Abstract
Human immunodeficiency virus type 1 (HIV-1) strain LAV-1 (HIV-1LAV-1) particles were collected by ultracentrifugation, treated with subtilisin, and then purified by Sepharose CL-4B column chromatography to remove microvesicles. The lysate of the purified HIV-1LAV-1 particles was subjected to two-dimensional (2D) gel electrophoresis and stained. The 2D gel electrophoresis image suggested that 24 proteins can be identified inside the virion. Furthermore, the stained protein spots were excised and digested with trypsin. The resulting peptide fragments were characterized by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Peptide mass fingerprinting data suggested that two isoforms of cyclophilin A (CyPA), one with an isoelectric point (pI) of 6.40 and one with a pI of 6.53, are inside the viral membrane; that another isoform, with a pI of 6.88, is outside the viral membrane; and that the CyPA isoform with a pI of 6.53 is N acetylated. The mechanisms that permit the redistribution of CyPA on the viral surface have not yet been clarified, but it is surmised that the CyPA isoform with a pI of 6.88 may play a critical role in the attachment of virions to the surface of target cells and that both CyPA isoforms with pIs of 6.40 and 6.53 may regulate the conformation of the HIV-1 capsid protein.
The integrated form of human immunodeficiency virus (HIV) type 1 (HIV-1), also known as the provirus, is approximately 9.8 kb in length (17). The genes of HIV encode at least nine proteins that are divided into three classes: (i) the major structural proteins (Gag, Pol, and Env), (ii) the regulatory proteins (Tat and Rev), and (iii) the accessory proteins (Vpu, Vpr, Vif, and Nef) (8). These viral proteins function in the replication cycle of HIV-1, which includes the following steps: viral entry, reverse transcription, integration, gene expression, assembly, budding, and maturation. The actions of these viral proteins alone, however, cannot completely explain how HIV-1 can efficiently replicate in a susceptible host. To date, several studies of purified HIV-1 virions have shown that in addition to proteins encoded by the virus, cellular proteins from the host are found in these virions (18, 19). Cellular proteins, such as cyclophilin A (CyPA), are included in HIV-1 virions as a result of their interaction with Gag proteins during assembly and release (6, 19, 28). CyPA is an abundant cytosolic protein ubiquitously expressed in eukaryotic cells (21) and binds to the interaction sites that are located in the N-terminal domain of p24gag around Gly89-Pro90 for packaging and in the C-terminal domain of p24gag around Gly156-Pro157 and Gly223-Pro224 for destabilization of the capsid cone (5, 9). In addition, additional evidence of the role of CyPA in the HIV-1 life cycle has recently emerged. CyPA mediates HIV-1 attachment to target cells via heparans (22). The actions of cellular protein CyPA support HIV-1 replication.
Furthermore, some viral proteins with cotranslational or posttranslational modifications, such as myristoylation and phosphorylation, are also included in virions. N myristoylation of p17gag of HIV-1 is essential for structural assembly and replication (3, 26, 27). Phosphorylation of p17gag is related to its dissociation from the membrane during the early postentry step of HIV-1 (4, 7). These cotranslational or posttranslational modifications also support HIV-1 replication.
In this study, a purified HIV-1 strain LAV-1 (HIV-1LAV-1) preparation was analyzed by two-dimensional (2D) gel electrophoresis and matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (MS) to identify not only unknown cellular and viral proteins inside HIV-1LAV-1 that are indispensable for its replication but also unknown cotranslational or posttranslational modifications of viral and cellular proteins that are essential for the viral life cycle.
Virus purification and subtilisin treatment.
