Abstract
Human papillomavirus type 16 (HPV16) is the most common cause of cervical carcinoma. Cervical cancer develops from low-grade lesions that support the productive stages of the virus life cycle. The 16E1∧E4 protein is abundantly expressed in such lesions and can be detected in cells supporting vegetative viral genome amplification. Using an inducible mammalian expression system, we have shown that 16E1∧E4 arrests HeLa cervical epithelial cells in G2. 16E1∧E4 also caused a G2 arrest in SiHa, Saos-2 and Saccharomyces pombe cells and, as with HeLa cells, was found in the cytoplasm. However, whereas 16E1∧E4 is found on the keratin networks in HeLa and SiHa cells, in Saos-2 and S. pombe cells that lack keratins, 16E1∧E4 had a punctate distribution. Mutagenesis studies revealed a proline-rich region between amino acids 17 and 45 of 16E1∧E4 to be important for arrest. This region, which we have termed the “arrest domain,” contains a putative nuclear localization signal, a cyclin-binding motif, and a single cyclin-dependent kinase (Cdk) phosphorylation site. A single point mutation in the putative Cdk phosphorylation site (T23A) abolished 16E1∧E4-mediated G2 arrest. Arrest did not involve proteins regulating the phosphorylation state of Cdc2 and does not appear to involve the activation of the DNA damage or incomplete replication checkpoint. G2 arrest was also mediated by the E1∧E4 protein of HPV11, a low-risk mucosal HPV type that also causes cervical lesions. The E1∧E4 protein of HPV1, which is more distantly related to that of HPV16, did not cause G2 arrest. We conclude that, like other papillomavirus proteins, 16E1∧E4 affects cell cycle progression and that it targets a conserved component of the cell cycle machinery.
Papillomaviruses are small DNA viruses that infect the epithelial tissue of a wide range of vertebrates, including humans (47). Infection begins in cells of the basal layer, with the productive stages of the virus life cycle being initiated as these infected cells migrate toward the epithelial surface. In this way, the virus life cycle is tightly linked to the differentiation of the epithelium, which makes human papillomaviruses (HPVs) difficult to study in the laboratory (37). More than 200 papillomavirus types have been identified (12). These share a common organization of their ∼8-kb genomes but differ in the types of epithelium they infect and the pathologies that each virus causes (47). Infection occurs at cutaneous epithelial sites, i.e., the skin, for viruses such as HPV type 1 (HPV1) or at mucosal sites (e.g., the anogenital tract or the cervix) for viruses such as HPV11 and HPV16 (47). HPVs are additionally classified from low to high risk, with low-risk viruses such as HPV11 (and HPV1) being associated solely with benign lesions (often known as warts), whereas lesions caused by high-risk viruses such as HPV16 can progress to malignancy (47). HPV16 is the most prevalent high-risk virus and is the most common causative agent of cervical cancers (68).
Although the 16E1∧E4 protein is not expressed in lesions that have progressed to malignancy (K. Middleton, L. Morris, W. Peh, K. Sotlar, D. Jenkins, R. Seth, A. El-Sherif, C. Coleman, and J. Doorbar, Abstr. 19th Int. Papillomavirus Conf., abstr. P-21, 2001), in productive infections it is an abundant protein that is expressed in the upper layers of the epithelium. 16E1∧E4 is translated from a spliced transcript (20). The first five amino acids are derived from the E1 open reading frame (ORF), whereas the remainder are derived from the E4 ORF (19). The expression of 16E1∧E4 occurs late during infection and correlates with the onset of vegetative viral DNA amplification, prior to the expression of the viral structural proteins (10, 17). Despite the association with viral genome amplification, which takes place in the nucleus, 16E1∧E4 has a diffuse and filamentous distribution in epithelial cells and is located in the cytoplasm (17). This filamentous distribution is a result of an association between 16E1∧E4 and the cytokeratin networks and, at least in cell culture, this association can result in collapse of the keratin filament networks (16, 59). The 16E1∧E4 protein has also been found to bind a DEAD box RNA helicase (E4-DBP) and alter its ATPase activity (14). However, the significance for the virus of the 16E1∧E4 associations with keratins and E4-DBP is unclear, and the role of the E1∧E4 protein in the viral life cycle remains unknown (13).
To investigate the role of 16E1∧E4 during the virus life cycle, we constructed HPV16 mutant genomes that were unable to express the full-length 16E1∧E4 protein. Our preliminary experiments suggested that experimental lesions induced by these mutants were more proliferative than those induced by wild-type DNA, suggesting a possible role for 16E1∧E4 in cell cycle regulation. Here we show that 16E1∧E4 causes HeLa and Saos-2 cells to arrest in the G2 phase of the cell cycle. 16E1∧E4 caused a G2 arrest in Schizosaccharomyces pombe, enabling us to use this system as a model to investigate 16E1∧E4 function. The ability of 16E1∧E4 to induce G2 arrest was assayed in a variety of cell cycle mutant genetic backgrounds and in the presence of the drug pentoxifylline. Using a range of 16E1∧E4 mutants, residues in the N-terminal half of 16E1∧E4 were found to be important for G2 arrest. A single point mutation in a putative Cdk phosphorylation site within the “arrest domain” of 16E1∧E4 was sufficient to abolish the phenotype.
MATERIALS AND METHODS
Vectors, molecular subcloning, and mutagenesis.
The HPV 16E1∧E4 protein and green fluorescent protein (GFP) were expressed from the recombinant adenoviruses rAd.16E1∧E4 and rAd.GFP (14) under the control of the cytomegalovirus (CMV) early promoter. To prepare the yeast expression vectors, 0.3-kb fragments containing E1∧E4 wild-type and mutant sequences were PCR amplified from the plasmids pGBT9 (wild-type 16E1∧E4, Δ2-6, Δ12-16, Δ23-28, Δ27-32, Δ31-36, Δ36-41, Δ41-46, Δ46-51, Δ52-57, Δ63-68, Δ73-77, Δ80-83, Δ84-88, Δ86-92, and 16E1∧E4 mutants or wild-type 1E1∧E4) (14) and pGEX4T-1.11E1∧E4 (55a) with the primers shown in Table 1. The fragments were inserted into the NdeI-BamHI sites downstream of the inducible nmt1 (for “no message in thiamine”) promoter (51) in the plasmid pREP1 (52). Mutagenesis was performed by using the QuikChange site-directed mutagenesis kit (Stratagene) with the primers listed in Table 2. All mutations were confirmed by bidirectional sequencing. The HPV 16E1∧E4 sequences were also subcloned into the mammalian expression vector pMV11 (14) downstream of the CMV early promoter. Again, 0.3-kb fragments containing the HPV 16E1∧E4 wild-type and mutant sequences were PCR amplified from the above plasmids with the primers shown in Table 1 and inserted into the KpnI-EcoRI sites of pMV11. The plasmid pXJ.GFP has been described previously (71).
TABLE 1.
Primers used for subcloning by PCR
| Primer | Sequence (5′ to 3′) |
|---|---|
| 5′ Wild type, Δ12-16, Δ23-28, Δ27-32, Δ31-36, Δ36-41, Δ41-46, Δ46-51, Δ52-57, Δ63-68, Δ73-77, Δ80-83, Δ84-88, Δ86-92, and Δ51-92 16E1∧E4 into pREP1 | CTTGATCATATGGCTGATCCTGCAGCAGCAACG |
| 3′ Wild type, Δ2-6, Δ12-16, Δ23-28, Δ27-32, Δ31-36, Δ36-41, Δ41-46, Δ46-51, Δ52-57, Δ63-68, Δ73-77, Δ80-83, Δ1-49, and Δ1-16 16E1∧E4 into pREP1 | GTAACTGGATCCTTATGGGTGTAGTGTTACTATTA |
| 5′ Δ2-6 16E1∧E4 into pREP1 | AGAATTCATATGGCAGCAACGAAGTATCCTCTC |
| 3′ Δ84-88 16E1∧E4 into pREP1 | AGAAATGGATCCTTATGGGTGTAGTGTTCCGTCCT |
| 3′ Δ87-92 16E1∧E4 into pREP1 | AGAAATGGATCCTTAAGTTAATCCGTCCTTTGTGTG |
| 5′ Δ1-49 16E1∧E4 into pREP1 | AGAATTCATATGCAGACACCGGAAACCCCTGCC |
| 5′ Δ1-16 16E1∧E4 into pREP1 | AGAATTCATATGGGCAGCACTTGGCCAACCACC |
| 3′ Δ51-92 16E1∧E4 into pREP1 | GTAACTGGATCCTTACTGGCTCTGATCTTGGTC |
| 5′ 11E1∧E4 into pREP1 | AGATTCCATATGGCGGACGATTCAGCACTGTAC |
| 5′ 16E1∧E4 into pREP1 | AGAATTCATATGGCAGATAATAAAGCTCCCCAA |
| 3′ 11E1∧E4 into pREP1 | AGAATTGGATCCCTATAGGCGTAGCTGCACTGT |
| 3′ 1E1∧E4 into pREP1 | AGAATTAGATCTTTACACAGACCACGGGTGGAT |
| 5′ Wild type, T22A, T23A 16E1∧E4 into pMV11 | AGAATTGGTACCATGGCTGATCCTGCAGCAGCA |
| 3 Wild type, T22A, T23A 16E1∧E4 into pMV11 | AGAATTGAATTCTTATGGGTGTAGTGTTACTATTA |
TABLE 2.
