Abstract
Rhesus monkey rhadinovirus (RRV) is a close relative of Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8). RRV serves as an in vitro and an in vivo model for KSHV, and the mapping of its transcription program during lytic replication is significant since it represents de novo infection in the absence of stimulation with phorbol esters. Further, the RRV lytic system facilitates the making of recombinant viruses, and hence transcription profiling of the wild-type virus is important. Currently, the kinetics of lytic gene expression of RRV, the function of the RRV Orf50/Rta gene, and the presence of the RRV R8 and R8.1 genes are not known. This study details the transcription profile seen during RRV lytic replication and shows that RRV latency-associated nuclear antigen, viral FLIP (vFLIP), and vCyclin are transcribed during the RRV lytic phase. In addition, this study describes the identification of three new spliced products of the RRV Orf50, R8, and R8.1 genes, which are structural homologs of the KSHV Orf50, K8, and K8.1 genes, respectively. Characterization of the RRV Orf50 protein identifies it as a strong transcriptional transactivator capable of activating three early RRV promoters. Interestingly, the KSHV Orf50 transactivator can also activate these simian virus promoters, suggesting that there exists a conservation of gene function between the key transcription factors of KSHV and RRV.
Kaposi's sarcoma (KS) is a neoplastic vascular disorder occurring most frequently in patients with AIDS. Several lines of evidence strongly implicate Kaposi's sarcoma-associated herpesvirus (KSHV; human herpesvirus 8) as the causal agent of KS (15, 16, 23, 34, 35). KSHV was discovered in KS tissues by representative differential analysis (10) and was subsequently classified as a lymphotropic gamma-2 herpesvirus in the subfamily Rhadinovirinae. Since then, KSHV has been consistently identified in biopsy samples from patients with KS of all types including classical/Mediterranean (14), AIDS related (3), sporadic/endemic childhood (22), and renal transplant related (39). KSHV has also been identified in primary effusion lymphomas (7, 10), also known as body cavity-based lymphomas, as well as multicentric Castleman's disease (17, 47).
In 1997, the New England Regional Primate Research Center (NERPRC) isolated a gamma-2 herpesvirus from rhesus macaques that showed a high degree of sequence relatedness to human KSHV (12). This new virus was named rhesus monkey rhadinovirus (RRV), also called RV2-mac (11, 12) or MmuRHV-2. Sequence analysis of two distinct isolates by both the Oregon Regional Primate Research Center and NERPRC confirms that RRV is very closely related to KSHV by both sequence similarity and genomic organization (2, 46). Short stretches of related but phylogenetically distinct viral sequences have also been amplified from Macaca mulatta and Macaca nemestrina and are named RFHvMm and RFHvMn, respectively (6, 42, 45, 48). This second group of putative rhadinoviruses has not been cultured due to difficulty in finding a permissive lytic system. Hence, only a few viral genes have been sequenced, and these appear to be more closely related to KSHV than RRV. However, more detailed information on genomic organizations and sequence relatedness is needed. Short stretches of related sequence from African green monkeys (19) and from chimpanzees and gorillas (25) have also been amplified. Again, these viruses have not yet been cultured due to difficulty in establishing a permissive lytic system.
Manifestations of RRV infection parallel those of KSHV infection in humans. Wong et al. (55) have shown that rhesus macaques coinfected with simian immunodeficiency virus (SIV) and RRV (strain 17757) developed a hyperplastic lymphoproliferative disorder similar to Castleman's disease (55). Also, Mansfield et al. (32) demonstrated that experimental infection of macaques coinfected with RRV (strain 26-95) and SIV was associated with clinical lymphadenopathy characterized initially by paracortical hyperplasia and vascular hypertrophy/hyperplasia, which subsequently were replaced by marked follicular hyperplasia (32). In the most-severe cases, this follicular hyperplasia obliterated medullary sinuses and completely effaced the normal lymph node architecture. They also observed that three of four monkeys coinfected with RRV and SIV developed an arteriopathy. This arteriopathy appeared fundamentally similar to the vascular endothelial lesions observed in patients with KS and to the large-vessel arteritis induced in mice infected with murine gammaherpesvirus 68 (MHV-68).
Several laboratories have studied the kinetics of gene expression during KSHV reactivation using Northern hybridization analyses (44, 49, 50) and microarray technology (20, 37). These studies have aided our understanding of this virus's replication strategies. However, due to the unavailability of a naturally permissive lytic replication system for KSHV, these studies rely on the use of B-cell lines latently infected with KSHV (40). To activate the viral lytic cycle, these latent cell lines are artificially induced by phorbol esters such as 12-O-tetradecanoylphorbol-13-acetate (TPA) or n-butyrate (33, 40) and so lytic activity is a measure of viral reactivation rather than viral replication. Since typically only 25 to 30% of all latently infected B cells stimulated with TPA undergo lytic reactivation (8) and since it is estimated that only 5 to 10% of stimulated cells produce virus (40), there is always a background of latent gene expression. Conversely, in studies of latent transcription, there is a background of lytic transcripts due to the fact that 2 to 5% of these cells undergo spontaneous lytic reactivation (33).
The availability of the entire RRV genomic sequence, the ability to grow the virus lytically and to high titers, and the ability to infect rhesus macaques with RRV make RRV an excellent model system to study the contribution of individual genes to viral replication and persistence. Further, the RRV lytic system facilitates the making of recombinant viruses and is more amenable to the genetic engineering of mutant viruses than the KSHV system. Currently the kinetics of gene expression of RRV is unknown. The mapping of the RRV transcription program during lytic replication is relevant because it represents de novo infection in the absence of stimulation with phorbol esters, and transcription profiling of RRV can serve to corroborate the transcription seen with the artificially induced KSHV reactivation system. This paper reports the characterization of the kinetics of gene expression of RRV strain 26-95 following de novo infection of rhesus fibroblasts and the classification of genes into immediate-early, early, and late classes. We also report the identification of three new, previously unidentified, full-length cDNAs for the RRV Orf50, R8, and R8.1 genes. Whereas RRV Orf50 and R8 proteins are localized in the nucleus, R8.1 is localized in the cytoplasm. Furthermore, we demonstrate that RRV Orf50 is a potent transcriptional activator of several RRV promoters including the RRV R8 and Orf57 promoters, which can also be transactivated by KSHV Orf50.
MATERIALS AND METHODS
Cell culture.
An immortalized rhesus macaque skin fibroblast cell line was established by transduction of primary cells with a defective retrovirus that expresses human telomerase reverse transcriptase (5) and puromycin resistance. The retrovirus was generated from plasmid pLPChTERT (Clontech), and cells were infected as previously described (1). Transduced cells were selected by several passages in media containing 1 μg of puromycin (Sigma)/ml. Cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle medium-H with Gluta-max supplemented with 10% fetal bovine serum and 1 μg of puromycin/μl.
Virus and infections.
RRV isolate H26-95 (12) was used for infection at a multiplicity of infection of approximately 5. Infections were carried out for 0 (mock), 6, 12, 24, 36, 48, and 72 h. For the cycloheximide experiments, cycloheximide (Sigma; C-7698) was used at a concentration of 50 to 100 μg/ml, infections were carried out for 12 or 24 h, and cells were incubated simultaneously with cycloheximide and virus. For the phosphonoacetic acid (PAA) experiments, PAA (Sigma; P-6909) was used at a concentration of 50 to 100 μg/ml and infections were carried out for 30, 36, or 40 h. Initial experiments were carried out with PAA at a concentration of 200 μg/ml, and similar results were obtained.
RNA preparations and Northern hybridizations.