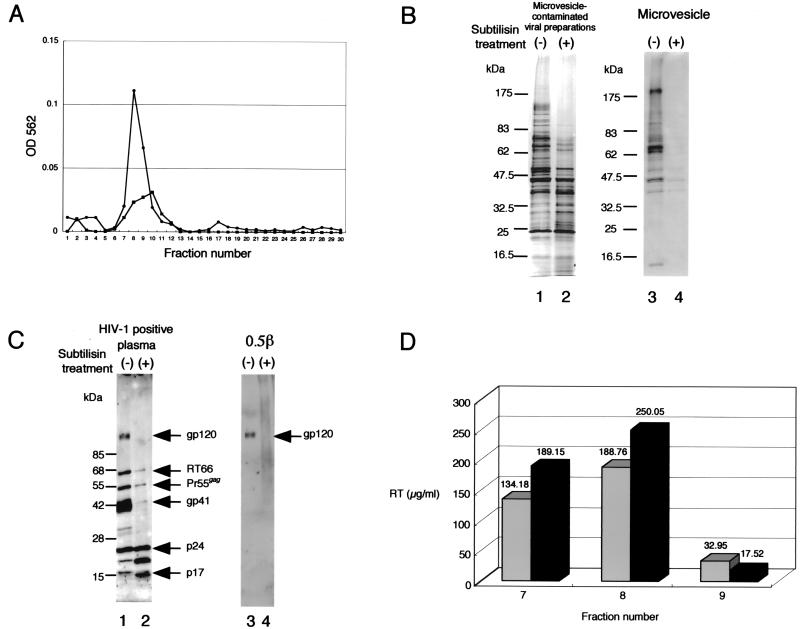
The study of proteins inside HIV-1 was complicated by the potential contamination of the HIV-1 preparation with nonviral particles called microvesicles (1, 11, 19). In this study, a microvesicle-contaminated HIV-1LAV-1 preparation (from the culture medium of chronically HIV-1LAV-1-infected T-cell line CEM) and microvesicles from uninfected T-cell line CEM were prepared and then purified with Sepharose CL-4B according to the protocol of McGrath et al. (16). As shown in Fig. 1A, both the microvesicle-contaminated HIV-1 preparation and the microvesicles alone eluted at the same positions as fractions 7, 8, and 9 on a Sepharose CL-4B gel with calcium- and magnesium-free phosphate-buffered saline [PBS(−)]. The microvesicle-contaminated HIV-1LAV-1 preparation and the microvesicles alone were digested with subtilisin (ICN Biomedicals Inc., Costa Mesa, Calif.) to remove microvesicles by using the protocol of Ott et al. (19, 20). The subtilisin-treated HIV-1LAV-1 preparation and the microvesicles alone were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (15) (Multigel 4/20; Daiichi Pure Chemicals Co., Ltd., Tokyo, Japan), and the separated proteins were subsequently subjected to Western immunoblot analysis with HIV-1-positive plasma and antibody 0.5β (a kind gift from Shuzo Matsushita, AIDS Research Institute, Kumamoto University, Kumamoto, Japan).
FIG. 1.
Elution profiles of both the microvesicle-contaminated HIV-1LAV-1 preparation and microvesicles alone, protein profiles of the subtilisin-treated HIV-1LAV-1 preparation and microvesicles alone, and HIV-1 reverse transcriptase activity assay results. (A) Elution profiles of the microvesicle-contaminated HIV-1LAV-1 preparation (circles) and microvesicles alone (squares) determined with a 10-ml column of Sepharose CL-4B. OD, optical density. (B) SDS-PAGE of the microvesicle-contaminated HIV-1LAV-1 preparation and microvesicles alone derived from CEM cells. Lanes containing non-subtilisin-treated and subtilisin-treated materials are denoted by minus and plus signs, respectively. (C) Western immunoblot analysis of the subtilisin-treated HIV-1LAV-1 preparation. Antibodies used for analyses are displayed above the blots. The bands were visualized by means of chemiluminescence detection (NEN Life Science Products, Inc., Boston, Mass.). Lanes containing non-subtilisin-treated and subtilisin-treated HIV-1LAV-1 materials are denoted by minus and plus signs, respectively. RT66, reverse transcriptase. SDS-PAGE and Western immunoblot analyses of digests were done with samples containing 1 μg of starting total protein. (D) Reverse transcriptase (RT) activity of each fraction containing non-subtilisin-treated (gray bars) or subtilisin-treated (black bars) HIV-1LAV-1, determined in accordance with the manufacturer’s instructions.
Typical results for the subtilisin-treated HIV-1LAV-1 preparation and the microvesicles alone are shown in Fig. 1B and C, respectively. As expected, more than 95% of microvesicle-associated proteins were removed by the subtilisin treatment (Fig. 1B, lane 4). In contrast, proteins inside the virion, such as p17gag and p24gag, were not digested, although gp120 was completely digested (Fig. 1C, lanes 2 and 4). In addition, HIV-1 reverse transcriptase activity, measured by using a reverse transcriptase assay nonradioactive kit (Roche Diagnostics Corp., Tokyo, Japan) in accordance with the manufacturer's instructions, was not decreased by the subtilisin treatment (Fig. 1D). These results suggest that the proteins inside the virion were not digested by the subtilisin treatment because they were protected by the virion membrane. Therefore, the subtilisin digestion procedure was used to identify unknown cellular and viral proteins inside HIV-1LAV-1. Finally, the subtilisin-treated HIV-1LAV-1 preparation was repurified by column chromatography with a 10-ml Sepharose CL-4B column with PBS(−) prior to proteome analysis.
The microvesicle-contaminated HIV-1LAV-1 preparation, the microvesicle-free HIV-1LAV-1 preparation, and the microvesicles alone were pooled and centrifuged at 100,000 × g for 1 h at 4°C. Each resulting pellet was boiled for 1 min and lysed in 200 μl of lysis buffer, which consisted of 8 M urea and 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) in 2% immobilized pH gradient (IPG) buffer (pH 6 to 11).
2D gel separation of HIV-1LAV-1 lysates and characterization of proteins inside the virion.