Primers used for mutagenesis reactions
| Mutant | Primer sequences (5′ to 3′) |
|---|---|
| K37A,K38A,H39A | GCCGTCGCCTTGGGCACCGGCGGCAGCCAGACGACTATCCAGCGACC |
| GGTCGCTGGATAGTCGTCTGGCTGCCGCCGGTGCCCAAGGCGACGGC | |
| S43A,S44A | CGAAGAAACACAGACGACTAGCCGCCGACCAAGATCAGAGCCAG |
| CTGGCTCTGATCTTGGTCGGCGGCTAGTCGTCTGTGTTTCTTCG | |
| R40A,L42A | GGGCACCGAAGAAACACGCACGAGCATCCAGCGACCAAGATC |
| GATCTTGGTCGCTGGATGCTCGTGCGTGTTTCTTCGGTGCCC | |
| T23A,P24A,P25A,P27A | CAGCACTTGGCCAACCGCCGCGGCGCGAGCGATACCAAAGCCGTCG |
| CGACGGCTTTGGTATCGCTCGCGCCGCGGCGGTTGGCCAAGTGCTG | |
| T23A | CTTGGCCAACCGCCCCGCCGCGACC |
| GGTCGCGGCGGGGCGGTTGGCCAAG | |
| T23D | CTTGGCCAACCGACCCGCCGCGACC |
| GGTCGCGGCGGGTCGGTTGGCCAAG | |
| T23E | CTTGGCCAACCGAGCCGCCGCGACC |
| GGTCGCGGCGGCTCGGTTGGCCAAG | |
| T22A,T23A | CAGCACTTGGCCAGCCGCCCCGCCGCGACCC |
| GGGTCGCGGCGGGGCGGCTGGCCAAGTGCTG |
Generation of HeLa Tet-on cell line for the stable expression of 16E1∧E4.
A 0.3-kb BamHI-EcoRI fragment containing HPV 16E1∧E4 was excised from the plasmid pMV11.16E1∧E4 (14), and the 3′-overhangs were filled in with Klenow polymerase. The fragment was subcloned into the unique BamHI-PvuII sites in the plasmid pTRE2pur (Clontech) to generate pTRE2pur.16E1∧E4, a plasmid expressing 16E1∧E4 under the control of the reverse tetracycline-controlled transactivator (rtTA). To generate the HeLa-16E1∧E4-Tet-on cell line, pTRE2pur-16E1∧E4 was transfected into HeLa Tet-on cells (Clontech), a HeLa-derived cell line which expresses the chimeric rtTA. The cells were cultured in Dulbecco modified Eagle medium (DMEM), supplemented with 10% tetracycline-free fetal calf serum and 0.1 mg of G418 ml−1. Puromycin (0.5 mg ml−1) was added to select for the pTRE2pur.16E1∧E4 plasmid. After induction with the tetracycline derivative doxycycline (2 μg ml−1), the HeLa-16E1∧E4-Tet-on positive clones were detected by immunofluorescence with TVG402 (15) Alexa-fluor 488-conjugated antibody and a clonal line was derived from a single positive cell. Populations of 16E1∧E4-expressing cells were obtained by incubating the cells with doxycycline (3 μg ml−1) for 24 h.
Synchronization and recombinant adenovirus infection of mammalian cells.
Saos-2 cells were synchronized at G1/S and infected with recombinant adenovirus by incubating them in 2 mM thymidine in DMEM for 17 h, washing the cells twice with DMEM to remove the thymidine, incubating them with rAd.GFP or rAd.16E1∧E4 (at a multiplicity of infection of 15) for 8 h, and finally incubating them again in 2 mM thymidine in DMEM for 17 h. The block was released by washing the cells twice with DMEM and replacing them with 10% fetal calf serum in DMEM. SiHa and HeLa cells were synchronized at the G1/S border with thymidine (as described above) or with low serum-aphidicolin as described by Liu et al. (45). At 6 h prior to release from the block, SiHa cells were infected with 100 rAd particles per cell. For HeLa cells, infection was carried out 3 h prior to release with 200 rAd particles per cell.
S. pombe strains and media.
S. pombe strains (Table 3) were maintained in Edinburgh minimal medium (EMM) supplemented with adenine, histidine, uracil, and in the absence of plasmid, leucine (each at 0.1 mg ml−1). S. pombe were normally grown at 30°C except for temperature-sensitive strains, which were grown at the permissive temperature of 23°C and shifted to the restrictive temperature of 35°C to induce the phenotype. To repress the nmt promoter, thiamine was added to the medium to a final concentration of 75 μg ml−1. When required, pentoxifylline (Sigma) was added to the growth medium at a final concentration of 5 mM. To induce expression from the nmt promoter, cells were washed twice with EMM (minus thiamine) and reinoculated into minus-thiamine EMM. S. pombe were transformed with plasmid DNA by electroporation (1.5 kV, 25 μF, 200 Ω, and 0.2 cm) with a GenePulser (Bio-Rad).
TABLE 3.
S. pombe strains used in this study
| Name | Strain | Genotype | Description | G2 arresta |
|---|---|---|---|---|
| Wild type | PR109 | leu1-32 ura4-D18 h− | Normal cell cycle | + |
| cdc2-1w | JM305 | cdc2-1w leu1-32 ura4-D18 h+ | Constitutively active Cdc2, insensitive to Y15 phosphorylation by Wee1 | + |
| cdc2-3w | JM125 | cdc2-3w leu1-32 ura4-D18 h− | Cdc2 active independently of Cdc25 | + |
| cdc2-3w Δcdc25 | JM192 | cdc2-3w cdc25::ura4 leu1-32 ura4-D18 h | As for cdc2-3w plus no expression of Cdc25 | + |
| wee1-50 | JM604 | wee1-50 leu1-32 ura4D-18 h− | Temperature-sensitive mutant of Wee1 | + |
| wee1-50 Δmik1 | PR337 | wee1-50 mik1::ura4 leu1.32 ura4-D18 h− | Same as for wee1-50 plus no expression of Mik1 | + |
| wee1-50 cdc25-22 | JM1657 | wee1-50 cdc25-22 leu1-32 ura4-D18 h− | Same as for wee1-50 plus temperature-sensitive mutant of Cdc25 | + |
| Δrad24 | JM1172 | rad24::ura4 leu1-32 ura4-D18 ade6-704 h− | No expression of Rad24 | + |
| Δrad25 | JM1173 | rad25::ura4 leu1-32 ura4-D18 ade6-704 h− | No expression of Rad25 | + |
| cdc10-129 | JM195 | cdc10-129 leu1.32 h− | Temperature-sensitive arrest in G1 | + |
+, positive for G2 arrest.
Immunofluorescence staining and microscopy.
Approximately 5 × 107 S. pombe cells were fixed with −20°C methanol and resuspended in PEM buffer (100 mM pipes, pH 6.9; 1 mM EGTA; 1 mM magnesium sulfate). The cell wall was digested with 250 to 1,000 U of lyticase in PEMS buffer (1.2 M sorbitol, PEM) for 7 min at 37°C, and the cells were permeabilized with 1% Triton X-100 in PEMS for 30 s. The cells were incubated overnight with TVG402 (15) Alexa-fluor 488-conjugated antibody in PEMBAL (1% bovine serum albumin, 0.1% sodium azide, 0.1 M l-lysine, PEM), and the DNA was stained with DAPI (4′,6′-diamidino-2-phenylindole; 1 μg ml−1). Mammalian cells were fixed with 5% formaldehyde for 5 min and blocked with 2% bovine serum albumin-0.01% sodium azide in phosphate-buffered saline (PBS) for 30 min. Cells were incubated with TVG402 Alexa-fluor 488-conjugated antibody and DAPI (1 μg ml−1) in 0.1% fetal calf serum in PBS. After five 5-min washes in PBS, the cells were examined by using a fluorescent Labophot II microscope (Nikon). Digital images were captured with a SenSys monochrome camera and IP Lab imaging software (Roper Scientific).
Western blot analysis.
S. pombe cells from mid-log-phase cultures were lysed in extraction buffer (2% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol, 50 mM Tris-HCl [pH 7.0]) by vortexing them with an equal volume of 425- to 600-μm-diameter glass beads. Mammalian cells (HeLa-16E1∧E4-Tet-on, Saos-2, and Cos-7) were lysed in extraction buffer by gentle pipetting and incubation at 95°C for 5 min. Debris from both S. pombe and mammalian cell extracts was pelleted (10,000 × g, 10 min), and the protein concentration in the supernatant estimated by using the detergent-compatible protein assay (Bio-Rad) according to the manufacturer's instructions. Proteins were separated by SDS-polyacrylamide gel electrophoresis on 15% gels and transferred to Immobilon membrane (Millipore) by wet blotting. 16E1∧E4 was detected by using TVG402 hybridoma supernatant (15) horseradish peroxidase-conjugated anti-mouse immunoglobulin (Amersham Pharmacia Biotech) and enhanced chemiluminescence (Amersham Pharmacia Biotech).
Flow cytometric analysis.
Approximately 106 human cells were fixed with 70% ethanol, incubated with Alexa-fluor 488-conjugated TVG402 antibody (15) for 30 min, treated with RNase A (0.1 mg ml−1)-0.1% NP-40 in PBS for 20 min, and stained with propidium iodide (12.5 μg ml−1) for 30 min. Approximately 0.3 × 107 S. pombe cells were fixed with methanol, treated with RNase A (0.1 mg ml−1) in 50 mM sodium citrate for 16 h, and stained with propidium iodide (2 μg ml−1) for 30 min. Fluorescence was analyzed on a FACScan (Becton Dickinson) and analyzed with CellQuest software (Becton Dickinson).
Mitotic index analysis.
Cos-7 cells were seeded at 3 × 104 cells per well and 24 h later were transfected with pXJ.GFP, pMV11.16E1∧E4, or pMV11.T22A,T23A,16E1∧E4 by using Effectene (Qiagen) according to the manufacturer's instructions. At 24 h after transfection, nocodazole was added to a final concentration of 0.1 μg ml−1 for a further 24 h. The cells were harvested by using 0.1 mM EDTA in PBS and applied to a poly-l-lysine-coated slide by cytospinning. The cells were fixed with 5% formaldehyde for 5 min and stained with TVG402 (15) Alexa-fluor 488-conjugated antibody and DAPI (1 μg ml−1). The percentages of cells positive for transgene expression that had condensed chromosomes were determined by microscopy.
RESULTS
HPV16 E1∧E4 protein induces G2 cell cycle arrest.
