Abstract
Moderation of hepatitis delta virus (HDV) replication is a likely prerequisite in the establishment of chronic infections and is thought to be mediated by the intracellular accumulation of large hepatitis delta antigen (L-HDAg). The regulatory role of this protein was suggested from several studies showing that cotransfection of plasmid cDNAs expressing both L-HDAg and HDV RNA results in a potent inhibition of HDV RNA replication. However, since this approach differs significantly from natural HDV infections, where HDV RNA replication is initiated from an RNA template, and L-HDAg appears only late in the replication cycle, it remains unclear whether L-HDAg can modulate HDV RNA replication in the natural HDV replication cycle. In this study, we investigated the effect of L-HDAg, produced as a result of the natural HDV RNA editing event, on HDV RNA replication. The results showed that following cDNA-free HDV RNA transfection, a steady-state level of RNA was established at 3 to 4 days posttransfection. The same level of HDV RNA was reached when a mutant HDV genome unable to make L-HDAg was used, suggesting that L-HDAg did not play a role. The rates of HDV RNA synthesis, as measured by metabolic labeling experiments, were identical at 4 and 8 days posttransfection and in the wild type and the L-HDAg-deficient mutant. We further examined the effect of overexpression of L-HDAg at various stages of the HDV replication cycle, showing that HDV RNA synthesis was resistant to L-HDAg when it was overexpressed 3 days after HDV RNA replication had initiated. Finally, we showed that, contrary to conventional thinking, L-HDAg alone, at a certain molar ratio with HDV RNA, can initiate HDV RNA replication. Thus, L-HDAg does not inherently inhibit HDV RNA synthesis. Taken together, these results indicated that L-HDAg affects neither the rate of HDV RNA synthesis nor the final steady-state level of HDV RNA and that L-HDAg is unlikely to act as an inhibitor of HDV RNA replication in the natural HDV replication cycle.
Hepatitis delta virus (HDV) is a small RNA virus that is associated with severe acute and chronic liver disease in humans. The HDV virion comprises an inner core structure (28) containing a small, single-stranded, circular RNA molecule of about 1.7 kb that is associated with the only known virus-encoded protein, hepatitis delta antigen (HDAg), and an outer envelope of hepatitis B surface antigen (2). In infected cells, three HDV-specific RNA species are detected. The most abundant is the full-length (1.7-kb) genomic strand, followed by the complementary strand (antigenomic HDV RNA), which is present at 10- to 20-fold lower levels. The least abundant (approximately 1/500th of the genomic species) is a subgenomic 0.8-kb mRNA species, also of antigenomic sense, that encodes HDAg (7). HDV RNA replication proceeds via a double rolling-circle mechanism, in which the genomic and antigenomic full-length species serve as the respective templates for the synthesis of the other strand (3, 20). Based on the different sensitivities to α-amanitin, the synthesis of genomic HDV RNA appears to be carried out by host cellular RNA polymerase II (pol II) (20, 23, 24), whereas the synthesis of antigenomic HDV RNA requires a different, as-yet-unidentified, RNA polymerase (20, 23). The genomic strand also serves as a template for synthesis of the HDAg-encoding mRNA by a pol II-dependent mechanism (23). HDAg occurs as two species, small HDAg (S-HDAg) (p24) and large HDAg (L-HDAg) (p27). L-HDAg is synthesized only late in the viral replication cycle, as a result of an RNA editing event which alters the termination codon of the open reading frame for S-HDAg (16). The two HDAg species have different roles in HDV replication. S-HDAg is an essential activator of HDV RNA replication (13), whereas L-HDAg is essential for virion assembly (4). HDV RNA is synthesized in the nucleus (9, 19) and, like S- and L-HDAg (27), was thought to reside primarily in the nuclei of HDV-infected hepatocytes (9). However, our recent studies showed that HDV RNA, particularly the genomic strand, was also detected in the cytoplasm (19).
The need of HDV particles for a hepatitis B surface antigen envelope dictates that natural HDV infections occur as coinfections with hepatitis B virus or superinfections of hepatitis B virus carriers. In the latter scenario, infection typically leads to severe acute hepatitis with a short incubation span, which progresses to a chronic HDV infection in the majority of cases. The development of the chronic state is associated with a decrease in the level of circulating virus and hepatic markers of infection (31). This moderation of HDV RNA replication is a likely prerequisite for the establishment of persistent HDV infections. The intracellular accumulation of L-HDAg is generally thought to play a major role in the down-regulation of HDV RNA replication. This was concluded from a number of studies in which cotransfection of plasmids expressing HDV RNA and L-HDAg resulted in potent inhibition of HDV RNA replication (5, 6, 8). However, in these experiments, L-HDAg was always introduced early in HDV RNA replication, a situation that does not reflect the delayed appearance of this protein in natural infections. Moreover, these studies have invariably relied on transcription from an HDV cDNA template to initiate HDV RNA replication. Recently, using an HDV cDNA-free RNA transfection system, we showed that only the synthesis of genomic-sense and not that of antigenomic-sense HDV RNA was sensitive to inhibition by L-HDAg (21). This finding explains how HDV RNA replication can be established despite the fact that HDV particles contain equal amounts of L- and S-HDAg (1). However, the strong inhibitory effect of genomic HDV RNA synthesis by L-HDAg when the antigenomic strand was transfected is puzzling, since the accumulation of antigenomic HDV RNA after genomic RNA transfection will require several rounds of RNA-to-RNA replication. We would have expected that both genomic and antigenomic HDV RNA accumulation following genomic RNA transfection would be inhibited by L-HDAg. With this reasoning, we hypothesized that the sensitivity of genomic HDV RNA synthesis to inhibition by L-HDAg may occur only at the initiation of HDV RNA synthesis from the artificially introduced antigenomic HDV RNA. Since L-HDAg appears only late in the natural HDV life cycle, L-HDAg may not inhibit HDV RNA replication in natural infection, as previously believed.