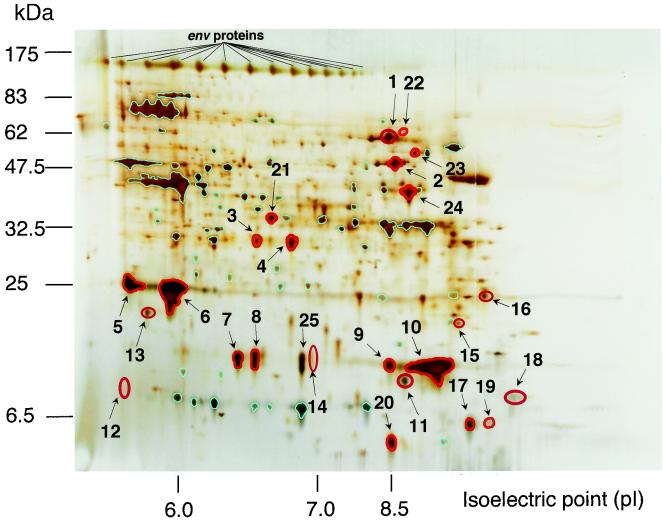
To create a 2D image of HIV-1 virion proteins, the nontreated HIV-1LAV-1 preparation, the subtilisin-treated HIV-1LAV-1 preparation, and the microvesicles alone were individually analyzed by 2D PAGE under the same conditions by using the protocol of Görg et al. (12). Each sample was loaded on the gel by anodic cup loading with an 18-cm Immobiline Drystrip (pH 6 to 11) (Amersham Biosciences Corp., Buckinghamshire, United Kingdom). Each gel was run in the gradient mode: the voltage was raised from 500 to 3,500 V over 8 h and then was maintained at 3,500 V for 25 h. After the 2D electrophoresis (12 to 14% ExcelGel XL SDS; Amersham Biosciences Corp.), the gel was silver stained by using the protocols of Shevchenko et al. (25) and Wilm et al. (31). The 2D gel was analyzed by using Bio-Rad Melanie II software (Nippon Bio-Rad Laboratories, Yokohama, Japan). Spots in the protein profile of the nontreated HIV-1LAV-1 preparation that overlapped those in the profile of the subtilisin-treated HIV-1LAV-1 preparation are emphasized in red, except for the spots newly produced by the subtilisin treatment (Fig. 2). The results suggested that 24 spots can be identified as proteins inside the virion (Fig. 2). In addition, spots in the protein profile of the non-subtilisin-treated HIV-1LAV-1 preparation that overlapped those in the profile of the microvesicles alone are emphasized in light blue (Fig. 2). Sixty-one spots were identified as proteins derived from microvesicles. The other spots were identified as proteins outside the virion.
FIG. 2.
2D gel image of HIV-1LAV-1. The horizontal axis shows protein separation by isoelectric focusing. The vertical axis shows protein separation by molecular mass (in kilodaltons). The gel was silver stained. 2D SDS-PAGE standards (Nippon Bio-Rad Laboratories) were used for the reproducibility of 2D experiments. The most striking advantage of these standards is that they allow valid comparisons of 2D electrophoresis patterns of different samples. The 2D gel pattern shows the results obtained after superimposition of the patterns of the subtilisin-treated HIV-1LAV-1 preparation and microvesicles alone on that of the non-subtilisin-treated HIV-1LAV-1 preparation. Spots of proteins inside the virion are emphasized in red. Spots derived from microvesicles are emphasized in light blue.
The spots corresponding to the proteins inside the virion were excised and destained prior to enzymatic digestion by using the protocol of Gharahdaghi et al. (10). These pieces of gel were immersed in 100 μl of acetonitrile, dried under vacuum centrifugation for 60 min, rehydrated in 50 μl of trypsin solution (25 ng of trypsin/μl in 100 mM ammonium bicarbonate), and then incubated for 45 min on ice. Unabsorbed trypsin solution was removed, and the gel pieces were immersed in 20 μl of 100 mM ammonium bicarbonate (pH 8.0) and further incubated for 12 h at 37°C. The resulting tryptic peptides were purified by using ZipTip C18 in accordance with the manufacturer's instructions (literature number TN 224; Nihon Millipore Ltd., Tokyo, Japan) and then analyzed by MALDI-TOF MS with the FAST accessory for post-source decay (Burker Daltonik GmbH, Bremen, Germany) and α-cyano-4-hydroxycinnamic acid as the matrix.