Papillomaviruses modulate the cell cycle in order to create a cellular environment that allows the replication of viral genomes and the production of infectious virions. With the exception of the E4 ORF, genes contained in the early region of HPV16 (E7, E6, E1, E2, and E5) have been shown to encode proteins that modulate the cell cycle or which associate with components of the cell cycle machinery in order to facilitate viral DNA replication (3, 27, 28, 44, 49). To determine whether the product of the HPV16 E4 ORF also affects cell cycle progression, a tetracycline-inducible HeLa cell line expressing 16E1∧E4 was generated (HeLa-16E1∧E4-Tet-on). To induce expression of 16E1∧E4, cells were treated with doxycycline (a tetracycline derivative) for 24 and 48 h. The cells were stained with propidium iodide and anti-16E1∧E4 antibodies (Alexa-fluor 488-conjugated) in order to identify cells expressing the 16E1∧E4 protein. It was necessary to include the 16E1∧E4 immunostain since despite being a clonal cell line, only 30% of the cells expressed high levels of 16E1∧E4. The cell cycle distribution of the cells was then determined by measuring the DNA content by using flow cytometry. The results show that in the 16E1∧E4-expressing population the cells arrested with an approximately 4N DNA content (Fig. 1A). Some degree of endoreduplication was also apparent, as evidenced by the presence of cells containing a DNA content of greater than 4N. This is in contrast to the non-16E1∧E4-expressing cells which, like the cells not treated with doxycycline, showed a typical HeLa cell cycle profile, i.e., with the majority of the cells being in G1 (Fig. 1A). Microscopic analysis of large populations of 16E1∧E4-expressing cells showed that they had not arrested with condensed chromatin (data not shown). These data suggest that 16E1∧E4 expression induced the HeLa cell line to arrest predominantly in G2, i.e., with a 4N DNA content. It was not possible, based on the profiles shown in Fig. 1A, to establish whether E1∧E4 expression also retarded progression through S phase or whether some cells were arresting with less than a 4N DNA content. To examine this more closely, 16E1∧E4 was transiently expressed in SiHa cells after synchronization in G1. E1∧E4 was expressed from a recombinant adenovirus (rAd.16E1∧E4 [14]), and cell cycle progression was compared at 4-h intervals to that seen after expression of the cellular protein Myt1 (rAd.Myt), which is known to arrest cells in G2 (45), or β-galactosidase (rAd.β-gal), which has no affect the cell cycle (45). No discernible difference could be detected in the rate of exit from G1 or the rate of progression through S-phase in cells expressing 16E1∧E4, Myt1, or β-galactosidase (see 0-, 4-, and 8-h time points in Fig. 1B). Accumulation of cells with a 4N DNA copy number was apparent in cells expressing both Myt1 and 16E1∧E4, however (see the 12-, 16-, and 20-h time points in Fig. 1B). This can be seen most clearly at the 20-h time point in cells expressing 16E1∧E4 (Fig. 1B). No significant difference was apparent in the spread of the G2 peak after expression of 16E1∧E4, Myt1 or β-galactosidase, suggesting that 16E1∧E4 does not markedly affect exit from S phase. Although the majority of the 16E1∧E4-expressing cells appeared to arrest with a 4N DNA content, some cells appeared to have a DNA content of greater than 4N, as would be expected if endoreduplication had taken place. This was apparent in cells expressing both 16E1∧E4 and Myt1, however. As with the HeLa-16E1∧E4-Tet-on cell line, microscopic analysis of large numbers of 16E1∧E4 expressing SiHa cells showed that they had not arrested with condensed chromatin (data not shown). We conclude from these results that 16E1∧E4, like Myt1, causes cell cycle arrest predominantly in G2, i.e., with a 4N DNA content but prior to chromosome condensation.
FIG. 1.
Expression of wild-type 16E1∧E4 in mammalian cells. (A) The HeLa-16E1∧E4-Tet-on cell line that can be induced to express 16E1∧E4 by the addition of doxycycline was treated for 0, 24, and 48 h with doxycycline. The cells were stained with propidium iodide and analyzed by flow cytometry. At 24 and 48 h the cells were additionally stained with TVG402 anti-16E1∧E4 antibody, and the results are shown for the 16E1∧E4-negative (−) and -positive (+) populations. The population of 16E1∧E4-expressing cells arrests with a 4N DNA content, whereas the 16E1∧E4-negative population continues to cycle. Histograms show the relative number of cells (on the y axis) with a particular DNA content (on the x axis). (B) SiHa cells were synchronized at G1 and infected with recombinant adenoviruses that express β-galactosidase (rAd.β-gal), 16E1∧E4 (rAd.16E1∧E4), or Myt1 (rAd.Myt1). The cells were harvested at 0, 4, 8, 12, 16, and 20 h after block release, stained with propidium iodide, and analyzed by flow cytometry as in panel A. Cells expressing 16E1∧E4 or Myt1 show a cell cycle arrest with a 4N DNA content. (C) Saos-2 cells were synchronized at G1 and infected with recombinant adenoviruses that express GFP (rAd.GFP) or 16E1∧E4 (rAd.16E1∧E4). The cells were harvested at 0, 12, and 24 h after block release, stained with propidium iodide, and analyzed by flow cytometry as in panel A. Cells infected with rAd.16E1∧E4 showed a cell cycle arrest with a 4N DNA content. (D) Total cell extracts from Saos-2 cells infected with rAd.GFP or rAd.16E1∧E4, from the HeLa-16E1∧E4-Tet-on cell line, and from SiHa cells infected with rAd.Myt1, rAd.16E1∧E4, or rAd.β-gal were separated on 15% polyacrylamide-SDS gels and Western blotted with TVG402 anti-16E1∧E4 antibody.
The HeLa and SiHa cell lines were derived from cervical cancers caused by HPV18 (42) and HPV16, respectively (23), and the proliferation of these cells is dependent on the continued expression of the E6 and E7 oncogenes. It was therefore possible that the 16E1∧E4 protein was inducing G2 arrest by affecting the function of another HPV protein (either E6 or E7). This occurs with the HPV E2, which when expressed in HPV transformed cell lines, induces G2 arrest and subsequent apoptosis by downregulating the expression of E6 and E7 (31, 69). To test whether 16E1∧E4 was functioning in a similar manner, it was transiently expressed from a recombinant adenovirus (14) in Saos-2 cells, a non-HPV transformed osteocarcoma cell line. The Saos-2 cells were synchronized in G1 and infected with recombinant adenoviruses expressing 16E1∧E4 (rAd.16E1∧E4) or the control protein GFP (rAd.GFP) (14). At 24 h postinfection, the cells were stained with anti-16E1∧E4, Alexa-fluor 488-conjugated antibodies to identify cells expressing 16E1∧E4 before being stained with propidium iodide. The cell cycle distribution of the cells was then determined by measuring the DNA content by using flow cytometry. The results showed that as with the HeLa-16E1∧E4-Tet-on cell line, expression of 16E1∧E4 in Saos-2 cells vastly increased the number of cells found in the G2 phase of the cell cycle (Fig. 1C). This confirms that 16E1∧E4 is not acting to induce G2 arrest via an effect on HPV proteins already present in the cell (Saos-2 cells do not express endogenous HPV proteins) and indicates that 16E1∧E4-induced arrest occurs independently of the 16E1∧E4 keratin association (Saos-2 cells also do not express cytokeratins). In the absence of cytokeratin filaments, 16E1∧E4 staining remains predominantly cytoplasmic but has a punctate staining pattern that may be the result of 16E1∧E4 self-association (data not shown).
The human immunodeficiency virus (HIV) Vpr protein and the HPV E2 protein, in addition to causing G2 arrest in mammalian cells, have also been shown to be capable of inducing G2 arrest in S. pombe (27, 72). To examine whether this is also the case for 16E1∧E4, the 16E1∧E4 cDNA was cloned into pREP1 under the control of the thiamine-repressible promoter, nmt1, and transformed into S. pombe. In the absence of thiamine, 16E1∧E4 expression was detected (Fig. 2A). Microscopic examination of the S. pombe cells revealed an elongated morphology, a finding typical of a cdc phenotype (Fig. 2B). This phenotype occurs when growth continues in the absence of cell division and is indicative of a cell cycle block. To determine whether the 16E1∧E4-induced block was occurring at G1 or G2, the S. pombe cells were analyzed by flow cytometry (Fig. 2C). In the presence of thiamine, the S. pombe cells do not express 16E1∧E4 and are found predominantly in G2, as is normal for an asynchronous population of S. pombe cells since the G1 phase of the cell cycle in S. pombe is extremely short compared to G2. In the absence of thiamine, the elongated S. pombe cells expressing 16E1∧E4 remain in G2. (The increase in DNA content beyond the standard G2 peak is due to continued replication of mitochondrial DNA and is a normal feature of G2-arrested S. pombe cells.) This is in contrast to the cdc10-129 mutant (included as a control), which at the restrictive temperature arrests in G1. These data suggest that, as in mammalian cells, the 16E1∧E4 protein does not retard S-phase progression but causes S. pombe cells to arrest predominantly in G2, making expression of 16E1∧E4 in S. pombe a suitable model system in which to study 16E1∧E4-induced G2 arrest. This observation provides further support for the idea that arrest is independent of the 16E1∧E4-keratin association, since S. pombe cells also do not contain keratins. As with Saos-2 cells, despite the fact that the cells lack keratins, 16E1∧E4 immunostaining is not observed in the nucleus of S. pombe (Fig. 2D).
FIG. 2.
Expression of wild-type 16E1∧E4 in S. pombe cells. S. pombe cells transformed with pREP1.16E1∧E4 were grown for 20 h under repressing (+Thiamine) and inducing (−Thiamine) conditions. (A) Total cell extracts were separated on 15% polyacrylamide-SDS gels and Western blotted with TVG402 anti-16E1∧E4 antibody. (B) Cells were stained with DAPI and observed by fluorescent microscopy. Bar, 20 μm. Cells induced to express 16E1∧E4 show the cdc phenotype, i.e., they are elongated, a characteristic of cell cycle arrest. (C) Cells were stained with propidium iodide and analyzed by flow cytometry. A reference G1 peak was provided by the S. pombe strain cdc10-129 that was grown for 4 h at the restrictive temperature of 37°C prior to propidium iodide staining. Histograms show the number of cells (on the y axis) with a particular DNA content (on the x axis). The cells induced to express 16E1∧E4 are mainly found to contain 2C DNA content, indicating that arrest is occurring in G2. (D) Cells expressing 16E1∧E4 were stained with DAPI (blue) and TVG402 anti-16E1∧E4 antibody (green) and observed by fluorescent microscopy. 16E1∧E4 immunostaining is found at apical foci in the cytoplasm of S. pombe. Bar, 10 μm.
