experiment were low (<10 SFC/106 PBMCs). The results indicate that peptide pools elicited IFN-γ ELISPOT responses equivalent (in magnitude) to those stimulated with naturally processed antigen provided by vac-gag infection of target cells. It is noteworthy that the quality of the spots was generally better when the peptide pool rather than vac-gag was used for restimulation. Responses to the vaccinia virus control (vac-sc) were usually low; however, a few individual monkeys had high background responses to vac-sc. Also, autologous B-cell lines infected with vac-gag were found to be poor targets for eliciting a response to this ELISPOT assay since they resulted in high background responses and poor-quality spots (data not shown).
TABLE 1.
Equivalent IFN-γ ELISPOT responses to peptide pools and naturally processed Gag delivered with a vaccinia virus vectora
| Rhesus macaque no. | IFN-γ ELISPOT response after stimulation with:
|
|||
|---|---|---|---|---|
| Gag peptide pool | vac-gag | vac-sc (control) | Medium control | |
| 930147 | 78 | 80 | 3 | 5 |
| 920078 | 666 | 569 | 8 | 9 |
| 94R013 | 348 | 413 | 1 | 6 |
| 94R031 | 135 | 94 | 3 | 6 |
| 940218 | 95 | 59 | 0 | 5 |
PBMCs from monkeys immunized i.m. with 1 to 5 mg of HIV gag DNA vaccine at weeks 0, 4, 8, and 24 were assayed at week 36. The results are expressed as SFC per 106 input PBMCs.
IFN-γ ELISPOT response of rhesus monkeys immunized with HIV gag DNA vaccines.
Candidate HIV gag DNA vaccines were constructed by altering the DNA sequence of the gag gene (but not the resultant amino acid sequence) such that it contained codons more commonly recognized by mammalian tRNAs instead of wild-type viral codons, many of which are recognized inefficiently by rare tRNAs. The expression vector, V1Jns, was derived from a pUC19 plasmid containing the human cytomegalovirus immediate-early promoter with its intron A sequence, multiple restriction sites (including BglII) for cloning, the bovine growth hormone polyadenylation signal sequence, and the kanamycin antibiotic resistance gene (16). The vaccine was constructed by inserting a full-length p55 gag sequence immediately downstream from the cytomegalovirus promoter. In some experiments, an alternative vector, containing the signal peptide sequence from human tissue-specific plasminogen activator inserted into the beginning of the gag sequence, was used. Plasmid DNA used for immunization was purified from Escherichia coli DH5 cells by a modified alkaline lysis procedure, and DNA was further purified by banding via two sequential equilibrium density gradient centrifugations, using cesium chloride-ethidium bromide gradients (16). Plasmid DNA expressing HIV gag with or without the tissue plasminogen activator (tPA) leader sequence was injected intramuscularly (i.m.) into rhesus monkeys at 0, 4, and 8 weeks. In experiment 1 (Table 2), the monkeys were boosted at week 20. At periodic intervals after vaccination, PBMCs were seeded into 96-well ELISPOT plates at 4 × 105 cells per well and stimulated with a pool of 20-mer peptides encompassing the Gag protein. Cells were incubated for ∼20 h, and then the plates were processed for development of ELISPOTs as described above. The results shown in Table 2 are the net responses after subtraction of spots formed in medium control wells, which were generally less than 10% of the response to the peptide pool. The results from experiment 1 indicate that there was not a marked dose-response effect, since approximately equivalent responses were elicited with the 1- and 5-mg doses of vaccine, and there was no apparent advantage to incorporating the tPA leader sequence into the vaccine. Of note, responses elicited by the DNA vaccine were durable; they were shown to persist for at least 18 months after administration of the final (fourth) dose of the vaccine (experiment 1).
TABLE 2.
