Abstract
The human immunodeficiency virus type 1 (HIV-1) gp120 exterior envelope glycoprotein is conformationally flexible. Upon binding to the host cell receptor CD4, gp120 assumes a conformation that is recognized by the second receptor, CCR5 and/or CXCR4, and by the CD4-induced (CD4i) antibodies. Guided by the X-ray crystal structure of a gp120-CD4-CD4i antibody complex, we introduced changes into gp120 that were designed to stabilize or disrupt this conformation. One mutant, 375 S/W, in which the tryptophan indole group is predicted to occupy the Phe 43 cavity in the gp120 interior, apparently favors a gp120 conformation closer to that of the CD4-bound state. The 375 S/W mutant was recognized as well as or better than wild-type gp120 by CD4 and CD4i antibodies, and the large decrease in entropy observed when wild-type gp120 bound CD4 was reduced for the 375 S/W mutant. The recognition of the 375 S/W mutant by CD4BS antibodies, which are directed against the CD4-binding region of gp120, was markedly reduced compared with that of the wild-type gp120. Compared with the wild-type virus, viruses with the 375 S/W envelope glycoproteins were resistant to neutralization by IgG1b12, a CD4BS antibody, were slightly more sensitive to soluble CD4 neutralization and were neutralized more efficiently by the 2G12 antibody. Another mutant, 423 I/P, in which the gp120 bridging sheet was disrupted, did not bind CD4, CCR5, or CD4i antibodies, even though recognition by CD4BS antibodies was efficient. These results indicate that CD4BS antibodies recognize conformations of gp120 different from that recognized by CD4 and CD4i antibodies.
Over 35 million people are currently infected with human immunodeficiency virus type 1 (HIV-1), the major cause of AIDS (4, 24). The development of a preventive vaccine, which optimally should elicit both virus-neutralizing antibodies and cellular immune responses, is of high priority and urgency (25, 32).
Neutralizing antibodies must bind the HIV-1 envelope glycoproteins, which mediate the entry of the virus into the target cell (97). The trimeric envelope glycoprotein complex is anchored in the host cell or viral membrane by the gp41 transmembrane glycoprotein, which is noncovalently attached to the gp120 exterior envelope glycoprotein. Most of the surface-exposed elements of the trimeric envelope glycoprotein complex are contained on the gp120 exterior envelope glycoprotein (52). Comparison of the gp120 glycoproteins from different HIV-1 strains reveals regions of conservation interrupted by long regions of variability (V1 to V5) (46, 80). Intramolecular disulfide bonds in the gp120 glycoprotein result in the incorporation of the first four variable regions (V1 to V4) into large, surface-exposed loops (43, 52). The conserved gp120 regions fold into a core, which contains many of the gp120 elements important for receptor binding (6, 65, 98).
CD4 and the chemokine receptors CCR5 and CXCR4 serve as HIV-1 receptors (14, 17-19, 22, 31). The binding of the HIV-1 gp120 glycoprotein to CD4 contributes to the attachment of the virus to the target cell and also triggers conformational changes in gp120 that allow high-affinity binding to the chemokine receptor (2, 76, 87, 89, 94). Much of our current understanding of gp120-CD4 interaction is based on mutagenic analyses (3, 7, 16, 40, 47, 60, 64), X-ray crystal structures (38, 39, 73, 91, 93, 95), and thermodynamic studies (56). The gp120 core consists of an inner domain, which is thought to contact the gp41 ectodomain, an outer domain thought to face outward on the assembled trimer, and a bridging sheet (38, 39, 95). In binding CD4, all three gp120 elements contact the most amino-terminal of the four immunoglobulin-like domains of CD4. Large areas of the gp120 and CD4 surface are occluded by their interaction, and two cavities exist within the interface of these proteins. A large, shallow cavity (approximately 300 Å3) is bounded equally by gp120 and CD4 and is filled with water molecules. Another cavity (approximately 150 Å3) is deeper, extends into the interior of gp120, and is bounded by well-conserved residues derived from all three gp120 core elements. Phenylalanine 43 of CD4, which is critical for gp120 binding, also contacts this cavity and plugs the opening of what otherwise would be a deep pocket on the gp120 surface. Mutagenic analyses (16, 40, 60) suggest that most of the gp120-CD4 contacts that contribute to the efficiency of the interaction occur in a spatially localized “hot spot” near the Phe 43 cavity.
The induction of chemokine receptor binding by the gp120-CD4 interaction has been suggested to involve conformational changes in the gp120 variable loops and in the conserved core. The V2 variable loop apparently masks the chemokine receptor-binding site on gp120 and is moved out of the way by CD4 binding (96). In some cases, HIV-1 gp120 glycoproteins with deletions or alterations in the V2 loop exhibit the ability to bind chemokine receptor in the absence of CD4; viruses with these altered envelope glycoproteins can infect CD4-negative cells that express the appropriate chemokine receptor (34, 35).
The conserved core of HIV-1 gp120 is also conformationally changed by CD4 binding, as evidenced by the unusually large changes in gp120 entropy documented by isothermal titration calorimetry. These studies (56) suggest that both full-length gp120 and the gp120 core are flexible proteins that are conformationally fixed by CD4 binding. A structural interpretation of these data is that, in the absence of CD4, gp120 exhibits interdomain flexibility. Upon CD4 binding, many contacts between the gp120 inner and outer domains are created and the bridging sheet is formed. The possibility of CD4-induced formation of the bridging sheet is particularly important in light of the contribution of this gp120 element to chemokine receptor binding (71, 72).
The HIV-1 gp120 V3 loop is the major genetic determinant of chemokine receptor choice (5, 12, 14, 15, 26, 58, 77, 79). The V3 loop apparently cooperates with a highly conserved sequence in the β19 strand of gp120 to form a high-affinity binding site for the chemokine receptors (71, 72). Chemokine receptor binding is thought to trigger additional conformational changes in the HIV-1 envelope glycoproteins that lead to the fusion of the viral membrane with the target cell membrane (13, 45, 92).
The HIV-1 envelope glycoproteins elicit antibody responses during the course of natural infection and have been extensively employed as immunogens in vaccine studies (9-11, 44, 48, 63, 97). Many elicited antibodies do not recognize the functional envelope glycoprotein trimers efficiently and thus fail to neutralize virus infection (23, 49, 62, 75, 84). Neutralizing antibodies are directed against both variable and conserved elements of the gp120 glycoprotein (52, 54). The conserved gp120 neutralization epitopes are discontinuous elements dependent on the native conformation of gp120 (50, 82, 95, 97). These epitopes include those near the CD4 binding site (CD4BS epitopes) (67, 86, 88), those induced by CD4 binding (the CD4-induced [CD4i] epitopes) (87), and the carbohydrate-dependent 2G12 epitope (90).
The locations of these epitopes on the crystallized gp120 core have been mapped by mutagenesis (95). The CD4BS antibodies can compete with CD4 for gp120 binding (54, 67). The CD4i antibodies bind a conserved HIV-1 gp120 structure that is induced by CD4 binding and has been implicated in binding the CCR5 chemokine receptor (71, 72, 87). The epitopes for CD4i antibodies are located near the bridging sheet of the HIV-1 gp120 core (39, 95). The 2G12 antibody binds the carbohydrate-rich gp120 outer domain (90, 95).
Because of the chronic nature of HIV-1 infection, the viral envelope glycoproteins have evolved to minimize the potential impact of neutralizing antibodies on virus infection. This adaptation includes an inefficiency in the elicitation of desirable neutralizing antibodies and a resistance, particularly among primary HIV-1 isolates, to neutralization by antibodies (53, 85). The conserved receptor-binding regions of gp120, which must be at least partly exposed on the virion surface, are potentially vulnerable to antibody-mediated neutralization. Overlying variable loops and adjacent N-linked carbohydrates can restrict the access of potentially neutralizing antibodies to the gp120 receptor-binding sites (51, 55, 57, 83). The conformational flexibility of gp120 may render the presentation of discontinuous receptor-binding regions to the immune system inefficient and creates an entropic barrier that must be overcome or bypassed by antibodies targeting receptor-binding regions (P. D. Kwong , M. Doyle, D. Casper, C. Cicala, S. Leavitt, S. Majeed, T. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. Parren, J. Robinson, L. Wang, D. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos, submitted for publication). The “shedding” of gp120 from the HIV-1 envelope glycoprotein complex results in the elicitation of high titers of antibodies to gp120 and gp41 epitopes not accessible on the functional trimer (78, 95, 100, 101). Thus, many nonneutralizing antibodies are elicited by these and other nonfunctional forms of the envelope glycoproteins that act as “decoy molecules.” An appreciation of the conformations available to the HIV-1 gp120 glycoprotein in the context of a free monomer and the functional envelope glycoprotein complex may suggest ways to circumvent viral strategies for immune evasion.
