Abstract
Vaccinia virus, a member of the poxvirus family, contains a conserved J1R open reading frame that encodes a late protein of 17.8 kDa. The 18-kDa J1R protein is associated mainly with the membrane fraction of intracellular mature virus particles. This study examines the biological function of J1R protein in the vaccinia virus life cycle. A recombinant vaccinia virus was constructed to conditionally express J1R protein in an isopropyl-β-d-galactopyranoside (IPTG)-inducible manner. When J1R is not expressed during vaccinia virus infection, the virus titer is reduced approximately 100-fold. In contrast, J1R protein is not required for viral gene expression, as indicated by protein pulse-labeling. J1R protein is also not required for DNA processing, as the resolution of the concatemer junctions of replicated viral DNA was detected without IPTG. A deficiency of J1R protein caused a severe delay in the processing of p4a and p4b into mature core proteins 4a and 4b, indicating that J1R protein participates in virion morphogenesis. Infected cells grown in the absence of IPTG contained very few intracellular mature virions in the cytoplasm, and enlarged viroplasm structures accumulated with viral crescents attached at the periphery. Abundant intermediate membrane structures of abnormal shapes were observed, and many immature virions were either empty or partially filled, indicating that J1R protein is important for DNA packaging into immature virions. J1R protein also coimmunoprecipited with A45R protein in infected cells. In summary, these results indicate that vaccinia virus J1R is a membrane protein that is required for virus growth and plaque formation. J1R protein interacts with A45R protein and performs an important role during immature virion formation in cultured cells.
Vaccinia virus is the prototypical member of the poxvirus family; it contains double-stranded DNA and replicates in the cytoplasm of infected cells (37). Poxviruses are difficult to study because of the large sizes of their genomes and their complex structures and compositions. Most poxviruses infect several types of cells and produce multiple distinct infectious particles during the virus life cycle (16, 36). Virion morphogenesis is highly complex and evolves with multiple stages at precise intracellular locations in the infected cell (60). For example, intracellular mature virions (IMV) are formed at the intermediate compartment after viral late gene expression (58). IMV are converted to particles of intracellular enveloped viruses (IEV) when they are enveloped with membrane structures derived from Golgi cisternae (53). IEV are transported to the cell periphery by interactions with microtubules (18, 21, 43, 51, 66, 70, 71). IEV fuse with the plasma membrane and remain cell-associated virions or are released from the cells as extracellular enveloped viruses (9, 10).
Different forms of the virion are important for different routes of virus infection (25). IMV are very stable, resistant to environmental stress, and therefore well suited for transmission between hosts. Cell-associated virions are derived from IEV at the cell periphery, where they acquire actin-based motility that facilitates transmission between cells (10, 21, 67). Extracellular enveloped viruses are released from infected cells and play a role in transmission of the virus through the host bloodstream (38).
As expected, the different forms of virions have unique biochemical structures and compositions. Previously, two-dimensional (2D) gel electrophoresis experiments identified 11 viral proteins associated with IMV-containing membranes (27). Some of these proteins have been studied. For example, antibodies to A27L, D8L, and L1R proteins neutralize vaccinia virus infection, indicating that they are important for IMV infection (22, 26, 35, 80). In addition, soluble recombinant forms of the extracellular domains of A27L, D8L, and H3L proteins recognize cell surface glycosaminoglycans and facilitate virus attachment (22, 23, 33). Experiments with electron microscopy of virus mutants revealed that A17L, A14L, A4L, and L1R proteins play important roles in virion morphogenesis (40, 45, 46, 48, 64, 78, 79). G4L protein, on the other hand, is an enzyme that plays a role in protein disulfide bond formation (56, 73, 74).
Due to sensitivity limitations, there were other, minor 2D protein spots in membrane fractions whose identities remained unknown (27). However, another approach based on published viral genomic sequences helped identify several novel membrane-associated viral proteins, and their roles in virion morphogenesis were subsequently revealed (8, 55, 63, 81).
This report investigates the role of vaccinia virus J1R protein in the virus life cycle. Antibodies recognizing J1R protein identified its association with IMV membranes. A recombinant vaccinia virus that conditionally expresses J1R protein in an isopropyl-β-d-galactopyranoside (IPTG)-inducible manner was constructed. The results indicate that J1R plays a role in virion morphogenesis.
(This work was done by W.-L. Chiu in partial fulfillment of the requirements for a Ph.D. degree from the Graduate Institute of Life Science of the National Defense University.)
MATERIALS AND METHODS
Reagents, cells, and viruses.
Mycophenolic acid, hypoxanthine, and xanthine were purchased from Sigma Inc., dissolved in 0.1 N NaOH at 10 mg/ml, and stored at −20°C. Cytosine β-d-arabinofuranoside was purchased from Sigma. BSC40 cells were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% calf serum (CS). Vaccinia virus wild-type strain WR was supplied by S. Pennathur. vT7lacOI was obtained from B. Moss (72). vT7lacOI expresses T7 RNA polymerase under the regulation of IPTG to allow high-level expression of the target gene, which is inserted at the nonessential A56R gene locus. All viruses were grown in BSC40 cells. IMV were purified by 20 to 40% sucrose gradient centrifugation and stored at −70°C as described previously (29). Rabbit antibodies were generated for vaccinia virus D8L, A27L, H3L, and L1R proteins, and some were described previously (22, 33). Antibodies against A13L, A14L, A17L-N′, and viral cores were provided by J. Krijnse Locker (27, 39, 50). A monoclonal antibody against A45R protein was provided by G. Smith (4).
Antigen preparation and rabbit immunization.
Full-length recombinant J1R protein was expressed in bacteria and used as an antigen to generate antibodies in rabbits. The J1R gene was amplified by PCR with the following two primers (restriction sites are underlined): 5′ primer, 5′-TATGAATTCATGGATCACAACCAGTATCTC-3′, and 3′ primer, 5′-CCCAAGCTTATTATTGTTCACTTTATT-3′. The PCR product was digested with EcoRI and HindIII and cloned into pET21a (Novagen). The resulting plasmid, pET21a-J1R, expressed J1R protein with a T7 tag peptide at the N terminus for detection and hexahistidine sequences at the C terminus for purification as described previously (23). In brief, the plasmid was transformed into Escherichia coli BL21(DE3), and cultures were induced with 0.2 mM IPTG for 30 min at 37°C and harvested. Cell lysates were sonicated and loaded onto a nickel affinity column, and recombinant J1R protein was eluted with 0.3 M imidazole. Purified J1R protein was loaded onto a sodium dodecyl sulfate (SDS)-12% polyacrylamide gel to remove minor contaminants. Gel slices containing 500 μg of J1R protein were excised and injected intramuscularly into each New Zealand White rabbit (F5 and F6). The rabbits were boosted with 250 μg of recombinant J1R protein prepared as described above five times at 2-week intervals and bled 1 week after the third boost. The antibody titer was determined by immunoblot analysis. The anti-J1R antibody (F6) recognized J1R protein in both Western blot (1:1,000) and immunoprecipitation (1:500) experiments.
Generation of viJ1R virus, which expresses J1R protein upon IPTG induction. (i) Plasmid construction.
To construct pMITEO-J1R with an inducible copy of the J1R gene, the full-length J1R open reading frame (ORF) was generated by PCR with the following two primers (NcoI and BamHI restriction sites are underlined): 5′-AAACCATGGATCACAACCAGTATCTC-3′ and 5′-CCCGGATCCTTAATTATTGTTCACTTT-3′. Vaccinia virus genomic DNA (strain WR) was used as the template. The PCR product was digested with NcoI and BamHI and cloned into pMITEOlac.20/3 to produce pMITEO-J1R (24).