HPV16 E1∧E4-induced cell cycle arrest occurs independently of Wee1, Cdc25, and Mik1.
In both mammalian and S. pombe cells, G2 arrest can occur in response to DNA damage (55) or incomplete replication (5) via the “checkpoint” pathways. To determine whether 16E1∧E4 was inducing G2 arrest via a checkpoint pathway, 16E1∧E4 was expressed in a range of S. pombe mutants that lack expression or responsiveness to proteins in these pathways (Table 3).
The checkpoint pathways converge on the Cdc2-cyclin B complex by inhibiting its activity via phosphorylation of the Tyr15 residue of the cyclin-dependent kinase (Cdk), Cdc2 (Fig. 3). Cdc2 Tyr15 phosphorylation is catalyzed by Wee1 and to a lesser extent by Mik1 (48). In the cdc2-1w strain, Cdc2 is insensitive to phosphorylation by Wee1, and in the wee1-50 strain the expression of Wee1 is prevented. In both of these strains expression of 16E1∧E4 still resulted in G2 arrest, suggesting that 16E1∧E4 is not acting solely on Wee1. Cdc25 is the phosphatase responsible for dephosphorylating Cdc2 Tyr15 (53). In the cdc2-3w strain, Cdc2 is insensitive to Cdc25 activity and in the cdc2-3w cdc25::ura4 strain, Cdc25 is not expressed. Again, in both cases 16E1∧E4 was still able to induce arrest, suggesting that it is not acting solely to prevent dephosphorylation of Cdc2 Tyr15. The ability of 16E1∧E4 to induce arrest in the wee1-50 cdc25-22 strain shows that it is unlikely that 16E1∧E4 could act degenerately on both Wee1 and Cdc25. Lack of Wee1 in the wee1-50 strain is compensated for by the presence of Mik1; however, 16E1∧E4 was still able to induce G2 arrest in the wee1-50 Δmik1 strain. This finding confirms that 16E1∧E4 is not acting to upregulate the kinases that phosphorylate Cdc2 Tyr15.
FIG. 3.
Proteins involved at the S. pombe G2/M cell cycle boundary. As in higher eukaryotes, the ultimate determinant of entry into mitosis in S. pombe is the kinase activity of the Cdc2-cyclin B complex. Levels of this complex increase throughout G2, but premature activity is prevented by phosphorylation on Y15, catalyzed by Wee1 and Mik1. In late G2, the inhibitory phosphorylation is removed by Cdc25, which creates an active complex that can promote entry into mitosis. Cdc25 can itself become phosphorylated, and this targets it for cytoplasmic sequestration by the Rad24/25 proteins, thereby preventing access to Cdc2-cyclin B in the nucleus.
S. pombe strains Δrad24 and Δrad25 lack the genes for two 14-3-3 proteins (26) that act in a degenerate manner to sequester Cdc25 in the cytoplasm, thereby preventing its access to Cdc2 and hence inhibiting mitotic entry (46). The 16E1∧E4 G2 arrest phenotype is maintained in Δrad24 and Δrad25 strains, suggesting that the Rad24 and Rad25 14-3-3 proteins are not involved. This is supported by the fact that 16E1∧E4-induced arrest is independent of Cdc25, the downstream target of Rad24 and Rad25.
Together, the data obtained from expression of 16E1∧E4 in S. pombe cell cycle mutants suggest that arrest does not involve the kinases and phosphatase controlling the phosphorylation state of Cdc2 Tyr15 and is thus not occurring as a result of DNA damage or incomplete replication. These data also show that the mechanism of action of 16E1∧E4 differs from that of HIV Vpr, since Vpr-induced cell cycle arrest has been shown to be mediated by hyperphosphorylation of Cdc2 Tyr15 (33) and involves Wee1, Rad24, and Cdc25 (24, 50).
It has been shown in a number of situations in which G2 arrest is normally induced, e.g., exposure to DNA-damaging agents or expression of the HIV Vpr protein, that addition of the drug pentoxifylline abolishes the arrest (57). Pentoxifylline is a methylxanthine similar to caffeine. To test whether the 16E1∧E4-induced G2 arrest was sensitive to pentoxifylline, S. pombe cells transformed with pREP1.16E1∧E4 were treated with 5 mM pentoxifylline, induced to express 16E1∧E4, and analyzed by microscopy. The uninduced S. pombe cells were unaffected by the pentoxifylline, whereas the S. pombe cells induced to express only showed the cdc phenotype when not treated with pentoxifylline (Fig. 4A). It had already been shown that in the case of Vpr, the negative effect of pentoxifylline on G2 arrest was not mediated by decreasing the expression level of Vpr (57). To check that this was also true for 16E1∧E4, expression levels of 16E1∧E4 were compared in the absence or presence of pentoxifylline, and it was found that the expression level of 16E1∧E4 was not reduced in the presence of pentoxifylline (Fig. 4B). These data show that the 16E1∧E4-induced G2 can be abolished by pentoxifylline and that this is not mediated by affects on 16E1∧E4 expression levels. However, it is not clear how pentoxifylline is acting to prevent 16E1∧E4-mediated G2 arrest. In some situations, e.g., in response to Vpr expression, abrogation of G2 arrest by pentoxifylline is associated with decreasing the levels of hyperphosphorylated Cdc2 Tyr15 (57). However, since 16E1∧E4-induced G2 arrest does not appear to involve Cdc2 Tyr15, it seems unlikely that pentoxifylline is acting in this way in 16E1∧E4-expressing cells. It may be that, as with another methylxanthine drug, caffeine, the effects of pentoxifylline are pleiotropic, making the exact mechanism of action difficult to interpret.
FIG. 4.
Effect of pentoxifylline on 16E1∧E4-induced S. pombe G2 arrest. S. pombe transformed with pREP1.16E1∧E4 was cultured for 16 h under repressing (+Thiamine) and inducing (−Thiamine) conditions, both in the presence or in the absence of 5 mM pentoxifylline (+ or − PTX). (A) Cells were stained with DAPI and observed by fluorescent microscopy. Bar, 20 μm. In the presence of PTX, 16E1∧E4-induced G2 arrest is abolished. (B) Total cell extracts from 16E1∧E4-expressing cells were separated on 15% polyacrylamide-SDS gels and Western blotted with TVG402 anti-16E1∧E4 antibodies. The levels of 16E1∧E4 expression were equivalent in the absence or presence of pentoxifylline.
The region responsible for arrest lies in the N-terminal half of 16E1∧E4.
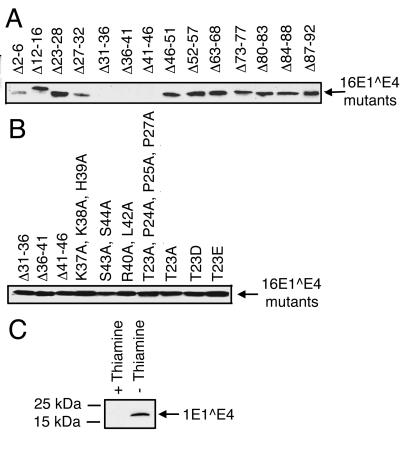
To determine the region of 16E1∧E4 required for G2 arrest, a panel of approximately five amino acid deletion mutations of 16E1∧E4 were analyzed in S. pombe for their ability to induce G2 arrest (Table 4). The mutated 16E1∧E4 sequences were subcloned into pREP1 and transformed into S. pombe. The cells were induced to express the mutated 16E1∧E4 proteins and were analyzed microscopically for G2 arrest. The expression level of each mutant was established by Western blotting (Fig. 5).
TABLE 4.
Ability of wild-type and mutant E1∧E4 proteins to induce arrest in S. pombe
| E1∧E4 protein | G2 arresta |
|---|---|
| Type 16 E1∧E4 wild type | + |
| Type 1 E1∧E4 wild type | − |
| Type 11 E1∧E4 wild type | + |
| Δ2-6 | + |
| Δ12-16 | + |
| Δ23-28 | − |
| Δ27-32 | − |
| Δ31-36 | − |
| Δ36-41 | − |
| Δ41-46 | − |
| Δ46-51 | + |
| Δ52-57 | + |
| Δ63-68 | + |
| Δ73-77 | + |
| Δ80-83 | + |
| Δ84-88 | + |
| Δ87-92 | + |
| Δ1-49 | − |
| Δ1-16 | + |
| Δ51-92 | + |
| K37A,K38A,H39A | + |
| S43A,S44A | + |
| R40A,L42A | + |
| T23A,P24A,P25A,P27A | − |
| T23A | − |
| T23D | − |
| T23E | − |
+, Positive for G2 arrest; −, no G2 arrest.
FIG. 5.
Expression of wild-type and mutant E1∧E4 sequences in S. pombe. S. pombe total cell extracts were separated on 15% polyacrylamide-SDS gels and Western blotted with anti-16E1∧E4 antibodies. (A) 16E1∧E4 mutants were detected with TVG402 anti-16E1∧E4 monoclonal antibody, which has an epitope between amino acids 31 and 46. (B) 16E1∧E4 mutants between amino acids 31 and 46 were detected with polyclonal rabbit anti-16E1∧E4 antibody. (C) Wild-type 1E1∧E4 was detected with 4.37 anti-1E1∧E4 monoclonal antibody.