In the present study, we examined the effect of L-HDAg on the steady-state level of viral RNA and the rate of HDV RNA synthesis. We also investigated the effect of the introduction of this protein at different stages of the HDV replication cycle. Our results indicate that the inhibitory activity of L-HDAg observed in previous studies likely will not occur in the natural HDV replication cycle.
MATERIALS AND METHODS
Cell culture and transfection.
The human hepatoma cell line HuH7 (25) was cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, and streptomycin and incubated at 37°C with 5% CO2. For transfection studies, cell cultures were seeded overnight either in six-well plates or, for immunofluorescence analysis, in eight-chamber CultureSlides (Falcon). Transfection mixtures comprised of 1 to 5 μg of nucleic acids (see figure legends for specific details), and transfection was performed using DMRIE-C reagent (GibcoBRL) according to the manufacturer's directions. Following transfection, the cultures were incubated overnight. Subsequently, the medium was changed and the cultures were incubated for up to an additional 12 days.
Plasmids and cloning.
Plasmids pBSδ1.2G and pBSδ1.2G(2xS) (in vitro transcription templates for the 1.2 × genome-length wild-type and L-HDAg− genomic HDV RNAs, respectively) and pBSδ1.2AG and pBSδ1.2AG(2xS) (in vitro transcription templates for the 1.2X genome-length wild-type and L-HDAg− antigenomic HDV RNAs, respectively) have been described elsewhere (19). Mammalian expression plasmids pCδ1.2G and PCδ1.2AG were used to express 1.2X genome-length transcripts of genomic and antigenomic HDV RNAs, respectively, under the control of the cytomegalovirus immediate-early promoter. These plasmids contain the same HDV cDNA insert as pBSδ1.2G and pBSδ1.2AG, respectively, cloned between the EcoRV and XbaI sites of plasmid pCDNA3 (Clontech). Plasmids pX9-1/II (22) and pB1-3-I/II (21) were used for in vitro transcription of S- and L-HDAg mRNAs, respectively, and pTMδSalB (17) and pBSδHX (20) were used for the generation of 32P-labeled probes to detect genomic and antigenomic HDV RNA, respectively. pSV24, used for constitutive expression of S-HDAg, was constructed by blunt-end cloning of the 898-nucleotide (nt) HindIII-XbaI fragment (HDV nt 1679 to 781 according to the numbering of Wang et al. [30]) from plasmid pTMδ3 (18) into the SmaI site of pSVL (Amersham Pharmacia Biotech). pSV27, used for constitutive expression of L-HDAg, has been described previously (26). The tetracycline-inducible L-HDAg expression plasmid pTet27 was constructed by blunt-end cloning of an 800-nt fragment containing the L-HDAg-coding region from pSV27 (released by double digestion with restriction enzymes BamHI and XbaI) into the EcoRV site of pTet-Splice (Life Technologies). Expression of L-HDAg from pTet27 requires the cotransfection of the tetracycline regulator plasmid pTet-tTak (Life Technologies).
In vitro transcription.
1.2X genome-length HDV RNAs were transcribed from plasmids pBSδ1.2G, pBSδ1.2AG, pBSδ1.2G(2xS), and pBSδ1.2AG(2xS) by using T7 MEGAscript kits (Ambion) after linearization with restriction enzyme NotI. Capped mRNA for HDAg was transcribed from plasmids pX9-I/II and pB1-3-I/II after linearization with restriction enzyme HindIII by using a T7 m-Message m-Machine kit (Ambion). The method for generation of HDV-specific 32P-labeled riboprobes has been described elsewhere (17).
Northern blot hybridization analysis.
RNA was extracted from intact cells by using Tri-Reagent (Molecular Research Center, Inc) according to the manufacturer's protocol. RNA samples were separated by electrophoresis through MOPS (morpholinepropanesulfonic acid)-formaldehyde-containing 1.2% agarose gels, transferred to a membrane, hybridized, and washed as described previously (19). Detection of genomic and antigenomic HDV RNAs was performed with in vitro-transcribed 32P-labeled probes generated from pTMδSalB and pBSδHX, respectively. The washed membrane was exposed to Biomax MR or MS X-ray film (Kodak). Quantitation was performed by phosphorimaging analysis with ImageQuant version 1.11 software (Molecular Dynamics). Detection of ChoA mRNA (10) was performed as described previously (23).
[32P]orthophosphate metabolic labeling.
Metabolic labeling with [32P]orthophosphate was performed for 4 h at 37°C in the presence of 50 μg of actinomycin D (Fisher Biotech) per ml as described in a recent study (20).
Immunoblot and immunofluorescence analysis for HDAg.