Three DNA fragments were used to replace the endogenous J1R gene with a lacZ expression cassette. The 5′-flanking sequence of a 618-bp DNA fragment upstream of the J1R ORF was generated by PCR with the following two primers: primer A, 5′-GGGCTCGAGGTCCTGAATGTGTATTCT-3′ (XhoI site is underlined), and primer B, 5′-CCAGTCGACGAATCATCATCTGCGAAG-3′ (SalI site is underlined). vT7lacOI genomic DNA was used as the template. The 3′-flanking fragment of 1,228 bp downstream of the J1R ORF was generated by PCR with the following two primers: primer E, 5′-GGGGGATCCATCGCATTTTCTAACGTG-3′ (BamHI site is underlined), and primer F, 5′-AAAGCGGCCGCGCACTGGCTGGTCAATGG-3′ (NotI site is underlined). vT7lacOI genomic DNA was used as the template. The 3-kb p11k-lacZ gene expression cassette was isolated from pSC11-5t by SalI and PstI digestion (13). pSC11-5t was derived from pSC11 by removing the p7.5K promoter and inserting a multiple cloning site with a SalI site; this made it possible to isolate the p11k-lacZ cassette as a 3-kb SalI-PstI fragment (13). The 5′-flanking DNA fragment was cloned into the pBluescript KS(−) vector (Stratagene) to create pL4L5, which was ligated to the lacZ cassette to create pL4L5/lacZ. Finally, the 3′-flanking sequence was inserted into pL4L5/lacZ to create pL4L5/lacZ/J2T7. The sequences of PCR fragments were confirmed by DNA sequencing.
(ii) Construction of recombinant viJ1R virus.
Recombinant viJ1R virus was constructed by established protocols as described previously (79). In brief, 5 × 105 CV-1 cells were seeded, incubated for 1 day, and infected with vT7lacOI at a multiplicity of infection (MOI) of 5 PFU per cell for 1 h at 37°C. The cells were then washed three times with DMEM and transfected with 6 μg of pMITEO-J1R in 60 μl of Lipofectamine (Gibco-BRL, Inc.). After 5 h, the transfection mixtures were removed and replaced with complete DMEM containing 10% fetal bovine serum. The lysates were harvested at 2 days postinfection (p.i.) and used to infect BSC40 cell monolayers in the presence of mycophenolic acid (25 μg/ml), xanthine (250 μg/ml), and hypoxanthine (15 μg/ml) to select for plaques formed by the intermediate virus, vJ1R/iJ1R, which expresses xanthine-guanine phosphoribosyltransferase, as described previously (79). Pure recombinant vJ1R/iJ1R virus was obtained after three rounds of plaque purification. The insertion of xanthine-guanine phosphoribosyltransferase and inducible J1R genes into the viral A56R gene locus was confirmed by PCR. vJ1R/iJ1R virus was used to generate viJ1R virus as follows.
CV-1 cells were infected with the intermediate virus, vJ1R/iJ1R, at an MOI of 5 PFU per cell and transfected with 6 μg of pL4L5/lacZ/J2T7 as described above. The lysates were harvested at 2 days p.i. and used to infect BSC40 cell monolayers in the presence of 50 μM IPTG. The infected cells were overlaid with agar, incubated for 2 days at 37°C, and then overlaid with a second layer of agar containing 150 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/ml. Two blue plaques were isolated independently after three consecutive rounds of plaque purification. Recombinant viJ1R viruses obtained from these two plaques behaved identically in our experiments.
One-step virus growth curve analysis.
BSC40 cell monolayers were infected with vaccinia virus at an MOI of 5 PFU per cell for 1 h at 37°C. The cells were then incubated in complete DMEM containing 10% CS with or without 50 μM IPTG and harvested at various times (0, 1, 2, 4, 6, 8, 12, 16, 18, and 24 h) after infection. The infected cells were subjected to three freeze-thaw cycles and sonicated, and virus titers were determined by plaque assays in the presence of 50 μM IPTG. The experiments were repeated twice, and the averages are presented.
Immunoblot analysis.
Viral proteins from purified IMV or extracts from virus-infected cells were fractionated by SDS-12% polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes. Membranes were blocked by incubation with 3% nonfat milk in 0.5% Tween 20-20 mM Tris-HCl (pH 7.4)-0.5 M NaCl and then incubated with primary antibody to viral proteins. Alkaline phosphatase-conjugated secondary antibody was used for detection by a chemiluminescence method according to the manufacturer's protocol (Tropix). Blots were stripped with stripping buffer (0.7% β-mercaptoethanol, 2% SDS, 62.5 mM Tris-HCl [pH 6.8]) at 50°C for 30 min and two washes in phosphate-buffered saline (PBS)-0.5%Tween 20 of 10 min each and then reused for subsequent probing.
Membrane protein extraction from IMV.
Vaccinia virus IMV were extracted with detergent to separate membrane and core fractions essentially as described previously (15, 55, 63, 74, 81). In brief, purified IMV (108 PFU) were incubated for 1 h at 37°C in 1% NP-40-50 mM Tris-HCl (pH 7.5) with or without 50 mM dithiothreitol (DTT). The insoluble and soluble fractions were separated by centrifugation at 14,000 × g for 30 min at 4°C. Proteins from the pellet and supernatant were analyzed by SDS-12% PAGE and transferred to nitrocellulose for Western blot analyses with various antibodies as described above.
[35S]methionine protein labeling.
BSC40 cells were infected with vT7lacOI or viJ1R virus at an MOI of 10 PFU per cell for 1 h at 37°C. The infected cells were incubated with complete DMEM containing 10% CS with or without 50 μM IPTG. The infected cells were pulse-labeled for 15 min with [35S]methionine (50 μCi/ml) at 1, 2, 4, 6, 8, 12, and 24 h p.i. Labeled proteins were harvested in SDS-containing sample buffer and analyzed by SDS-10% PAGE.
To monitor p4a and p4b core protein processing, cells were pulse-labeled with [35S]methionine (50 μCi/ml) for 30 min at 8 h p.i.; the pulse was followed by incubation in growth medium lacking [35S]methionine for 0, 0.25, 0.5, 1, 2, 4, 12, or 24 h. The cells were harvested in SDS-containing sample buffer, and proteins were analyzed by SDS-10% PAGE. After electrophoresis, the gel was fixed, dried, and analyzed by autoradiography as described previously (68).
Viral DNA analysis.
BSC40 cells were infected with viJ1R virus at an MOI of 10 PFU per cell for 1 h at 37°C. The infected cells were incubated with complete DMEM containing 10% CS with or without 50 μM IPTG. The cells were harvested at 2, 4, 8, and 24 h p.i.; viral DNA was extracted, digested with BstEII, separated on a 1% agarose gel, and transferred to nitrocellulose paper for hybridization with a 32P-end-labeled 70-mer oligonucleotide as described previously (6, 12). The 70-mer oligonucleotide sequence represents vaccinia virus tandem repeats and is close to the terminal loop: 5′-TTTTTGTGAGACCATCGAAGAGAGAAAGAGATAAAACTTTTTTACGACTCCATCAGAAAGAGGTTTAATA-3′. The blot was washed in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C for 60 min, air dried, and autoradiographed.
Electron microscopy of virion morphogenesis.
BSC40 cells were seeded on round coverslips and infected at an MOI of 20 PFU per cell. These cells were directly fixed on coverslips at 24 h p.i. with 2.5% glutaraldehyde in 0.1 M PBS (pH 7.0) at room temperature for 1 h and then rinsed in three 15-min changes of 0.1 M sodium phosphate buffer (pH 7.0). The cells were treated with 1% OsO4 in 0.1 M sodium phosphate (pH 7.0) at room temperature for 60 min and washed three times in 0.1 M sodium phosphate (pH 7.0). The cells were dehydrated with an ethanol series from 30 to 100% ethanol, and Spurr's resin was used for infiltration and embedding as described previously (61). After embedding, the cells were separated from coverslips and thin sectioned with an Ultracut Eultramicrotome. Thin sections of 90 nm were stained with uranyl acetate and lead citrate and analyzed under a Zeiss 902 transmission electron microscope (41).
Immunoprecipitation.
BSC40 cells were infected with viruses at an MOI of 5 PFU per cell and incubated for 24 h. The cells were rapidly chilled on ice, washed with ice-cold PBS, and lysed in lysis buffer (20 mM Tris-HCl [pH 8.0], 20 mM EDTA, 80 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride) (46). Insoluble material was removed by centrifugation (10,000 × g, 10 min, 4°C). Cell lysates were incubated with anti-A45R monoclonal antibody (1:500) or anti-J1R antibody (1:500) for 2 h at 4°C. Immune complexes were incubated with protein A/G-Sepharose (Santa Cruz), washed five times with lysis buffer, and separated by SDS-12% PAGE. After electrophoresis, the gel was subjected to immunoblot analysis with anti-J1R (1:1,000) or anti-A45R (1:5,000) antibody.