MATERIALS AND METHODS
Modeling HIV-1 gp120 mutants.
Conformations of the substituted amino acid residues were modeled by using the X-ray structure of the ternary complex consisting of the gp120 core from the YU2 primary HIV-1 strain, two-domain CD4, and the antigen-binding fragment of the human antibody 17b (PDB accession code 1G9N). Models were visually analyzed with the TOM/FRODO interactive graphics program (Alberta/Caltech version 3.0 [Mark Isreal, Arthur Chirino, David Schuller, and T. Alwyn Jones]). Briefly, the side chain of the amino acid was replaced, and the allowed classes of rotamer conformations, as specified by Ponder and Richards (66), were checked to minimize steric clashes. Generally, steric considerations permitted only one class of rotamer conformations. For each rotamer class, the chi1 and chi2 angles of the side chain were varied within 5 standard deviations of the most probable observed rotamer position to minimize steric clashes and enhance potential van der Waals or hydrogen bond-stabilizing effects.
Cells.
293T cells were cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and penicillin-streptomycin. Peripheral blood mononuclear cells (PBMC) from humans were stimulated with phytohemagglutinin for 5 days and cultured in medium containing interleukin-2.
HIV-1 gp120 mutants.
Site-directed mutagenesis was used to introduce amino acid changes into the wtΔ protein, derived from the YU2 HIV-1 strain (71, 72). The wtΔ protein contains deletions of gp120 residues 31 to 81 and 128 to 194, which represent the N terminus and V1/V2 variable loops, respectively. Numbering of gp120 amino acid residues is based on the sequence of the prototypic HXBc2 strain of HIV-1, according to current convention (37).
Transient expression of HIV-1 envelope glycoproteins.
293T cells grown to 70% confluence in 100-mm dishes were transfected with 2 μg of an envelope protein-expressing plasmid and 1 μg of an HIV-1 Tat-expressing plasmid with the Effectene transfection reagent (Qiagen). One day later, the medium was removed, the cells were washed once with 10 ml of phosphate-buffered saline (PBS), and labeling medium (4.5 ml of Dulbecco's modified Eagle's medium, 0.5 ml of heat-inactivated, dialyzed fetal bovine serum, 50 μl of penicillin-streptomycin solution (10 μg/ml), and 20 μl [∼230 μCi] each of [35S]cysteine and [35S]methionine) was added. The cells were incubated at 37°C for another day. The medium containing radiolabeled wtΔ or mutant gp120 protein was collected, cleared by centrifugation, and stored at 4°C.
For experiments in which 293T cells were transfected with plasmids expressing full-length HIV-1 envelope glycoproteins, radiolabeled cells were washed twice with PBS and lysed with NP-40 lysis buffer. The lysates were vortexed for 1 min and cleared by centrifugation in an Eppendorf centrifuge for 5 min. The supernatants were used for immunoprecipitation.
Immunoprecipitation of envelope glycoproteins.
For precipitation of radiolabeled HIV-1 envelope glycoproteins, 400 μl of medium containing the labeled proteins was mixed with 100 μl of 10% protein A-Sepharose (Pharmacia), 50 μl of 4% bovine serum albumin, and either 1 or 2 μg of monoclonal antibody, sCD4 plus the T45 anti-CD4 antibody, or 4 μl of a mixture of sera from HIV-1-infected individuals. PBS was added to bring the total volume to 1 ml. The samples were rocked at 4°C overnight or at room temperature for 2 h. The Sepharose beads were then washed twice with 1 ml of 0.5 M NaCl in PBS and once with 1 ml of PBS. The beads were mixed with 2× gel loading buffer and boiled for 3 min. Following removal of the beads by centrifugation, the supernatants were loaded on a sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) gel. The gel was enhanced with Autoflour (National Diagnostic) for 45 min before being dried at 80°C for 2 h and exposed to X-ray film. The gel was also used for PhosphorImager (Molecular Dynamics) analysis.
Virus infection assay.
Recombinant HIV-1 containing the firefly luciferase gene was produced by transfection of 293T cells with the pCMV Gag-Pol packaging plasmid, the pHIV-luc vector, and the pSVIIIenv plasmid expressing the wild-type or 375 S/W mutant YU2 HIV-1 envelope glycoprotein. Three days after transfection, the cell supernatants were harvested, assayed for virus by reverse transcriptase measurement, and frozen in aliquots. For infection of Cf2Th/CD4/CCR5 cells, 1 × 104 to 3 × 104 reverse transcriptase units of virus per well were incubated with the cells at 37°C overnight in 24-well plates. For PBMC infections, 105 reverse transcriptase units of virus was used. The next day, the medium of the Cf2Th/CD4/CCR5 cells was changed completely, and 1.5 ml of fresh medium was added to the PBMC cultures. Cells were cultured for an additional 2 days, after which the luciferase activity in the cell lysate was determined.
Virus neutralization assay.
Recombinant HIV-1 infection of Cf2Th/CD4/CCR5 cells, as described above, was used to examine the sensitivity of the viruses to neutralization by sCD4 and antibodies. Viruses were incubated with sCD4 or antibodies at different concentrations for 90 min at 37°C. The virus-ligand mixtures were then transferred to wells containing the target Cf2Th/CD4/CCR5 cells. After overnight incubation, 1.5 ml of fresh medium was added to each well, and the cells were cultured for 2 more days. The cells were then lysed and assayed for luciferase (Tuner 20; Promega).
Isothermal titration calorimetry.
Isothermal titration calorimetry experiments were conducted with a Microcal (Amherst, Mass.) VP-ITC microcalorimeter. The proteins were dialyzed against 10 mM sodium phosphate (pH 7.4)-200 mM sodium chloride-0.2 mM EDTA. Concentrations were determined from UV-visible spectra with molar extinction coefficients at 280 nm of 60,200 M−1 (sCD4), 75,100 M−1 (wild-type YU2 gp120), and 80,600 M−1 (375 S/W YU2 gp120), which were calculated from amino acid sequences (61). Data were analyzed with Microcal Origin software version 5.0 according to a single-binding-site model. Data for the wild-type gp120 and the 375 S/W mutant were measured on the same day with the same solution of sCD4 to optimize our ability to compare the enthalpy changes associated with CD4 binding.
RESULTS
Modeling HIV-1 gp120 mutants.
The high entropy of gp120 (56) in conjunction with structural studies (38, 39, 95) suggested the possibility that, in the absence of CD4, the spatial relationship of the gp120 inner and outer domains was flexible and that the bridging sheet assumed a conformation different than that observed in the CD4-bound state. Several programs designed to predict the secondary structures of proteins indicate a strong propensity of the β20 and β21 strands of the bridging sheet of HIV-1 gp120 to form an α-helix (39). Furthermore, many of the gp120 residues shown by mutagenic analysis to be important for the binding of CD4BS antibodies were not exposed on the surface of CD4-bound gp120 (95), suggesting that these antibodies might recognize distinct conformations of gp120.
To investigate these possibilities, we sought to alter gp120 so that the free molecule assumed a conformation that more closely resembled the CD4-bound state. Stabilization of the CD4-bound conformation was attempted both by filling the water-filled cavities observed in the CD4:gp120 X-ray structure and by stabilizing the interface between the inner and outer gp120 domains (Fig. 1 and Table 1). Many previous studies have documented the favorable effects of filling internal cavities on the stabilization of native protein conformation (1, 8, 20, 21, 27-30, 33, 36, 41, 42, 59, 68, 69, 81, 99). In the case of the HIV-1 gp120 glycoprotein, cavity-filling substitutions were chosen to increase side chain volume and to promote stabilizing hydrophobic interactions. A second strategy, attempting to stabilize the CD4-bound conformation of gp120, involved modification of the interface of the gp120 inner and outer domains. Hydrophilic residues surrounded by hydrophobic residues in this interface were altered in an attempt to increase hydrophobic interactions while avoiding steric clashes and maintaining opportunities for hydrogen bonding. For example, arginine 273 reaches across the inner domain-outer domain interface to make a hydrogen bond with carbonyl 233. Replacement of this arginine with tryptophan allowed maintenance of a hydrogen bond with the Trp Nɛ while adding stacking interactions with tyrosine 484 across the domain interface.