Immunoblot analysis for HDAg was performed as described in an earlier study (21). For detection of HDAg by immunostaining, HuH7 cells grown on eight-chamber CultureSlides were first fixed in a solution of 2% formaldehyde in phosphate-buffered saline (PBS) for 30 min at room temperature and then permeabilized in acetone at 4°C for 5 min. Following rehydration in PBS, the slides were incubated with one or two of three primary HDAg-specific antibodies (see figure legends for details) for 60 min at 37°C, washed in PBS, and then incubated for a further 40 min at 37°C with the appropriate fluorescein isothiocyanate (FITC)- or rhodamine-labeled secondary antibodies (see figure legends). After a final wash in PBS, the slides were mounted in ProLong antifade reagent (Molecular Probes) and examined by UV microscopy.
RESULTS
L-HDAg potently inhibits HDV RNA synthesis when L-HDAg is introduced early in replication by cDNA transfection.
Most of the previous reports showing that L-HDAg inhibits HDV RNA replication were based on cotransfection studies using HDV cDNA plasmids, where L-HDAg was produced continuously from the time of transfection (5, 6, 8). To reproduce this finding, HuH7 cells were cotransfected with either plasmid pCδ1.2G or pCδ1.2AG, which express 1.2X genome-length wild-type genomic or antigenomic HDV RNA, respectively (Fig. 1, HDV-G and HDV-AG), and a 1/10 amount of plasmid pSV27, which expresses L-HDAg (Fig. 1). At 6 days posttransfection, total cellular RNA was examined by Northern blot hybridization to detect genomic and antigenomic HDV RNAs. The results showed that the presence of 1/10 amount of pSV27 led to an approximately 10-fold decrease in the amounts of both genomic and antigenomic HDV RNAs compared to the control cultures cotransfected with vector DNA (Fig. 1, lanes + and −, respectively). We also used an HDAg− mutant HDV cDNA genome (pCδ1.2m), which does not express a functional S-HDAg. In this case, the HDV cDNA was cotransfected with a plasmid expressing S-HDAg (pSV24). When pSV24 and pSV27 were present at a 10:1 ratio, HDV RNA synthesis (both genomic and antigenomic) was again inhibited more than 10-fold (data not shown). These results are consistent with the previous reports showing that L-HDAg was a potent inhibitor of HDV RNA replication when L-HDAg was expressed from cDNA at the time of transfection (5, 6, 8). As we have shown previously, when L-HDAg was expressed from mRNA by using RNA transfection methods, only genomic, and not antigenomic, HDV RNA synthesis was inhibited (21).
FIG. 1.
L-HDAg potently inhibits HDV RNA synthesis following HDV cDNA transfection. Northern blot hybridization was used to detect HDV RNA. RNA was extracted from HuH7 cells 6 days after cotransfection with 4.5 μg of either plasmid pCδ1.2G (HDV-G) or pCδ1.2AG (HDV-AG) and 0.5 μg of either vector DNA (lanes −) or L-HDAg expression plasmid pSV27 (lanes +). Lane δ9, positive control from H1δ9 cells, a cell line that stably replicates HDV RNA (17). Lane M, RNA from mock-transfected HuH7 cells. All lanes except δ9 (0.5 μg) were loaded with 10 μg of total cellular RNA and hybridized with 32P-labeled RNA probes specific for genomic or antigenomic HDV RNA derived from plasmids pTMδSalB (17) and pBSδHX (19), respectively.
L-HDAg does not affect the steady-state level of HDV RNA.
In the above-described work and previously published reports, L-HDAg was expressed continually from the time of transfection. This does not reflect the kinetics of L-HDAg appearance in the natural HDV replication cycle, where, apart from a small amount that is carried with the input infecting virion, L-HDAg appears only after HDV RNA replication has been established. To investigate the possible effect of L-HDAg that appears as a result of the natural editing event on HDV RNA replication, we examined the kinetics and steady-state levels of HDV RNA from day 3 to 13 following transfection of HuH7 cells with genomic HDV RNA and an mRNA encoding S-HDAg (Fig. 2A). Unexpectedly, similar levels of antigenomic HDV RNA were detected from days 3 to 13 (Fig. 2A, middle panel) even though L-HDAg could be detected from day 4 onwards by Western blotting (Fig. 2A, lower panel). Identical results were also obtained when the level of genomic HDV RNA was examined in HuH7 cells at various time points posttransfection with antigenomic HDV RNA (Fig. 2A, top panel). This result suggests that HDV RNA replication had reached an equilibrium between RNA synthesis and RNA degradation by day 3 to 4 posttransfection.
FIG. 2.
Effect of L-HDAg on steady-state levels of HDV RNA. HuH7 cells were cotransfected with 2.5 μg of either wild-type [HDV RNA (WT)] (A) or L-HDAg− mutant [HDV RNA (2XS)] (B and C) 1.2X genome-length genomic- or antigenomic-sense HDV RNA and 2.5 μg of an mRNA expressing S-HDAg. RNA and protein samples collected at various times posttransfection (PT) were analyzed for the opposite-sense HDV RNA by Northern blotting (A and B, top panels, and C) and for HDAg by immunoblotting (A and B, bottom panels). Lanes δ9 and Mock, total RNA and protein extracted from H1δ9 and mock-transfected HuH7 cells, respectively. Also shown is a 29-kDa protein marker.