RESULTS
The conserved vaccinia virus J1R gene encodes a late viral membrane protein associated with IMV particles.
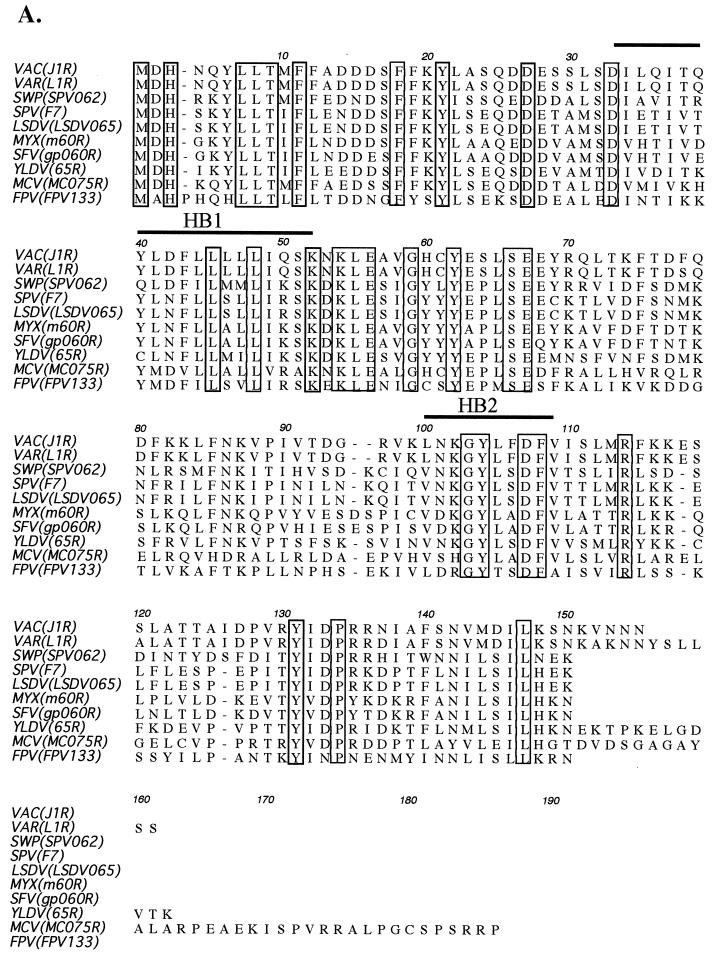
The vaccinia virus J1R gene encodes a polypeptide of 153 amino acids with a predicted molecular mass of 17.8 kDa (19). A multiple sequence alignment of the vaccinia virus J1R protein with its orthologues in the poxvirus family is shown in Fig. 1A. This alignment reveals a high level of homology among these proteins (>58% conserved residues), suggesting that J1R protein may play an important function in the poxvirus life cycle. The C-terminal region of J1R protein is less well conserved than the N-terminal region, and the length of the C-terminal region is variable.
FIG. 1.
Vaccinia virus J1R gene encodes a late protein. (A) Alignment of deduced amino acid sequences of vaccinia virus J1R and J1R orthologues in other poxviruses. Accession numbers are from GenBank. VAC, vaccinia virus (strain WR; accession no. P07616); VAR, variola virus (India-1967/isolate IND3; accession no. NP-042122); SWP, swine poxvirus (accession no. NP-570222); SPV, sheep poxvirus (accession no. P19746); LSDV, lumpy skin disease virus (accession no. AAK85026); MYX, myxoma virus (accession no. NP-051774); SFV, Shope fibroma virus (accession no. NP-051949); YLDV, Yaba-like disease virus (accession no. NP-073450); MCV, molluscum contagiosum virus subtype 1 (accession no. NP-044026); FPV, fowl poxvirus (accession no. NP-039096). The boxed sequences are identical amino acids, and the thick lines indicate hydrophobic sequences (HB). (B) Hydropathy plot of J1R protein. a.a., amino acid. (C) Expression of J1R protein in infected cells and IMV. BSC40 cells were infected with wild-type vaccinia virus at an MOI of 5 PFU per cell and harvested at the indicated times. Lysates were separated by SDS-12% PAGE and transferred to nitrocellulose for Western blotting with rabbit antibody against J1R protein. araC, cytosine β-d-arabinofuranoside; hp.i, hours postinfection; V, purified IMV particles.
The nucleotide sequence 5′ to the initiation codon of J1R, AATAA, matches the viral late promoter consensus sequence, indicating that J1R protein is likely to be expressed during the late phase of virus infection (49). A hydropathy analysis indicated that J1R protein has two hydrophobic domains (designated HB) spanning residues 34 to 52 and residues 100 to 109 (Fig. 1A and B) (30). Viral late proteins A17L, A14L, and L1R also have multiple hydrophobic regions which subsequently were shown to be membrane-spanning regions and components of IMV that play a role in virion morphogenesis or virus entry (26, 40, 45-48, 64, 79, 80). We postulate that J1R protein may be similar to these proteins in having a structural or morphogenic involvement with IMV particles.
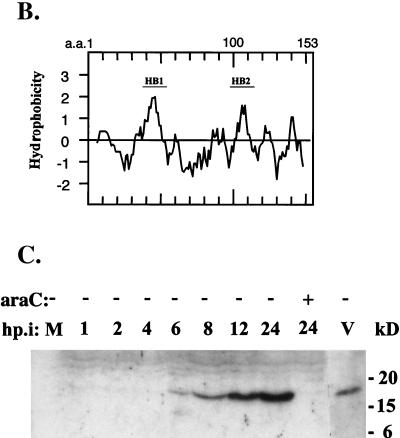
Anti-J1R antibody was used to study the localization of J1R protein during vaccinia virus infection. Rabbits were immunized with recombinant J1R protein purified from a prokaryotic expression system (see Materials and Methods). This rabbit serum was tested by Western blotting with lysates from virus-infected cells (Fig. 1C). The serum did not recognize any protein in mock-infected cells; however, it recognized an 18-kDa protein in virus-infected cells that was first detected at 6 h p.i. and increased in abundance until 24 h p.i. The 18-kDa protein was not detected in virus-infected cells treated with cytosine β-d-arabinofuranoside, which inhibits viral DNA replication and blocks expression from late viral promoters. The J1R antiserum also detected an 18-kDa protein in purified IMV (Fig. 1C).
The localization of J1R protein in IMV was also studied. Purified IMV were extracted with 1% NP-40 and 50 mM DTT. Virion membranes dissolved into the soluble detergent phase (supernatant), which was separated from insoluble core components (pellet) by centrifugation (4, 15, 39, 47, 55, 63, 74, 81). As shown in Fig. 2, J1R protein was extracted from virions and partially released into the supernatant in buffer containing 1% NP-40; J1R protein was extracted into the supernatant more completely in buffer containing 1% NP-40 and 50 mM DTT. The results indicated that J1R protein is associated with the membrane. Other IMV membrane proteins, including L1R, A17L, A27L, A14L, D8L, H3L, and A13L, behaved similarly and were extracted into the supernatant fraction (Fig. 2, lower panel). Viral core proteins 4a and 4b, A45R, and F17R were more resistant to detergent extraction and were associated with the insoluble pellet fraction (Fig. 2, upper panel) (4, 15, 39, 47, 50).
FIG. 2.
Membrane and core proteins after NP-40-DTT extraction of vaccinia virus IMV. Sucrose-purified vaccinia virus IMV were incubated with buffer containing 1% NP-40 and 50 mM DTT or no DTT. After centrifugation, the supernatant (S) and the insoluble pellet (P) were analyzed with antibodies as described in Materials and Methods. c, core; m, membrane.
Jensen et al. (27) carried out 2D gel electrophoresis of detergent-extracted IMV and identified 11 membrane-associated viral proteins in the IMV. However, several additional membrane proteins appeared on the 2D gel as minor spots that could not be identified because of their low abundance (27). J1R protein could be a low-abundance membrane-associated component of IMV that might have been overlooked in prior studies.