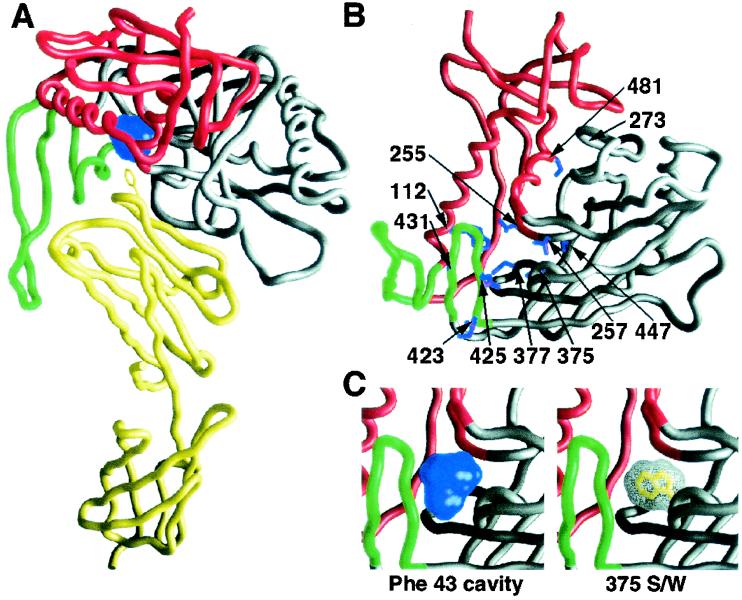
FIG. 1.
Locations of changes introduced into the HIV-1 YU2 gp120 glycoprotein. (A) Cα tracing of the HIV-1 YU2 gp120 core complexed with two-domain CD4 (yellow) (38). The gp120 inner domain is red, the outer domain is gray, and the bridging sheet is green. The Phe 43 cavity is colored blue. The side chain of phenylalanine 43 of CD4 is shown. (B) View of gp120 from the perspective of CD4, rotated 90° on the horizontal axis from the view in panel A. The gp120 domains are colored as in panel A. The gp120 residues altered in this study are shown. (C) Enlarged view of gp120, in the same orientation as that shown in panel B. The image at the left shows the Phe 43 interfacial cavity (blue). The image at the right shows the molecular surface of the modeled 375 S/W mutant, with the substituted tryptophan side chain occupying the Phe 43 cavity.
TABLE 1.
Modeling parametersa
| gp120 position | Introduced changeb | Δ volc (Å3) | Modeled orientationd
|
Comment | ||
|---|---|---|---|---|---|---|
| chi1, degrees (rmsd) | chi2, degrees (rmsd) | Observed % (optimal angles) | ||||
| 112 | W/A | −97 | Removes stabilizing interaction with Trp 427 | |||
| 255 | V/W | 58 | 110 (3.5) | −89 (0.0) | 20.7 (65°, −89°) | 2.6-Å close contact with Trp 112 ring |
| 257 | T/A | −26 | Removes stabilizing H-bond with 370 carbonyl | |||
| 273 | R/W | 15 | −58 (2.3) | −73 (1.9) | 6.9 (−73°, −88°) | Clashes with Gln 287, which can be easily resolved; ring makes H-bond with 233 carbonyl and stacking interactions with Tyr 484 across domain interface |
| 375 | S/W | 90 | 98 (2.5) | −84 (1.0) | 20.7 (65°, −84°) | 2.5-Å clash with Thr 257 side chain |
| S/F | 62 | 110 (3.7) | 91 (0.0) | 21.3 (66°, 91°) | ||
| S/Y | 68 | 110 (5.0) | 89 (0.0) | 15.0 (63°, 89°) | Reducing chi1 results in clash with Thr 257 | |
| 377 | N/L | 28 | −165 (0.0) | 133 (1.0) | 4.8 (−165°, 168°) | Good van der Waals contacts made with Phe 210 and Leu 116, stabilizing domain interface |
| 423 | I/P | −34 | Backbone phi needs to change roughly 30° to accommodate | |||
| 423 | I/M | 0 | Triple mutation enhances calculated helix-forming | |||
| 425 | N/K | 39 | propensity of β20 strand | |||
| 431 | G/E | 61 | ||||
| 447 | S/I | 51 | 75 (2.6) | −131 (4.0) | 16.1 (62°, 164°) | van der Waals contacts with Ile 261 and 262 sugar |
| 481 | S/F | 62 | −173 (0.7) | 79 (0.0) | 25.0 (−179°, 79°) | Clash with Gln 287, which can easily move; van der Waals contacts with Ile 285 and aliphatic base of 287 across domain interface |
The modeling was carried out with the YU2 gp120 core:D1D2 CD4:17b Fab complex (PDB accession code 1G9N). Residue numbers correspond to those of the prototype HXBc2 HIV-1 strain, as per current convention (37).
The amino acids are given in single-letter code. The amino acid residue in the wild-type YU2 gp120 is written first, followed by the substituted amino acid residue.
The change in volume associated with the amino acid substitution was calculated based on the volume enclosed by the van der Waals radii, using the atomic volumes provided by F. M. Richards (70).
Modeling was performed as described in Materials and Methods, using the experimentally determined side chain rotamer classes specified by Ponder and Richards (66). Modeled rotamers are specified by the percentage of the observed side chains that fall into a particular rotamer class (observed percent) and the optimal chi1/chi2 angles which indicated the observed optimal rotamer orientation for that particular class. The chi1 and chi2 values listed in the table are those that resulted in minimal steric clash while being close to a particular rotamer optimum. The root mean square deviation (rmsd) values represent the deviation of the modeled chi1/chi2 angles from the observed optimum values.
We also attempted to stabilize a gp120 conformation recognized by CD4BS antibodies. These attempts were based on the hypothesis that the β20 strand of gp120 in the CD4-bound conformation assumes an α-helical conformation when complexed to a CD4BS antibody. Stabilization of an α-helical conformation of the β20 region was attempted both by substitution of amino acids with increased helical propensity (e.g., 423 I/M + 425 N/K + 431 G/E) and by replacement of isoleucine 423 at the N terminus of the potential helix with a proline, placing conformational strain on the β-strand. Other gp120 substitutions attempted to destabilize the CD4-bound conformation and thus promote increased occupancy of other conformations. Residues away from the CD4 surface that performed conformationally sensitive functions were chosen. For example, tryptophan 112 stabilizes tryptophan 427, which is crucial for CD4 binding and is located at the end of the β20 strand (38, 39). Tryptophan 112 was changed to alanine in an attempt to destabilize the CD4-bound conformation.
Ligand binding of HIV-1 envelope glycoprotein mutants.
The changes described above were introduced into the wtΔ protein, which is derived from the YU2 primary R5 strain of HIV-1. The wtΔ protein lacks 52 N-terminal residues and the V1/V2 variable loops of the mature gp120 glycoprotein (72). The use of wtΔ eliminates possible indirect effects of amino acid changes on the conformation of the V1/V2 variable loops, which can mask binding sites for receptors and antibodies. The wtΔ and mutant derivatives were transiently produced by transfection of 293T cells, which were metabolically labeled with [35S]cysteine-methionine. The labeled wtΔ glycoprotein variants in the cell supernatants were precipitated by sCD4 in combination with an anti-CD4 antibody and by CD4i and CD4BS antibodies. The relative amounts of wtΔ and mutant glycoproteins in the cell supernatants were determined by precipitation with a mixture of sera from HIV-1-infected individuals, which recognizes multiple epitopes on the HIV-1 envelope glycoproteins. These ratios were used to normalize the relative amounts of wtΔ and mutant glycoprotein precipitated by the sCD4 and monoclonal antibodies.
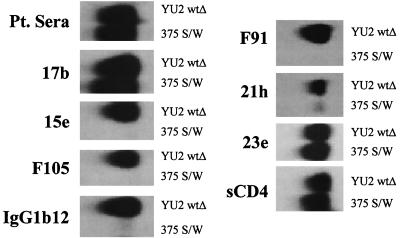
The abilities of the mutant glycoproteins to bind sCD4 and monoclonal antibodies relative to that of the wtΔ protein are shown in Table 2. The most dramatic phenotypes were seen for the mutants with aromatic amino acid substitutions at serine 375, which were designed to fill the Phe 43 cavity. These mutants bound sCD4 at least as well as the wtΔ protein and also were recognized efficiently by the CD4i antibody 17b. Recognition of the 375 S/W mutant by four other CD4i antibodies (48d, 23e, 49e, and 21c) (98a) was 75 to 100% of that seen for the wild-type gp120 glycoprotein (data not shown). Notably, recognition of the serine 375 mutants by the CD4BS antibodies was generally lower than recognition of the wtΔ protein. Of the three mutants in which serine 375 was altered, the tryptophan substitution, which is predicted to fill the Phe 43 cavity most efficiently, exhibited the most dramatic phenotype (Fig. 2). None of the mutants containing interdomain substitutions exhibited such a profound phenotype with respect to maintenance of sCD4 binding and loss of recognition by CD4BS antibodies.