To determine if this was due to the production of L-HDAg, we repeated the above-described experiment using a mutant HDV RNA genome (2XS) which is unable to express L-HDAg (Fig. 2B, lower panel). Following transfection of the genomic form of this mutant, the amounts of antigenomic HDV RNA were examined from day 1 to 8 posttransfection (Fig. 2B). HDV RNA was detected faintly at day 1 posttransfection (Fig. 2B, upper panel). Following a rapid increase during the first 3 days, the levels of antigenomic HDV RNA became stable, suggesting that a moderation of HDV RNA replication had occurred. Similar results were also obtained when genomic HDV RNA synthesis was examined after transfection with antigenomic HDV RNA (Fig. 2C). If L-HDAg were responsible for moderating HDV replication, the absence of this protein would have led to a continuous increase of HDV RNA. However, the kinetics of appearance and the final steady-state levels of HDV RNA detected in cells transfected with either the wild-type or the 2XS mutant genomes were roughly equivalent, suggesting that L-HDAg does not affect the synthesis or maintenance of HDV RNA.
To rule out the possibility that unregulated HDV RNA replication had caused the death and consequential loss of some HDV RNA-replicating cells, thus obscuring the effect of L-HDAg on HDV RNA levels, we examined cells transfected with the wild-type and 2XS mutant HDV RNAs by immunofluorescence analysis at days 4 and 8 posttransfection (Fig. 3), using an FITC-conjugated human antibody specific for both L- and S-HDAg. At day 4 posttransfection (Fig. 3A to F), a similar proportion of HDAg-positive cells (∼90%) was detected following transfection with either wild-type or 2XS antigenomic HDV RNA (compare Fig. 3D and F), while no staining was observed in mock-transfected cells (Fig. 3B). By day 8 posttransfection (Fig. 3G to L), similar numbers of HDAg-positive cells were again detected for the wild-type and the L-HDAg mutant (compare Fig. 3H and K), indicating that L-HDAg did not influence the rate of cell death. The same results were obtained following transfection of genomic-sense wild-type and 2XS HDV RNAs (data not shown). Moreover, when cultures transfected with wild-type HDV RNA were costained with a rabbit antibody (LP3, a kind gift from Stan Lemon) specific for L-HDAg (rhodamine) at day 8 posttransfection, virtually all FITC-positive cells were also positive for L-HDAg (compare Fig. 3H and I). As expected, no L-HDAg was detected in cells transfected with 2XS HDV RNA (Fig. 3L). These results indicate that the expression of S-HDAg and/or L-HDAg did not affect cell survival.
FIG. 3.
Immunofluorescent-antibody detection of HDAg in HDV RNA-transfected HuH7 cells. HuH7 cells were cotransfected with 0.5 μg of either wild-type [HDV-AG (WT)] or L-HDAg− mutant [HDV-AG (2XS)] 1.2X genome-length antigenomic-sense HDV RNA and 0.5 μg of an mRNA expressing S-HDAg. At days 4 (A to F) and 8 (G to L) posttransfection, cultures were stained for total HDAg (B, D, F, H, and K) with a 1:400 dilution of an FITC-conjugated human polyclonal anti-HDAg antibody. At day 8 posttransfection, cultures were also stained with a 1:100 dilution of a rabbit polyclonal antibody specific for L-HDAg (LP3) (29) (I and L), followed by a 1:100 dilution of a goat anti-rabbit serum conjugated to rhodamine (Jackson Immunoresearch). Fluorescent and phase-contrast images were captured with a Zeiss AxioCam digital camera and processed with Adobe Photoshop version 5.0.
The rate of HDV RNA synthesis is not affected by the presence of L-HDAg.
The constant levels of HDV RNA observed after day 3 to 4 posttransfection in the experiments described above could represent the persistence of previously synthesized HDV RNA, while no new RNA was synthesized; alternatively, the rates of HDV RNA synthesis and degradation may have reached an equilibrium. To distinguish between these possibilities, we subjected wild-type or 2XS antigenomic HDV RNA-transfected HuH7 cells to metabolic labeling with [32P]orthophosphate in the presence of actinomycin D at days 4 and 8 posttransfection (Fig. 4A). Total RNA was analyzed by agarose gel electrophoresis. As established in a recent study (20), in cells transfected with HDV RNA (Fig. 4A, lanes 4 to 7), two distinct RNA species representing HDV monomer and dimer RNAs were labeled (Fig. 4A, 1× and 2×, respectively). The dimer species appear as a doublet and likely represent linear and circular HDV RNA (Fig. 4A, 2×). Strikingly, the level of metabolically labeled monomeric and dimeric HDV RNA species was identical at both harvest points. In fact, there appeared to be slightly more 32P-labeled HDV RNA at day 8 than at day 4. Most importantly, the wild type and the 2XS mutant, which cannot make L-HDAg, yielded the same amounts of metabolically labeled HDV RNA, indicating that L-HDAg did not affect the rate of RNA synthesis. Analysis of duplicate transfected cultures by Northern blotting also revealed identical levels of both genomic and antigenomic HDV RNAs for both wild-type and 2XS HDV RNA transfections at both days 4 and 8 posttransfection (Fig. 4B, first two panels from top; compare lanes 4 to 7). Examination of cellular ChoA mRNA (10) in the same samples confirmed similar RNA loadings in all lanes (Fig. 4B, third panel). Finally, as expected, Western blot analysis of HDAg revealed only S-HDAg in cells transfected with 2XS HDV RNA at both day 4 and day 8 but a significant amount of L-HDAg at day 8 posttransfection with the wild-type HDV RNA (Fig. 4B, bottom panel). Taken together, the results from Fig. 2 to 4 indicate that L-HDAg neither inhibits HDV RNA synthesis nor affects the steady-state level of intracellular HDV RNA. This steady state is most likely maintained by a balance between HDV RNA synthesis and degradation. Thus, the previous observations of inhibition of HDV RNA synthesis by L-HDAg may have been the result of the artificial introduction of L-HDAg at the time of transfection.