Construction of a recombinant vaccinia virus expressing the inducible J1R gene under the control of the lac operator.
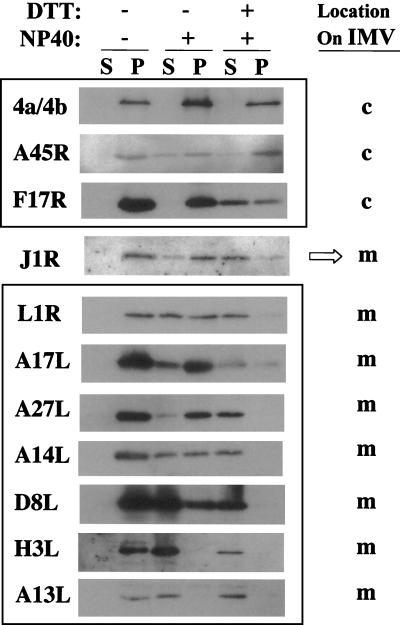
The role played by J1R protein during the vaccinia virus life cycle in cell cultures was explored by using a recombinant vaccinia virus, viJ1R, that conditionally expresses J1R protein in an IPTG-regulated manner. viJ1R was generated from parental virus vT7lacOI (Fig. 3A) (72). First, an inducible copy of J1R was inserted into the A56R (hemagglutinin) locus to produce vJ1R/iJ1R, an intermediate virus that retains the endogenous J1R locus. Second, the endogenous J1R locus of vJ1R/iJ1R was replaced with a lacZ marker gene by homologous recombination to produce viJ1R. Blue plaques were isolated in the presence of 50 μM IPTG and purified after three rounds of plaque purification.
FIG. 3.
Construction of viJ1R for conditional expression of J1R protein. (A) Schematic diagram of the parental virus vT7lacOI, the intermediate virus vJ1R/iJ1R, and the final mutant virus viJ1R. The J1R, J2R (thymidine kinase [TK]), and A56R (hemagglutinin) loci are indicated. DNA fragments inserted into these loci are shown below the lines. Abbreviations: T7 pol, bacteriophage T7 RNA polymerase gene; lacO, E. coli lac operator; PL, viral late promoter; PE/L, viral early and late promoters; lacI, E. coli lac repressor; EMC, encephalomyocarditis virus cap-independent translation enhancer element; gpt, E. coli guanine phophoribosyltransferase gene; P7.5 and P11, viral promoters; PT7, promoter for T7 RNA polymerase. (B) Expression of J1R protein in cells infected with viJ1R. Cells were infected with vT7lacOI or viJ1R at an MOI of 5 PFU per cell and harvested for Western blotting with antibody against J1R protein. M, mock-infected cells.
BSC40 cells were infected with viJ1R, cultured in medium with or without IPTG for 24 h, and harvested for immunoblot analysis (Fig. 3B). Parental virus vT7lacOI expressed comparable amounts of J1R protein in infected cells 24 h p.i. in the presence and absence of IPTG. Recombinant virus viJ1R expressed abundant J1R protein only in the presence of IPTG, and leaky expression of J1R protein was not detected in the absence of IPTG. These results are consistent with those of previous studies in which vT7lacOI was used to produce recombinant viruses that are tightly regulated by IPTG; thus, this system is useful for gene expression studies (12, 17, 55, 63, 74, 78, 79, 81).
J1R protein is required for plaque formation and IMV production in cell cultures.
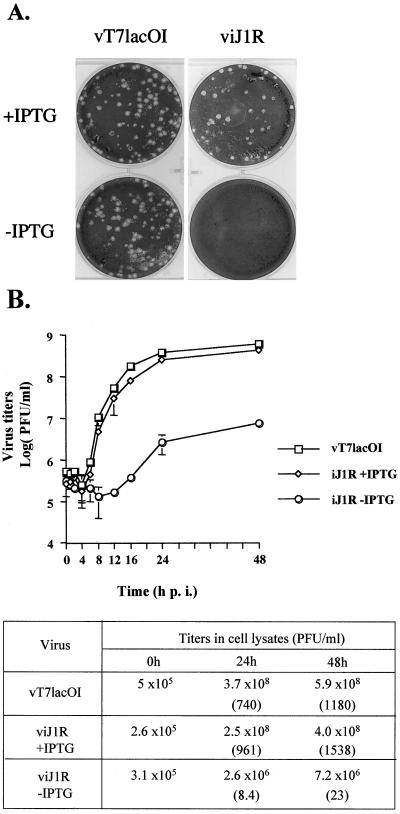
The role played by J1R protein during virus infection was examined by using BSC40 cells infected with viJ1R in the presence or absence of IPTG, which turns on or turns off J1R expression, respectively (Fig. 4A). Cells were also infected with parental virus vT7lacOI as a control. vT7lacOI formed plaques 3 days p.i in the presence or absence of IPTG. However, viJ1R formed plaques only in the presence of IPTG, indicating that the expression of J1R is required for plaque formation. viJ1R formed plaques similar in size to those of the parental virus, indicating that the inducible J1R gene has a nearly wild-type function. The infected cells were incubated in the absence of IPTG for up to 7 days, and no detectable plaques were produced (data not shown).
FIG. 4.
Characterization of viJ1R. (A) viJ1R mutant virus does not form plaques on BSC40 cells. BSC40 cells were infected with vT7lacOI or viJ1R, incubated in medium for 3 days, fixed, stained with crystal violet, and photographed. (B) One-step growth curve analysis of viJ1R virus. BSC40 cells were infected with parental vT7lacOI or viJ1R at an MOI of 5 PFU per cell; incubated in normal medium or medium with IPTG; and harvested at 0, 1, 2, 4, 6, 8, 12, 16, 24, and 48 h p.i. Error bars indicate standard deviations. Virus titers in the lysates were determined by plaque formation assays with BSC40 cells and are listed in the table. Numbers in parentheses are the fold increase in virus titer, determined as the virus titer at 24 or 48 h p.i. divided by the virus titer at 0 h.
viJ1R titers in cell cultures were measured by one-step growth analysis. BSC40 cells were infected with viJ1R at an MOI of 5 PFU per cell and cultured in the presence or absence of IPTG. Cell lysates were collected, and virus titers were determined (Fig. 4B). Parental virus vT7lacOI produced averages of 740- and 1,180-fold increases in virus titers in cell lysates at 24 and 48 h p.i., respectively (Fig. 4B, table). Recombinant virus viJ1R grew poorly in the absence of IPTG, increasing virus titers 8.4- and 23-fold at 24 and 48 h p.i., respectively. In contrast, viJ1R increased virus titers 961- and 1,538-fold in the presence of IPTG. These results show that viJ1R has a wild-type titer when J1R expression is induced with IPTG. In addition, a deficiency of J1R protein severely impairs virus growth and reduces the virus titer 114-fold in 24 h. Thus, J1R protein is important for vaccinia virus growth in cell cultures.
J1R protein is not required for viral protein synthesis but is required for processing of viral core proteins 4a and 4b.