The peptide masses obtained were subjected to a search to determine their matches in the SWISS-PROT and TrEMBL databases by using the program PeptIdent (http://expasy.proteome.org.au/tools/peptident.html), and protein matches were assigned by referring to theoretical fingerprints derived from published data. The remaining, nonmatching peptide masses were subjected to a search by using the FindMod program (http://www.expasy.ch/tools/findmod) in order to identify probable cotranslational or posttranslational modifications (30). Consequently, 9 of the 24 spots were identified as the major viral structural proteins (spots 1 to 6 and 9 to 11), and 2 of the 24 spots were identified as cellular proteins (spots 7 and 8) (Table 1). Interestingly, proteins corresponding to spots 7 and 8 (with pIs of 6.40 and 6.53, respectively) were assigned to two isoforms of CyPA inside the virion which we designated CyPA6.40 and CyPA6.53, respectively (Table 2). Only Env was assigned based on the 2D image by Western immunoblot analysis of the non-subtilisin-treated HIV-1LAV-1 preparation with antibody 0.5β because proteins outside the virion were completely digested by the subtilisin treatment (Fig. 2). Characterization of the other proteins inside the virion (spots 12 to 24 in Fig. 2) is ongoing.
TABLE 1.
Data for protein spots excised from the 2D gel
| Spota | Protein name | Accession no.e for gene | %Seqb | Gel featurec
|
Predicted datad
|
||
|---|---|---|---|---|---|---|---|
| pI | Mass(kDa) | pI | Mass (kDa) | ||||
| 1 | Reverse transcriptase/RNase H (fragment) | O40174 | 10.7 | 8.40 | 58 | 8.63 | 65 |
| 2 | POL (fragment) | Q9IDF2 | 27.2 | 8.59 | 49 | 8.64 | 60 |
| 3 | Integrase (fragment) | O92844 | 21.9 | 6.54 | 30 | 8.16 | 32 |
| 4 | Integrase (fragment) | O92844 | 28.1 | 6.80 | 30 | 7.75 | 32 |
| 5 | Chain 2: core protein p24 (core antigen) | P03348 | 11.3 | 5.69 | 24 | 6.26 | 26 |
| 6 | Chain 2: core protein p24 (core antigen) | P03348 | 54.5 | 5.94 | 24 | 6.26 | 26 |
| 7 | Peptidyl-prolyl cis-trans isomerase A (CyPA) | P05092 | 40.2 | 6.40 | 18 | 7.82 | 18 |
| 8 | Peptidyl-prolyl cis-trans isomerase A (CyPA) | P05092 | 41.5 | 6.53 | 18 | 7.82 | 18 |
| 9 | Chain 1: core protein p17 (matrix protein) | P03348 | 25.2 | 8.41 | 17 | 9.28 | 15 |
| 10 | Chain 1: core protein p17 (matrix protein) | P03348 | 35.9 | 9.72 | 17 | 9.28 | 15 |
| 11 | Chain 1: core protein p17 (matrix protein) | P03348 | 58.8 | 8.85 | 12 | 9.28 | 15 |
| 25 | Peptidyl-prolyl cis-trans isomerase A (CyPA) | P05092 | 48.2 | 6.88 | 18 | 7.82 | 17.9 |
Key to spot labels in Fig. 2.
Ratio of the length of peptide sequences actually detected to the total sequence length of the theoretical gene product, expressed as a percentage.
Calculated directly from the 2D gel.
Predicted for the theoretical gene product.
SWISS PROT/TrEMBL database.
TABLE 2.
Comparison of theoretical masses and observed masses derived from tryptic digests of spots 7.
| Spot | Tryptic peptide (amino acids) | Mass (m/z)
|
Corresponding sequence | No. of missed cleavages | |
|---|---|---|---|---|---|
| Theoretical | Observed | ||||
| 7 | T1 (1-18) | 1,946.00 | 1,946.30 | VNPTVFFDIAVDGEPLGR | 0 |
| T2 (19-30) | 1,379.76 | 1,380.00 | VSFELFADKVPK | 1 | |
| T3 (31-36) | 737.36 | 737.35 | TAENFR | 0 | |
| T4 (76-90) | 1,831.91 | 1,832.20 | SIYGEKFEDENFILK | 1 | |