TABLE 2.
Ligand-binding characteristics of wild-type YU2 and mutant derivativesa
| Envelope protein | Relative binding
|
||||||
|---|---|---|---|---|---|---|---|
| sCD4 | CD4i antibody 17b | CD4BS antibodies
|
|||||
| F105 | 15e | IgG1b12 | 21h | F91 | |||
| Wild type | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 112 W/A | 0.59 | 0.94 | 0.22 | 0.54 | 0.97 | 0.83 | 0.91 |
| 255 V/W | 0.38 | 0.88 | 0.00 | 0.06 | 0.51 | 0.80 | 0.76 |
| 257 T/A | 0.69 | 0.84 | 0.00 | 0.16 | 0.77 | 1.10 | 0.81 |
| 273 R/W | 0.78 | 0.91 | 0.72 | 0.90 | 0.46 | 0.78 | 0.76 |
| 375 S/W | 1.74 | 0.90 | 0.00 | 0.00 | 0.02 | 0.32 | 0.00 |
| 375 S/F | 1.10 | 0.70 | 0.00 | 0.22 | 0.57 | 0.72 | 0.31 |
| 375 S/Y | 1.13 | 1.08 | 0.00 | 0.68 | 0.90 | 0.91 | 0.45 |
| 377 N/L | 0.95 | 1.02 | 1.12 | 0.98 | 1.01 | 0.81 | 0.85 |
| 423 I/P | 0.09 | 0.05 | 1.01 | 0.85 | 1.27 | 0.83 | 0.86 |
| 423 I/M + 425 N/K + 431 G/E | 0.00 | 0.00 | 1.25 | 0.56 | 1.18 | 1.25 | 0.65 |
| 447 S/I | 0.00 | 0.75 | 0.00 | 0.76 | 0.58 | 0.84 | 0.38 |
| 481 S/F | 0.69 | 0.80 | 0.46 | 0.84 | 0.73 | 0.73 | 0.73 |
The indicated residue changes were introduced into the YU2 wtΔ protein. The radiolabeled proteins were expressed in 293T cells and precipitated by a polyclonal mixture of sera from HIV-1-infected individuals, by a combination of sCD4 and the T45 anti-CD4 antibody, or by monoclonal antibodies. The precipitated proteins were resolved by SDS-PAGE and autoradiography. The amounts of precipitated proteins were determined by PhosphorImager analysis (Molecular Dynamics). The relative binding value shown was calculated as follows: (mutant protein/wtΔ protein)ligand × (wtΔ protein/mutant protein)serum mixture. The values represent the average obtained from at least two independent experiments; relative binding in the experiments exhibited less than 25% variation from the values reported.
FIG. 2.
Precipitation of wtΔ and 375 S/W proteins by monoclonal antibodies. Equivalent amounts of metabolically labeled wtΔ and 375 S/W glycoproteins of the YU2 strain of HIV-1 were produced in 293T cell supernatants. The proteins were precipitated by a polyclonal pool of sera from HIV-1-infected individuals (Pt. Sera) or by the indicated ligands. The 17b and 23e monoclonal antibodies are CD4i antibodies, whereas the F105, 15e, IgG1b12, 21h, and F91 antibodies are CD4BS antibodies. The precipitated proteins were resolved by SDS-PAGE and autoradiography.
Two mutants, 423 I/P and 423 I/M + 425 N/K + 431 G/E, which were designed with the intention of altering the gp120 β20 strand, exhibited severe deficits in binding sCD4 and the 17b CD4i antibody (Table 2). However, recognition of these mutants by the panel of CD4BS antibodies was efficient.
These results indicate that gp120 mutants exist that exhibit distinct binding preferences for the CD4 and CD4i antibodies on the one hand and CD4BS antibodies on the other.
Binding of gp120 mutants to the CCR5 chemokine receptor.
Theoretically, if the 375 S/W mutant exhibited a preference for the CD4-bound conformation, it might be expected to bind CCR5 better than the wild-type protein in the absence of CD4. The influence of the 375 S/W change in the context of V1/V2 loop-deleted and full-length YU2 gp120 glycoproteins on CCR5 binding in the absence and presence of sCD4 was studied (Fig. 3). The 375 S/W mutants consistently demonstrated a slightly greater ability to bind CCR5 in the absence of sCD4 compared with their wild-type counterparts. These data are consistent with the 375 S/W mutants exhibiting a moderate preference for the CD4-bound state; it is clear, however, that the conformation of 375 S/W is not fully equivalent to that of gp120 bound to CD4. Although all of the proteins demonstrated substantial increases in CCR5 binding in the presence of sCD4, the binding of the 375 S/W mutants was slightly lower than that of the wild-type proteins in this context.
FIG. 3.
CCR5 binding of HIV-1 YU2 gp120 mutants. The ability of HIV-1 YU2 gp120 mutants to bind CCR5 was assessed in two separate experiments. In experiment 1, equivalent amounts of the ΔV1/V2 or ΔV1/V2 375 S/W mutants were incubated with either CCR5-negative or CCR5-positive Cf2Th cells in the absence of sCD4. In experiment 2, equivalent amounts of metabolically labeled YU2 gp120 variants were incubated with Cf2ThsynCCR5 cells, which express human CCR5, in the absence or presence of sCD4. The bound proteins were precipitated by a mixture of sera from HIV-1-infected individuals and resolved by SDS-PAGE. The positions of molecular size markers (lane M) are shown on the left (in kilodaltons).
Thermodynamic studies of 375 S/W gp120 binding to sCD4.
The binding of wild-type HIV-1 gp120 and sCD4 results in unusually large, balanced changes in enthalpy (ΔHobs) and entropy (ΔS) (56). Because the CD4 glycoprotein has been shown not to undergo substantial conformational changes upon gp120 binding (38, 39, 73, 91), by deduction most of these enthalpic and entropic changes derive from the gp120 glycoprotein. A gp120 molecule that exhibits a preference for the CD4-bound conformation would be expected to exhibit less of an entropic change and, probably, less of an enthalpic change upon binding CD4. To examine this, full-length wild-type and 375 S/W gp120 glycoproteins were produced in Drosophila cells, purified, and used for isothermal titration calorimetry with sCD4 (Fig. 4). The results in Table 3 indicate that the enthalpic change observed when the 375 S/W mutant bound sCD4 (ΔHobs = −35.5 kcal/mol of sCD4) was substantially less than that seen for the wild-type gp120 (ΔHobs = −52.1 kcal/mol of sCD4). The calculated entropies (−TΔS) for the 375 S/W mutant and wild-type gp120 were 23.9 and 41.6 kcal/mol of sCD4, respectively. The 375 S/W mutant exhibited an approximately sixfold increase in affinity for sCD4 compared with the wild-type gp120. These results strongly support the idea that the 375 S/W mutant exhibits a preference for a conformation closer to that of the CD4-bound gp120 glycoprotein.
FIG. 4.
Isothermal titration calorimetry data for the interaction of sCD4 with gp120 glycoproteins. The data on the left were generated with sCD4 and wild-type YU2 gp120, and the data on the right were generated with 375 S/W YU2 gp120. The top panel in each case shows the data generated following the sequential injection of 7 μl of 75 μM sCD4 into the calorimeter cell containing 5 μM gp120. Integration of the binding peaks yielded the heat generated per mole of sCD4 injected, as shown in the bottom panels. Best-fit curves for a single-site-binding model are shown with the data points. The conditions for the assay were 10 mM sodium phosphate, 200 mM sodium chloride, 0.2 mM EDTA, pH 7.4, and 37°C.
TABLE 3.
Thermodynamic parameters of the gp120-sCD4 interaction at 37°Ca
| Proteins | kd (nM) | ΔG (kcal/mol of sCD4) | ΔHobs (kcal/mol of sCD4) | −TΔS (kcal/mol of sCD4) |
|---|---|---|---|---|
| Wild-type YU2 gp120 + sCD4 | 38 ± 2 | −10.52 ± 0.03 | −52.1 ± 0.2 | 41.6 ± 0.2 |
| 375 S/W gp120 + sCD4 | 6.4 ± 0.9 | −11.62 ± 0.08 | −35.5 ± 0.2 | 23.9 ± 0.2 |
Isothermal titration calorimetry was performed with sCD4 and either the wild-type HIV-1 YU2 gp120 or the 375 S/W YU2 gp120 mutant, as described in Materials and Methods. The results, shown in Fig. 4, allowed determination of the kd values and the observed binding enthalpy changes (ΔHobs). The free energy changes (ΔG) and entropy changes (−TΔS) were calculated based on the ΔG and ΔHobs values. The molar ratios of sCD4 to gp120 were 1.037 ± 0.003 for the wild-type YU2 gp120 and 0.694 ± 0.002 for the 375 S/W mutant.