FIG. 4.
Effect of L-HDAg on rate of HDV RNA synthesis. (A) Huh7 cells were cotransfected with 2.5 μg of either wild-type (WT) (lanes 4 and 5) or L-HDAg− mutant (2XS) (lanes 6 and 7), 1.2X genome-length antigenomic-sense HDV RNA, and 2.5 μg of an mRNA expressing S-HDAg. At days 4 and 8 posttransfection (PT), the cultures were labeled for 4 h with [32P]orthophosphate in the presence of 50 μg of actinomycin D per ml, and the total RNA was analyzed on a 1.5% denaturing agarose gel. Lane 1, 32P-labeled RNA marker of approximately 1.75 kb. 1× and 2×, positions of monomer and dimer HDV RNAs, respectively; prRNA, 28S, and 18S, cellular rRNA precursor and 28S and 18S rRNA species, respectively. (B) Upper panel, Northern blot analysis of HuH7 cell RNA transfected as for panel A at days 4 and 8 posttransfection for genomic (G) and antigenomic (AG) HDV RNAs and HDAg. ChoA mRNA was used as loading control. Bottom panel, Western blot analysis for L-and S-HDAg.
HDV RNA synthesis is not inhibited by the overexpression of L-HDAg late in the replication cycle.
To further confirm this conclusion, we attempted to overexpress L-HDAg at various time points after RNA transfection. For this purpose, we needed first to devise a system in which we could express L-HDAg in a majority of HDV RNA-replicating cells at various time points, so that the possible effect of L-HDAg on HDV RNA replication could be observed. We used a tetracycline-inducible expression plasmid to produce L-HDAg and a constitutively expressing plasmid to produce S-HDAg. These plasmids were cotransfected with 1.2X genome-length HDV RNA. In trial experiments, we found that RNA transfection efficiencies (using our cDNA-free transfection system) were generally three to four times higher than those using DNA. We first determined whether L-HDAg was expressed in all of the cells undergoing HDV RNA replication.
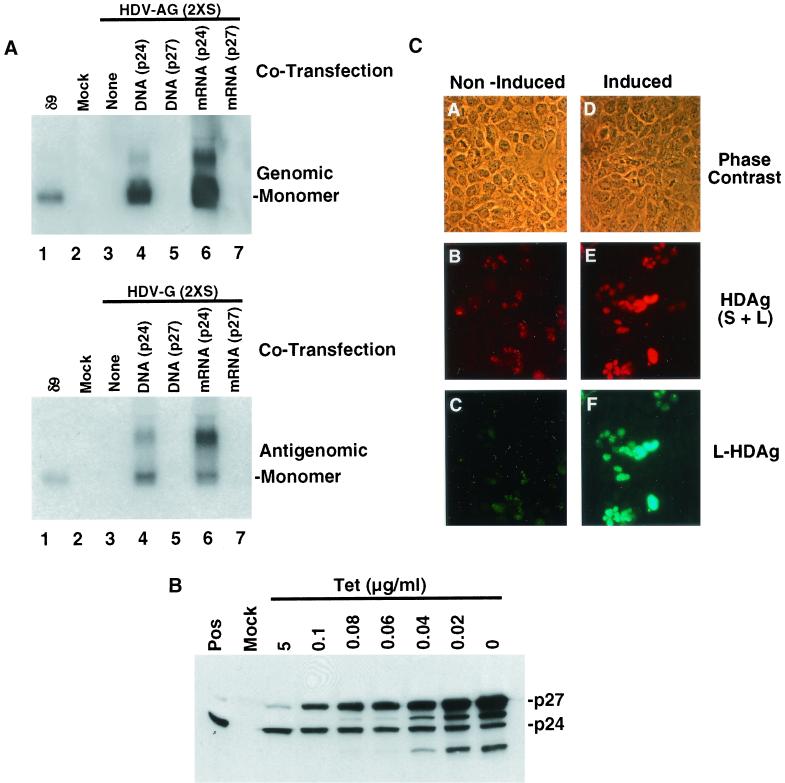
HuH7 cells were cotransfected with either 2XS antigenomic (Fig. 5A, upper panel) or 2XS genomic (Fig. 5A, lower panel) RNA and either plasmids or mRNAs expressing S- or L-HDAg. As expected, when HuH7 cells were transfected with HDV RNA alone, no evidence of RNA replication was detected (Fig. 5A, lanes 3). However, when cells were cotransfected with a plasmid (pSV24) expressing S-HDAg, HDV RNA replication could be detected, although at a lower efficiency than seen when they were cotransfected with S-HDAg mRNA (Fig. 5, lanes 4 and 6). No evidence of replication was observed when HDV RNA was cotransfected with either a plasmid (pSV27) or mRNA expressing L-HDAg (Fig. 5A, lanes 5 and 7, respectively). These results indicated that HDV RNA synthesis was absolutely dependent on the expression of S-HDAg and therefore must be restricted to cells that take up plasmid pSV24.
FIG. 5.