Persistent IFN-γ ELISPOT responses of rhesus monkeys immunized with HIV gag DNA vaccinesa
| Expt | Vaccine | Dose (mg) | Gag-specific IFN-γ ELISPOT response (SFC per 106 PBMCs) at wk:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4 | 8 | 12/13 | 20 | 24 | 28 | 33 | 99 | |||
| 1 | gag DNA | 5 | 0 | 19 | 45 | 122 | 65 | 337 | 280 | 303 | 176 |
| 0 | 42 | 53 | 181 | 125 | 275 | 294 | 100 | 119 | |||
| 2 | 60 | 102 | 114 | 90 | 134 | 74 | 15 | 107 | |||
| 0.7 | 40.3 | 66.7 | 139.0 | 93.3 | 248.7 | 216.0 | 139.3 | 134.0 | |||
| 1 | 0 | 8 | 59 | 79 | 118 | 223 | 125 | 112 | 57 | ||
| 3 | 48 | 34 | 89 | 73 | 109 | 90 | 175 | 45 | |||
| 4 | 50 | 15 | 213 | 89 | 40 | 116 | 38 | 35 | |||
| 2.3 | 35.3 | 36.0 | 127.0 | 93.3 | 124.0 | 110.3 | 108.3 | 45.7 | |||
| tPA-gag DNA | 5 | 0 | 194 | 101 | 57 | 48 | 68 | 41 | 172 | ||
| 5 | 3 | 49 | 38 | 9 | 5 | 3 | 30 | ||||
| 0 | 264 | 43 | 118 | 208 | 205 | 103 | 255 | ||||
| 1.7 | 153.7 | 64.3 | 71.0 | 88.3 | 92.7 | 49.0 | 152.3 | ||||
| 2 | gag DNA | 5 | 1 | 208 | 441 | 358 | 144 | 161 | |||
| 0 | 148 | 121 | 266 | 337 | 85 | ||||||
| 3 | 44 | 48 | 41 | 66 | 116 | ||||||
| 1.3 | 133.3 | 203.3 | 221.7 | 182.3 | 120.7 | ||||||
| 1 | 6 | 4 | 68 | 36 | 10 | 66 | |||||
| 0 | 46 | 135 | 101 | 81 | 111 | ||||||
| 1 | 98 | 349 | 499 | 273 | 568 | ||||||
| 2.3 | 49.3 | 184.0 | 212.0 | 121.3 | 248.3 | ||||||
Plasmid DNA containing a modified codon sequence of the HIV gag gene with or without the tPA leader sequence was injected i.m. into rhesus monkeys at 0, 4, and 8 weeks. In experiment 1, the monkeys were boosted at week 20. At the indicated times, PBMCs were seeded into 96-well ELISPOT plates at 4 × 105 cells per well and stimulated with a pool of overlapping 20-mer peptides (offset by 10 amino acids) encompassing the Gag protein. Cells were incubated for ∼20 h, and then the plates were processed for development of ELISPOTs. Results shown are net responses after subtraction of spots formed in medium control wells. The mean response for each group is shown in bold typeface for each time point.
IFN-γ ELISPOT response of rhesus monkeys to an HIV gag DNA-CRL8623 vaccine.
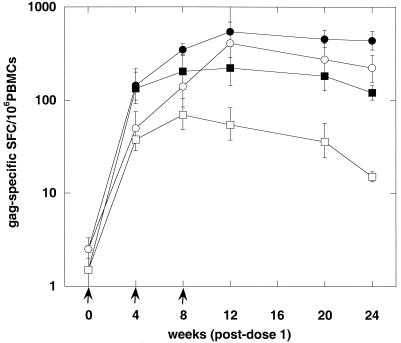
Recently, we documented the potent adjuvant effect for induction of CMI by using a simian immunodeficiency virus (SIV) gag DNA vaccine (23). In the present study, we compared the T-cell phenotype of rhesus macaques immunized with HIV gag formulated with CRL8623, a nonionic block copolymer adjuvant similar to CRL1005, with that of monkeys immunized with non-adjuvant-associated HIV gag DNA for the induction of IFN-γ ELISPOT responses. The DNA-CRL8623 mixture was kept at −70°C and then slowly warmed to room temperature prior to injection. Figure 1 shows the results for groups of three monkeys assayed at six time points after vaccination with 5 mg of HIV gag DNA, formulated either with saline or with 7.5 mg of CRL8623, at weeks 0, 4, and 8. ELISPOT responses were determined using PBMCs that were or were not depleted of CD4 T cells prior to in vitro culture. The results indicated that the DNA-CRL8623 formulation induced a more than twofold-higher ELISPOT response than did non-adjuvant-associated DNA. CD4 depletion experiments suggested that the DNA, with or without the adjuvant, was able to induce both CD4+ and CD8+ T cells. It would appear, with the limited cohort size, that adjuvant-associated DNA tended to induce a higher fraction of antigen-specific T cells that were CD8+ than the non-adjuvant-associated form.