Total cellular RNA was extracted by using the RNA Stat-60 protocol (Tel-Test, Inc.), electrophoresed in 1.0 to 1.5% agarose-formaldehyde gels, and transferred to nylon membranes (Hybond N+; Amersham Pharmacia) by standard protocols. Probes were generated by random-primed oligonucleotide labeling (Random Primed DNA-labeling kit, catalog no. 1004760; Roche) of linearized plasmids or PCR products with [α-32P]dCTP (New England Nuclear). Hybridizations were carried out for 5 to 15 h at 68°C in QuikHyb (Stratagene) solution; hybridization temperature was lowered to 60°C for RNA probes. Membranes were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) for 15 min at room temperature and then washed once with 0.1× SSC-0.1% SDS for 30 min at 60°C and analyzed with a PhosphorImager (Molecular Dynamics).
Plasmids.
RRV Orf50, Orf65, Orf62, Orf73, and R1 genes were all cloned into pQE30 (Qiagen) between BamHI and SacI sites. The viral interleukin-6 (vIL-6) gene was cloned into pEF1-HisA (Invitrogen) between BamHI and XbaI sites, and the Orf57 gene was cloned into pEF1-HisC (Invitrogen) between BamHI and EcoRI sites. vCyclin, vFLIP, glycoprotein B (gB), pol, and viral interferon regulatory factor (vIRF) genes were all cloned with Topo-TA (Invitrogen) into pCDNA3. The Orf73 gene was subsequently subcloned into pSP72 (Promega) between BamHI and HindIII sites for in vitro transcription probe generation (see below). Human GAPDH (glyceraldehyde-3-phosphate dehydrogenase) control plasmid pHcGAP was used (52). For promoter activation studies, the promoter regions of five RRV genes were cloned into pGL2-Basic as follows. A 193-bp sequence (nucleotides [nt] 65155 to 65347) upstream of the RRV Orf50 gene was cloned by using KpnI and SacI, a 240-bp (nt 75324 to 75563) sequence upstream of the Orf57 gene was cloned by using KpnI and SacI, a 172-bp (nt 78671 to 78500) sequence upstream of the vIRF-1 gene was cloned by using KpnI and SacI, a 383-bp (nt 9300 to 9682) sequence upstream of the gB gene was cloned by using XhoI and HindIII, and, after rapid amplification of cDNA ends (RACE) experiments identified RRV R8, a 316-bp region (nt 68025 to 68340) upstream of the R8 gene was cloned by using KpnI and SacI. RRV Orf50 cDNA was cloned into pcDNA3 with the pcDNA3 TOPO kit (Invitrogen) to create pcDNA3-RRVOrf50. N-terminal green fluorescent protein (GFP) fusion proteins were created for RRV Orf50, R8, and R8.1 by cloning the cDNA for the genes in frame with the enhanced GFP (EGFP) gene from plasmid pEGFP-N1 (Clontech).
In vitro transcription.
The Orf73 gene was subcloned from pQE30-RRV73 into plasmid pSP72 between the BamHI and HindIII restriction enzyme sites. In vitro transcription was carried out with T7 RNA polymerase for the antisense strand-specific probe and with SP6 polymerase for the sense strand-specific probe by using the Riboprobe system (Promega) and [α-32P]dUTP.
5′ and 3′ RACE.
Poly(A) RNA was isolated from rhesus fibroblasts infected with RRV H26-95 for 48 h with an mRNA-Stat 30 kit (Tel-Test, Inc.). 5′ and 3′ RACE was carried out with the Smart RACE cDNA amplification kit (Clontech). Refer to Table 1 for exact genomic locations of the primers used. 5′ and 3′ RACE PCR products were then cloned into pCR2.1-TOPO (Invitrogen), and individual colonies were sequenced. For cDNA cloning of the individual messages, poly(A) mRNA was isolated, first-strand cDNA was made by reverse transcription, and primers specific to each message were used to amplify the specific cDNA clone by PCR.
TABLE 1.
Primers used for 5′ and 3′ RACE for mapping the RRV Orf50, R8, and R8.1 transcripts
| Primer no. | RACE | RRV genomic location (nt) | Sites mapped (nt) |
|---|---|---|---|
| 1 | 5′ | 66760 | Orf50 transcription start site (68285), Orf50 exon 1 (65348-65368), Orf50 exon 2 (66312) |
| 2 | 3′ | 67941 | R8 exon 1 (68341-68787), R8 exon 2 (68902-69024), R8 exon 3 (69146-69280), Orf50 transcription stop (68171) |
| 3 | 5′ | 68400 | R8 exon 1 (68341), Orf50 exon 2 (68024) |
| 4 | 3′ | 68342 | R8 exon 1 (68787), R8β exon 1 (69024), R8 exon 2 (68902-69024), R8 exon 3 (69146-69280) |
| 5 | 5′ | 68972 | R8 exon 2 (68902), R8 exon 1 (68341-68787) |
| 6 | 3′ | 69200 | R8 exon 3 (69280), R8.1 exon 1 (69423-69865), R8.1 exon 2 (69951-70366) |
| 7 | 5′ | 70359 | R8.1 exon 1 (69423), R8 exon 3 (69146-69280), R8 exon 2 (69024) |
| 8 | 3′ | 69423 | R8.1 exon 2 (70366) |
In vitro transcription and translation.
The cDNA sequences for RRV Orf50, R8, and R8.1 were cloned into the pcDNA3 expression vector under the control of the cytomegalovirus (CMV) and T7 promoters. The T7 in vitro transcription and translation kit from Promega was used along with [35S]methionine to radiolabel the in vitro-synthesized proteins. The manufacturer's protocol was followed.
Proteins from each sample were then electrophoresed in a 10% SDS-polyacrylamide gel, dried, and exposed to a phosphorimager.
Western blotting.
The cDNA sequences for RRV Orf50, R8, and R8.1 were tagged with the coding sequence for the AU1 epitope (Covance) at the C-terminal end for Orf50 and at the N-terminal end for R8 and R8.1 (after the putative signal peptide) by PCR. The PCR products were cloned into the pcDNA3 expression vector downstream of the CMV promoter. Cos-1 cells were transfected with 10 μg of each plasmid as well as a pEGFPN1 vector control that has a CMV promoter upstream of the EGFP gene with Superfect (Qiagen). Cells were harvested 48 h posttransfection with radioimmunoprecipitation assay buffer (150 mM NaCl, 1% NP-40, 50 mM Tris [pH 8], 0.5% sodium deoxycholate, 0.1% SDS) and subjected to SDS-polyacrylamide gel electrophoresis (PAGE). The gels were transferred to nitrocellulose, and the Western blots were probed with an anti-AU1 antibody. Immunoblot detection was performed with the enhanced chemiluminescence ECL kit (Amersham).
Promoter activation studies.
The cDNA for RRV Orf50 was tagged with the coding sequence for the AU1 epitope (Covance) at the C-terminal end and cloned into pCDNA3.1. The promoters for five RRV genes were cloned into pGL2-Basic as described above. Three micrograms of each promoter construct was transfected into 293 cells by the SuperFect protocol (Qiagen) with 3 μg of either pcDNA3, pcDNA3-RRVOrf50, or pcDNA3-KSHVOrf50. One microgram of a β-galactosidase (β-Gal)-expressing plasmid was cotransfected with each sample for transfection efficiency normalization. Each promoter was analyzed in triplicate in two separate experiments. For comparison of levels of promoter activation, each transactivator or empty pcDNA3 was transfected with pGL2-Basic. At 48 h posttransfection, cells were harvested by scraping and lysed using 1× reporter lysis buffer (Promega) and freeze-thawed. Samples were centrifuged at 13,000 × g for 3 min, and supernatants were collected and analyzed for luciferase activity and β-Gal activity with the luciferase assay system (Promega) and Galacto-Star kits (Tropix).
RESULTS
Immediate-early transcripts of RRV: Orf50 and Orf57 genes.