Optimization of the transfection systems for analysis of the effects of L-HDAg. (A) Northern blot analysis of RNA from HuH7 cells cotransfected with 2.5 μg of mutant L-HDAg− HDV RNA of either genomic [HDV-G (2XS)] or antigenomic [HDV-AG (2XS)] polarity and 2.5 μg of either vector DNA (None) or plasmid DNA or mRNA expressing S-HDAg (p24) or L-HDAg (p27). RNA was extracted at 6 days posttransfection. The monomeric HDV RNA species is indicated. (B) Tetracycline induction of L-HDAg. HuH7 cells were cotransfected with 1 μg of S-HDAg expression plasmid pSV24, 1 μg of vector DNA, 1.5 μg of tetracycline-regulated L-HDAg expression plasmid pTet27, and 1.5 μg of the tetracycline regulator plasmid pTet-tTak in the presence of medium containing 5 μg of tetracycline per ml. At 1 day posttransfection, the medium was replaced with medium containing various concentrations of tetracycline (Tet), and the cultures were harvested and analyzed for HDAg expression 24 h later. Lanes Pos and Mock, HDAg expression in H1δ9 and mock-transfected HuH7 cells, respectively. p24 and p27, positions of S- and L-HDAg, respectively. (C) Immunofluorescence analysis to detect HDAg in HuH7 cells transfected as described for panel B. L-HDAg was either noninduced (5 μg of tetracycline per ml) or induced (0.08 μg of tetracycline per ml) at day 3 posttransfection, and the cultures were harvested for staining 3 days later. Detection of total HDAg (panels B and E) was performed with a 1:50 dilution of a mixture of three HDAg-specific mouse monoclonal antibodies (11) followed by a 1:100 dilution of a rhodamine-conjugated goat anti-mouse serum (American Qualex). L-HDAg was detected (panels C and F) with a 1:100 dilution of antibody LP3 and a 1:100 dilution of an FITC-conjugated goat anti-rabbit serum (American Qualex).
The efficiency of tetracycline-regulated L-HDAg production was then evaluated (Fig. 5B). HuH7 cells were cotransfected with pSV24, pTet27, and the tetracycline-regulator plasmid pTet-tTak in the presence of 5 μg of tetracycline per ml, which suppresses the expression of L-HDAg. At 1 day posttransfection, the medium was replaced with fresh medium containing various concentrations of tetracycline, and the level of L-HDAg was measured 24 h later by Western blotting (Fig. 5B). As expected, similar levels of S-HDAg were seen in all transfections. In the presence of 5 μg of tetracycline per ml, a trace amount of L-HDAg could be detected, which increased as the tetracycline concentration was reduced. However, at very low tetracycline concentrations (0 to 0.04 μg/ml) (Fig. 5B), a substantial amount of smaller products, which likely represented breakdown products of L-HDAg or the results of mistranslation, were also detected. To minimize these smaller products, we selected a concentration of 0.08 μg of tetracycline per ml, at which the highest level of L-HDAg production with the minimum degree of breakdown products was detected, for all subsequent experiments. To determine whether L- and S-HDAg were expressed in the same cells, we transfected HuH7 cells with pSV24, pTet-tTak, and pTet27 in the presence of 5 μg of tetracycline per ml. At 3 days posttransfection, the tetracycline concentration was reduced to 0.08 μg/ml, and after an additional 3 days, the cultures were costained with a monoclonal antibody specific for both L- and S-HDAg (rhodamine) and a rabbit antibody (LP3) specific for L-HDAg only (FITC). In the presence of 5 μg of tetracycline per ml (noninduced) (Fig. 5C, panels A to C), approximately 20% of the cells were rhodamine positive; about 40% of these cells also showed a weak FITC signal, indicating that there was a slight leakiness of L-HDAg expression. In the presence of 0.08 μg of tetracycline per ml, a similar number of cells stained positive for rhodamine (Fig. 5C, panels D to F); however, almost all of these cells were also strongly positive for FITC staining, indicating that almost all S-HDAg-positive cells were also positive for L-HDAg. Since HDV RNA synthesis occurs only in cells expressing S-HDAg, this result indicates that L-HDAg will be expressed in almost all of the cells undergoing HDV RNA replication. Therefore, this system enables the study of the effect of L-HDAg on HDV RNA replication.
The effect of early and late L-HDAg production on subsequent HDV RNA synthesis was then examined (Fig. 6). The 2XS mutant HDV RNA genome, which does not make L-HDAg, was used so that the expression level of L-HDAg would not be affected by HDV RNA replication. When L-HDAg was expressed at the time of transfection (at a 1:1 ratio for S- and L-HDAg), genomic HDV RNA synthesis was virtually completely inhibited at day 3 posttransfection (Fig. 6B, lane 4), while antigenomic HDV RNA synthesis was inhibited to a smaller extent (Fig. 6A, lane 4), in agreement with the previous result obtained with mRNAs for S- and L-HDAg for transfection (21). The production of almost equimolar levels of L- and S-HDAg was demonstrated by immunoblotting (Fig. 6, lower panels, lanes 4). In contrast, when L-HDAg production was induced at day 3 posttransfection and HDV RNA synthesis was examined at day 6 posttransfection, no differences in the level of antigenomic or genomic HDV RNA between the uninduced and L-HDAg-induced cultures were observed (Fig. 6, lanes 6 and 7). As a comparison, when the same cultures were harvested at day 3 before induction of L-HDAg (Fig. 6, lanes 5), the amounts of both antigenomic and genomic HDV RNAs were smaller than those seen in cultures expressing S-HDAg alone (Fig. 6, compare lanes 3 and 5), indicating that even the leaky expression of L-HDAg resulted in significant inhibition of HDV RNA replication early in the viral replication cycle (Fig. 6, lower panels, lanes 5). Strikingly, the levels of HDV RNA at day 6 were much higher than those in the uninduced cultures at day 3, regardless of the presence or absence of L-HDAg. Taken together, these results clearly demonstrate that while L-HDAg can inhibit HDV RNA synthesis when provided at the time of HDV RNA (or cDNA) transfection, it has no effect at later points of the HDV replication cycle.