| T5 (82-90) | 1,154.57 | 1,153.58 | FEDENFILK | 0 | |
| T6 (118-124) | 848.41 | 848.35 | TEWLDGK | 0 | |
| T7 (118-130) | 1,515.80 | 1,515.57 | TEWLDGKHVVFGK | 1 | |
| T8 (118-132) | 1,742.96 | 1,742.91 | TEWLDGKHVVFGKVK | 2 | |
| 8 | T1 (1-18) | 1,988.01 | 1,988.26 | Acetyl-VNPTVFFDIAVDGEPLGR | 0 |
| T2 (31-36) | 737.36 | 737.37 | TAENFR | 0 | |
| T3 (37-43) | 705.38 | 705.42 | ALSTGEK | 0 | |
| T4 (55-68) | 1,541.72 | 1,542.00 | IIPGFMCQGGDFTR | 0 | |
| T5 (76-90) | 1,831.91 | 1,832.20 | SIYGEKFEDENFILK | 1 | |
| T6 (82-90) | 1,154.57 | 1,153.58 | FEDENFILK | 0 | |
| T7 (118-124) | 848.41 | 848.35 | TEWLDGK | 0 | |
| T8 (118-130) | 1,515.80 | 1,515.57 | TEWLDGKHVVFGK | 1 | |
| T9 (131-143) | 1,505.75 | 1,506.00 | VKEGMNIVEAMER | 1 | |
| T10 (133-143) | 1,278.58 | 1,278.80 | EGMNIVEAMER | 0 | |
| 25 | T1 (1-18) | 1,946.00 | 1,945.70 | VNPTVFFDIAVDGEPLGR | 0 |
| T2 (31-36) | 737.36 | 737.46 | TAENFR | 0 | |
| T3 (55-68) | 1,541.72 | 1,541.50 | IIPGFMCQGGDFTR | 0 | |
| T4 (76-90) | 1,831.91 | 1,832.29 | SIYGEKFEDENFILK | 1 | |
| T5 (82-90) | 1,154.57 | 1,153.75 | FEDENFILK | 0 | |
| T6 (118-124) | 848.41 | 848.36 | TEWLDGK | 0 | |
| T7 (118-130) | 1,515.80 | 1,515.58 | TEWLDGKHVVFGK | 1 | |
| T8 (131-143) | 1,505.75 | 1,505.50 | VKEGMNIVEAMER | 1 | |
| T9 (133-143) | 1,278.58 | 1,278.40 | EGMNIVEAMER | 0 | |
Posttranslational modification.
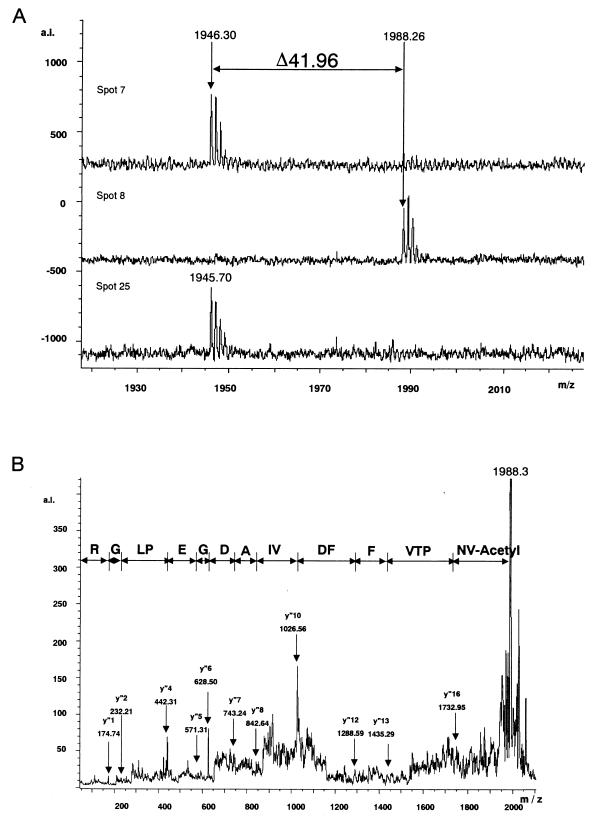
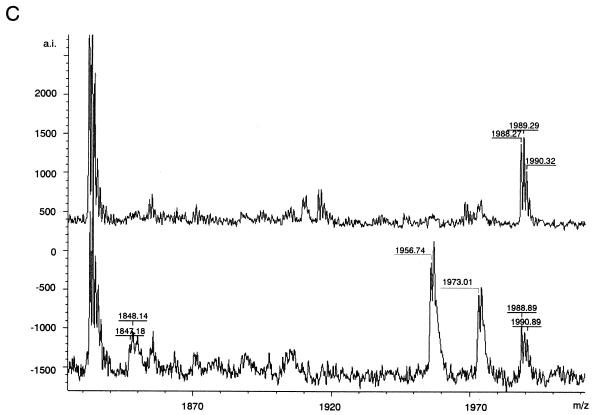
Proteins corresponding to 9 of the 11 identified spots (spots 3 to 11) were estimated to be the products of three genes, suggesting the existence of multiple protein isoforms resulting from cotranslational or posttranslational modifications of the gene products. Peptide mass fingerprinting data were used to identify probable cotranslational or posttranslational modifications. Consequently, the acetylation of N-terminal tryptic peptide Val1-Arg18 of CyPA6.53 was estimated by using the FindMod program because the mass spectrum corresponding to N-terminal tryptic peptide Val1-Arg18 exhibited a peak at m/z 1,946.30 in spot 7 and the mass spectrum corresponding to the acetylated form exhibited a peak at m/z 1,988.26 in spot 8 (Fig. 3A, upper and middle spectra). The acetylation was confirmed by sequential cleavages with MS determination of the peptide sequence fragment (Fig. 3B) and liberation of N-acetylated Val1 from the amino-terminal peptide of CyPA6.53 (Fig. 3C). The fragment ions (y" series) at m/z 174.74, 232.21, 442.31, 571.31, 628.50, 743.24, 842.64, 1,026.56, 1,288.59, 1,435.29, and 1,732.95 were defined according to the nomenclature of Biemann (2) and provided sufficient information on the formation of acetylated Val1 (Fig. 3B). Furthermore, the peak of (M + H)+ at m/z 1,847.18 corresponded to the peptide (Asn2-Arg18) whose N-acetylated Val1 was cleaved away by an N-acylamino acid-releasing enzyme (29) (Takara Shuzo Co., Ltd., Tokyo, Japan) in accordance with the manufacturer's instructions (Fig. 3C, lower spectrum). These findings demonstrated for the first time that N-acetylated CyPA is actually present in the virion, although the N acetylation of CyPA has been presumed because there were previous cases where the N-terminal amino acid sequence of CyPA was not obtained (14, 19), and that N-acetylation can be considered one of the possible causes of the difference in pI between CyPA6.40 and CyPA6.53.