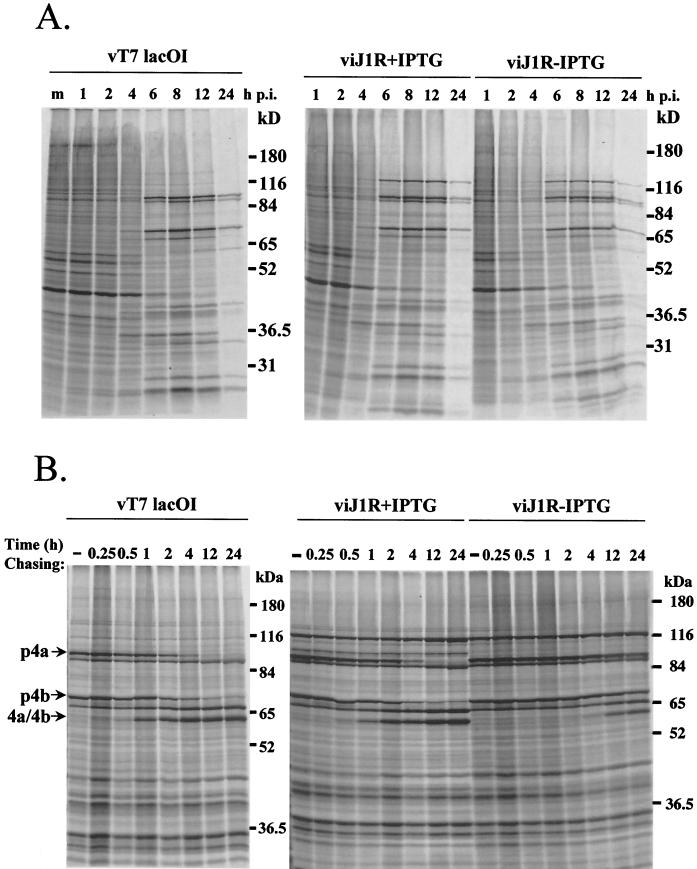
Experiments were carried out to determine more precisely when and where J1R protein is required during the vaccinia virus life cycle. Viral protein synthesis was monitored by a pulse-labeling experiment. BSC40 cells were pulse-labeled with [35S]methionine at various times after virus infection (Fig. 5A). In BSC40 cells infected with vT7lacOI, the synthesis of host proteins was gradually shut off and the synthesis of viral intermediate and late proteins was predominant from 4 to 12 h p.i. (37, 42). The patterns of viral protein synthesis were similar in cells infected with viJ1R in the presence and absence of IPTG. These results demonstrate that J1R protein is not required for viral gene expression.
FIG. 5.
Viral protein synthesis in cells infected by viJ1R. (A) Pulse-labeling of viral proteins. BSC40 cells were infected with vT7lacOI or viJ1R at an MOI of 10 PFU per cell in the presence (+) or absence (−) of 50 μM IPTG. The cells were labeled with [35S]methionine (50 μCi/ml) for 15 min at 1, 2, 4, 6, 8, 12, and 24 h p.i. m, mock infected. Immediately after labeling, the cells were washed and lysed, and the labeled proteins were separated by SDS-10% PAGE. (B) Pulse-chase analysis of precursor p4a and p4b processing. BSC40 cells were infected with either vT7lacOI or viJ1R in the presence (+) or absence (−) of 50 μM IPTG. At 8 h p.i., the cells were pulse-labeled with [35S]methionine for 30 min and either immediately harvested (−) or chased with normal medium for 0.25, 0.5, 1, 2, 4, 12, or 24 h. Proteins were denatured and analyzed by SDS-10% PAGE followed by autoradiography. The mobilities of p4a and p4b and their mature processed forms, 4a and 4b, are shown at the left (68, 69).
Protein processing plays an important role during the vaccinia virus life cycle (68, 69, 75, 76). For example, p4a and p4b are major precursors of vaccinia virus core proteins that are proteolytically processed to become mature core proteins 4a and 4b (68, 69). This processing occurs during the late phase of viral infection and can be used as a diagnostic marker for the conversion of immature virions (IV) to IMV (68). If 4a and 4b processing is delayed or absent, then it is likely that IMV formation will be blocked (12, 32, 33, 40, 45, 48, 63, 74, 76, 79, 81, 83; B. Moss and E. N. Rosenblum, Letter, J. Mol. Biol. 81:267-269, 1973). Experiments were performed to determine whether J1R protein is important for p4a and p4b processing (Fig. 5B). BSC40 cells were infected with vT7lacOI or viJ1R, and viral protein synthesis was pulse-labeled for 30 min starting at 8 h p.i. and chased for various times up to 24 h p.i. In cells infected with vT7lacOI, p4a and p4b were synthesized and processed to mature core proteins 4a and 4b with a half-life (t1/2) of 4 h (68). In cells infected with viJ1R and cultured in the presence of IPTG, a similar pattern of p4a and p4b processing was observed. However, when cells were infected with viJ1R and cultured in the absence of IPTG, the processing of p4a and p4b was significantly delayed, and only minor processing was observed after 12 to 24 h, with a t1/2 of 24 h. These results strongly suggest that a deficiency of J1R protein blocks the processing of p4a and p4b. The processing of other viral proteins, such as A17L protein, was also inhibited in virus infections that did not express J1R (data not shown).
Abnormal empty IV associated with enlarged dense viroplasm structures accumulate during infections lacking J1R protein.
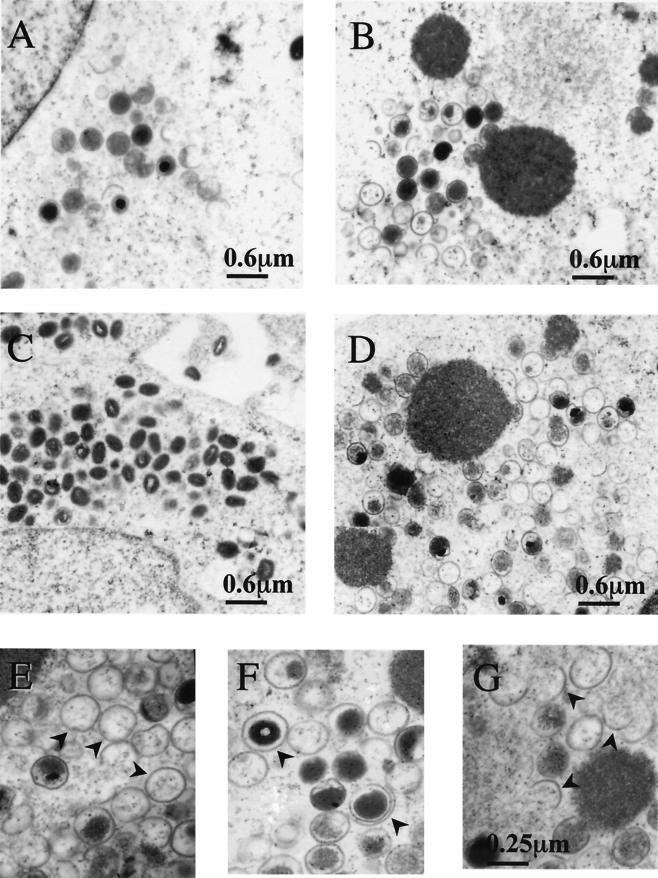
BSC40 cells were infected with viJ1R in the presence or absence of IPTG, and cells were analyzed at 12 and 24 h p.i. by electron microscopy (Fig. 6). At 12 h p.i., IV and other intermediate membrane structures were detected in cells infected with viJ1R in the presence of IPTG (Fig. 6A). Similar viral structures were also detected in infected cells in the absence of IPTG. In addition, electron-dense viroplasm structures were also present in these infected cells (Fig. 6B). At 24 h p.i., a large number of dense IMV particles were detected in the cytoplasm of cells infected with viJ1R in the presence of IPTG (Fig. 6C). However, in cells infected with viJ1R in the absence of IPTG, large dense viroplasm structures accumulated more and were often associated with abnormal membrane structures (Fig. 6D). Viral crescents formed around the edges of the enlarged viroplasm structures; half-filled crescents were also observed separate from the dense viroplasm structures. Many abnormal IV particles were detected in the cytoplasm, including “electron-lucent” or empty double layers and half-circle and open-circle structures (Fig. 6E to G). The internal dense content of the IV appeared to be separate from the inner membrane of the IV, and the condensed nucleoid inside the IV was minimal. Mature brick-shaped IMV were rare in these infected cells. This analysis suggested that the initial step of crescent formation is not affected by the lack of J1R protein; however, J1R protein is required for the formation of IV. When J1R protein is absent, the IV membrane does not enclose much viroplasm, resulting in electron-lucent IV particles or partially packaged IV particles.
FIG. 6.
Electron micrographs of vaccinia virion morphogenesis in cells infected with viJ1R. BSC40 cells were infected with viJ1R virus at an MOI of 20 PFU per cell either in the presence (A and C) or in the absence (B and D to G) of IPTG and fixed at 12 h (A and B) or 24 h (C to G) p.i. for electron microscopy. Photos were taken at magnifications of ×12,000 (A to D) and ×30,000 (E to G). Arrowheads in panels E to G represent aberrant membrane structures, such as empty IV (E), double-layer membranes (F), and half-circle membranes (G).