375 S/W change in the context of functional envelope glycoproteins.
The 375 serine-to-tryptophan change was introduced into the full-length YU2 envelope glycoproteins, which were expressed transiently along with the wild-type YU2 envelope glycoproteins in 293T cells. The wild-type and 375 S/W envelope glycoprotein precursors were proteolytically processed with comparable efficiency (Fig. 5A). Similar levels of the gp120 glycoprotein were observed in the cell supernatants for both the wild-type and 375 S/W glycoproteins. It appears that the folding, transport, processing, and subunit association of the wild-type and 375 S/W envelope glycoproteins are roughly equivalent.
FIG. 5.
Complete YU2 envelope glycoproteins with the 375 S/W change. (A) The full-length YU2 HIV-1 envelope glycoproteins, either wild-type (wt) or 375 S/W, were expressed in 293T cells and radiolabeled. Cell lysates and supernatants were precipitated by a mixture of sera from HIV-1-infected individuals, and precipitated proteins were resolved by SDS-PAGE. (B) An env-defective HIV-1 provirus expressing firefly luciferase was complemented with plasmids expressing wild-type YU2 envelope glycoproteins or mutant envelope glycoprotein 375 S/W or 375 S/W + 369 P/Q. Recombinant viruses were incubated with Cf2Th canine thymocytes expressing human CD4 and CCR5 (left panel) or human CCR5 only (right panel). Luciferase activity in the target cells was measured and normalized to that found in Cf2Th CCR5+ CD4+ cells exposed to viruses with the wild-type YU2 envelope glycoproteins.
To determine whether the 375 S/W mutant envelope glycoproteins would support HIV-1 entry into cells, a plasmid expressing either wild-type or 375 S/W YU2 envelope glycoproteins was used to complement an env-defective HIV-1 provirus encoding firefly luciferase. Recombinant virions produced in 293T cells were incubated with Cf2Th cells stably expressing CD4 and CCR5, and the luciferase activity in the target cells was measured. Figure 5B shows that the 375 S/W envelope glycoproteins supported infection of the Cf2Th/CD4/CCR5 cells at a level only 10% of that of the wild-type YU2 glycoproteins. This was significantly higher than the background of the assay, which was determined by using an HIV-1 envelope glycoprotein containing a large deletion of most of gp120 or with target cells lacking appropriate receptors. In these instances, the luciferase activity in the target cells was less than 1% of that seen when the wild-type HIV-1 envelope glycoproteins were used (data not shown). The ability of the 375 S/W mutant to complement HIV-1 entry into human PBMC was also low compared with that of the wild-type YU2 envelope glycoproteins but was detectable (data not shown).
During the course of these studies, we discovered that a second change (369 P/Q) inadvertently introduced into some of the clones during the PCR mutagenesis facilitated the infection of Cf2Th/CD4/CCR5 cells by envelope glycoproteins with the 375 S/W change (Fig. 5B). None of the envelope glycoproteins tested detectably supported the infection of CD4-negative Cf2ThsynCCR5 cells. We conclude that the 375 S/W mutant can support HIV-1 infection but at a reduced efficiency compared with the wild-type envelope glycoproteins.
Neutralization sensitivity of viruses with 375 S/W envelope glycoproteins.
The sensitivity of recombinant viruses with the wild-type YU2 or 375 S/W envelope glycoproteins to neutralization by sCD4 and monoclonal antibodies was examined. Viruses with the 375 S/W envelope glycoproteins were slightly more sensitive to neutralization by sCD4 than viruses with wild-type YU2 envelope glycoproteins (Fig. 6), consistent with the higher CD4 affinity of the 375 S/W mutant. By contrast, viruses with the 375 S/W envelope glycoproteins were completely resistant to neutralization by a CD4BS antibody, IgG1b12; viruses with the wild-type YU2 envelope glycoproteins were neutralized by IgG1b12 with a 50% inhibitory concentration of approximately 5 μg/ml. This result is consistent with the data indicating reduced recognition of the 375 S/W glycoprotein by IgG1b12 and other CD4BS antibodies.
FIG. 6.
Neutralization of viruses with wild-type and 375 S/W envelope glycoproteins. Recombinant viruses expressing firefly luciferase and containing either the wild-type (wt) YU2 (○) or 375 S/W (•) envelope glycoproteins were incubated with the indicated concentrations of antibody or sCD4 for 1 h at 37°C. The viruses were then added to Cf2Th cells expressing CD4 and CCR5. Forty-eight hours later, the cells were lysed, and luciferase activity was measured. Values were normalized to those observed for the viruses with wild-type YU2 envelope glycoproteins in the absence of added antibody. The results shown are from a single experiment that was repeated with comparable results.
The CD4i antibodies, 17b and 48d, did not neutralize viruses with the wild-type or 375S/W envelope glycoproteins at antibody concentrations of up to 20 μg/ml (data not shown).
The 2G12 antibody, which recognizes a carbohydrate-dependent epitope on the gp120 outer domain (90), exhibited minimal ability to neutralize the viruses with wild-type YU2 envelope glycoproteins. The viruses with 375 S/W glycoproteins were more sensitive to neutralization by the 2G12 antibody. To investigate the basis for this observation, we examined 2G12 recognition of the wild-type YU2 and 375 S/W soluble and shed gp120, as well as the gp160 envelope glycoprotein precursor in cell lysates. In all three contexts, the 2G12 antibody precipitated the 375 S/W mutants more efficiently than the wild-type glycoproteins. Apparently, the 375 S/W change can subtly influence the glycosylation of the envelope glycoproteins and increase recognition by the 2G12 antibody.
The 2F5 antibody, which recognizes a gp41 epitope, did not efficiently neutralize viruses with either wild-type or 375 S/W envelope glycoproteins. The poor neutralization of these viruses by the 2F5 antibody probably results from polymorphism of the linear gp41 epitope in the YU2 HIV-1 strain.
DISCUSSION
Here we identified two groups of HIV-1 gp120 mutants and used the ligand-binding phenotypes of these mutants to reach the conclusion that the gp120 glycoprotein can assume at least two distinct conformations. This conclusion necessitates that we distinguish among global misfolding, changes in conformational state, and local alterations of epitopes as consequences of the amino acid changes studied. One group of mutants, exemplified by the 375 S/W glycoprotein, bound CD4 and CD4i antibodies well; however, these mutants were inefficiently recognized by CD4BS antibodies. The second group, exemplified by the 423 I/P mutant, bound CD4BS antibodies but not CD4 or CD4i antibodies. The ability of each of these mutants to bind one of these sets of conformation-dependent ligands rules out global misfolding of the altered glycoproteins.
Several pieces of evidence support the explanation that an alteration in gp120 conformational state accounts for the observed phenotype of the 375 S/W mutant. The indole ring of the substituted tryptophan residue in this mutant is expected to fill the Phe 43 cavity and to increase the propensity of gp120 to assume a conformation close to that of the CD4-bound state. The microcalorimetry studies confirmed that both the enthalpic and entropic changes associated with the binding of 375 S/W gp120 and sCD4 are significantly decreased compared with those seen for wild-type gp120 and sCD4. The entropic gains associated with the 375 S/W change more than compensate for the unfavorable enthalpic changes, resulting in a sixfold increase in CD4-binding affinity. Multiple ligands, particularly those interacting with the receptor-binding surfaces of gp120, induce large compensating changes in enthalpy and entropy upon binding gp120 (Kwong et al., submitted). Thus, it is virtually certain that the source of these changes is the gp120 glycoprotein.
Free gp120 is thought to exhibit interdomain flexibility and thus to sample many conformations. Ligands like CD4 that bind across the gp120 domains decrease the entropy of gp120 and promote the formation of energetically favorable interdomain bonds. The tryptophan substitution at residue 375 results in similar changes, strongly suggesting that this substitution favors the sampling by free gp120 of conformations closer to the CD4-bound state. The small but reproducible increase in CCR5 binding of the 375 S/W mutant in the absence of sCD4 is consistent with this model.