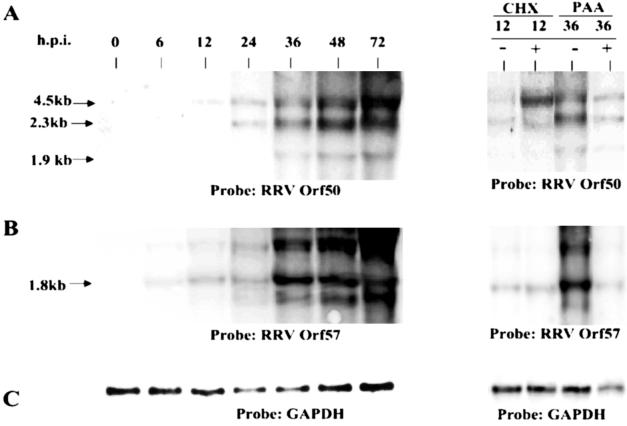
We infected rhesus fibroblast cells with RRV at a multiplicity of infection of 5 and harvested RNA at different time points postinfection (p.i.). The RNA samples were electrophoresed through 1.0% agarose gels, transferred to nylon membranes, and hybridized with a radioactive double-stranded DNA probe to specific genes. The KSHV Orf50 gene (Rta/ART/LytA) has been identified as an immediate-early gene in Northern analyses (50, 57) and microarray studies (20, 37). Further, Orf50 has been shown to be a viral transactivator sufficient for reactivation from latency in KSHV (18, 29, 30, 49) and in MHV-68 (56). Northern hybridization analysis demonstrated that the RRV Orf50 gene transcript is expressed between 6 and 12 h p.i. (Fig. 1A). The largest transcript of the Orf50 gene, approximately 4 to 5 kb, was the first RNA species to appear at 12 h p.i. This is likely a polycistronic message encoding Orf50, R8, and R8.1, as has been shown for KSHV (57). Two slightly higher-mobility species at approximately 2.3 and 1.9 kb appeared at 24 and 36 h, respectively (Fig. 1A). These likely represent messages for Orf50 alone as well as for Orf50 and R8 since we have isolated cDNA clones with these two different messages. Transcription of the Orf50 gene was not inhibited by the presence of cycloheximide (100 μg/ml) or PAA treatment (Fig. 1A). Three different time points were used for the PAA experiments (30, 36, and 40 h p.i.) with no visible difference in the results. Thus, the Orf50 gene is an immediate-early gene of RRV, and transcription of the Orf50 gene likely does not terminate by 72 h p.i.
FIG. 1.
Northern analysis of immediate-early gene expression. Total RNA from infected cells was prepared at the indicated times after infection and electrophoresed through 1.0% agarose gels, transferred to nylon membranes, and hybridized with a radioactive double-stranded DNA probe containing either the Orf50 (A) or the Orf57 (B) gene sequence. To ensure equivalent loading, membranes were also hybridized with a GAPDH probe (C). Orf50 and Orf57 gene expression was analyzed at various times after infection in the absence of metabolic inhibitors and at 12 h p.i. in the presence or absence of cycloheximide (CHX) or at 36 h p.i. in the presence or absence of PAA. Arrows indicate sizes of the transcripts that would be predicted from the sequence of the specific RRV gene.
Another immediate-early gene transcribed during the lytic phase is the RRV Orf57 gene. Here, the 1.8-kb RRV Orf57 gene transcript was first detected at 6 h p.i. (Fig. 1B), appears to precede the RRV Orf50 gene transcript, and was also not inhibited by cycloheximide. Similar to what was found for the RRV Orf50 gene, we do not observe a termination in RRV Orf57 gene transcription before 72 h p.i. We find that, in the RRV system, unlike the KSHV system, the immediate-early Orf57 gene transcript appears to precede the immediate-early Orf50 gene message since Northern analyses were performed on the identical RNA samples but with different probes. Although this needs to be studied in further detail, it may imply that there is a potential role for Orf57 in reactivation of the viral lytic cycle in conjunction with Orf50 or that the Orf50 mRNA is transcribed first but, because of low abundance or instability, is not detected first. Also, since Orf57 has been shown to be a posttranscriptional regulator (24), it might serve to increase Orf50 mRNA levels at the very onset of viral replication.
Identification of the 5′ and 3′ ends of the RRV Orf50, R8, and R8.1 gene transcripts by RACE.
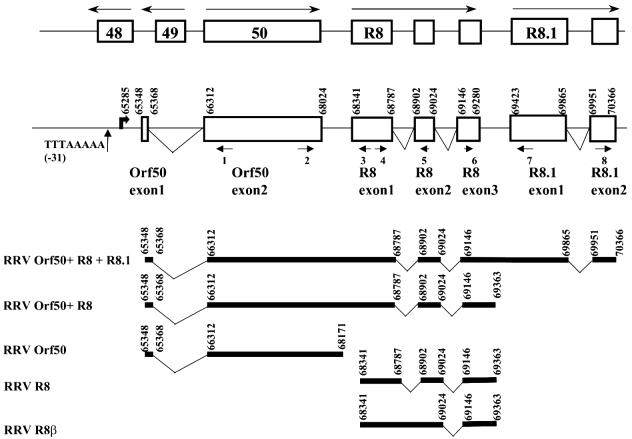
To define the 5′ end of the Orf50 gene message, we employed 5′ RACE using three separate primers internal to the Orf50 gene sequence (Fig. 2). Agarose separation of RACE PCR products confirmed that a single species was present in each reaction mixture and that no products were found when reverse transcriptase was left out of the reaction mixture (data not shown). Sequence analysis of the products of all three reactions showed the existence of a small exon 5′ to the genomic sequence (Fig. 2). Exon 1 of the Orf50 gene spans nt 65348 to 65368, and exon 2 begins at nt 66312. The intron between these two exons is large and contains the entire coding region of the Orf49 gene (2). This is similar to the situation in KSHV, where the KSHV Orf50 gene is also encoded by two exons and the first exon is similarly located upstream of the Orf49 gene (49). With the newly discovered exon included, an amino acid homology comparison between KSHV and RRV Orf50 proteins revealed 39% identity and 55% similarity, which is a significant improvement over the original genomic assignment for the RRV Orf50 sequence, which showed 36% identity and 51% similarity to the KSHV sequence.
FIG. 2.
Splicing pattern of the RRV Orf50, R8, and R8.1 transcripts. The genomic locations of the Orf50, R8, and R8.1 genes for RRV are shown as boxes. Nucleotide numbers for the RRV gene splice donor and splice acceptor sites are shown. Small arrows under genes, locations and numbers of primers used for 5′ and 3′ RACE. The exact locations of the primers are listed in Table 1. A weak putative TATA box is present at a position 31 bp upstream of the Orf50 gene transcription start site. The multiply spliced transcripts of all three genes are also shown as a schematic diagram. Long arrows above the boxes represent the direction of gene transcription.
When we used 3′ RACE (Table 1) to identify the 3′ end of the Orf50 gene, we discovered two new spliced gene transcripts. One of these was a new, previously unknown transcript, which we have named R8, since its product shows homology to KSHV K8 (also known as KbZip) at the amino acid level (28, 57). The R8 gene is multiply spliced, and the splicing pattern appears similar to that of the KSHV K8 gene and is shown in Fig 2. cDNA cloning revealed that the R8 gene was transcribed by itself as well as bicistronically on a transcript encoding Orf50 (Fig. 2). Since we found transcripts that only contained the R8 gene and not the Orf50 gene, this suggests that a promoter for the R8 gene may be located between the Orf50 and R8 genes. A second R8β gene transcript, similar to the KSHV K8β gene transcript (28, 57), was also identified (Fig. 2). The R8 and K8 proteins show 39% identity and 53% similarity at the amino acid level.
We also discovered a second new spliced transcript (Fig. 2) of another previously unknown gene, which we have named R8.1, since it shows homology to the KSHV K8.1 glycoprotein gene (9, 27). The R8.1 and K8.1 proteins show 26% identity and 43% similarity at the amino acid level. A more detailed characterization of the proteins encoded by the Orf50, R8, and R8.1 transcripts is provided below.
Early transcripts of RRV.