Viral DNA processing occurs normally in infected cells lacking J1R protein.
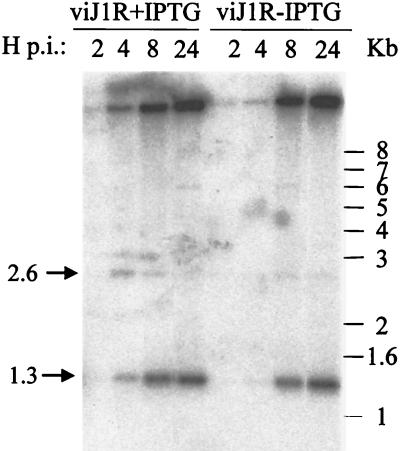
The presence of empty and partially packaged IV particles indicates that J1R protein is required for viral DNA packaging during IV formation. Alternatively, J1R protein could be involved in the processing of viral DNA concatemers into monomers, and the failure of the resolution of viral DNA ends in the absence of J1R protein could lead to the same phenotype as that of a DNA packaging mutant.
In order to differentiate the above two possibilities, viral DNA was isolated from cells infected with viJ1R in the presence or absence of IPTG. Because BstEII cleaves at a distance of 1.3 kb from each end of the unit-length mature genome, fragments of 2.6 kb are formed by enzyme cleavage of concatemeric genomic molecules (12). With a 70-oligomer probe marking the DNA ends, replicated viral DNA accumulated at 8 to 24 h p.i., and a predominant band of 1.3 kb was observed, indicating that the efficient formation of unit-length genomes occurs with or without J1R protein (Fig. 7). The 2.6-kb fragment was difficult to detect, consistent with normal rapid processing of viral DNA concatemers (6). We therefore concluded that J1R protein is not required for viral DNA processing and, hence, that the defect in morphogenesis resides in a DNA packaging step.
FIG. 7.
Processing of viral DNA. BSC40 cells were infected with viJ1R at an MOI of 10 PFU per cell and incubated with complete DMEM containing 10% CS with (+) or without (−) 50 μM IPTG. Cells were harvested at 2, 4, 8, and 24 h p.i. Viral DNA was extracted, digested with BstEII, separated on an 1% agarose gel, and transferred to nitrocellulose paper for hybridization with a 32P-end-labeled 70-mer oligonucleotide as described previously (12). The arrow labeled 1.3 marks the position of the diagnostic 1.3-kb DNA fragment, which indicates the resolution of concatemers to monomeric DNA ends.
Vaccinia virus J1R protein interacts with A45R protein in virus-infected cells.
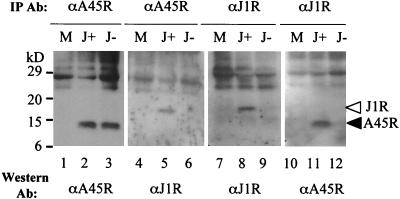
A previous study with a yeast two-hybrid screen suggested that vaccinia virus J1R interacts with itself and with A45R (34). However, it was not known whether such interactions occur in infected cells. A45R protein is a 13.5-kDa viral core protein which has a superoxide dismutase-like motif and which is not essential for vaccinia virus growth in cells (4). J1R protein, as reported here, is essential for virus growth in cell cultures. To investigate whether J1R protein interacts with A45R protein in virus-infected cells, J1R protein was immunoprecipitated from cell lysates harvested at 24 h p.i. and analyzed by SDS-PAGE and Western blotting (Fig. 8). Anti-A45R antibody immunoprecipitated similar amounts of A45R protein from infected cells with and without IPTG (Fig. 8, lanes 2 and 3). Anti-A45R antibody also immunoprecipitated the 18-kDa J1R protein from lysates harvested in the presence of IPTG (Fig. 8, lane 5). Since anti-A45R antibody does not cross-react with J1R protein, these results indicate that A45R protein interacts with J1R protein. The interaction is specific to J1R protein and was not detected in infected cells grown in the absence of IPTG (Fig. 8, lane 6). Furthermore, A45R protein was also detected when immunoprecipitation was performed with anti-J1R antibody and lysates from cells incubated with IPTG (Fig. 8, lanes 11 and 12). These results indicate that J1R protein interacts with A45R protein in virus-infected cells.
FIG. 8.
Specific interaction between A45R and J1R proteins. BSC40 cells were either mock infected (M) or infected with viJ1R at an MOI of 5 PFU per cell and then incubated in medium with IPTG (J+) or without IPTG (J−). Cell lysates were harvested at 24 h p.i. and immunoprecipitated (IP) with antibody. Immune complexes were separated by SDS-12% PAGE and analyzed by Western blotting with anti-A45R (1:5,000) or anti-J1R (1:1,000) antibody.
DISCUSSION
The vaccinia virus J1R gene encodes a conserved protein that belongs to a family of homologues in poxviruses with an average of 37% identical amino acid residues. The J1R gene occurs widely in the chordopoxvirinae, but it is not present in the entomopoxvirinae subfamily of poxviruses (1-3, 5, 7, 11, 19, 28, 31, 54, 57, 65, 77). Amino acid sequences in the N-terminal region of J1R protein are more highly conserved than sequences in the C-terminal region. The residues between hydrophobic regions 1 and 2, i.e., residues 66 to 101, are 30% identical to residues within the von Willebrand factor type A domain of cellular adhesion molecule Lu-ECAM-1 (GenBank accession no. T02152). Residues 91 to 138 of J1R protein are homologous to the phosphotriesterase-like domain of a hypothetical protein encoded by Mycoplasma pulmonis (14). The phosphotriesterase activity was first identified in soil bacteria and appears to have evolved the ability to hydrolyze the insecticide paraoxon (52). The significance of this homology to J1R protein is not known.
The J1R gene has a typical late transcription initiation site upstream of the translation initiation ATG codon. J1R protein is detected as a 18-kDa protein during late viral gene expression. J1R has two putative hydrophobic regions, indicating a possible association with the virion membrane. This suggestion is consistent with the fact that J1R can be efficiently extracted from purified IMV by detergent in the presence of DTT. This behavior has also been observed with other vaccinia virus membrane proteins, such as L1R, A17L, A27L, and A14L (39, 44, 47, 63). Although J1R has two hydrophobic regions that may serve as transmembrane regions, computer programs such as TMpred have suggested otherwise. Alternatively, J1R could be embedded in the membrane. Anti-J1R antiserum failed to specifically recognize native J1R protein in electron microscopy or confocal microscopy, probably because the antiserum mainly recognizes epitopes on denatured J1R protein (data not shown). The topology of J1R protein in the IMV membrane remains unknown and is currently under investigation.
The role of J1R protein in the vaccinia virus life cycle was examined with a recombinant virus (viJ1R) that expresses J1R protein in an IPTG-inducible manner. This system utilizes parental virus vT7lacOI, which has been widely used for numerous recombinant virus constructions (12, 17, 55, 63, 74, 78, 79, 81). The expression of J1R protein in viJ1R was tightly regulated by IPTG, and no leaky protein expression was detected in the absence of IPTG. Furthermore, when J1R protein was not expressed, viJ1R grew very poorly, with a 100-fold reduction in IMV progeny, indicating that J1R protein plays an important role during vaccinia virus infection. The minimal growth of viJ1R in the absence of IPTG may indicate that J1R protein plays a regulatory role to enhance viral growth. Alternatively, it remains possible that a trace amount of J1R protein was expressed in the absence of IPTG and aided virion assembly under nonpermissive conditions.