Compared to the wild-type gp120 glycoprotein, the 375 S/W mutant was precipitated inefficiently by the CD4BS antibodies. The preferred conformation of the 375 S/W mutant is apparently not suitable for binding the CD4BS antibodies. This interpretation is supported by the limited solvent accessibility in the CD4-bound state of many of the gp120 residues implicated by mutagenesis in the binding of the CD4BS antibodies (39, 95). A direct effect of the tryptophan substitution on the CD4BS epitopes is not possible if gp120 maintains a CD4-bound conformation, because residue 375 is not solvent accessible in this case (39). Thus, whether the effect of the tryptophan substitution at serine 375 on the binding of CD4BS antibodies is mediated by conformational alteration or epitope disruption, gp120 must assume different conformations when binding CD4 and CD4BS antibodies.
The argument that CD4 and CD4BS antibodies recognize distinct gp120 conformations is further supported by the phenotypes of the 423 I/P and 423 I/M + 425 N/K + 431 G/E mutants. The substitutions in these mutants were designed to alter the architecture of the β20 and β21 gp120 strands, two critical components of the bridging sheet and CD4-binding region (38, 39). As expected, these mutants were markedly defective in binding CD4. That the effect of the 423 I/P change on CD4 binding is secondary to conformational disruption is supported by the observations that isoleucine 423 does not contact CD4 (38, 39) and that a 423 I/S change does not affect CD4 binding (72).
Binding of the 423 I/P and 423 I/M + 425 N/K + 431 G/E mutants to the 17b CD4i antibody was also poor, as expected from the contribution of the bridging sheet to contacts with this antibody (38, 39). In the available X-ray crystal structures, isoleucine 423 directly contacts the 17b Fab fragment, and another change in this gp120 residue, 423 I/S, has previously been shown to eliminate 17b binding (72). The 423 I/S change also eliminates the binding of another CD4i antibody, 48d, and neither 17b nor 48d binding can be restored by incubation of the mutant gp120 glycoprotein with sCD4 (72). These observations suggest that the negative effects of the 423 I/P change on 17b and 48d binding probably result from alteration of a side chain that is critical for antibody contact. By contrast, the negative effects of the 423 I/P change on the binding of another subset of CD4i antibodies appear to be mediated through conformational disruption. Three newly described CD4i antibodies (23e, 21c, and 49e) failed to recognize the 423 I/P mutant but, in contrast to the results seen for the 17b and 48d antibodies, precipitated the mutant following incubation with high concentrations of sCD4 (data not shown). Moreover, the 23e, 49e, and 21c antibodies efficiently recognized the 423 I/S mutant even in the absence of sCD4 (98a).
Despite the major disruption of the epitopes for both CD4 and CD4i antibodies, the recognition of the 423 I/P and 423 I/M + 425 N/K + 431 G/E mutants by the CD4BS antibodies was efficient and, in the case of the IgG1b12 antibody, even increased relative to that of the wild-type protein. The epitope of the IgG1b12 antibody has been modeled by gp120 mutagenesis and structural analysis of the free antibody (74). These studies have suggested extensive IgG1b12 contacts with the outer domain of gp120, a model consistent with our results. We recently observed that IgG1b12 binding does not lead to large reductions in gp120 entropy, in contrast to the binding of most CD4BS antibodies (Kwong et al., submitted). An IgG1b12 epitope centered on the outer gp120 domain and not reliant on contacts across gp120 domains would explain this observation. The available data suggest that, although the epitopes of various CD4BS epitopes differ, they are all relatively insensitive to changes that disrupt the conformational integrity of the bridging sheet. In this respect, CD4BS epitopes differ from the binding sites for CD4, CD4i antibodies, and CCR5, which are thought to bind similar or identical conformations of gp120.
The 375 S/W envelope glycoproteins were able to support HIV-1 infection, albeit at a reduced level compared with that of the wild-type envelope glycoproteins. This result suggests that the Phe 43 cavity is not absolutely required for envelope glycoprotein function. Examination of primate immunodeficiency virus sequences reveals that, although most HIV-1 strains have a serine residue at position 375, group O HIV-1 strains generally have a histidine and chimpanzee strains a methionine at this position. A tryptophan residue is found at this position in most HIV-2 and simian immunodeficiency virus (SIV) isolates. One of the Phe 43 cavity-lining residues, tryptophan 112 in HIV-1, is a phenylalanine in HIV-2/SIV gp120 glycoproteins, perhaps allowing tryptophan 375 to be accommodated. The highly conserved nature of the other gp120 residues contacting the Phe 43 cavity (38, 39) suggests that gp120 architecture in this region is similar among the primate immunodeficiency viruses. Thus, tryptophan 375 in the HIV-2 and SIV gp120 molecules probably represents a cavity-filling residue and might contribute to some of the properties of these viruses that differ from those of HIV-1. For example, SIV strains often exhibit some degree of CD4 independence. HIV-2 and SIV rarely, if ever, elicit CD4BS antibodies, a property that might be explained by preferred gp120 conformations approximating the CD4-bound state and not recognized by CD4BS antibodies.
The 375 S/W change influences the sensitivity of the virus to neutralization. Compared with the wild-type virus, viruses with 375 S/W envelope glycoproteins exhibited a significant increase in sensitivity to the 2G12 antibody, a slight increase in sensitivity to sCD4, and a marked resistance to the CD4BS antibody IgG1b12. These neutralization phenotypes can be explained by the observed alterations in the affinity of monomeric 375 S/W gp120 preparations for 2G12, sCD4, and IgG1b12. The observed alterations in neutralization sensitivity imply that different conformations can be assumed by the gp120 molecule in the context of the wild-type HIV-1 envelope glycoprotein trimer, at least in the presence of particular ligands. More studies will be required to assess the degree of gp120 conformational flexibility on the free envelope glycoprotein trimer.
The conformational flexibility of the HIV-1 envelope glycoproteins is important to the function of these molecules in mediating virus entry and in evading the humoral immune response. Therefore, understanding the range of conformations available to these glycoproteins is important for an appreciation of their role in HIV-1 replication and for guiding attempts at intervention. The feasibility of targeting the conserved receptor-binding regions of HIV-1 gp120 with drugs or antibodies will no doubt be influenced by the conformational variation of these structures. Limiting the conformational heterogeneity of the gp120 core, as we have begun to do with the 375 S/W change, might increase the efficiency with which antibodies directed against receptor-binding surfaces are generated. Although the 375 S/W mutant does not fully mimic the CD4-bound state, the phenotypes observed and the approaches used herein will be useful in guiding efforts to modify the HIV-1 envelope glycoprotein further to achieve that end. Stabilization of other conformational states of the gp120 glycoprotein, such as that recognized by CD4BS antibodies, would also be a desirable goal.
Acknowledgments
This study was supported by grants from the National Institutes of Health (AI24755, AI31783, AI41851, and AI24030), by a Center for AIDS Research grant (AI42848), and by gifts from the G. Harold and Leila Y. Mathers Charitable Foundation, the Bristol-Myers Squibb Foundation, the Friends 10, the late William F. McCarty-Cooper, and Douglas and Judith Krupp. S.-H. Xiang was supported by an Australian National Health and Medical Research Council HIV/AIDS fellowship (987185). Peter D. Kwong was a recipient of a Burroughs Wellcome Career Development Award.