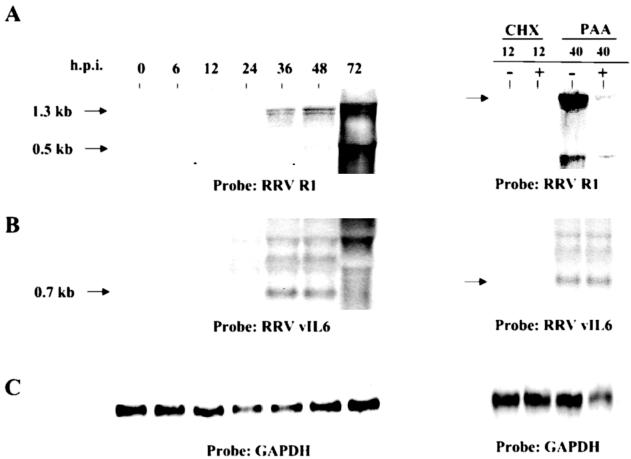
The RRV R1 gene has been previously studied in the context of cellular transformation and lymphocyte signaling (2, 11). Northern hybridizations for the R1 gene revealed that it was first transcribed between 12 and 24 h p.i. (Fig. 3A). Based on its time of appearance, which is after immediate-early gene transcription, and sensitivity to cycloheximide at 12 (Fig. 3A) and 20 h (not shown) and the fact that PAA fails to completely abolish its expression at 40 h p.i., we characterize it as an early gene (Fig. 3A). Transcript sizes for the R1 gene are 1.3 and 0.5 kb, and the R1 gene sequence is 1,272 bp, so the 1.3-kb transcript likely represents the full-length mRNA.
FIG. 3.
Northern analysis of early gene expression. Total RNA from infected cells was prepared at the indicated times after infection and electrophoresed through 1.0% agarose gels, transferred to nylon membranes, and hybridized with a radioactive double-stranded DNA probe containing either the RRV R1 (A) or the RRV vIL-6 (B) gene sequence (B). To ensure equivalent loading, membranes were also hybridized with a GAPDH probe (C). R1 and vIL-6 gene expression was analyzed at various times after infection in the absence of metabolic inhibitors and at 12 h p.i. in the presence or absence of cycloheximide (CHX) or at 40 h p.i. in the presence or absence of PAA. Arrows indicate sizes of transcripts that would be predicted from the sequences of the specific RRV genes.
Like KSHV, RRV carries many genes encoding proteins with cellular homologs including vIL-6. RRV vIL-6 has been shown to have IL-6-like activity in cultured cells (2, 21) and presumably has many of the same properties as KSHV vIL-6. The expression patterns of RRV vIL-6 reveal that transcription initiates between 12 and 24 h p.i (Fig. 3B), with the major transcript being approximately 700 bp long (likely corresponding to the 623-bp genomic sequence). Based on its time of appearance and sensitivity to cycloheximide at 12 (Fig. 3B) and 20 h (not shown) and the fact that PAA fails to completely abolish its expression at 40 h p.i., we characterize it as an early gene (Fig. 3B). We have also observed that the RRV DNA polymerase gene is expressed as an early lytic gene by Northern blot analyses (data not shown).
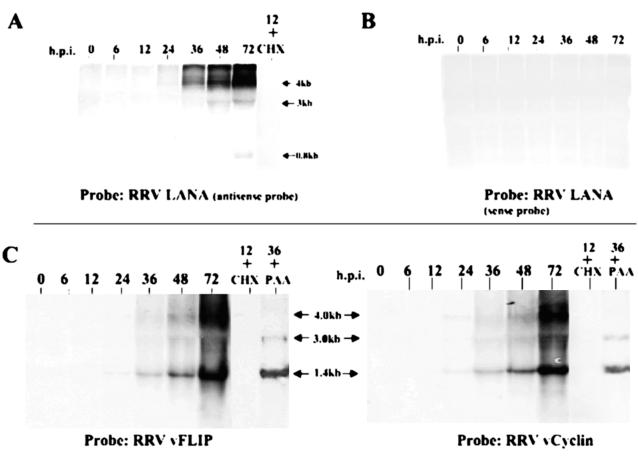
The Orf73 gene of KSHV and RRV encodes the latency-associated nuclear antigen (LANA). While RRV LANA is relatively uncharacterized, KSHV LANA has been extensively studied (13, 38). Commonly used as a serological marker, LANA has a number of functions in the viral life cycle. The LANA protein is known to maintain latency by preserving the viral episome during cell division by being tethered to the mitotic chromosomes (4). Expression of the KSHV LANA gene, which is classically described as a latent gene, has been shown by Paulose-Murphy et al. to be induced during reactivation (37). We sought to investigate if transcription of the LANA gene can be detected during lytic RRV replication. Northern hybridizations were performed with both sense and antisense riboprobes (Fig. 4A and B). The results depicted in Fig. 4A demonstrated that only the antisense riboprobe produced a signal and that hybridization to both 3.0- and 4.0-kb transcripts occurred, suggesting that both these transcripts represent sense strand mRNAs capable of expressing LANA. These transcripts were sensitive to cycloheximide and insensitive to PAA (data not shown). We also performed Northern hybridizations probing for RRV vFLIP and vCyclin genes (Fig. 4C) and noticed patterns very similar to that seen with KSHV vFLIP and vCyclin genes, possibly because these genes are expressed on bicistronic or tricistronic transcripts (including the Orf73 gene), as has been demonstrated for an analogous transcription unit in KSHV (13, 44, 51). Transcription in RRV was initiated by 24 h p.i. with a predominant RNA species at 1.4 kb and minor species of 3.0 and 4.0 kb. The combined genomic sequence size for these two genes is 1,347 bp, which corresponds well with the 1.4-kb transcript. The time of appearance of the vCyclin and vFLIP gene transcripts and their sensitivity to cycloheximide at 12 h and insensitivity to the presence of PAA at three different time points (30, 36, and 40 h p.i.) (Fig. 4C) classify them as early or delayed-early genes in the lytic replication cycle. An assessment of the cycloheximide sensitivity of these transcripts at a time point after 24 h could not be performed due to the cell toxicity effects of this drug. Increases in the amounts of RRV LANA, vFLIP, and vCyclin gene transcripts transcribed during de novo infection of rhesus fibroblasts may represent a characteristic of viral replication in rhesus fibroblasts, which may differ from the kinetics of gene expression observed during KSHV viral reactivation in B cells.
FIG. 4.
Northern analysis of LANA, vFLIP, and vCyclin gene expression. Total RNA from infected cells was prepared at the indicated times after infection and electrophoresed through 1.0% agarose gels, transferred to nylon membranes, and probed as described in Materials and Methods. To ensure equivalent loading, membranes were stripped and rehybridized with a GAPDH probe (data not shown). (A) Orf73 gene expression was analyzed at various times after infection in the absence of metabolic inhibitors. The membrane was hybridized with a radioactive sense-specific riboprobe. (B) An identical membrane was hybridized with an antisense-specific riboprobe. (C) vFLIP gene expression was analyzed at various times after infection in the absence of metabolic inhibitors and at 12 and 36 h p.i. in the presence of cycloheximide (CHX) or PAA, respectively. vCyclin gene expression was analyzed at various times after infection in the absence of metabolic inhibitors and at 12 and 36 h p.i. in the presence of cycloheximide or PAA, respectively. Arrows indicate sizes of the transcripts that would be predicted from the sequences of the specific RRV genes.
Late transcripts of RRV.