FIG. 1.
IFN-γ ELISPOT response of rhesus monkeys to an HIV gag DNA vaccine formulated with CRL8623 adjuvant. Groups of three monkeys were injected i.m. with 5 mg of HIV gag DNA formulated with CRL8623 (circles) or with saline (squares) at 0, 4, and 8 weeks. ELISPOT responses were enumerated at the indicated times postvaccination, using PBMCs that were unfractionated (closed symbols) or depleted of CD4 T cells (open symbols).
ELISPOT response profile to individual peptides from HIV Gag.
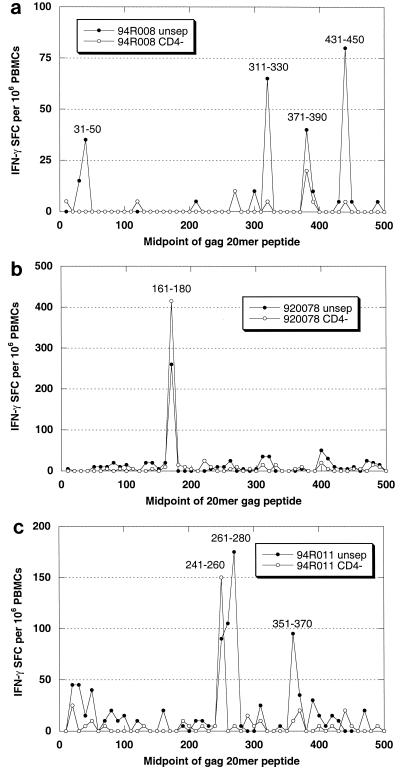
An advantage of using peptide pools to monitor the response to vaccination is that once a responder has been identified, the fine specificity of the response can be ascertained by restimulation with individual peptides. Figure 2 contains examples of peptide profiles representative of IFN-γ ELISPOT responses of rhesus monkeys to HIV gag DNA vaccines. Rhesus monkeys were injected i.m. with 5 mg of HIV gag plasmid DNA vaccine at weeks 0, 4, 8, and 24. PBMCs collected at week 51 were tested for their response to individual 20-mer peptides representing the HIV Gag sequence. Unseparated and CD4-depleted PBMCs were tested in parallel for reactivity to each of 50 peptides. Figure 2a shows an example of a CD4-dominated response in which four distinct peptides were recognized. Figure 2b shows the results for a monkey that responded to a single dominant peptide. Note that the response increased following CD4 depletion, indicating that it was predominantly (or entirely) CD8 mediated. Figure 2c provides an example of a mixed-phenotype response; a strong CD8 response to one peptide was observed, while an equally vigorous CD4 response was directed to two distinct peptides. In each example, robust responses were detected 6 months following administration of the final vaccine dose.
FIG. 2.
Response profiles representative of IFN-γ ELISPOT responses of rhesus monkeys to HIV gag DNA vaccines. Rhesus monkeys were injected i.m. with 5 mg of HIV gag plasmid DNA vaccine at weeks 0, 4, 8, and 24. PBMCs collected at week 51 were tested for their response to individual 20-mer peptides comprising the HIV Gag sequence. Unseparated (unsep) and CD4-depleted (CD−) PBMCs were tested in parallel for reactivity to each of 50 peptides. (a) Example of a CD4-dominated response in which four distinct peptides were recognized. (b) Results for a monkey that responded to a single peptide. Note that the magnitude of the response increased following CD4 depletion, indicating that the response was predominantly CD8 mediated. (c) Example of a mixed-phenotype response yielding a strong CD8 response to one peptide and an equally vigorous CD4 response directed to two distinct peptides.