REFERENCES
- 1.Akasako, A., M. Haruki, M. Oobatake, and S. Kanaya. 1997. Conformational stabilities of Escherichia coli RNase HI variants with a series of amino acid substitutions at a cavity within the hydrophobic core. J. Biol. Chem. 272:18686-18693. [DOI] [PubMed] [Google Scholar]
- 2.Allan, J. S. 1991. Receptor-mediated activation of immunodeficiency viruses in viral fusion. Science 252:1322-1323. [DOI] [PubMed] [Google Scholar]
- 3.Arthos, J., K. C. Deen, M. A. Chaikin, J. A. Fornwald, G. Sathe, Q. J. Sattentau, P. R. Clapham, R. A. Weiss, J. S. McDougal, C. Pietropaolo, et al. 1989. Identification of the residues in human CD4 critical for the binding of HIV. Cell 57:469-481. [DOI] [PubMed] [Google Scholar]
- 4.Barre-Sinoussi, F., J. C. Chermann, F. Rey, M. T. Nugeyre, S. Chamaret, J. Gruest, C. Dauguet, C. Axler-Blin, F. Vezinet-Brun, C. Rouzioux, W. Rozenbaum, and L. Montagnier. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., R. A. Fridell, I. Aramori, S. S. Ferguson, M. G. Caron, and B. R. Cullen. 1997. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 coreceptor. EMBO J. 16:2599-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binley, J. M., R. Wyatt, E. Desjardins, P. D. Kwong, W. Hendrickson, J. P. Moore, and J. Sodroski. 1998. Analysis of the interaction of antibodies with a conserved enzymatically deglycosylated core of the HIV type 1 envelope glycoprotein 120. AIDS Res. Hum. Retrovir. 14:191-198. [DOI] [PubMed] [Google Scholar]
- 7.Brodsky, M. H., M. Warton, R. M. Myers, and D. R. Littman. 1990. Analysis of the site in CD4 that binds to the HIV envelope glycoprotein. J. Immunol. 144:3078-3086. [PubMed] [Google Scholar]
- 8.Buckle, A. M., P. Cramer, and A. R. Fersht. 1996. Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities. Biochemistry 35:4298-4305. [DOI] [PubMed] [Google Scholar]
- 9.Burton, D. R. 1997. A vaccine for HIV type 1: the antibody perspective. Proc. Natl. Acad. Sci. USA 94:10018-10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton, D. R., and D. C. Montefiori. 1997. The antibody response in HIV-1 infection. AIDS 11:S87-S98. [PubMed] [Google Scholar]
- 11.Burton, D. R., and J. P. Moore. 1998. Why do we not have an HIV vaccine and how can we make one? Nat. Med. 4:495-498. [DOI] [PubMed] [Google Scholar]
- 12.Carrillo, A., D. B. Trowbridge, P. Westervelt, and L. Ratner. 1993. Identification of HIV1 determinants for T lymphoid cell line infection. Virology 197:817-824. [DOI] [PubMed] [Google Scholar]
- 13.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 14.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 15.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 16.Cordonnier, A., L. Montagnier, and M. Emerman. 1989. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature 340:571-574. [DOI] [PubMed] [Google Scholar]
- 17.Dalgleish, A. G., P. C. Beverley, P. R. Clapham, D. H. Crawford, M. F. Greaves, and R. A. Weiss. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763-767. [DOI] [PubMed] [Google Scholar]
- 18.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 19.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 20.Durr, E., and I. Jelesarov. 2000. Thermodynamic analysis of cavity creating mutations in an engineered leucine zipper and energetics of glycerol-induced coiled coil stabilization. Biochemistry 39:4472-4482. [DOI] [PubMed] [Google Scholar]
- 21.Eriksson, A. E., W. A. Baase, and B. W. Matthews. 1993. Similar hydrophobic replacements of Leu99 and Phe153 within the core of T4 lysozyme have different structural and thermodynamic consequences. J. Mol. Biol. 229:747-769. [DOI] [PubMed] [Google Scholar]
- 22.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 23.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, et al. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 25.Heeney, J. L., and B. H. Hahn. 2000. Vaccines and immunology: elucidating immunity to HIV-1 and current prospects for AIDS vaccine development. AIDS 14:S125-S127. [PubMed] [Google Scholar]
- 26.Hwang, S. S., T. J. Boyle, H. K. Lyerly, and B. R. Cullen. 1991. Identification of the envelope V3 loop as the primary determinant of cell tropism in HIV-1. Science 253:71-74. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa, K., H. Nakamura, K. Morikawa, and S. Kanaya. 1993. Stabilization of Escherichia coli ribonuclease HI by cavity-filling mutations within a hydrophobic core. Biochemistry 32:6171-6178. [PubMed] [Google Scholar]
- 28.Izard, J., M. W. Parker, M. Chartier, D. Duche, and D. Baty. 1994. A single amino acid substitution can restore the solubility of aggregated colicin A mutants in Escherichia coli. Protein Eng. 7:1495-1500. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, S. E., M. Moracci, N. elMasry, C. M. Johnson, and A. R. Fersht. 1993. Effect of cavity-creating mutations in the hydrophobic core of chymotrypsin inhibitor 2. Biochemistry 32:11259-11269. [DOI] [PubMed] [Google Scholar]
- 30.Kellis, J. T., Jr., K. Nyberg, D. Sali, and A. R. Fersht. 1988. Contribution of hydrophobic interactions to protein stability. Nature 333:784-786. [DOI] [PubMed] [Google Scholar]
- 31.Klatzmann, D., E. Champagne, S. Chamaret, J. Gruest, D. Guetard, T. Hercend, J. C. Gluckman, and L. Montagnier. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767-768. [DOI] [PubMed] [Google Scholar]
- 32.Klein, E., and R. Ho. 2000. Challenges in the development of an effective HIV vaccine: current approaches and future directions. Clin. Ther. 22:295-314. [DOI] [PubMed] [Google Scholar]
- 33.Kocher, J. P., M. Prevost, S. J. Wodak, and B. Lee. 1996. Properties of the protein matrix revealed by the free energy of cavity formation. Structure 4:1517-1529. [DOI] [PubMed] [Google Scholar]
- 34.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolchinsky, P., T. Mirzabekov, M. Farzan, E. Kiprilov, M. Cayabyab, L. J. Mooney, H. Choe, and J. Sodroski. 1999. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J. Virol. 73:8120-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kono, H., M. Saito, and A. Sarai. 2000. Stability analysis for the cavity-filling mutations of the Myb DNA-binding domain utilizing free-energy calculations. Proteins 38:197-209. [DOI] [PubMed] [Google Scholar]
- 37.Korber, B., F. Foley, C. Kuiken, S. Pillai, and J. Sodroski. 1998. Numbering positions in HIV relative to HXBc2, p. III-102-III-103. In B. Korber, C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 38.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold. Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 39.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lasky, L. A., G. Nakamura, D. H. Smith, C. Fennie, C. Shimasaki, E. Patzer, P. Berman, T. Gregory, and D. J. Capon. 1987. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell 50:975-985. [DOI] [PubMed] [Google Scholar]
- 41.Lassalle, M. W., H. Yamada, H. Morii, K. Ogata, A. Sarai, and K. Akasaka. 2001. Filling a cavity dramatically increases pressure stability of the c-Myb R2 subdomain. Proteins 45:96-101. [DOI] [PubMed] [Google Scholar]
- 42.Lee, J., K. Lee, and S. Shin. 2000. Theoretical studies of the response of a protein structure to cavity-creating mutations. Biophys. J. 78:1665-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leonard, C. K., M. W. Spellman, L. Riddle, R. J. Harris, J. N. Thomas, and T. J. Gregory. 1990. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J. Biol. Chem. 265:10373-10382. [PubMed] [Google Scholar]
- 44.Letvin, N. L. 1998. Progress in the development of an HIV-1 vaccine. Science 280:1875-1880. [DOI] [PubMed] [Google Scholar]
- 45.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:465-471. [DOI] [PubMed] [Google Scholar]
- 46.Modrow, S., B. H. Hahn, G. M. Shaw, R. C. Gallo, F. Wong-Staal, and H. Wolf. 1987. Computer-assisted analysis of envelope protein sequences of seven human immunodeficiency virus isolates: prediction of antigenic epitopes in conserved and variable regions. J. Virol. 61:570-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moebius, U., L. K. Clayton, S. Abraham, S. C. Harrison, and E. L. Reinherz. 1992. The human immunodeficiency virus gp120 binding site on CD4: delineation by quantitative equilibrium and kinetic binding studies of mutants in conjunction with a high-resolution CD4 atomic structure. J. Exp. Med. 176:507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montefiori, D. C., and T. G. Evans. 1999. Toward an HIV type 1 vaccine that generates potent, broadly cross-reactive neutralizing antibodies. AIDS Res. Hum. Retrovir. 15:689-698. [DOI] [PubMed] [Google Scholar]
- 49.Moore, J. P., Y. Cao, L. Qing, Q. J. Sattentau, J. Pyati, R. Koduri, J. Robinson, C. F. Barbas III, D. R. Burton, and D. D. Ho. 1995. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 69:101-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moore, J. P., and D. D. Ho. 1993. Antibodies to discontinuous or conformationally sensitive epitopes on the gp120 glycoprotein of human immunodeficiency virus type 1 are highly prevalent in sera of infected humans. J. Virol. 67:863-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by with monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moore, J. P., Q. J. Sattentau, R. Wyatt, and J. Sodroski. 1994. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J. Virol. 68:469-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore, J. P., Q. J. Sattentau, H. Yoshiyama, M. Thali, M. Charles, N. Sullivan, S. W. Poon, M. S. Fung, F. Traincard, M. Pinkus, et al. 1993. Probing the structure of the V2 domain of human immunodeficiency virus type 1 surface glycoprotein gp120 with a panel of eight monoclonal antibodies: human immune response to the V1 and V2 domains. J. Virol. 67:6136-6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morikita, T., Y. Maeda, S. Fujii, S. Matsushita, K. Obaru, and K. Takatsuki. 1997. The V1/V2 region of human immunodeficiency virus type 1 modulates the sensitivity to neutralization by soluble CD4 and cellular tropism. AIDS Res. Hum. Retrovir. 13:1291-1299. [DOI] [PubMed] [Google Scholar]