Only mutations Δ23-28, Δ27-32, Δ31-36, Δ36-41, and Δ41-46 when expressed in S. pombe did not result in the cdc phenotype (Table 4). The deletions made outside this region had no affect on elongation. All of the deletion mutants were expressed at similar levels (Fig. 5). To confirm that the region required for arrest lay in the N-terminal half of 16E1∧E4, two truncation mutations were expressed in S. pombe. The first lacked the C-terminal half of 16E1∧E4 (Δ51-92) but was still able to cause arrest. However, the second, which lacked the N-terminal half of 16E1∧E4 (Δ1-49), was unable to cause arrest. A further N-terminal truncation mutation was constructed that lacked amino acids 1 to 16 (Δ1-16). This mutated sequence was able to induce arrest, suggesting that residues before amino acid 17 are not involved in G2 arrest. These data suggest that the arrest domain lies within amino acids 17 to 45 (Fig. 6). This region will now be referred to as the “arrest domain.” In addition to indicating that the region of 16E1∧E4 responsible for arrest lies in the N terminus of 16E1∧E4, these data suggest that the DEAD box helicase protein identified to bind the C terminus of 16E1∧E4, is not involved in 16E1∧E4-mediated G2 arrest. They also support the theory that the keratins are not involved, since the region of 16E1∧E4 known to associate with keratins lies within amino acids 1 to 16.
FIG. 6.
16E1∧E4 arrest region. Mutation analysis of 16E1∧E4 expression in S. pombe identified the region responsible for arrest as lying between residues 17 and 45. Mutations outside of this region did not abolish arrest. A number of interesting features are found within the arrest region. These include putative nuclear localization and cyclin-binding motifs and an abundance of proline residues, some of which constitute part of a Cdk consensus phosphorylation motif.
The ability to induce G2 arrest in S. pombe is not a feature of all HPV E1∧E4 proteins.
To determine whether the ability to induce G2 arrest is a general feature of all E1∧E4 proteins, the HPV1 and HPV11 E1∧E4 proteins were cloned into pREP1 and expressed in S. pombe. Like that of HPV16, HPV11 E1∧E4 caused a G2 arrest. In contrast, the E1∧E4 protein of HPV1 did not cause the cells to elongate. This was not a result of the failure of the 1E1∧E4 protein to be expressed (Fig. 5C). This finding shows that the 1E1∧E4 protein does not induce G2 arrest in S. pombe and suggests that the G2 arrest phenotype may not be a general feature of all HPV E1∧E4 proteins.
HPV16 E1∧E4-induced G2 arrest is independent of the potential NLS, adjacent phosphorylation sites, and putative cyclin-binding motif.
The arrest domain contains a number of interesting features that warranted further investigation. To achieve this, individual or small groups of amino acids were mutated and analyzed microscopically in S. pombe. 16E1∧E4 contains a stretch of amino acids fitting the criteria for a classical nuclear localization signal (NLS), i.e., a cluster of five basic residues (Fig. 6). Although 16E1∧E4 immunostaining shows a cytoplasmic localization, it is possible that the 16E1∧E4 protein shuttles in and out of the nucleus but spends the majority of the time in the cytoplasm. Since most proteins that affect the cell cycle are found at least transiently in the nucleus, the potential relevance of the 16E1∧E4 sequence corresponding to the NLS was investigated. Adjacent to the NLS motif are two serine residues and, since phosphorylation of nearby residues is a common mechanism used for regulating the activity of the NLS, it was thought that these might be important. A mutated 16E1∧E4 with a disrupted NLS sequence (K37A K38A H39A) was generated. The final two basic residues of the putative NLS were left undisturbed to preserve the cyclin-binding motif (described below). The K37A K38A H39A mutation was still able to induce G2 arrest. This was also the case for the 16E1∧E4 mutation that lacked the two serine residues adjacent to the NLS sequence (S43A and S44A). Both mutants were expressed at levels comparable to that of wild-type 16E1∧E4 (Fig. 5). This suggests that any potential of this region of 16E1∧E4 to act as an NLS is not significant for G2 arrest.
Also present in the arrest domain is a sequence, HRRL, that corresponds to a putative cyclin-binding motif, i.e., RXL, where “X” is basic and the arginine is preceded by a basic amino acid or cysteine (Fig. 6). Such motifs are found in a variety of proteins that are both substrates and inhibitors of Cdk complexes, key regulators of the cell cycle. An interaction between 16E1∧E4 and a Cdk complex could provide a mechanism for affecting entry into mitosis either by direct inhibition of kinase activity or by blocking access of potential substrates to the Cdk complex. To test the significance of the 16E1∧E4 putative cyclin-binding motif, a mutated 16E1∧E4 sequence that lacked the RXL consensus (R40A L42A) was expressed in S. pombe (Fig. 5). Again, this mutant still showed the cdc phenotype, suggesting that the putative cyclin-binding motif is also not required for G2 arrest.
HPV16 E1∧E4-induced G2 arrest is dependent on residues within the proline-rich region.
The arrest domain is particularly rich in proline residues, an amino acid noted for its unique effects on protein conformation and its importance in many protein-protein interactions. Proline-rich regions are also frequently the target of phosphorylation and contained within the 16E1∧E4 arrest domain is a sequence that corresponds to the consensus site for phosphorylation by Cdc2, S/TPXP+, where “X” is any amino acid and “+” is a positively charged residue (Fig. 6). To determine whether this sequence was important for 16E1∧E4-mediated arrest, a mutated 16E1∧E4 sequence that lacked the Cdc2 consensus phosphorylation site (T23A P24A P25A P27A) was expressed in S. pombe and assayed microscopically for arrest. Despite successfully expressing the T23A P24A P25A P27A 16E1∧E4 protein (Fig. 5), the S. pombe cells did not elongate, indicating that this 16E1∧E4 mutant fails to induce G2 arrest. Because of the gross changes to protein structure that can occur as a result of the mutation of proline residues, a more subtle mutation was constructed in this region, in which the threonine residue alone was converted to alanine (T23A). Again, although adequately expressed (Fig. 5), the T23A 16E1∧E4 protein fails to induce G2 arrest in S. pombe. This single amino acid change is relatively innocuous and is unlikely to have drastic affects on protein structure. This suggests that the threonine residue itself is important for 16E1∧E4-mediated G2 arrest.
One potential explanation for the importance of the threonine residue is that it could be phosphorylated. One method by which this possibility can be addressed is by converting the residue to aspartic or glutamic acid. The negatively charged carboxyl groups contained within the side chains of these amino acids are thought to mimic phosphate. Two mutants were constructed in which the threonine residue was replaced by aspartic or glutamic acid (T23D and T23E, respectively). These were expressed in S. pombe and assayed microscopically for arrest. In both cases, although the mutant proteins were adequately expressed, the S. pombe cells did not elongate, indicating that these mutants were not able to induce arrest (Fig. 5). Although this finding does not support the theory that phosphorylation of residue 23 is important for 16E1∧E4-induced G2 arrest, it does not rule it out either since aspartic or glutamic acid substitutions are not always adequate mimics of phosphorylated amino acids (43).
Analysis of 16E1∧E4 mutants in mammalian cells.
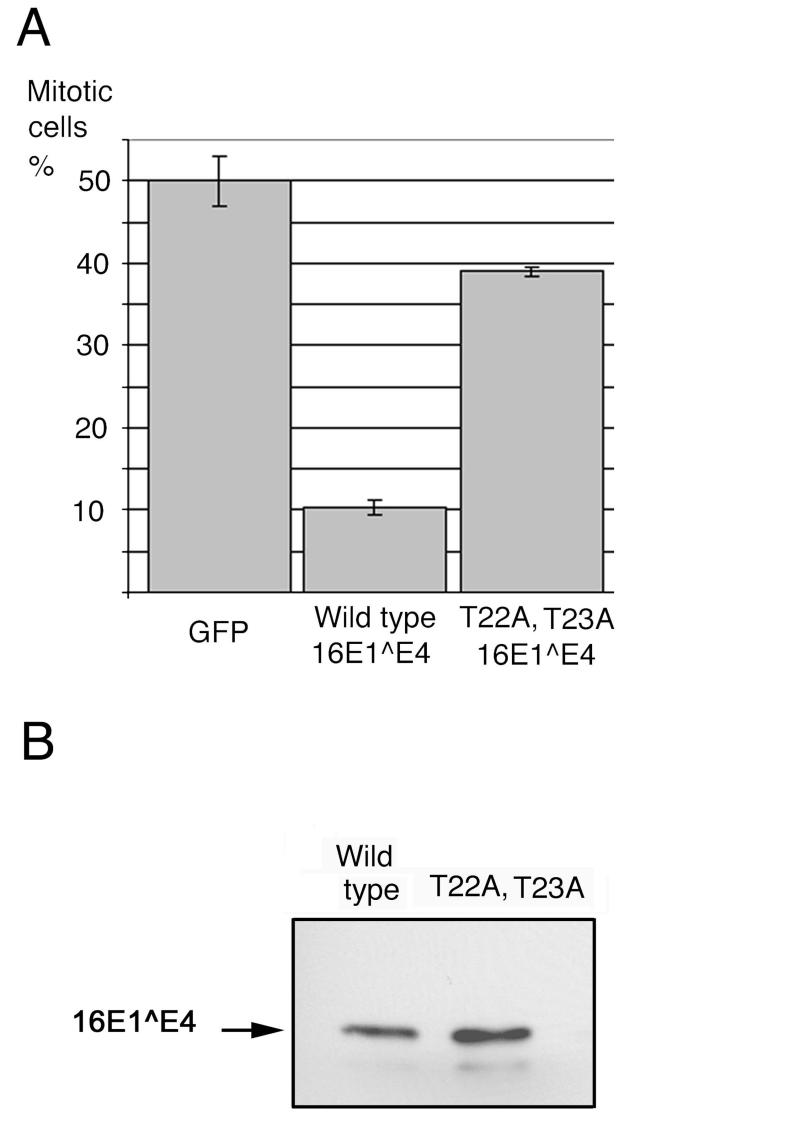
To determine whether the observations made in S. pombe with the 16E1∧E4 mutants were also applicable in mammalian cells, the T22A,T23A 16E1∧E4 mutant was transfected into Cos-7 cells and analyzed for its ability to prevent entry into mitosis. Cos-7 cells were chosen because they have previously been used extensively for studies of E1∧E4 function (14, 16, 59) and are readily identifiable when mitotic. At 24 h posttransfection, the cells were treated for a further 24 h with nocodazole, which arrests the cells in mitosis by interfering with the formation of the mitotic spindle. The cells were then stained with anti-16E1∧E4, Alexa-fluor 488-conjugated antibodies, and the DNA stain DAPI before being analyzed microscopically to determine the mitotic index (percentage of cells in mitosis). In this assay, any G2 arrest affect is manifested as a decrease in the number of cells that are mitotic since they are unable to reach this stage of the cell cycle. The results showed that only 10% of cells expressing wild-type 16E1∧E4 reach mitosis compared to 50% of cells expressing the control protein GFP (Fig. 7A). Analysis of the 16E1∧E4 mutant T22A,T23A showed that cells expressing this mutants were far more likely to be mitotic than those expressing wild-type 16E1∧E4. In fact, the proportion of cells reaching mitosis in cells expressing this mutant (ca. 40%) approaches the value obtained for cells expressing GFP (50%). This was not a result of failure to adequately express this mutant 16E1∧E4 protein (Fig. 7B). These data suggest that, as for S. pombe, sequences in the N-terminal half of the 16E1∧E4 protein are important for inducing G2 arrest in mammalian epithelial cells.