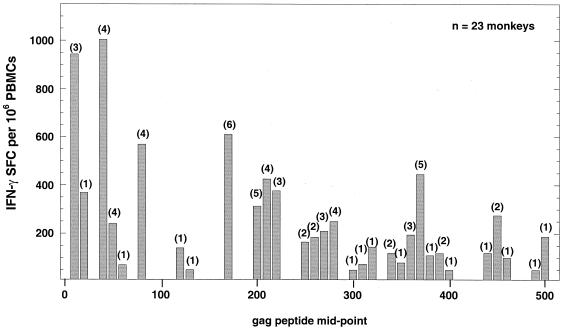
We went on to examine the fine specificity of the IFN-γ ELISPOT responses of PBMCs from 23 monkeys immunized with three or four doses of HIV gag DNA vaccine (formulated without adjuvant). Cells were cultured at a density of 2 × 105 per well with each of 50 overlapping 20-mer peptides (offset by 10 amino acids). Results (Fig. 3) are expressed as the total IFN-γ ELISPOT response (adjusted per 106 PBMCs) to each peptide for the 23 monkeys tested. The number in parentheses above each column in Fig. 3 indicates the number of monkeys that responded to the corresponding peptide at a level of ≥50 SFC per 106 PBMCs. The results show that, collectively, the 23 monkeys responded to 31 of a possible 50 peptides. It should be noted, however, that epitopes may be contained in more than one peptide since the individual peptides overlap by 10 amino acids. If one makes the conservative assumption that responses of an individual to adjacent peptides result, in every case, from a response to the overlapping amino acids, then a minimum of 25 different epitopes were recognized by the 23 monkeys. The number of epitopes recognized by individual monkeys ranged from one to five, with an average of 2.7 peptides being recognized per monkey.
FIG. 3.
Profile of ELISPOT responses to individual 20-mer peptides from HIV Gag. PBMCs from 23 monkeys immunized with three or four doses of HIV gag DNA vaccine were cultured at a density of 2 × 105 cells per well with each of 50 overlapping 20-mer peptides (offset by 10 amino acids). Results are expressed as the total IFN-γ ELISPOT response (adjusted per million PBMCs) to each peptide for all 23 monkeys. The number in parentheses above each column indicates the number of monkeys that responded to the corresponding peptide at a level of ≥50 SFC per 106 PBMCs.
Considerable effort has gone into identifying epitopes and determining their MHC restriction in subjects infected with HIV, as well as in animal models of HIV and SIV, thereby providing a rationale for the construction of novel HIV vaccine candidates comprising a string of CTL epitopes that can be delivered as plasmid DNA (10) or in viral vectors (9). Such vaccines have the potential advantage of incorporating epitopes from divergent HIV clades, but they are necessarily limited to containing only those epitopes that have been previously identified, either through studies of infected individuals or through motif scanning to detect binding of peptide to individual MHC molecules (2). This approach also raises the possibility of unintentionally creating novel epitopes that overlap the defined epitopes in such multiepitope vaccines. These artificial epitopes could have unknown and potentially deleterious effects, such as the induction of autoimmune responses. The results presented here suggest that it may be unnecessary, and perhaps counterproductive, to limit a vaccine to a restricted set of epitopes. Our data show that the native peptide sequence encoded by the HIV gag gene, when delivered as a DNA vaccine to a small number of monkeys (n = 23), elicited T-cell responses to at least 25 different epitopes. These animals recognized an average of 2.7 epitopes per monkey; however, 7 of 23 monkeys recognized only a single epitope. This may be important in light of the recent observation that breakthrough disease in a gag DNA-vaccinated monkey resulted from a mutation in a single T-cell epitope (3). The use of adenovirus vectors to deliver HIV gene sequences either alone or in a mixed vaccine regimen may overcome this problem, since these vaccines elicit much more potent CMI responses (23).