- 56.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oberlin, E., A. Amara, F. Bachelerie, C. Bessia, J. L. Virelizier, F. Arenzana-Seisdedos, O. Schwartz, J. M. Heard, I. Clark-Lewis, D. F. Legler, M. Loetscher, M. Baggiolini, and B. Moser. 1996. The CXC chemokine SDF-1 is the ligand for LESTR/fusin and prevents infection by T-cell-line-adapted HIV-1. Nature 382:833-835. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien, W. A., Y. Koyanagi, A. Namazie, J. Q. Zhao, A. Diagne, K. Idler, J. A. Zack, and I. S. Chen. 1990. HIV-1 tropism for mononuclear phagocytes can be determined by regions of gp120 outside the CD4-binding domain. Nature 348:69-73. [DOI] [PubMed] [Google Scholar]
- 59.Ohmura, T., T. Ueda, K. Ootsuka, M. Saito, and T. Imoto. 2001. Stabilization of hen egg white lysozyme by a cavity-filling mutation. Protein Sci. 10:313-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Olshevsky, U., E. Helseth, C. Furman, J. Li, W. Haseltine, and J. Sodroski. 1990. Identification of individual human immunodeficiency virus type 1 gp120 amino acids important for CD4 receptor binding. J. Virol. 64:5701-5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parren, P. W., I. Mondor, D. Naniche, H. J. Ditzel, P. J. Klasse, D. R. Burton, and Q. J. Sattentau. 1998. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 72:3512-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Parren, P. W., J. P. Moore, D. R. Burton, and Q. J. Sattentau. 1999. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS 13:S137-S162. [PubMed] [Google Scholar]
- 64.Peterson, A., and B. Seed. 1988. Genetic analysis of monoclonal antibody and HIV binding sites on the human lymphocyte antigen CD4. Cell 54:65-72. [DOI] [PubMed] [Google Scholar]
- 65.Pollard, S. R., M. D. Rosa, J. J. Rosa, and D. C. Wiley. 1992. Truncated variants of gp120 bind CD4 with high affinity and suggest a minimum CD4 binding region. EMBO J. 11:585-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ponder, J. W., and F. M. Richards. 1987. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J. Mol. Biol. 193:775-791. [DOI] [PubMed] [Google Scholar]
- 67.Posner, M. R., T. Hideshima, T. Cannon, M. Mukherjee, K. H. Mayer, and R. A. Byrn. 1991. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. J. Immunol. 146:4325-4332. [PubMed] [Google Scholar]
- 68.Rashin, A. A., B. H. Rashin, A. Rashin, and R. Abagyan. 1997. Evaluating the energetics of empty cavities and internal mutations in proteins. Protein Sci. 6:2143-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ratnaparkhi, G. S., and R. Varadarajan. 2000. Thermodynamic and structural studies of cavity formation in proteins suggest that loss of packing interactions rather than the hydrophobic effect dominates the observed energetics. Biochemistry 39:12365-12374. [DOI] [PubMed] [Google Scholar]
- 70.Richards, F. M. 1974. The interpretation of protein structures: total volume, group volume distributions and packing density. J. Mol. Biol. 82:1-14. [DOI] [PubMed] [Google Scholar]
- 71.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 72.Rizzuto, C. D., R. Wyatt, N. Hernandez-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 73.Ryu, S. E., P. D. Kwong, A. Truneh, T. G. Porter, J. Arthos, M. Rosenberg, X. P. Dai, N. H. Xuong, R. Axel, R. W. Sweet, et al. 1990. Crystal structure of an HIV-binding recombinant fragment of human CD4. Nature 348:419-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saphire, E. O., P. W. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton, and I. A. Wilson. 2001. Crystal structure of a neutralizing human IgG against HIV-1: a template for vaccine design. Science 293:1155-1159. [DOI] [PubMed] [Google Scholar]
- 75.Sattentau, Q. J., and J. P. Moore. 1995. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 182:185-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sattentau, Q. J., J. P. Moore, F. Vignaux, F. Traincard, and P. Poignard. 1993. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J. Virol. 67:7383-7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shioda, T., J. A. Levy, and C. Cheng-Mayer. 1992. Small amino acid changes in the V3 hypervariable region of gp120 can affect the T-cell-line and macrophage tropism of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 89:9434-9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Si, Z., M. Cayabyab, and J. Sodroski. 2001. Envelope glycoprotein determinants of neutralization resistance in a simian-human immunodeficiency virus (SHIV-HXBc2P 3.2) derived by passage in monkeys. J. Virol. 75:4208-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Starcich, B. R., B. H. Hahn, G. M. Shaw, P. D. McNeely, S. Modrow, H. Wolf, E. S. Parks, W. P. Parks, S. F. Josephs, R. C. Gallo, et al. 1986. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell 45:637-648. [DOI] [PubMed] [Google Scholar]
- 81.Steif, C., H. J. Hinz, and G. Cesareni. 1995. Effects of cavity-creating mutations on conformational stability and structure of the dimeric 4-alpha-helical protein ROP: thermal unfolding studies. Proteins 23:83-96. [DOI] [PubMed] [Google Scholar]
- 82.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to gp120. Science 254:105-108. [DOI] [PubMed] [Google Scholar]
- 83.Sullivan, N., Y. Sun, J. Binley, J. Lee, C. F. Barbas III, P. W. Parren, D. R. Burton, and J. Sodroski. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J. Virol. 72:6332-6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sullivan, N., Y. Sun, J. Li, W. Hofmann, and J. Sodroski. 1995. Replicative function and neutralization sensitivity of envelope glycoproteins from primary and T-cell-line-passaged human immunodeficiency virus type 1 isolates. J. Virol. 69:4413-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sullivan, N., M. Thali, C. Furman, D. D. Ho, and J. Sodroski. 1993. Effect of amino acid changes in the V1/V2 region of the human immunodeficiency virus type 1 gp120 glycoprotein on subunit association, syncytium formation, and recognition by a neutralizing antibody. J. Virol. 67:3674-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thali, M., C. Furman, D. D. Ho, J. Robinson, S. Tilley, A. Pinter, and J. Sodroski. 1992. Discontinuous, conserved neutralization epitopes overlapping the CD4-binding region of human immunodeficiency virus type 1 gp120 envelope glycoprotein. J. Virol. 66:5635-5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 90.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang, J. H., Y. W. Yan, T. P. Garrett, J. H. Liu, D. W. Rodgers, R. L. Garlick, G. E. Tarr, Y. Husain, E. L. Reinherz, and S. C. Harrison. 1990. Atomic structure of a fragment of human CD4 containing two immunoglobulin-like domains. Nature 348:411-418. [DOI] [PubMed] [Google Scholar]
- 92.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 93.Wu, H., P. D. Kwong, and W. A. Hendrickson. 1997. Dimeric association and segmental variability in the structure of human CD4. Nature 387:527-530. [DOI] [PubMed] [Google Scholar]
- 94.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 95.Wyatt, R., P. D. Kwong, E. Desjardins, R. W. Sweet, J. Robinson, W. A. Hendrickson, and J. G. Sodroski. 1998. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature 393:705-711. [DOI] [PubMed] [Google Scholar]
- 96.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 98.Wyatt, R., N. Sullivan, M. Thali, H. Repke, D. Ho, J. Robinson, M. Posner, and J. Sodroski. 1993. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J. Virol. 67:4557-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98a.Xiang, S.-H., N. Doka, R. Choudhary, J. Sodroski, and J. Robinson. Characterization of CD4-induced epitopes on the HIV-1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir., in press. [DOI] [PubMed]
- 99.Xu, B., Q. X. Hua, S. H. Nakagawa, W. Jia, Y. C. Chu, P. G. Katsoyannis, and M. A. Weiss. 2002. A cavity-forming mutation in insulin induces segmental unfolding of a surrounding alpha-helix. Protein Sci. 11:104-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yang, Z. N., T. C. Mueser, J. Kaufman, S. J. Stahl, P. T. Wingfield, and C. C. Hyde. 1999. The crystal structure of the SIV gp41 ectodomain at 1.47 Å resolution. J. Struct. Biol. 126:131-144. [DOI] [PubMed] [Google Scholar]
- 101.Ye, Y., Z. H. Si, J. P. Moore, and J. Sodroski. 2000. Association of structural changes in the V2 and V3 loops of the gp120 envelope glycoprotein with acquisition of neutralization resistance in a simian-human immunodeficiency virus passaged in vivo. J. Virol. 74:11955-11962. [DOI] [PMC free article] [PubMed] [Google Scholar]