FIG. 3.
Spectra of N-terminal tryptic peptides derived from CyPA and corresponding to spots 7, 8, and 25; postsource decay spectrum; and N-acylamino acid-releasing enzyme treatment of an N-terminal tryptic peptide derived from the spot 8 protein, N-acetyl-VNPTVFFDIAVDGEPLGR. (A) MALDI-TOF mass spectra of N-terminal tryptic peptides derived from CyPA and corresponding to spots 7, 8, and 25. As shown in the top and bottom spectra, the peaks of (M + H)+ at m/z 1,946.30 and 1,945.70 represent N-terminal tryptic peptide Val1-Arg18 of CyPA. As shown in the middle spectrum, the peak of (M + H)+ at m/z 1,988.26 could be assigned to the acetylated form of N-terminal tryptic peptide Val1-Arg18. The fragment with a molecular mass of 41.96 and corresponding to the acetyl group was deleted. a.i., absolute intensity. (B) MS determination of the partial peptide sequence of the N-terminal tryptic peptide derived from the spot 8 protein. y" series ions were defined according to the nomenclature of Biemann (2) (C) Liberation of N-acetylated Val1 from the N-terminal peptide derived from the spot 8 protein. As shown in the lower spectrum, the peak at m/z 1,847.18 and corresponding to the peptide Asn2-Arg18 (theoretical mass, m/z 1,846.93) was detected after N-acylamino acid-releasing enzyme treatment. The peak at m/z 1,847.18 was not found before N-acylamino acid-releasing enzyme treatment (upper spectrum). The unknown peaks of (M + H)+ at m/z 1,956.74 and 1,973.01 were also found in control experiments under identical conditions, except for the omission of tryptic peptides derived from the spot 8 protein.
The other modified proteins (spots 3 to 6 and 9 to 11) included the products of the gag and pol genes; however, specific modifications could not be clearly established (Table 1). Although 11.3 to 58.8% of the total predicted sequence was recovered by proteome analysis, the remaining tryptic peptides might be either modified or not ionized well during MS.
Localization of CyPA.
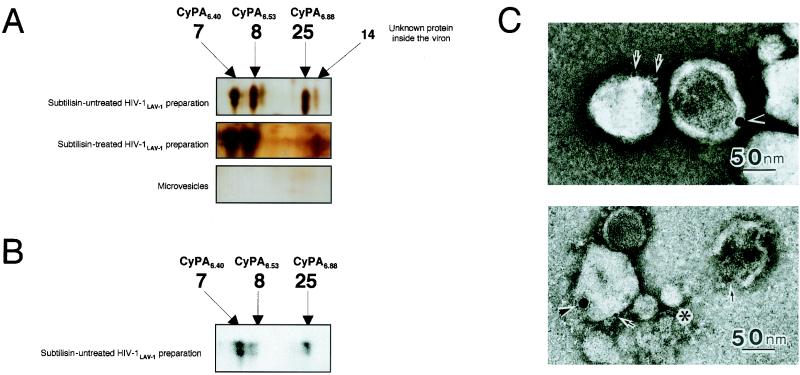
To identify proteins outside the virion, spots that did not overlap in the protein profile of the non-subtilisin-treated HIV-1LAV-1 preparation and that of the microvesicles alone were also investigated. Figure 4 shows an expanded view of the same area of interest in each 2D gel image. As shown in Fig. 4A, spot 25 was observed only in the protein profile of the non-subtilisin-treated HIV-1LAV-1 preparation that was then subjected to tryptic peptide mass fingerprinting by using MALDI-TOF MS. Interestingly, the protein corresponding to spot 25 (with a pI of 6.88) was assigned to a novel isoform of CyPA located outside the virion which we designated CyPA6.88 (Tables 1 and 2). Indeed, spot 25 on the 2D gel of the non-subtilisin-treated HIV-1LAV-1 preparation was also identified as CyPA by Western immunoblot analysis with anti-CyPA antibody (Upstate Biotechnology, Inc., Waltham, Mass.) (13) (Fig. 4B). Furthermore, negative-stain transmission electron microscopy demonstrated that CyPA is outside the virion but not outside the microvesicle (Fig. 4C).