FIG. 7.
Analysis of mutant 16E1∧E4 sequences in mammalian cells. Cos-7 cells were transfected with plasmids expressing GFP or wild-type or T22A,T23A 16E1∧E4 and treated for 24 h with nocodazole. (A) The cells were harvested, stained with TVG402 anti-16E1∧E4 antibody and DAPI, and analyzed microscopically for the percentage of cells expressing transgenes that were mitotic. The results shown are derived from a minimum of four replicate experiments and are displayed as the mean ± the standard error. (B) Total cell extracts were separated on 15% polyacrylamide-SDS gels and Western blotted with TVG402 anti-16E1∧E4 antibody.
DISCUSSION
HPV16 infects cervical epithelium and is the major cause of cervical cancer (68). The molecular basis for cancer progression is well understood and involves the integration of viral DNA into the host cell chromosome and the deregulated expression of the viral E6 and E7 oncogenes. By comparison, relatively little is known about the role of the viral proteins during productive infection. 16E1∧E4 is abundantly expressed during the late stages of the papillomavirus life cycle and is found in the cytoplasm of cells, supporting viral genome amplification (17, 61). Its role during the virus life cycle is unclear, although it is known to associate with keratin filaments (16) and to bind a putative RNA helicase (E4-DBP) (14). We have shown in the present study that 16E1∧E4 has a potent G2-arrest function that does not depend on its ability to bind keratins or E4-DBP. Furthermore, our finding that E1∧E4-mediated arrest occurs both in mammalian cells and S. pombe suggests that 16E1∧E4 targets a conserved component of the cell cycle machinery. The in vivo expression pattern of 16E1∧E4 and its ability to cause G2 arrest in HeLa cells suggests a possible role for E1∧E4 in antagonizing E7-mediated cell proliferation during productive stages of infection.
Several viruses, including baculovirus, parvovirus, reovirus, adenovirus, and HIV, have proteins that mediate G2/M arrest during the late stages of their life cycles (4, 54, 56, 70). The baculovirus ODV-EC27 protein is a cyclin B analogue. Like the host cyclin, ODV-EC27 associates with Cdk1 to create an active kinase, but the viral complex is not degraded and exit from mitosis is inhibited (2). Baculovirus DNA replication occurs in the absence of cellular replication and is thought to be enhanced after arrest in G2. The HIV Vpr protein induces G2 arrest during infection and when expressed in S. pombe (72) and acts by increasing the inhibitory phosphorylation on Cdc2 Tyr15 via its interaction with protein phosphatase 2A (38, 50). Vpr-induced G2 arrest prolongs the time that the long terminal repeat is active and gives rise to increased virus production (30). Interestingly, the HPV16 E2 protein also causes arrest in G2 and, although the precise mechanism is unknown, it does not involve its transactivation function (27, 28). 16E1∧E4 does not resemble a cyclin molecule and does not act on the proteins controlling phosphorylation of Cdc2 Tyr15, but it is possible that its G2 arrest activity in some way contributes to more efficient production of new viruses.
The E4 ORF is contained entirely within that of E2 and is expressed from the differentiation-dependent promoter late in infection (19). The overlap between these two ORFs and the finding that both proteins inhibit cell division suggests that they may cooperate during the virus life cycle. There are several examples of cooperation between viral proteins, and it is possible that E1∧E4 cooperates with E2 or with other viral gene products. This is the case for E7, which stimulates S-phase entry in differentiating epithelial cells. The coexpression of E6 along with E7 prevents an apoptotic response as a result of unscheduled DNA synthesis (29). The viral E1 and E2 proteins, which are involved in viral DNA replication, cooperate in the assembly of the initiation complex at the viral origin (63), whereas L2 and E2 may cooperate in the packaging of the replicated genomes into infectious virions (11, 34). The differentiation-dependent promoter directs expression of mRNAs encoding E1, E2, E4, and E5. The E1 and E1∧E4 proteins are expressed from mRNA species that share the same 5′ termini, whereas E5 is encoded by the abundant bicistronic transcript that also encodes E1∧E4 (37). In fact, our current knowledge of the papillomavirus life cycle suggests that E1∧E4 is first expressed in a replication-competent cell that is expressing E6 and E7, as well as the viral replication proteins E1, E2, and possibly also E5. The fact that the E4 ORF is embedded in a transcriptional unit that also contains E1 and E2 is intriguing given that the first appearance of the E1∧E4 protein correlates with the onset of vegetative viral genome amplification. The ability of 16E1∧E4 to antagonize E7-mediated cell proliferation in HeLa cells may reflect a requirement of 16E1∧E4 (and E2) to inhibit cell division during the normal virus life cycle.
Our flow cytometric analysis showed that 16E1∧E4 had the affect of arresting cells with a predominantly 4N DNA content. This indicated that most cells were arresting somewhere after DNA replication but before the completion of cytokinesis. The arrested cells were found to contain decondensed chromosomes, and mitotic index analysis showed that 16E1∧E4-expressing cells failed to reach mitosis. When considered together, these data show that 16E1∧E4 induces cell cycle arrest prior to mitosis. During natural infection by HPV16, the onset of viral DNA replication and the expression of E1∧E4 coincide closely (17). Although cellular DNA replication arrests at 4N after the expression of 16E1∧E4 in mammalian cells and S. pombe, we hypothesize that viral DNA replication may continue. Such a situation would arise if E2F-inducible genes continued to be expressed after S phase (as a result of E7 expression) or if S-phase entry was restimulated in the upper epithelial layers by E5, which can reactivate cellular DNA replication in quiescent cells (3, 67) and which is encoded on the same abundant mRNA that encodes E1∧E4 (20). The molecular mechanisms that inhibit the rereplication of cellular DNA, which involve the phosphorylation (and inactivation) of cellular initiation factors (e.g., MCM and Cdc6 [25, 40]) would not necessarily be expected to inhibit the replication of viral DNA. Papillomaviruses encode their own replication initiation proteins (E1 and E2) and are not thought to depend on cellular factors for the initiation of viral DNA synthesis (8, 49). The G2 arrest seen here may be manifested in vivo as arrest in a pseudo S-phase state in which viral but not cellular DNA replication can proceed.
The use of S. pombe as a model system in which to analyze 16E1∧E4 function proved very beneficial. First, it allowed us to rule out a number of potential cellular pathways as being important for G2 arrest, in particular those involving the kinase and phosphatases controlling the phosphorylation state of Cdc2 Tyr15. Second, it defined the region of 16E1∧E4 (amino acids 17 to 45) required for G2 arrest and identified a single point mutation (T23A) that abrogated the phenotype. Since mutation of amino acids in this region also prevented the arrest of mammalian cells, it seems likely that whatever the mechanism used by 16E1∧E4, it is conserved between S. pombe and higher eukaryotes. The ability to cause G2 arrest extended to the E1∧E4 protein of HPV11 but did not appear to be a characteristic of the full-length E1∧E4 protein of HPV1. HPV11 infects mucosal epithelium like that of HPV16, whereas HPV1 never causes lesions at these sites (22). HPV1 is the primary cause of plantar warts and is evolutionarily distinct from the human mucosal papillomavirus types that cause genital lesions (6). This is reflected in the degree of divergence of the E1∧E4 sequences of HPV1, HPV11, and HPV16 (Fig. 8). HPV1 is not transmitted by intimate physical contact, and lesions caused by this virus are hugely productive (1, 18). A similar function may thus be expected for the E1∧E4 protein of HPV1, and its inability to cause G2 arrest in S. pombe may reflect a failure of the protein to be correctly processed in yeast (e.g., by proteolytic cleavage [J. Doorbar, I. Coneron, and P. H. Gallimore, Abstr. 7th Int. Papillomavirus Workshop, p. 150, 1988] or phosphorylation [32]) or that this activity is encoded by another viral gene product in HPV1 (27). Differences in the activity of viral proteins among HPV types have already been shown for E6 and E7, which vary in their ability to stimulate cell proliferation (7) and to degrade p53 (9, 62, 64).
FIG. 8.
Sequence alignments of fragments of 16E1∧E4, 11E1∧E4, and 1E1∧E4 proteins corresponding to the arrest domain. The arrest domain of 16E1∧E4 was aligned with the corresponding regions of 11E1∧E4 and 1E1∧E4 by using CLUSTAL V software (36). Conserved residues are highlighted. Sequences corresponding to the consensus for a Cdk phosphorylation motif are underlined.
The mechanism of 16E1∧E4-mediated G2 arrest does not appear to involve known E1∧E4-binding proteins such as E4-DBP, since deletions affecting the last 27 amino acids of 16E1∧E4 abolish binding to E4-DBP (14) but do not prevent cell cycle arrest. Neither does arrest involve keratins, since deletion of the leucine cluster Δ12-16 did not abolish arrest even though keratin binding is abolished (58, 60). That the interaction with keratins is not essential for G2 arrest in S. pombe is not surprising since S. pombe cells do not express keratins. However, this also appears to be the case in human cells since 16E1∧E4 can induce G2 arrest in the Saos-2 cell line, which was derived from an osteosarcoma and does not contain keratins. Interestingly, 16E1∧E4 was predominantly cytoplasmic in both Saos-2 cells and S. pombe, indicating that the association with keratin filaments is not essential to maintain the protein in the cytoplasm.