The observation that the phenotype of the T-cell response to our HIV gag DNA vaccine can be manipulated by use of certain adjuvants may have implications for vaccine efficacy in human clinical trials. The DNA vaccines were able to elicit mixed CD8+ and CD4+ T-cell responses; formulation with CRL8623 appeared to induce a larger fraction of the antigen-specific T cells that were CD8+. The importance of CD8 T cells in prevention of AIDS has been inferred from studies of sex workers in Nairobi (19) as well as studies using the rhesus macaque-SIV model (21). On the other hand, Rosenberg et al. (17) showed that a robust HIV-specific CD4 T-cell response was associated with control of viremia in individuals with AIDS. This suggests that memory T cells may be resistant to infection with HIV, perhaps through rapid induction of a chemokine or cytokine response to virus-infected cells. Because CD4 T cells are necessary for the persistence of long-term CD8 T-cell responses, an HIV vaccine that induces both of these T-cell phenotypes is desirable. Our finding that long-term (>18-month) memory T-cell responses can be elicited to multiple epitopes in a relevant animal model provides hope that such vaccines may be effective in preventing or moderating HIV infection. The ELISPOT assay described herein provides a useful tool suitable for testing such vaccines in large-scale clinical trials.
Acknowledgments
We thank R. Druilhet at the New Iberia Research Center for help with bleeding of and preparation of cells from rhesus monkeys and W. McClements and H. Joseph for helpful comments on the manuscript.
REFERENCES
- 1.Akridge, R., F. Hladik, J. Markee, C. Alef, H. Kelley, A. Collier, and M. J. McElrath. 1999. Cellular immunity and target cell susceptibility in persons with repeated HIV-1 exposure. Immunol. Lett. 66:15-19. [DOI] [PubMed] [Google Scholar]
- 2.Allen, T. M., B. R. Mothé, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A∗01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 4.Boyer, J. D., K. E. Ugen, B. Wang, M. Agadjanyan, L. Gilbert, M. L. Bagarazzi, M. Chattergoon, P. Frost, A. Javadian, W. V. Williams, Y. Refaili, R. Ciccarelli, D. McCallus, L. Coney, and D. B. Weiner. 1997. Protection of chimpanzees from high-dose heterologous HIV-1 challenge by DNA vaccination. Nat. Med. 3:526-532. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, J. 1999. AIDS vaccines. Glimmerings of hope from the bottom of the well. Science 285:656-657. [DOI] [PubMed] [Google Scholar]
- 6.Evans, T. G., M. C. Keefer, K. J. Weinhold, M. Wolff, D. Montefiori, G. J. Gorse, B. S. Graham, M. J. McElrath, M. L. Clements-Mann, M. J. Mulligan, P. Fast, M. C. Walker, J. L. Excler, A. M. Duliege, and J. Tartaglia. 1999. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes given alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J. Infect. Dis. 180:290-298. [DOI] [PubMed] [Google Scholar]
- 7.Forsthuber, T., H. C. Yip, and P. V. Lehmann. 1996. Induction of TH1 and TH2 immunity in neonatal mice. Science 271:1728-1730. [DOI] [PubMed] [Google Scholar]
- 8.Goulder, P. J., Y. Tang, C. Brander, M. R. Betts, M. Altfeld, K. Annamalai, A. Trocha, S. He, E. S. Rosenberg, G. Ogg, C. A. O'Callaghan, S. A. Kalams, R. E. McKinney, Jr., K. Mayer, R. A. Koup, S. I. Pelton, S. K. Burchett, K. McIntosh, and B. D. Walker. 2000. Functionally inert HIV-specific cytotoxic T lymphocytes do not play a major role in chronically infected adults and children. J. Exp. Med. 192:1819-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanke, T., T. J. Blanchard, J. Schneider, C. M. Hannon, M. Becker, S. C. Gilbert, A. V. S. Hill, G. L. Smith, and A. McMichael. 1998. Enhancement of MHC class I-restricted peptide-specific T cell induction by a DNA prime/MVA boost vaccination regime. Vaccine 16:439-445. [DOI] [PubMed] [Google Scholar]