FIG. 4.
2D gel image analysis, Western immunoblot analysis with anti-CyPA antibody, and immunoelectron microscopic analysis. (A) After 2D gel electrophoresis and protein visualization by silver staining, the 2D gel images of the non-subtilisin-treated HIV-1LAV-1 preparation, the subtilisin-treated HIV-1LAV-1 preparation, and microvesicles alone were expanded. The expanded image of the non-subtilisin-treated HIV-1LAV-1 preparation shows four spots (spots 7, 8, 14, and 25). Spot 25 completely disappeared on subtilisin treatment. This result suggests that CyPA6.88 exists on the viral surface. Spots 7, 8, 14, and 25 were not detected in the image of the microvesicles. The spot numbers are in agreement with those in Fig. 2. (B) Spots 7, 8, and 25 on the 2D gel of the non-subtilisin-treated HIV-1LAV-1 preparation were subjected to Western immunoblot analysis with anti-CyPA antibody. (C) A microvesicle-contaminated HIV-1 preparation was fixed with 0.1 M sodium phosphate buffer (pH 7.4) containing 4% paraformaldehyde and 0.25% glutaraldehyde at 4°C. After brief washing, the samples were mounted on carbon-coated nickel grids. Double immunolabeling techniques were used as follows. The first step was rabbit anti-CyPA antibody (Upstate Biotechnology) and then 15-nm-diameter gold-labeled secondary antibody, and the second step was murine anti-gp120 (strain IIIB) monoclonal antibody (ImmunoDiagnostics, Inc., Woburn, Mass.) and then 5-nm-diameter gold-labeled secondary antibody. The immunolabeled samples were negatively stained with 3% uranyl acetate and examined with a transmission electron microscope. The asterisk indicates a microvesicle; arrows and arrowheads indicate gp120 and CyPA, respectively.
The cause of the difference in pI between CyPA6.40 and CyPA6.88 is unknown, but the difference may be due to other types of posttranslational modifications because the N-terminal residue of CyPA6.88 is free (Table 2).
Recently, Sherry et al. (24) and Saphire et al. (22) proposed that CyPA is located outside the virion and plays a direct role in the attachment of the virus to target cells. Our observations seem to be consistent with their model. However, our observation that the outside of CyPA is completely cleaved away by the subtilisin treatment (Fig. 4A) seems to be different from that in a previous study which showed that only a small part of CyPA would be removed upon proteolytic cleavage (22). In order to reconcile this finding with our own observation, we propose that two isoforms of CyPA, namely, CyPA6.40 and CyPA6.53, are inside the viral membrane; that another isoform, CyPA6.88, is found on the viral surface; and that one of the isoforms inside the virion may change to CyPA6.88 to penetrate the viral membrane. Although the mechanisms that permit the redistribution of CyPA on the viral surface have not yet been clarified, CyPA itself may have the characteristic of membrane penetration because CyPA has been reported to be a secreted growth factor induced by oxidative stress (32) and a proinflammatory secretory product of activated macrophages (23). Taken together, these data suggest that CyPA6.88 may play a critical role in the attachment of virions to the surface of target cells and that both CyPA6.40 and CyPA6.53 may regulate the conformation of the HIV-1 capsid protein.
Acknowledgments
We thank S. Matsushita (AIDS Research Institute, Kumamoto University, Kumamoto, Japan) for providing anti-gp120 antibody 0.5β.