Although 16E1∧E4 is a cytoplasmic protein, it contains a classical NLS (35) similar to that present in simian virus 40 T antigen (PKKHRRL in E1∧E4 and PKKKRKV in T antigen [41]). Although this motif lies in the “arrest domain” of 16E1∧E4, it did not appear necessary for 16E1∧E4-mediated arrest in S. pombe, and its role during virus infection is unclear. G2 arrest was also induced in the absence of the adjacent potential phosphorylation sites. Although the loss of an NLS at one site can sometimes be compensated for by sequences at other sites, 16E1∧E4 does not appear to have additional sequences that could fulfill this function. In fact, if transient nuclear entry of 16E1∧E4 is required for G2 arrest, then given its small size it should be able to enter by passive diffusion through the nucleopore and thus would not need an NLS. Given the importance of cyclin molecules to the cell cycle, it was perhaps surprising that the cyclin-binding motif, RXL, identified in 16E1∧E4 was not involved in the G2 cell cycle arrest. RXL motifs have not been shown to be involved in the targeting of S. pombe proteins to Cdk-cyclin complexes, and it may be that the RXL motif is important for some other function that occurs only in mammalian cells. Why the threonine residue found at position 23 of 16E1∧E4 should be so important for arrest is not fully clear, although it does form part of a Cdk consensus phosphorylation site (66). A similar consensus is found in the sequence of 16E1∧E4 corresponding to the arrest domain but is not found in the corresponding sequence of 1E1∧E4 (Fig. 8). Attempts to mimic phosphorylation by using acidic residues failed to produce a G2 arrest, and this could indicate either inadequate mimicry or that phosphorylation is not required for G2 arrest. If this latter case is true, then it may be that the threonine is important for some other reasons, such as the formation of hydrogen bonds with an interacting protein.
A major function of viral proteins is to modify the cell cycle to ensure efficient virus synthesis. E7 and E6 drive cells into S phase by interfering with the normal function of Rb and p53. E5 affects the function of cell surface receptors (39) and is mitogenic (65), whereas E2 has a negative affect on proliferation, and acts both to downregulate E7/E6 expression (21) and to prevent entry into mitosis (27). It now seems that E1∧E4, like many of the other HPV proteins, also acts to perturb the host cell cycle during the normal virus life cycle. Future experiments should reveal the mechanism by which 16E1∧E4 acts and its affect on productive infection.
Acknowledgments
C.E.D. was funded by a BBSRC/Roche CASE award to J.D. D.J.J., J.B.A.M., and Q.W. are supported by the United Kingdom Medical Research Council. N.F.F. was an MRC-funded sandwich student. S.A.S. was supported by an Association for International Cancer Research joint award to J.D. and J.B.A.M. K.R. was supported by grants from Cancer Research Switzerland and the Swiss National Science Foundation to P. Beard (Institut Suisse de Recherches Experimentales sur le Cancer, Epalinges, Switzerland). J.D. was supported by the Royal Society and the United Kingdom Medical Research Council.
We thank John Skehel and Jonathan Stoye for their support and encouragement during the course of this study. We thank S. Roberts (University of Birmingham, Birmingham, United Kingdom) for the gift of the 16E1∧E4 deletion mutants; K. Zumbach (DKFZ, Heidelberg, Germany) for advice regarding 16E1∧E4-mediated G2 arrest in yeast; and J. Dickson, J. Saldanha, and V. Buck for technical assistance.
REFERENCES
- 1.Barrera-Oro, J. G., K. O. Smith, and J. L. Melnick. 1962. Quantitation of papova virus particles in human warts. J. Natl. Cancer Inst. 29:583-595. [PubMed] [Google Scholar]
- 2.Belyavskyi, M., S. C. Braunagel, and M. D. Summers. 1998. The structural protein ODV-EC27 of Autographa californica nucleopolyhedrovirus is a multifunctional viral cyclin. Proc. Natl. Acad. Sci. USA 95:11205-11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 4.Brand, K., R. Klocke, A. Possling, D. Paul, and M. Strauss. 1999. Induction of apoptosis and G2/M arrest by infection with replication-deficient adenovirus at high multiplicity of infection. Gene Ther. 6:1054-1063. [DOI] [PubMed] [Google Scholar]
- 5.Canman, C. E. 2001. Replication checkpoint: preventing mitotic catastrophe. Curr. Biol. 11:R121-R124. [DOI] [PubMed] [Google Scholar]
- 6.Chan, S.-Y., H. Delius, A. L. Halpern, and H.-U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: uniting typing, phylogeny, and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciccolini, F., G. Di Pasquale, F. Carlotti, L. Crawford, and M. Tommasino. 1994. Functional studies of E7 proteins from different HPV types. Oncogene 9:2633-2638. [PubMed] [Google Scholar]
- 8.Conger, K. L., J. S. Liu, S. R. Kuo, L. T. Chow, and T. S. Wang. 1999. Human papillomavirus DNA replication: interactions between the viral E1 protein and two subunits of human DNA polymerase alpha/primase. J. Biol. Chem. 274:2696-2705. [DOI] [PubMed] [Google Scholar]
- 9.Crook, T., J. A. Tidy, and K. H. Vousden. 1991. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and transactivation. Cell 67:547-556. [DOI] [PubMed] [Google Scholar]
- 10.Crum, C. P., S. Barber, M. Symbula, K. Snyder, A. M. Saleh, and J. K. Roche. 1990. Coexpression of the human papillomavirus type 16 E4 and L1 open reading frames in early cervical neoplasia. Virology 178:238-246. [DOI] [PubMed] [Google Scholar]
- 11.Day, P. M., R. B. S. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Villiers, E. M. 1999. Human papillomavirus: introduction. Semin. Cancer Biol. 9:377. [DOI] [PubMed] [Google Scholar]
- 13.Doorbar, J. 1998. Late stages of the papillomavirus life cycle. Papillomavirus Rep. 9:119-123. [Google Scholar]
- 14.Doorbar, J., R. Elston, S. Napthine, K. Raj, E. Medcalf, D. Jackson, N. Coleman, H. Griffin, P. Masterson, S. Stacey, Y. Mengitsu, and J. Dunlop. 2000. The E1∧E4 protein of human papillomavirus type 16 associates with a putative RNA helicase through sequences in its C terminus. J. Virol. 74:10081-10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doorbar, J., S. Ely, N. Coleman, M. Hibma, D. H. Davies, and L. Crawford. 1992. Epitope-mapped monoclonal antibodies against the HPV16 E1∧E4 protein. Virology 187:353-359. [DOI] [PubMed] [Google Scholar]
- 16.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV16 16E1∧E4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 17.Doorbar, J., C. Foo, N. Coleman, E. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterisation of events during the late stages of HPV16 infection in vivo using high-affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 18.Doorbar, J., and P. H. Gallimore. 1987. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus type 1a. J. Virol. 61:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doorbar, J., and G. Myers. 1996. The E4 protein, p. 58-80. In G. Myers, H. Delius, J. Icenogel, H.-U. Bernard, C. Baker, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses 1996, vol. III. Los Alamos National Laboratory, Los Alamos, N.Mex. [Google Scholar]
- 20.Doorbar, J., A. Parton, K. Hartley, L. Banks, T. Crook, M. Stanley, and L. Crawford. 1990. Detection of novel splicing patterns in a HPV16-containing keratinocyte cell line. Virology 178:254-262. [DOI] [PubMed] [Google Scholar]
- 21.Dowhanick, J. J., A. A. McBride, and P. M. Howley. 1995. Suppression of cellular proliferation by the papillomavirus E2 protein. J. Virol. 69:7791-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egawa, K., Y. Inaba, K. Yoshimura, and T. Ono. 1993. Varied clinical morphology of HPV1-induced warts, depending on anatomical factors. Br. J. Dermatol. 128:271-276. [DOI] [PubMed] [Google Scholar]
- 23.El Awady, M. K., J. B. Kaplan, S. J. O'Brien, and R. D. Burk. 1987. Molecular analysis of integrated human papillomavirus16 sequences in the cervical cancer cell line SiHa. Virology 159:389-398. [DOI] [PubMed] [Google Scholar]
- 24.Elder, R. T., M. Yu, M. Chen, X. Zhu, M. Yanagida, and Y. Zhao. 2001. HIV-1 Vpr induces cell cycle G2 arrest in fission yeast (Schizosaccharomyces pombe) through a pathway involving regulatory and catalytic subunits of PP2A and acting on both Wee1 and Cdc25. Virology 287:359-370. [DOI] [PubMed] [Google Scholar]
- 25.Findeisen, M., M. El-Denary, T. Kapitza, R. Graf, and U. Strausfeld. 1999. Cyclin A-dependent kinase activity affects chromatin binding of ORC, Cdc6, and MCM in egg extracts of Xenopus laevis. Eur. J. Biochem. 264:415-426. [DOI] [PubMed] [Google Scholar]
- 26.Ford, J. C., F. al-Khodairy, E. Fotou, K. S. Sheldrick, D. J. Griffiths, and A. M. Carr. 1994. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265:533-535. [DOI] [PubMed] [Google Scholar]
- 27.Fournier, N., K. Raj, P. Saudan, S. Utzig, R. Sahli, V. Simanis, and P. Beard. 1999. Expression of human papillomavirus 16 E2 protein in Schizosaccharomyces pombe delays the initiation of mitosis. Oncogene 18:4015-4021. [DOI] [PubMed] [Google Scholar]
- 28.Frattini, M. G., S. D. Hurst, H. B. Lim, S. Swaminathan, and L. Laimins. 1997. Abrogation of a mitotic checkpoint by E2 proteins from oncogenic human papillomaviruses correlates with increased turnover of the p53 tumour suppressor protein. EMBO J. 16:318-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galloway, D. A., and J. K. McDougall. 1996. The disruption of cell cycle checkpoints by papillomavirus oncoproteins contributes to anogenital neoplasia. Semin. Cancer Biol. 7:309-315. [DOI] [PubMed] [Google Scholar]