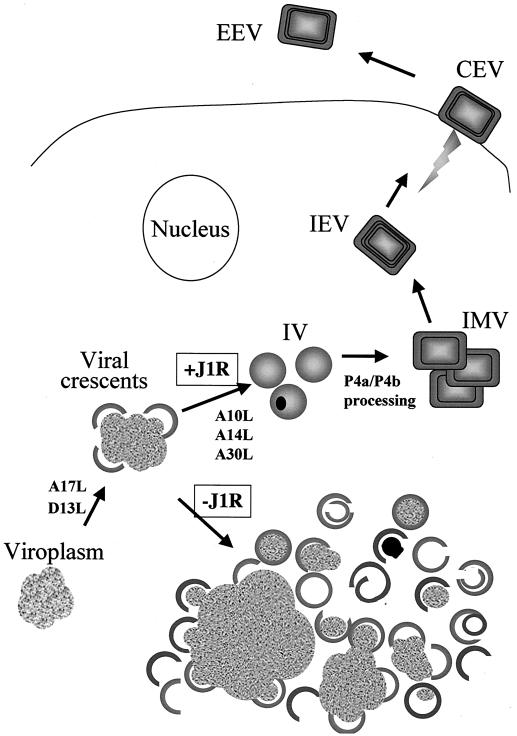
J1R protein is not required for viral protein synthesis in infected cells. However, the proteolytic processing of core protein precursors p4a and p4b was severely delayed, indicating that virion assembly was interrupted at or prior to the conversion of IV to IMV. Not only a pulse-chase experiment but also immunoblot analysis of cell lysates with an anti-core antibody recognized abundant p4a precursor accumulation and the absence of mature 4a protein when J1R was not expressed (data not shown). Electron micrographs of cells infected with viJ1R in the absence of IPTG demonstrated a dramatic accumulation of enlarged electron-dense viroplasm structures that remained associated with viral crescents. Thus, J1R protein is not required for crescent formation, in contrast to the A17L and D13L mutants (45, 47, 59, 79). A more striking defect caused by J1R deficiency was the presence of numerous membrane structures that contained half-circles, open circles, abnormal shapes, or other structures reminiscent of empty or partially filled IV. Viral DNA processing appeared normal in the absence of J1R protein. IV with electron-dense contents or IV with condensed nucleoids were very rare during infections lacking J1R. These results indicate that J1R is required for DNA packaging during IV formation (Fig. 9).
FIG. 9.
J1R protein is required for DNA packaging in IV formation during vaccinia virion morphogenesis. Schematic drawing of the stages of vaccinia virus morphogenesis, including crescent formation, IV formation, and IMV formation. IMV could be enveloped to become IEV. J1R protein is not required for crescent formation; instead, it plays a role in DNA packaging in IV formation. Also shown are several other viral proteins (A10L, A14L, A17L, A30L, and D13L) in which a mutation interferes with virion morphogenesis before or during the formation of IV (20, 45, 47, 48, 59, 63, 79, 82). CEV, cell-associated virions; EEV, extracellular enveloped viruses.
Several other vaccinia virus mutants that are also defective at IV formation during morphogenesis were identified, such as A10L, A14L, and A30L (20, 48, 63, 64). The phenotypes of these mutants are not identical to that of the J1R mutant. For example, mutations in A14L resulted in the accumulation of dense viroplasm structures and no IV formation in cells. However, the viral membrane structures observed in these cells were more fragmented and tubular-vesicular, indicating an additional role for A14L in crescent formation (48, 64). Mutations in A10L produced DNA bundles that were not incorporated into IMV, resulting in empty spherical viral particles similar to those observed with the J1R mutant (20). However, the organization of electron-dense viroplasm structures into regularly spaced bands that accumulated in the cytoplasm of cells was observed only in cells infected with the A10L mutant virus and not with the J1R mutant virus.
Abnormal membrane structures and empty IV were also described for the A30L mutant virus (62, 63). However, certain features remain distinct from those of the J1R mutant virus. First, enlarged masses of granular viroplasm were not associated with viral membranes in the A30L mutant, whereas a close association of viral crescents and viroplasm structures was common in the J1R mutant. Holes in the dense viroplasm observed in cells infected with the A30 mutant were not observed in cells infected with the J1R mutant.
Although the J1R mutant does not exhibit the same phenotypes as the A14L, A10L, and A30L mutants, mutations in each of the genes result in a blockage of IV maturation (Fig. 9). It will be interesting to determine whether J1R protein interacts with any of these proteins to facilitate efficient virion assembly.
J1R protein interacts with A45R protein in infected cells, consistent with previous results obtained with a yeast two-hybrid screen (34). A deficiency of J1R expression blocks virion assembly, whereas a deficiency of A45R expression does not alter vaccinia virus growth in cells or animals; these findings indicate that J1R protein may bind to other proteins besides A45R protein (4). The binding of J1R protein to A45R protein still occurred in infected cells treated with rifampin, indicating that the interaction occurs prior to virion assembly (data not shown). Since A45R protein was also reported to interact with A4L protein, it is possible that the stability of a ternary complex composed of A45R, A4L, and J1R proteins is important for further virion morphogenesis (34). This possibility will be investigated in the future.
Acknowledgments
We thank Sue-Ping Lee for excellent technical support in electron microscopy. We also thank J. Krijnse Locker and G. Smith for providing antibodies.
This work was supported by grants from Academia Sinica and the National Science Council (NSC91-2311-B-001-015) of R.O.C.
REFERENCES
- 1.Afonso, C. L., E. R. Tulman, Z. Lu, E. Oma, G. F. Kutish, and D. L. Rock. 1999. The genome of Melanoplus sanguinipes entomopoxvirus. J. Virol. 73:533-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2000. The genome of fowlpox virus. J. Virol. 74:3815-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, F. A. Osorio, C. Balinsky, G. F. Kutish, and D. L. Rock. 2002. The genome of swinepox virus. J. Virol. 76:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Almazan, F., D. C. Tscharke, and G. L. Smith. 2001. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J. Virol. 75:7018-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 6.Baroudy, B. M., S. Venkatesan, and B. Moss. 1983. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47:723-729. [DOI] [PubMed] [Google Scholar]
- 7.Bawden, A. L., K. J. Glassberg, J. Diggans, R. Shaw, W. Farmerie, and R. W. Moyer. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120-139. [DOI] [PubMed] [Google Scholar]
- 8.Betakova, T., E. J. Wolffe, and B. Moss. 2000. The vaccinia virus A14.5L gene encodes a hydrophobic 53-amino-acid virion membrane protein that enhances virulence in mice and is conserved among vertebrate poxviruses. J. Virol. 74:4085-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cameron, C., S. Hota-Mitchell, L. Chen, J. Barrett, J. X. Cao, C. Macaulay, D. Willer, D. Evans, and G. McFadden. 1999. The complete DNA sequence of myxoma virus. Virology 264:298-318. [DOI] [PubMed] [Google Scholar]
- 12.Cassetti, M. C., M. Merchlinsky, E. J. Wolffe, A. S. Weisberg, and B. Moss. 1998. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J. Virol. 72:5769-5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakrabarti, S., K. Brechling, and B. Moss. 1985. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol. Cell. Biol. 5:3403-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chambaud, I., R. Heilig, S. Ferris, V. Barbe, D. Samson, F. Galisson, I. Moszer, K. Dybvig, H. Wroblewski, A. Viari, E. P. Rocha, and A. Blanchard. 2001. The complete genome sequence of the murine respiratory pathogen Mycoplasma pulmonis. Nucleic Acids Res. 29:2145-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.da Fonseca, F. G., E. J. Wolffe, A. Weisberg, and B. Moss. 2000. Characterization of the vaccinia virus H3L envelope protein: topology and posttranslational membrane insertion via the C-terminal hydrophobic tail. J. Virol. 74:7508-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earles L stain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia, A. D., and B. Moss. 2001. Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes. J. Virol. 75:6460-6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 19.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266, 517-563. [DOI] [PubMed] [Google Scholar]
- 20.Heljasvaara, R., D. Rodriguez, C. Risco, J. L. Carrascosa, M. Esteban, and J. R. Rodriguez. 2001. The major core protein P4a (A10L gene) of vaccinia virus is essential for correct assembly of viral DNA into the nucleoprotein complex to form immature viral particles. J. Virol. 75:5778-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsiao, J.-C., C.-S. Chung, and W. Chang. 1999. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates intracellular mature virion adsorption to cells. J. Virol. 73:8750-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao, J. C., C. S. Chung, and W. Chang. 1998. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J. Virol. 72:8374-8379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu, X., L. J. Carroll, E. J. Wolffe, and B. Moss. 1996. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J. Virol. 70:7669-7677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ichihashi, Y. 1996. Extracellular enveloped vaccinia virus escapes neutralization. Virology 217:478-485. [DOI] [PubMed] [Google Scholar]
- 26.Ichihashi, Y., and M. Oie. 1996. Neutralizing epitope on penetration protein of vaccinia virus. Virology 220:491-494. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, T. Ashford, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson, G. P., S. J. Goebel, and E. Paoletti. 1993. An update on the vaccinia virus genome. Virology 196:381-401. [DOI] [PubMed] [Google Scholar]