This study was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 2.Biemann, K. 1990. Appendix 5. Nomenclature for peptide fragment ions (positive ions). Methods Enzymol. 193:886-887. [DOI] [PubMed] [Google Scholar]
- 3.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukrinskaya, A. G., A. Ghorpade, N. K. Heinzinger, T. E. Smithgall, R. E. Lewis, and M. Stevenson. 1996. Phosphorylation-dependent human immunodeficiency virus type 1 infection and nuclear targeting of viral DNA. Proc. Natl. Acad. Sci. USA 93:367-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endrich, M. M., P. Gehrig, and H. Gehring. 1999. Maturation-induced conformational changes of HIV-1 capsid protein and identification of two high affinity sites for cyclophilins in the C-terminal domain. J. Biol. Chem. 26:5326-5332. [DOI] [PubMed] [Google Scholar]
- 6.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 7.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 8.Gallo, R., F. Wong-Staal, L. Montagnier, W. A. Haseltine, and M. Yoshida. 1988. HIV/HTLV gene nomenclature. Nature 333:504. [DOI] [PubMed] [Google Scholar]
- 9.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 10.Gharahdaghi, F., C. R. Weinberg, D. A. Meagher, B. S. Imai, and S. M. Mische. 1999. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: a method for the removal of silver ions to enhance sensitivity. Electrophoresis 20:601-605. [DOI] [PubMed] [Google Scholar]
- 11.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 12.Görg, A., C. Obermaier, G. Boguth, A. Csordas, J. J. Diaz, and J. J. Madjar. 1997. Very alkaline immobilized pH gradients for two-dimensional electrophoresis of ribosomal and nuclear proteins. Electrophoresis 18:328-337. [DOI] [PubMed] [Google Scholar]
- 13.Haendler, B., R. Hofer-Warbinek, and E. Hofer. 1987. Complementary DNA for human T-cell cyclophilin. EMBO J. 6:947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieffer, L. J., T. Thalhammer, and R. E. Handschumacher. 1992. Isolation and characterization of a 40-kDa cyclophilin-related protein. J. Biol. Chem. 267:5503-5507. [PubMed] [Google Scholar]
- 15.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 16.McGrath, M., O. Witte, T. Pincus, and I. L. Weissman. 1978. Retrovirus purification: method that conserves envelope glycoprotein and maximizes infectivity. J. Virol. 25:923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muesing, M. A., D. H. Smith, C. D. Cabradilla, C. V. Benton, L. A. Lasky, and D. J. Capon. 1985. Nucleic acid structure and expression of the human AIDS/lymphadenopathy retrovirus. Nature 313:450-458. [DOI] [PubMed] [Google Scholar]
- 18.Ott, D. E. 1997. Cellular proteins in HIV virions. Rev. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 19.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder, I. I., L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 20.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder, I. I., Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 21.Ryffel, B., G. Woerly, B. Greiner, B. Haendler, M. J. Mihatsch, and B. M. Foxwell. 1991. Distribution of the cyclosporine binding protein cyclophilin in human tissues. Immunology 72:399-404. [PMC free article] [PubMed] [Google Scholar]
- 22.Saphire, A. C., M. D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry, B., N. Yarlett, A. Strupp, and, A., Cerami. 1992. Identification of cyclophilin as a proinflammatory secretory product of lipopolysaccharide-activated macrophages. Proc. Natl. Acad. Sci. USA 89:3511-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherry, B., G. Zybarth, M. Alfano, L. Dubrovsky, R. Mitchell, D. Rich, P. Ulrich, R. Bucala, A. Cerami, and M. Bukrinsky. 1998. Role of cyclophilin A in the uptake of HIV-1 by macrophages and T lymphocytes. Proc. Natl. Acad. Sci. USA 95:1758-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68:850-858. [DOI] [PubMed] [Google Scholar]
- 26.Shiraishi, T., S. Misumi, M. Takama, I. Takahashi, and S. Shoji. 2001. Myristoylation of human immunodeficiency virus type 1 gag protein is required for efficient env protein transportation to the surface of cells. Biochem. Biophys. Res. Commun. 282:1201-1205. [DOI] [PubMed] [Google Scholar]
- 27.Tashiro, A., S. Shoji, and Y. Kubota. 1989. Antimyristoylation of the gag proteins in the human immunodeficiency virus-infected cells with N-myristoyl glycinal diethylacetal resulted in inhibition of virus production. Biochem. Biophys. Res. Commun. 165:1145-1154. [DOI] [PubMed] [Google Scholar]
- 28.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Göttlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 29.Tsunasawa, S., and K. Narita. 1976. Acylamino acid-releasing enzyme from rat liver. Methods Enzymol. 45:552-561. [DOI] [PubMed] [Google Scholar]
- 30.Wilkins, M. R., E. Gasteiger, A. A. Gooley, B. R. Herbert, M. P. Molloy, P. A. Binz, K. Ou, J. C. Sanchez, A. Bairoch, K. L. Williams, and D. F. Hochstrasser. 1999. High-throughput mass spectrometric discovery of protein post-translational modifications. J. Mol. Biol. 289:645-657. [DOI] [PubMed] [Google Scholar]
- 31.Wilm, M., A. Shevchenko, T. Houthaeve, S. Breit, L. Schweigerer, T. Fotsis, and M. Mann. 1996. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature 379:466-469. [DOI] [PubMed] [Google Scholar]
- 32.Jin, Z. G., M. G. Melaragno, D. F. Liao, C. Yan, J. Haendeler, Y. A. Suh, J. D. Lambeth, and B. C. Berk. 2000. Cyclophilin A is a secreted growth factor induced by oxidative stress. Circ. Res. 87:789-796. [DOI] [PubMed] [Google Scholar]