FIG. 6.
Effect of L-HDAg on HDV RNA replication when induced at various time points. (A) Antigenomic HDV RNA and HDAg production following induction of L-HDAg at various times posttransfection. Lanes 1 and 2, extracts from H1δ9 and mock-transfected HuH7 cells, respectively. Lanes 3 to 7, extracts from HuH7 cells cotransfected with 1 μg each of pSV24 and genomic-sense 2XS HDV RNA either alone (lane 3) or together with either 1 μg of pSV27 (lane 4) or 1.5 μg each of pTet27 and pTet-tTak (lanes 5 to 7). Where necessary, all transfection mixtures were made up to 5 μg of total nucleic acid with vector DNA. Samples were harvested at days 3 and 6 posttransfection. L-HDAg was uninduced (lanes U) (5 μg of tetracycline per ml) or induced (lanes I) (0.08 μg of tetracycline per ml) at day 3 posttransfection. L and S, L- and S-HDAg, respectively; monomer, monomeric HDV RNA. ChoA mRNA was used as a loading control. (B) Same as for panel A except that cultures were transfected with antigenomic-sense 2XS HDV RNA, and genomic HDV RNA was measured by Northern blotting.
L-HDAg alone, at low molar ratios relative to HDV RNA, allows limited initiation of HDV RNA replication.
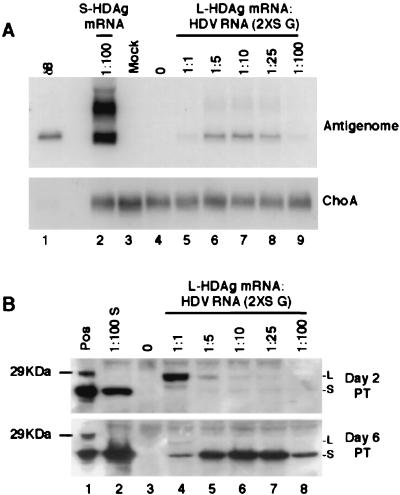
These results indicated that L-HDAg does not inhibit HDV RNA replication when it is expressed later in the HDV replication cycle. This conclusion raised a fundamental question regarding the differences observed previously between S- and L-HDAg in their role in HDV RNA replication. We hypothesize that there may not be fundamental differences, but only quantitative differences, between L- and S-HDAg in their ability to initiate HDV RNA replication under some conditions. Therefore, we examined whether L-HDAg mRNA alone could allow initiation of HDV RNA replication (Fig. 7). For this purpose, identical amounts of genomic-sense 2XS HDV RNA were cotransfected with different amounts of L-HDAg mRNA, and HDV RNA synthesis was examined at 6 days posttransfection (Fig. 7). At 1:1 ratio of L-HDAg mRNA to genomic HDV RNA, a trace amount of antigenomic HDV RNA was detected (Fig. 7A, lane 5). Surprisingly, as the relative ratio of L-HDAg mRNA to genomic HDV RNA was reduced to between 1:5 and 1:25, a significantly larger amount of antigenomic HDV RNA was detected (Fig. 7A, lanes 6 to 8). The amount of HDV RNA decreased as the ratio dropped to 1:100. These results indicate that initiation of antigenomic RNA synthesis occurred in the presence of L-HDAg only. Nevertheless, L-HDAg was clearly a poor substitute for S-HDAg in this process, as S-HDAg at a 1:100 ratio with genomic HDV RNA yielded at least 20 times more antigenomic HDV RNA than the largest amount of HDV RNA attainable with L-HDAg mRNA (Fig. 7A, compare lanes 2 with lanes 6 to 8). Antigenomic HDV RNA was not detected in the absence of any transfected HDAg mRNA (Fig. 7A, lane 4).
FIG. 7.
Initiation of HDV RNA replication in the presence of L-HDAg only. Antigenomic HDV RNA synthesis (A) and HDAg production (B) following cotransfection of six-well plate cultures of HuH7 cells with 2.5 μg of 2XS genomic-sense HDV RNA with either 2.5 μg (1:1), 500 ng (1:5), 250 ng (1:10), 100 ng (1:25), or 25 ng (1:100) of L-HDAg mRNA are shown. As controls, 2.5 μg of 2XS genomic-sense HDV RNA was either transfected alone (lanes 0) or cotransfected with 25 ng of S-HDAg mRNA (S-HDAg mRNA 1:100). Where necessary, the total amount of transfected RNA was adjusted to 5 μg with yeast tRNA. Samples were harvested at day 6 posttransfection (PT) for HDV RNA analysis and at days 2 and 6 posttransfection for HDAg. L and S, L- and S-HDAg, respectively. Also shown is a 29-kDa protein marker. ChoA mRNA was used as a loading control.
HDAg analysis at day 2 posttransfection revealed the presence of L-HDAg in all cotransfected samples (Fig. 7B, upper panel, lanes 4 to 8). The amounts were proportional to the amounts of L-HDAg mRNA transfected. Interestingly, small amounts of S-HDAg were also detected in those samples in which antigenomic HDV RNA synthesis was detected (Fig. 7A, lanes 5 to 7). By day 6 posttransfection, only S-HDAg was detected (Fig. 7B, bottom panel), and it was most abundant in those samples supporting HDV RNA synthesis. Since S-HDAg production is dependent on the input 2XS genomic HDV RNA, this result indicates that HDAg mRNA transcription can also occur in the presence of only L-HDAg.