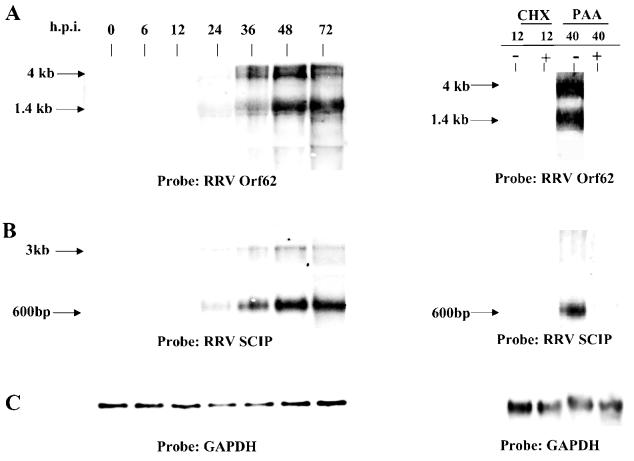
In the original sequencing of KSHV, Orf62 was described as a protein involved in assembly and DNA maturation based on the homology of this gene to other herpesvirus genes (43). However, recent structural studies have shown that KSHV Orf62 is a triplex member, a component of the capsid (36). The RRV Orf62 gene transcript first appeared at 24 h p.i. (Fig 5A) and was abolished by PAA, identifying the Orf62 gene as a late gene. There were two transcripts of 1.4 and 4 kb. The gene for Orf62 is 995 bp long, so the 1.4-kb transcript probably represents the Orf62 mRNA. A second gene, identified in structural studies as encoding a capsid protein, is the KSHV Orf65 gene. The Orf65 gene encodes a small capsomer-interacting protein (SCIP) (36), which is a homolog of the Epstein-Barr virus (EBV) small viral capsid antigen. Northern hybridizations for the RRV SCIP gene revealed two transcripts, beginning at 24 h p.i. (Fig. 5B). The major transcript was 600 bp, which corresponds well with the predicted coding sequence of 509 bp. Transcription was completely abolished when infected cells were treated with cycloheximide for 12 h (assessment after 24 h could not be performed due to the cell toxicity of the drug) and PAA (Fig. 5B), thus confirming that RRV SCIP is encoded by a late gene, similar to KSHV SCIP (36). In addition, we have observed that RRV vIRF and RRV gB genes are also expressed as late genes (data not shown).
FIG. 5.
Northern analysis of late gene expression. Total RNA from infected cells was prepared at the indicated times after infection and electrophoresed through 1.0% agarose gels, transferred to nylon membranes, and hybridized with a radioactive double-stranded DNA probe containing either the RRV Orf62 gene sequence (A) or the RRV SCIP gene sequence (B). To ensure equivalent loading, membranes were also hybridized with a GAPDH probe (C). Orf62 and SCIP gene expression was analyzed at various times after infection in the absence of metabolic inhibitors and at 12 and 40 h p.i. in the presence or absence of cycloheximide (CHX) or PAA, respectively. Arrows indicate sizes of the transcripts that would be predicted from the sequences of the specific RRV genes.
We have mapped the transcription profile of RRV upon de novo lytic infection of rhesus fibroblasts in the absence of TPA or n-butyrate. Although the transcription program of the KSHV lytic cycle was studied under artificially induced conditions, the RRV de novo lytic transcription program, with a few exceptions, is similar to that of KSHV. Thus, the experiments described here corroborate the data obtained with TPA induction and validate the use of the RRV system.
RRV Orf50 and RRV R8 proteins are localized in the nucleus, whereas RRV R8.1 is localized in the cytoplasm.
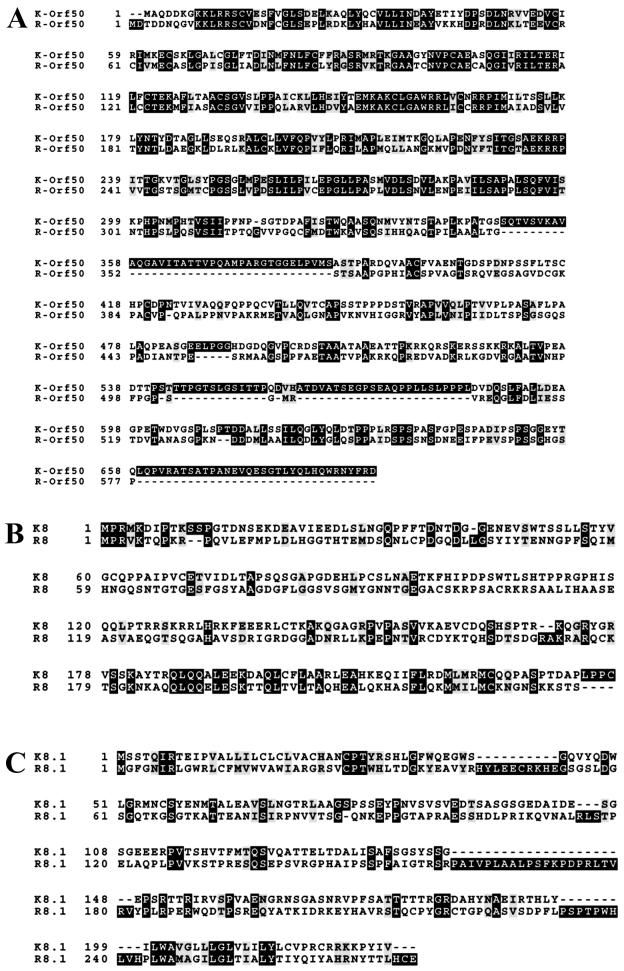
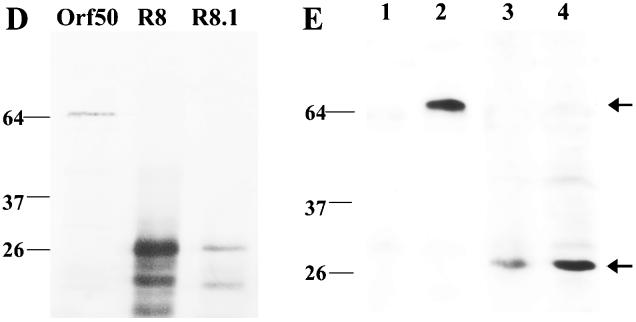
The three newly identified transcripts in Fig. 2 encode previously unidentified proteins RRV Orf50, RRV R8, and RRV R8.1, which show a high degree of sequence similarity to the Orf50 (Fig. 6A ), K8 (Fig. 6B), and K8.1 proteins of KSHV (Fig. 6C). Figure 6A to C show the alignments of the respective KSHV and RRV proteins by using the ClustalW alignment program. To determine the mobility of these newly identified RRV proteins, we added the coding sequence for the AU1 epitope tag to the cDNA of these proteins by PCR and cloned the cDNA into pCDNA3 vectors containing CMV and T7 promoters. All three proteins were synthesized with the in vitro transcription and translation kit from Promega along with [35S]methionine to radiolabel the proteins. The proteins were subjected to SDS-PAGE, and the gel was dried and exposed to a phosphorimager (Fig. 6D). In addition, each of the plasmids was transfected into Cos-1 cells, and cells were harvested 48 h posttransfection. Cell extracts were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with an anti-AU1 antibody as shown in Fig. 6E. The RRV Orf50 protein ran at a mobility of ∼64 kDa, which is close to its predicted size (Fig. 6D and E). Likewise, the RRV R8 and R8.1 proteins ran at a mobility of ∼30 kDa, which is close to the predicted sizes of these proteins (Fig. 6D and E).
FIG. 6.
Expression of RRV Orf50, R8, and R8.1. (A to C) Homology between the KSHV Orf50 and RRV Orf50 proteins (A), K8 and R8 proteins (B), and K8.1 and R8.1 proteins (C). The alignments of the respective KSHV and RRV proteins were performed with the ClustalW alignment program. Identical amino acids are highlighted in black, and similar amino acids are highlighted in grey. Dashes, amino acids missing in one protein sequence. (D) The RRV Orf50, R8, and R8.1 mRNAs were transcribed and translated in vitro with [35S]methionine, and the proteins were subjected to SDS-PAGE, dried, and exposed to a phosphorimager. RRV Orf50 ran at a mobility of ∼64 kDa, and R8 and R8.1 ran at a mobility of ∼30 kDa. (E) pCDNA3-RRV Orf50, pCDNA3-R8, pCDNA3-R8.1, and a pEGFPN1 control vector were each transfected into Cos-1 cells, and cells were harvested 48 h posttransfection. Cell extracts were subjected to SDS-PAGE, transferred to nitrocellulose, and probed with an anti-AU1 antibody. Lane 1, pEGFPN1 control-transfected cells; lane 2, RRV Orf50 protein; lane 3, RRV R8 protein; lane 4, RRV R8.1 protein. RRV Orf50 ran at a mobility of ∼64 kDa, and R8 and R8.1 ran at a mobility of ∼30 kDa.