- 10.Hanke, T., V. C. Neumann, T. J. Blanchard, P. Sweeney, A. V. Hill, G. L. Smith, and A. McMichael. 1999. Effective induction of HIV-specific CTL by multi-epitope using gene gun in a combined vaccination regime. Vaccine 17:589-596. [DOI] [PubMed] [Google Scholar]
- 11.Hay, C. M., D. J. Ruhl, N. O. Basgoz, C. C. Wilson, J. M. Billingsley, M. P. DePasquale, R. T. D'Aquila, S. M. Wolinsky, J. M. Crawford, D. C. Montefiori, and B. D. Walker. 1999. Lack of viral escape and defective in vivo activation of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes in rapidly progressive infection. J. Virol. 73:5509-5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lalvani, A., R. Brookes, S. Hambleton, W. J. Britton, A. V. S. Hill, and A. J. McMichael. 1997. Rapid effector function in CD8+ memory T cells. J. Exp. Med. 186:859-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larsson, M., X. Jin, B. Ramratnam, G. S. Ogg, J. Engelmayer, M. A. Demoitie, A. J. McMichael, W. I. Cox, R. M. Steinman, D. Nixon, and N. Bhardwaj. 1999. A recombinant vaccinia virus based ELISPOT assay detects high frequencies of Pol-specific CD8 T cells in HIV-1-positive individuals. AIDS 13:767-777. [DOI] [PubMed] [Google Scholar]
- 14.Letvin, N. L., D. C. Montefiori, Y. Yasutomi, H. C. Perry, M. E. Davies, C. Lekutis, M. Alroy, D. C. Freed, C. I. Lord, L. K. Handt, M. A. Liu, and J. W. Shiver. 1997. Potent, protective anti-HIV immune responses generated by bimodal HIV envelope DNA plus protein vaccination. Proc. Natl. Acad. Sci. USA 94:9378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lubeck, M. D., R. J. Natuk, M. Chengalvala, P. K. Chanda, K. K. Murthy, S. Murthy, S. Mizutani, S. G. Lee, M. S. Wade, B. M. Bhat, et al. 1994. Immunogenicity of recombinant adenovirus-human immunodeficiency virus vaccines in chimpanzees following intranasal administration. AIDS Res. Hum. Retrovir. 10:1443-1449. [DOI] [PubMed] [Google Scholar]
- 16.Montgomery, D. L., J. W. Shiver, K. R. Leander, H. C. Perry, A. Friedman, D. Martinez, J. B. Ulmer, J. J. Donnelly, and M. A. Liu. 1993. Heterologous and homologous protection against influenza A by DNA vaccination: optimization of DNA vectors. DNA Cell Biol. 12:777-783. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 18.Rowland-Jones, S., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, et al. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. (Erratum, 1:598.) [DOI] [PubMed] [Google Scholar]
- 19.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rowland-Jones, S. L., R. E. Phillips, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1992. Human immunodeficiency virus variants that escape cytotoxic T-cell recognition. AIDS Res. Hum. Retrovir. 8:1353-1354. [DOI] [PubMed] [Google Scholar]
- 21.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 22.Seth, A., I. Ourmanov, M. J. Kuroda, J. E. Schmitz, M. W. Carroll, L. S. Wyatt, B. Moss, M. A. Forman, V. M. Hirsch, and N. L. Letvin. 1998. Recombinant modified vaccinia virus Ankara-simian immunodeficiency virus gag pol elicits cytotoxic T lymphocytes in rhesus monkeys detected by a major histocompatibility complex class I/peptide tetramer. Proc. Natl. Acad. Sci. USA 95:10112-10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 24.van der Meide, P. H., R. J. Groenestein, M. C. de Labie, J. Heeney, P. Pala, and M. Slaoui. 1995. Enumeration of lymphokine-secreting cells as a quantitative measure for cellular immune responses in rhesus macaques. J. Med. Primatol. 24:271-281. [DOI] [PubMed] [Google Scholar]
- 25.Wilson, J. D., G. S. Ogg, R. L. Allen, P. J. Goulder, A. Kelleher, A. K. Sewell, C. A. O'Callaghan, S. L. Rowland-Jones, M. F. Callan, and A. J. McMichael. 1998. Oligoclonal expansions of CD8+ T cells in chronic HIV infection are antigen specific. J. Exp. Med. 188:785-790. [DOI] [PMC free article] [PubMed] [Google Scholar]