- 30.Goh, W. C., M. E. Rogel, C. M. Kinsey, S. F. Michael, P. N. Fultz, M. A. Nowak, B. H. Hahn, and M. Emerman. 1998. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 4:65-71. [DOI] [PubMed] [Google Scholar]
- 31.Goodwin, E. C., and D. DiMaio. 2000. Repression of human papillomavirus oncogenes in HeLa cervical carcinoma cells causes the orderly reactivation of dormant tumor suppressor pathways. Proc. Natl. Acad. Sci. USA 97:12513-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grand, R. J. A., J. Doorbar, K. J. Smith, I. Coneron, and P. H. Gallimore. 1989. Phosphorylation of the human papillomavirus type 1 E4 proteins in vivo and in vitro. Virology 170:201-213. [DOI] [PubMed] [Google Scholar]
- 33.He, J., S. Choe, R. Walker, P. Di Marzio, D. O. Morgan, and N. R. Landau. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J. Virol. 69:6705-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heino, P., J. Zhou, and P. F. Lambert. 2000. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304-314. [DOI] [PubMed] [Google Scholar]
- 35.Hicks, G. R., and N. V. Raikhel. 1995. Protein import into the nucleus: an integrated view. Annu. Rev. Cell. Dev. Biol. 11:155-188. [DOI] [PubMed] [Google Scholar]
- 36.Higgins, D. G., A. J. Bleasby, and R. Fuchs. 1992. CLUSTAL V: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 37.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 38.Hrimech, M., X. J. Yao, P. E. Branton, and E. A. Cohen. 2000. Human immunodeficiency virus type 1 Vpr-mediated G2 cell cycle arrest: Vpr interferes with cell cycle signaling cascades by interacting with the B subunit of serine/threonine protein phosphatase 2A. EMBO J. 19:3956-3967. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39.Hwang, E. S., T. Nottoli, and D. Dimaio. 1995. The HPV16 E5 protein: expression, detection, and stable complex formation with transmembrane proteins in COS cells. Virology 211:227-233. [DOI] [PubMed] [Google Scholar]
- 40.Ishimi, Y., and Y. Komamura-Kohno. 2001. Phosphorylation of Mcm4 at specific sites by cyclin-dependent kinase leads to loss of Mcm4,6,7 helicase activity. J. Biol. Chem. 276:34428-34433. [DOI] [PubMed] [Google Scholar]
- 41.Jans, D. A., M. J. Ackermann, J. R. Bischoff, D. H. Beach, and R. Peters. 1991. p34cdc2-mediated phosphorylation at T124 inhibits nuclear import of SV40 T antigen proteins. J. Cell Biol. 115:1203-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones, H. W., Jr., V. A. McKusick, P. S. Harper, and K. D. Wuu. 1971. George Otto Gey (1899-1970): the HeLa cell and a reappraisal of its origin. Obstet. Gynecol. 38:945-949. [PubMed] [Google Scholar]
- 43.Ku, N. O., J. Liao, and M. B. Omary. 1998. Phosphorylation of human keratin 18 serine 33 regulates binding to 14-3-3 proteins. EMBO J. 17:1892-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubbutat, M. H. G., and K. H. Vousden. 1996. Role of E6 and E7 oncoproteins in HPV-induced anogenital malignancies. Semin. Virol. 7:295-304. [Google Scholar]
- 45.Liu, F., C. Rothblum-Oviatt, C. E. Ryan, and H. Piwnica-Worms. 1999. Overproduction of human Myt1 kinase induces a G2 cell cycle delay by interfering with the intracellular trafficking of Cdc2-cyclin B1 complexes. Mol. Cell. Biol. 19:5113-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez-Girona, A., B. Furnari, O. Mondesert, and P. Russell. 1999. Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397:172-175. [DOI] [PubMed] [Google Scholar]
- 47.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, vol. 2. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa. [Google Scholar]
- 48.Lundgren, K., N. Walworth, R. Booher, M. Dembski, M. Kirschner, and D. Beach. 1991. mik1 and wee1 cooperate in the inhibitory tyrosine phosphorylation of cdc2. Cell 64:1111-1122. [DOI] [PubMed] [Google Scholar]
- 49.Ma, T., N. Zou, B. Y. Lin, L. T. Chow, and J. W. Harper. 1999. Interaction between cyclin-dependent kinases and human papillomavirus replication-initiation protein E1 is required for efficient viral replication. Proc. Natl. Acad. Sci. USA 96:382-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masuda, M., Y. Nagai, N. Oshima, K. Tanaka, H. Murakami, H. Igarashi, and H. Okayama. 2000. Genetic studies with the fission yeast Schizosaccharomyces pombe suggest involvement of wee1, ppa2, and rad24 in induction of cell cycle arrest by human immunodeficiency virus type 1 Vpr. J. Virol. 74:2636-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maundrell, K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857-10864. [PubMed] [Google Scholar]
- 52.Maundrell, K. 1993. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene 123:127-130. [DOI] [PubMed] [Google Scholar]
- 53.Millar, J. B., C. H. McGowan, G. Lenaers, R. Jones, and P. Russell. 1991. p80cdc25 mitotic inducer is the tyrosine phosphatase that activates p34cdc2 kinase in fission yeast. EMBO J. 10:4301-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morita, E., K. Tada, H. Chisaka, H. Asao, H. Sato, N. Yaegashi, and K. Sugamura. 2001. Human parvovirus B19 induces cell cycle arrest at G2 phase with accumulation of mitotic cyclins. J. Virol. 75:7555-7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Connell, M. J., N. C. Walworth, and A. M. Carr. 2000. The G2-phase DNA-damage checkpoint. Trends Cell Biol. 10:296-303. [DOI] [PubMed] [Google Scholar]
- 55a.Peh, W. L., K. Middleton, N. Christensen, P. Nicholls, K. Egawa, K. Sotlar, J. Brandsma, A. Percival, J. Lewis, W. J. Liu, and John Doorbar. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 56.Poggioli, G. J., T. S. Dermody, and K. L. Tyler. 2001. Reovirus-induced sigma1s-dependent G2/M phase cell cycle arrest is associated with inhibition of p34cdc2. J. Virol. 75:7429-7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poon, B., J. B. Jowett, S. A. Stewart, R. W. Armstrong, G. M. Rishton, and I. S. Chen. 1997. Human immunodeficiency virus type 1 vpr gene induces phenotypic effects similar to those of the DNA alkylating agent, nitrogen mustard. J. Virol. 71:3961-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roberts, S., I. Ashmole, L. Gibson, S. M. Rookes, G. J. Barton, and P. H. Gallimore. 1994. Mutational analysis of human papillomavirus E4 proteins: identification of structural features important in the formation of cytoplasmic E4/cytokeratin networks in epithelial cells. J. Virol. 68:6432-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roberts, S., I. Ashmole, G. D. Johnson, J. W. Kreider, and P. H. Gallimore. 1993. Cutaneous and mucosal human papillomavirus E4 proteins form intermediate filament-like structures in epithelial cells. Virology 197:176-187. [DOI] [PubMed] [Google Scholar]
- 60.Roberts, S., I. Ashmole, S. Rookes, and P. Gallimore. 1997. Mutational analysis of the human papillomavirus type 16 E1∧E4 protein shows that the C terminus is dispensable for keratin cytoskeleton association but is involved in inducing disruption of the keratin filaments. J. Virol. 71:3554-3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruesch, M. N., F. Stubenrauch, and L. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheffner, M., T. Takahashi, J. M. Huibregtse, J. D. Minna, and P. M. Howley. 1992. Interaction of the human papillomavirus type 16 E6 oncoprotein with wild-type and mutant human p53 proteins. J. Virol. 66:5100-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sedman, J., and A. Stenlund. 1995. Co-operative interaction between the initiator E1 and the transcriptional activator E2 is required for replicator specific DNA replication of bovine papillomavirus in vivo and in vitro. EMBO J. 14:6218-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Storey, A., M. Thomas, A. Kalita, C. Harwood, D. Gardiol, F. Mantovani, J. Breuer, I. M. Leigh, G. Matlashewski, and L. Banks. 1998. Role of a p53 polymorphism in the development of human papillomavirus-associated cancer. Nature 393:229-234. [DOI] [PubMed] [Google Scholar]
- 65.Straight, S., P. M. Hinckle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of HPV16 transforms fibroblasts and effects the downregulation of the EGF receptor in keratinocytes. J. Virol. 69:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeda, D. Y., J. A. Wohlschlegel, and A. Dutta. 2001. A bipartite substrate recognition motif for cyclin-dependent kinases. J. Biol. Chem. 276:1993-1997. [DOI] [PubMed] [Google Scholar]
- 67.Valle, G. F., and L. Banks. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239-1245. [DOI] [PubMed] [Google Scholar]
- 68.Walboomers, J., M. Jacobs, Manos, M. M., F. Bosch, J. Kummer, K. Shah, P. Snijders, J. Peto, C. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 69.Wells, S. I., D. A. Francis, A. Y. Karpova, J. J. Dowhanick, J. D. Benson, and P. M. Howley. 2000. Papillomavirus E2 induces senescence in HPV-positive cells via pRB- and p21CIP-dependent pathways. EMBO J. 19:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wersto, R. P., E. R. Rosenthal, P. K. Seth, N. T. Eissa, and R. E. Donahue. 1998. Recombinant, replication-defective adenovirus gene transfer vectors induce cell cycle dysregulation and inappropriate expression of cyclin proteins. J. Virol. 72:9491-9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xiao, J. H., I. Davidson, H. Matthes, J. M. Garnier, and P. Chambon. 1991. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell 65:551-568. [DOI] [PubMed] [Google Scholar]
- 72.Zhao, Y., J. Cao, M. R. O'Gorman, M. Yu, and R. Yogev. 1996. Effect of human immunodeficiency virus type 1 protein R (vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J. Virol. 70:5821-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]