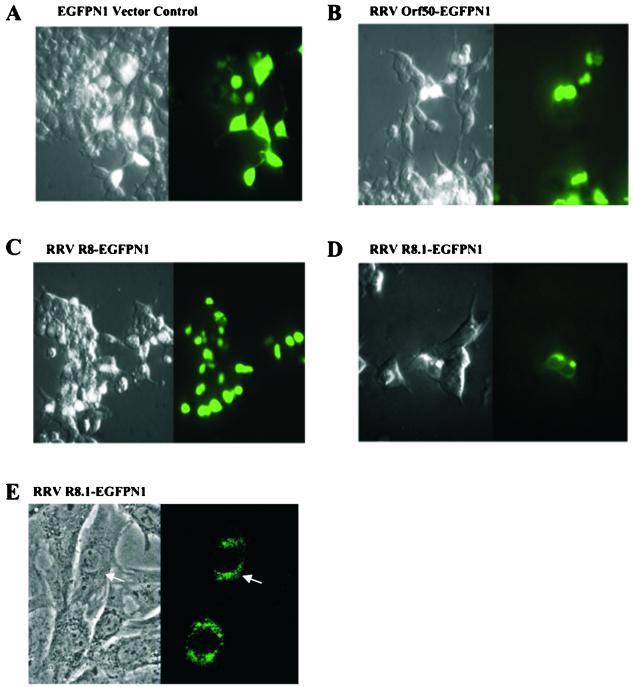
To determine the localization of the three newly identified proteins, we created N-terminal fusions of RRV Orf50, R8, and R8.1 to EGFP with the EGFPN1 vector. The constructs were transfected into 293 cells, and expression and localization of the fusion proteins were analyzed 48 h posttransfection. The RRV Orf50-EGFP (Fig. 7B) and RRV R8-EGFP (Fig. 7C) fusion proteins were localized in the nucleus, while the RRV R8.1-EGFP fusion protein was primarily seen in the periplasm and cytoplasm of the cell (Fig. 7D and E).
FIG. 7.
Cellular localization of RRV Orf50, R8, and R8.1 proteins. The Orf50, R8, and R8.1 cDNAs were fused in frame with the N terminus of GFP in the pEGFPN1 expression vector. 293 cells were transfected with either the EGFPN1 vector alone (A), the RRV Orf50-EGFPN1 fusion plasmid (B), the RRV R8-EGFPN1 fusion plasmid (C), or the RRV R8.1-EGFPN1 fusion plasmid (D). (E) Higher magnification (×40) of the RRV R8.1-EGFPN1 fusion plasmid expressed in rhesus fibroblasts. The EGFPN1 protein was expressed in both the nucleus and the cytoplasm, while the RRV Orf50-EGFPN1 and RRV R8-EGFPN1 proteins were targeted to the nucleus in contrast to the RRV R8.1-EGFPN1 protein, which was targeted to the periplasm and cytoplasm of the cell.
RRV Orf50 is a potent transcriptional activator of RRV promoters.
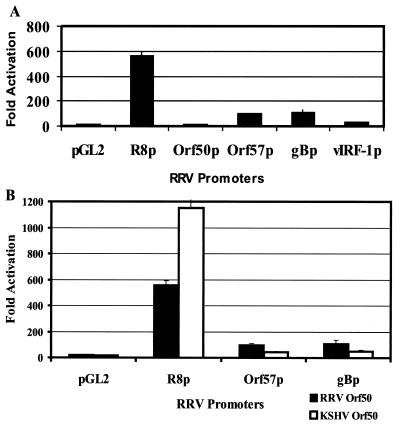
Since the RRV Orf50 gene is expressed as an immediate-early gene during RRV lytic replication and since RRV Orf50 shows a high degree of homology to the KSHV Orf50 transcriptional transactivator, we investigated whether RRV Orf50 could transactivate a set of five RRV promoters: Orf57p, Orf50p, R8p, gBp, and vIRF-1p. The cDNA for RRV Orf50 was tagged with the coding sequence for the AU1 epitope (Covance) at the C-terminal end and cloned into pCDNA3.1. The vector was transfected into 293 cells, and, at 48 h posttransfection, expression of the protein was analyzed by Western blotting of cell extracts using an AU1 antibody (Covance), which revealed that RRV Orf50 ran as an ∼64-kDa protein (data not shown), similar to its size when synthesized in vitro (Fig. 6D). The five promoter plasmids were cloned into the pGL2basic vector (Promega) upstream of a luciferase reporter gene. The pCDNA3-RRV Orf50 expression vector was cotransfected into 293 cells with the individual promoter plasmids and a β-Gal expression vector. Transfections were done in triplicate and normalized to β-Gal activity to control for transfection efficiency. As shown in Fig. 8A, RRV Orf50 is a potent transcriptional activator capable of activating the R8 promoter greater than 500-fold and of activating the Orf57 and gB promoters greater than 100-fold. The vIRF promoter was slightly activated, but the Orf50 promoter itself was not activated significantly. This is the first finding that shows that RRV Orf50 is a transcriptional activator.
FIG. 8.
Conservation of gene function between the human and rhesus Orf50 transcriptional transactivators. (A) RRV Orf50 is a transcriptional transactivator of the RRV Orf57, R8, and gB promoters. The promoters for five RRV genes, the Orf57, R8, gB, vIRF-1, and Orf50 genes, were cloned upstream of a luciferase reporter construct. The promoter plasmids were transfected with or without the pCDNA3-RRV Orf50 expression vector into 293 cells, and luciferase activity was assayed 48 h posttransfection. All assays were performed in triplicate and normalized to β-Gal to control for transfection efficiency. RRV Orf50 could transactivate the R8 promoter (R8p) greater than 500-fold and the Orf57 (Orf57p) and gB (gBp) promoters greater than 100-fold. It did not significantly activate its own promoter (Orf50p) or that of vIRF-1 (vIRF-1p). (B) KSHV Orf50 can transcriptionally transactivate RRV promoters. The RRV R8, Orf57, and gB promoter plasmids were transfected with or without the pCDNA3-RRV Orf50 expression vector into 293 cells, and luciferase activity was assayed 48 h posttransfection. All assays were performed in triplicate and normalized to β-Gal to control for transfection efficiency. These promoters were significantly transactivated by both the KSHV and RRV Orf50 proteins.
Since the RRV and KSHV Orf50 proteins have a high degree of homology at the amino acid level, we decided to investigate whether the KSHV Orf50 protein could activate the five RRV promoter plasmids tested in the assay described above. A pCDNA3-KSHV Orf50 expression construct was transfected into 293 cells along with the five promoter plasmids and β-Gal. Analogous to RRV Orf50, the KSHV Orf50 protein could transactivate the RRV R8, Orf57, and gB promoters significantly (Fig. 8B) but not the RRV Orf50 or vIRF promoters. Thus, there is a conservation of gene function that is shared between the human and the simian virus, which further validates the use of RRV as a model system to study events in the KSHV life cycle.
DISCUSSION
Studies on KSHV lytic gene transcription have used TPA to reactivate virus from latently infected cells in order to study the kinetics of gene expression during KSHV replication. Since only a small percentage of TPA-stimulated cells are typically reactivated under these conditions there is always a high background of latently expressed transcripts. In the RRV infection system, there is no background of latent infection, and we have monitored the RRV transcription program during de novo infection rather than reactivation. This system has enabled us to study the expression profiles of a number of lytically expressed genes from various transcriptional classes without the interference of latent gene expression.