- 29.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 30.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 31.Lee, H. J., K. Essani, and G. L. Smith. 2001. The genome sequence of Yaba-like disease virus, a yatapoxvirus. Virology 281:170-192. [DOI] [PubMed] [Google Scholar]
- 32.Lee, P., and D. E. Hruby. 1993. trans processing of vaccinia virus core proteins. J. Virol. 67:4252-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin, C. L., C. S. Chung, H. G. Heine, and W. Chang. 2000. Vaccinia virus envelope H3L protein binds to cell surface heparan sulfate and is important for intracellular mature virion morphogenesis and virus infection in vitro and in vivo. J. Virol. 74:3353-3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCraith, S., T. Holtzman, B. Moss, and S. Fields. 2000. Genome-wide analysis of vaccinia virus protein-protein interactions. Proc. Natl. Acad. Sci. USA 97:4879-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, H., N. Osterrieder, and C. P. Czerny. 1994. Identification of binding sites for neutralizing monoclonal antibodies on the 14-kDa fusion protein of orthopox viruses. Virology 200:778-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan, C., S. Ellison, H. Rose, and D. Moore. 1954. Structure and development of viruses observed in the electron microscope. II. Vaccinia and fowlpox viruses. J. Exp. Med. 100:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 38.Payne, L. G. 1980. Significance of extracellular enveloped virus in the in vitro and in vivo dissemination of vaccinia. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. K. Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravanello, M. P., and D. E. Hruby. 1994. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J. Virol. 68:6401-6410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds, E. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 55:541-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice, A. P., and B. E. Roberts. 1983. Vaccinia virus induces cellular mRNA degradation. J. Virol. 47:529-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 44.Risco, C., J. R. Rodriguez, W. Demkowicz, R. Heljasvaara, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1999. The vaccinia virus 39-kDa protein forms a stable complex with the p4a/4a major core protein early in morphogenesis. Virology 265:375-386. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez, D., M. Esteban, and J. R. Rodriguez. 1995. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J. Virol. 69:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez, D., C. Risco, J. R. Rodriguez, J. L. Carrascosa, and M. Esteban. 1996. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J. Virol. 70:7641-7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1997. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J. Virol. 71:1821-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez, J. R., C. Risco, J. L. Carrascosa, M. Esteban, and D. Rodriguez. 1998. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J. Virol. 72:1287-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosel, J. L., P. L. Earl, J. P. Weir, and B. Moss. 1986. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J. Virol. 60:436-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. K. Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanderson, C. M., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81:47-58. [DOI] [PubMed] [Google Scholar]
- 52.Scanlan, T. S., and R. C. Reid. 1995. Evolution in action. Chem. Biol. 2:71-75. [DOI] [PubMed] [Google Scholar]
- 53.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senkevich, T. G., E. V. Koonin, J. J. Bugert, G. Darai, and B. Moss. 1997. The genome of molluscum contagiosum virus: analysis and comparison with other poxviruses. Virology 233:19-42. [DOI] [PubMed] [Google Scholar]
- 55.Senkevich, T. G., A. S. Weisberg, and B. Moss. 2000. Vaccinia virus E10R protein is associated with the membranes of intracellular mature virions and has a role in morphogenesis. Virology 278:244-252. [DOI] [PubMed] [Google Scholar]
- 56.Senkevich, T. G., C. L. White, E. V. Koonin, and B. Moss. 2000. A viral member of the ERV1/ALR protein family participates in a cytoplasmic pathway of disulfide bond formation. Proc. Natl. Acad. Sci. USA 97:12068-12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shchelkunov, S. N., V. M. Blinov, S. M. Resenchuk, A. V. Totmenin, L. V. Olenina, G. B. Chirikova, and L. S. Sandakhchiev. 1994. Analysis of the nucleotide sequence of 53 kbp from the right terminus of the genome of variola major virus strain India-1967. Virus Res. 34:207-236. [DOI] [PubMed] [Google Scholar]
- 58.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van 't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sodeik, B., G. Griffiths, M. Ericsson, B. Moss, and R. W. Doms. 1994. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J. Virol. 68:1103-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sodeik, B., and J. Krijnse-Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 61.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 62.Szajner, P., A. S. Weisberg, and B. Moss. 2001. Unique temperature-sensitive defect in vaccinia virus morphogenesis maps to a single nucleotide substitution in the A30L gene. J. Virol. 75:11222-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Szajner, P., A. S. Weisberg, E. J. Wolffe, and B. Moss. 2001. Vaccinia virus A30L protein is required for association of viral membranes with dense viroplasm to form immature virions. J. Virol. 75:5752-5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Traktman, P., K. Liu, J. DeMasi, R. Rollins, S. Jesty, and B. Unger. 2000. Elucidating the essential role of the A14 phosphoprotein in vaccinia virus morphogenesis: construction and characterization of a tetracycline-inducible recombinant. J. Virol. 74:3682-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, G. F. Kutish, and D. L. Rock. 2001. Genome of lumpy skin disease virus. J. Virol. 75:7122-7130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 67.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26-36. [DOI] [PubMed] [Google Scholar]
- 68.VanSlyke, J. K., C. A. Franke, and D. E. Hruby. 1991. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J. Gen. Virol. 72:411-416. [DOI] [PubMed] [Google Scholar]
- 69.Vanslyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]
- 70.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ward, G. A., C. K. Stover, B. Moss, and T. R. Fuerst. 1995. Stringent chemical and thermal regulation of recombinant gene expression by vaccinia virus vectors in mammalian cells. Proc. Natl. Acad. Sci. USA 92:6773-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.White, C. L., T. G. Senkevich, and B. Moss. 2002. Vaccinia virus G4L glutaredoxin is an essential intermediate of a cytoplasmic disulfide bond pathway required for virion assembly. J. Virol. 76:467-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.White, C. L., A. S. Weisberg, and B. Moss. 2000. A glutaredoxin encoded by the G4L gene of vaccinia virus is essential for virion morphogenesis. J. Virol. 74:9175-9183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Whitehead, S. S., N. A. Bersani, and D. E. Hruby. 1995. Physical and molecular genetic analysis of the multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein. J. Gen. Virol. 76:717-721. [DOI] [PubMed] [Google Scholar]
- 76.Whitehead, S. S., and D. E. Hruby. 1994. Differential utilization of a conserved motif for the proteolytic maturation of vaccinia virus proteins. Virology 200:154-161. [DOI] [PubMed] [Google Scholar]
- 77.Willer, D. O., G. McFadden, and D. H. Evans. 1999. The complete genome sequence of shope (rabbit) fibroma virus. Virology 264:319-343. [DOI] [PubMed] [Google Scholar]
- 78.Williams, O., E. J. Wolffe, A. S. Weisberg, and M. Merchlinsky. 1999. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J. Virol. 73:4590-4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wolffe, E. J., D. M. Moore, P. J. Peters, and B. Moss. 1996. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J. Virol. 70:2797-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wolffe, E. J., S. Vijaya, and B. Moss. 1995. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology 211:53-63. [DOI] [PubMed] [Google Scholar]
- 81.Yeh, W. W., B. Moss, and E. J. Wolffe. 2000. The vaccinia virus A9L gene encodes a membrane protein required for an early step in virion morphogenesis. J. Virol. 74:9701-9711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang, Y., and B. Moss. 1992. Immature viral envelope formation is interrupted at the same stage by lac operator-mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology 187:643-653. [DOI] [PubMed] [Google Scholar]
- 83.Zhang, Y. F., and B. Moss. 1991. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J. Virol. 65:6101-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]