The immediate-early genes expressed during the RRV life cycle are the Orf50 and Orf57 genes, similar to what is seen with KSHV reactivation. Our findings with respect to early genes such as the vIL-6 and DNA polymerase genes are mostly consistent with previously published reports on KSHV, but we have also observed some differences. We observed that a cluster of genes defined as latent in KSHV, which contains Orf73 (LANA), vCyclin, and vFLIP genes, were expressed lytically in the RRV culture system. A polycistronic mRNA containing all three of these genes has been observed in KSHV (13, 44), and our results suggest that a similar transcript may exist in RRV. While the majority of reports on the KSHV LANA gene indicate that it is strictly a latent gene because there is no significant induction of expression following TPA treatment of latently infected B cells, a small number of publications have reported a slight induction of the LANA gene transcript following TPA stimulation (13, 20, 37, 51). Further, Sun et al. have reported, by using Northern analysis, that vFLIP (1.8-kb transcript) and LANA (4.5-kb transcript) genes are both up-regulated during the KSHV lytic cycle at 13 to 20 h after n-butyrate stimulation (50). In MHV-68, the Orf73 gene has been identified as a latent as well as an immediate-early lytic gene transcript (41) and the cyclin gene has also been shown to be expressed during the lytic cycle (53). Finally, in EBV, the EBNA-1 transcript is transcribed when EBV-transformed Burkitt's lymphoma cells enter the lytic cycle (26). Thus, reports on other closely related viruses suggest that LANA homologs can be expressed during the lytic cycle. While our experiments clearly demonstrate that latent genes, such as the LANA, vCyclin, and vFLIP genes, are expressed during the lytic cycle of RRV, we cannot rule out the possibility that, although RRV and KSHV are closely related viruses, they may show differences in their transcription programs or that the transcription program following de novo infection of fibroblasts differs from that seen during viral reactivation.
Late genes identified in this study include the vIRF-1, Orf62, Orf65, and gB genes. KSHV vIRF-1 is up-regulated 24 h after TPA induction (37). An earlier study showed that the K9/vIRF-1 gene was transcribed during both the latent and lytic cycles (44). Results presented here show that the RRV vIRF-1 gene is transcribed during the lytic cycle similarly to the KSHV vIRF gene (44). Our findings regarding the Orf62, Orf65, and gB genes are consistent with studies done on these genes in other herpesviruses. In agreement with recent structural studies by Nealon et al. (36), we find that, in RRV, Orf62 gene transcription is dependent on viral DNA replication, and Orf62 is thus a late gene.
We have reported the identification of three new spliced genes in RRV, which are the RRV Orf50, RRV R8, and RRV R8.1 genes. Discovery of a 5′ exon in the Orf50 gene is consistent with what is known about the splicing pattern of Orf50 mRNAs of other gamma-2 herpesviruses, including KSHV (57), MHV-68 (56), and herpesvirus saimiri (HVS) (54). The KSHV Orf50 mRNA has a 100-bp 5′ exon, followed by a second exon encoding the remaining 2,751 bp of the Orf50 gene sequence (57). Similarly in RRV, the Orf50 mRNA has an 83-bp upstream exon followed by a 1,861-bp second exon. Interestingly, in HVS, the Orf50 gene homolog is expressed in both spliced and unspliced forms (54), although no function has been ascribed to the unspliced form. The MHV-68 Orf50 transcript has a 35-bp first exon and a 1,712-bp second exon (56). In contrast gamma-1 herpesvirus EBV encodes an Orf50 homolog, BRLF1, whose cDNA is identical to that of the genomic open reading frame (31). We have demonstrated that RRV Orf50, similar to its gamma herpesvirus counterparts, is localized to the nucleus and is a potent transcriptional activator capable of activating three RRV promoters significantly. We have also shown that the functionality between KSHV Orf50 and RRV Orf50 is conserved because the KSHV Orf50 gene product can also activate these same RRV promoters to a significant extent as well. KSHV Orf50 significantly activates the RRV R8 promoter. However, it does not contain the KSHV Orf50 binding sequences reported for the KSHV K8 promoter (29, 30). Hence, further analysis of this promoter remains to be performed.
Finally we report the identification of two other new RRV proteins, R8 and R8.1, which had not been identified from the genomic sequence due to the fact that their messages are highly spliced. The R8 protein is localized in the nucleus and shows 39% identity and 53% similarity at the amino acid level to the K8 bZip protein of KSHV. The R8.1 protein is localized in the cytoplasm and shows 26% identity and 43% similarity at the amino acid level. The R8.1 protein has two potential glycosylation sites (NXS/T).
In conclusion, we have mapped the transcription program for RRV with respect to 12 genes, which all have corresponding homologs in KSHV. We find that transcription of the LANA, vCyclin, and vFLIP genes occurs in the context of this de novo lytic system. We have identified two new, previously unidentified, spliced transcripts of RRV and have named them R8 and R8.1. We have also determined the full-length cDNA for RRV Orf50, which includes a newly identified exon that is required to code for a functionally competent transcriptional transactivator. We show that the RRV Orf50 protein is nuclear and is a potent transcriptional transactivator and that there is conservation of gene function between the KSHV and RRV Orf50 genes. The elucidation of these spliced transcripts and proteins is useful for determining the contribution of individual genes to the viral life cycle and will further assist in the design of mutant and recombinant RRVs. We anticipate that these studies will further the understanding of gamma-2 herpesvirus lytic replication and help to substantiate and further develop RRV as a tractable model to study the KSHV viral life cycle.
Acknowledgments
We thank Stuart Krall for assistance with the viral infections. We are grateful to Ronald Desrosiers for giving us RRV strain 26-95 and rhesus fibroblasts. We thank members of the Damania lab for invaluable discussions and manuscript reading, R. Gosselin for technical assistance, and Nancy Raab-Traub for reagents and manuscript reading.
S. DeWire was supported by NIH training grant NIH5T32GM07092-26. This work was supported in part by grants from the V Foundation, UNC-CFAR, AHA, and NIH (R01CA096500) (to B.D.) and the A. D. Williams Fund of the Medical College of Virginia (to M.A.M.).
REFERENCES
- 1.Abbate, I., F. Dianzani, O. Turriziani, G. Antonelli, G. D'Offizi, V. Galati, M. Pierdominici, F. Pandolfi, and M. R. Capobianchi. 2001. Changes in host cell molecules acquired by circulating HIV-1 in patients treated with highly active antiretroviral therapy and interleukin-2. AIDS 15:11-16. [DOI] [PubMed] [Google Scholar]
- 2.Alexander, L., L. Denenkamp, A. Knapp, M. Auerbach, S. Czajak, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma herpesvirus and rhesus rhadinovirus isolate17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambroziak, J. A., D. J. Blackbourn, B. G. Herndier, R. G. Glogau, J. H. Gullett, A. R. McDonald, E. T. Lennette, and J. A. Levy. 1995. Herpes-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science 268:582-583. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Bodnar, A. G., M. Ouellette, M. Frolkis, S. E. Holt, C. P. Chiu, G. B. Morin, C. B. Harley, J. W. Shay, S. Lichtsteiner, and W. E. Wright. 1998. Extension of life-span by introduction of telomerase into normal human cells. Science 279:349-352. [DOI] [PubMed] [Google Scholar]
- 6.Bosch, M. L., E. Harper, A. Schmidt, K. B. Strand, S. Thormahlen, M. E. Thouless, and Y. Wang. 1999. Activation in vivo of retroperitoneal fibromatosis-associated herpesvirus, a simian homologue of human herpesvirus-8. J. Gen. Virol. 80:467-475. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. R., C. Bloomer, and B. Chandran. 1998. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology 240:118-126. [DOI] [PubMed] [Google Scholar]
- 9.Chandran, B., C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140-149. [DOI] [PubMed] [Google Scholar]
- 10.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 11.Damania, B., and R. C. Desrosiers. 2001. Simian homologues of human herpesvirus 8. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers, R. C., V. G. Sasseville, S. C. Czajak, X. Zhang, K. G. Mansfield, A. Kaur, R. P. Johnson, A. A. Lackner, and J. U. Jung. 1997. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J. Virol. 71:9764-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupin, N., M. Grandadam, V. Calvez, I. Gorin, J. T. Aubin, S. Havard, F. Lamy, M. Leibowitch, J. M. Huraux, J. P. Escande, et al. 1995. Herpesvirus-like DNA sequences in patients with Mediterranean Kaposi's sarcoma. Lancet 345:761-762. [DOI] [PubMed] [Google Scholar]
- 15.Ganem, D. 1998. Human herpesvirus 8 and its role in the genesis of Kaposi's sarcoma. Curr. Clin. Top. Infect. Dis. 18:237-251. [PubMed] [Google Scholar]
- 16.Ganem, D. 1997. KSHV and Kaposi's sarcoma: the end of the beginning? Cell 91:157-160. [DOI] [PubMed] [Google Scholar]
- 17.Gessain, A., A. Sudaka, J. Briere, N. Fouchard, M. A. Nicola, B. Rio, M. Arborio, X. Troussard, J. Audouin, J. Diebold, et al. 1996. Kaposi sarcoma-associated herpes-like virus (human herpesvirus type 8) DNA sequences in multicentric Castleman's disease: is there any relevant association in non-human immunodeficiency virus-infected patients? Blood 87:414-416. [PubMed] [Google Scholar]
- 18.Gradoville, L., J. Gerlach, E. Grogan, D. Shedd, S. Nikiforow, C. Metroka, and G. Miller. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J. Virol. 74:6207-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greensill, J., J. A. Sheldon, N. M. Renwick, B. E. Beer, S. Norley, J. Goudsmit, and T. F. Schulz. 2000. Two distinct gamma-2 herpesviruses in African green monkeys: a second gamma-2 herpesvirus lineage among Old World primates? J. Virol. 74:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaleeba, J. A., E. P. Bergquam, and S. W. Wong. 1999. A rhesus macaque rhadinovirus related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 encodes a functional homologue of interleukin-6. J. Virol. 73:6177-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasolo, F. C., E. Mpabalwani, and U. A. Gompels. 1997. Infection with AIDS-related herpesviruses in human immunodeficiency virus-negative infants and endemic childhood Kaposi's sarcoma in Africa. J. Gen. Virol. 78:847-855. [DOI] [PubMed] [Google Scholar]
- 23.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. [DOI] [PubMed] [Google Scholar]
- 24.Kirshner, J. R., D. M. Lukac, J. Chang, and D. Ganem. 2000. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 74:3586-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacoste, V., P. Mauclere, G. Dubreuil, J. Lewis, M. C. Georges-Courbot, and A. Gessain. 2000. KSHV-like herpesviruses in chimps and gorillas. Nature 407:151-152. [DOI] [PubMed] [Google Scholar]
- 26.Lear, A. L., M. Rowe, M. G. Kurilla, S. Lee, S. Henderson, E. Kieff, and A. B. Rickinson. 1992. The Epstein-Barr virus (EBV) nuclear antigen 1 BamHI F promoter is activated on entry of EBV-transformed B cells into the lytic cycle. J. Virol. 66:7461-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, M., J. MacKey, S. C. Czajak, R. C. Desrosiers, A. A. Lackner, and J. U. Jung. 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J. Virol. 73:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, S. F., D. R. Robinson, G. Miller, and H. J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lukac, D. M., J. R. Kirshner, and D. Ganem. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348-9361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukac, D. M., R. Renne, J. R. Kirshner, and D. Ganem. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304-312. [DOI] [PubMed] [Google Scholar]
- 31.Manet, E., H. Gruffat, M. C. Trescol-Biemont, N. Moreno, P. Chambard, J. F. Giot, and A. Sergeant. 1989. Epstein-Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 8:1819-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mansfield, K. G., S. V. Westmoreland, C. D. DeBakker, S. Czajak, A. A. Lackner, and R. C. Desrosiers. 1999. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J. Virol. 73:10320-10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, G., L. Heston, E. Grogan, L. Gradoville, M. Rigsby, R. Sun, D. Shedd, V. M. Kushnaryov, S. Grossberg, and Y. Chang. 1997. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J. Virol. 71:314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, G., M. O. Rigsby, L. Heston, E. Grogan, R. Sun, C. Metroka, J. A. Levy, S. J. Gao, Y. Chang, and P. Moore. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292-1297. [DOI] [PubMed] [Google Scholar]
- 35.Moore, P. S., and Y. Chang. 1995. Detection of herpesvirus-like DNA sequences in Kaposi's sarcoma in patients with and without HIV infection. N. Engl. J. Med. 332:1181-1185. [DOI] [PubMed] [Google Scholar]
- 36.Nealon, K., W. W. Newcomb, T. R. Pray, C. S. Craik, J. C. Brown, and D. H. Kedes. 2001. Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus. J. Virol. 75:2866-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Regamey, N., M. Tamm, M. Wernli, A. Witschi, G. Thiel, G. Cathomas, and P. Erb. 1998. Transmission of human herpesvirus 8 infection from renal-transplant donors to recipients. N. Engl. J. Med. 339:1358-1363. [DOI] [PubMed] [Google Scholar]
- 40.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 41.Rochford, R., M. L. Lutzke, R. S. Alfinito, A. Clavo, and R. D. Cardin. 2001. Kinetics of murine gammaherpesvirus 68 gene expression following infection of murine cells in culture and in mice. J. Virol. 75:4955-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose, T. M., K. B. Strand, E. R. Schultz, G. Schaefer, G. W. Rankin, Jr., M. E. Thouless, C. C. Tsai, and M. L. Bosch. 1997. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J. Virol. 71:4138-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schultz, E. R., G. W. Rankin, Jr., M. P. Blanc, B. W. Raden, C. C. Tsai, and T. M. Rose. 2000. Characterization of two divergent lineages of macaque rhadinoviruses related to Kaposi's sarcoma-associated herpesvirus. J. Virol. 74:4919-4928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Searles, R. P., E. P. Bergquam, M. K. Axthelm, and S. W. Wong. 1999. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J. Virol. 73:3040-3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 48.Strand, K., E. Harper, S. Thormahlen, M. E. Thouless, C. Tsai, T. Rose, and M. L. Bosch. 2000. Two distinct lineages of macaque gamma herpesviruses related to the Kaposi's sarcoma associated herpesvirus. J. Clin. Virol. 16:253-269. [DOI] [PubMed] [Google Scholar]
- 49.Sun, R., S. F. Lin, L. Gradoville, Y. Yuan, F. Zhu, and G. Miller. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 95:10866-10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 52.Tso, J. Y., X. H. Sun, T. H. Kao, K. S. Reece, and R. Wu. 1985. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res 13:2485-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Dyk, L. F., H. I. Virgin, and S. H. Speck. 2000. The murine gammaherpesvirus 68 v-cyclin is a critical regulator of reactivation from latency. J. Virol. 74:7451-7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol. 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wong, S. W., E. P. Bergquam, R. M. Swanson, F. W. Lee, S. M. Shiigi, N. A. Avery, J. W. Fanton, and M. K. Axthelm. 1999. Induction of B cell hyperplasia in simian immunodeficiency virus-infected rhesus macaques with the simian homologue of Kaposi's sarcoma-associated herpesvirus. J. Exp. Med. 190:827-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, T. T., E. J. Usherwood, J. P. Stewart, A. A. Nash, and R. Sun. 2000. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol. 74:3659-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu, F. X., T. Cusano, and Y. Yuan. 1999. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5556-5567. [DOI] [PMC free article] [PubMed] [Google Scholar]