We also performed similar analysis with antigenomic HDV RNA transfection. Genomic HDV RNA synthesis was not observed at any relative ratio of L-HDAg mRNA to antigenomic 2XS HDV RNA used for transfection (data not shown). Thus, L-HDAg appears to be able to initiate HDV RNA synthesis only from the genomic RNA strand.
DISCUSSION
The gradual intracellular accumulation of L-HDAg, as a result of an RNA editing event, has long been considered to play a key role in the moderation of HDV replication, a likely prerequisite in the establishment of chronic HDV infections. However, the only evidence supporting this hypothesis has come from cotransfection studies in which L-HDAg was expressed from plasmids at the beginning of HDV RNA replication (5, 6, 8). In these studies, L-HDAg was shown to potently inhibit HDV RNA replication when initiated from HDV cDNA. While we were able to confirm these observations, this approach has little in common with natural infections, where there are no cDNA intermediates and L-HDAg is produced only late in the replication cycle. Thus, in this study, we used RNA transfection to examine the effect of naturally derived L-HDAg on HDV RNA replication. Under these conditions, L-HDAg was shown to have no influence on the steady-state level of HDV RNA, as a wild-type HDV RNA genome and a mutant RNA unable to synthesize L-HDAg yielded the same steady-state level of HDV RNA. Moreover, metabolic labeling experiments showed similar quantities of 32P-labeled HDV RNA at days 4 and 8 posttransfection, indicating that L-HDAg also does not affect the rate of HDV RNA synthesis. Finally, we demonstrated that while HDV RNA synthesis could be inhibited by L-HDAg soon after transfection, overexpression of this protein late in the replication cycle had no effect.
The results presented in this study indicate that the role of L-HDAg in HDV replication needs to be reevaluated. The main function of L-HDAg appears to be for virus assembly (4). Thus, mature HDV particles package almost the same amounts of S- and L-HDAg (1). However, the second function previously proposed for L-HDAg, namely, inhibition of HDV RNA replication, now appears to be the result of a transfection artifact. Even though the present study showed that the presence of L-HDAg at the beginning of replication could influence HDV RNA synthesis, the amount of L-HDAg in the virion particles is not likely to inhibit the initiation of HDV replication, since antigenomic HDV RNA synthesis from genomic RNA is relatively resistant to L-HDAg (21), and by the time new L-HDAg appears (as a result of the natural editing event), HDV RNA replication is no longer sensitive to inhibition. Therefore, in the natural HDV replication cycle, L-HDAg will not have a significant effect on HDV replication. Since this protein shares many properties with S-HDAg, including the ability to stabilize HDV RNA (14) and enhance ribozyme activity (12), it is conceivable that L-HDAg generated after RNA editing may be able to act as a functional supporter, rather than an inhibitor, of HDV replication. Consistent with this hypothesis, we showed here that L-HDAg, at a certain molar ratio with HDV RNA, can initiate HDV RNA synthesis even in the absence of S-HDAg. Why, then, does L-HDAg so potently inhibit HDV RNA replication when overexpressed during the early stages of HDV replication (Fig. 1)? This may in part be related to the ability of L-HDAg to inhibit pol II-mediated, DNA-dependent transcription (15). Another possibility is that the large amount of L-HDAg artificially expressed at the early step of HDV RNA replication may trap the HDV RNA in a conformation or in a subcellular compartment unfavorable for RNA replication. Alternatively, inhibition of HDV RNA replication may be dependent on the stoichiometry of HDV RNA and L-HDAg. In previous cotransfection experiments, the number of HDV RNA molecules synthesized early posttransfection would be low compared to the high level of L-HDAg produced, thus leading to the inhibition of subsequent HDV RNA replication. In contrast, in the experiments presented here where L-HDAg was induced later, the ratio of HDV RNA to L-HDAg may then be much higher, allowing HDV RNA replication to continue. Nevertheless, in light of the results from this study, this inhibitory effect at early time points posttransfection may have little (if any) biological significance and most likely represents an artifact of the transfection systems used.
Regulation of replication is an important and highly evolved process in a number of virus systems and is critical for the establishment of chronic infections. Examination of the kinetics of HDV RNA synthesis following transfection with the 2XS mutant HDV genome revealed that following an initial exponential amplification during the first 3 days posttransfection, a steady-state level was reached. These results indicated that in the absence of L-HDAg, a moderation of HDV RNA synthesis still occurs. While the mechanism responsible for this moderation is unclear, it seems likely that this is a function of HDAg, since this is the only HDV-encoded protein. Previously we demonstrated that HDAg mRNA is produced throughout the HDV replication cycle (22), indicating a continual requirement for de novo-synthesized HDAg. It is possible that a mutual balance exists between HDAg and HDV RNA, such that the level of HDV RNA regulates HDAg production and the level of HDAg, in turn, controls the rate of HDV RNA synthesis. Evidence for this comes from kinetics studies in which HDV RNA production in the presence of large amounts of HDAg (following transfection into a cell line stably expressing S-HDAg) was found to be rapidly accelerated, whereas production of HDV RNA with limiting amounts of HDAg (by lowering the proportion of HDAg mRNA used for cotransfection) was delayed (unpublished observations). In both cases, however, the same steady-state level was reached, although after shorter or longer times, respectively, posttransfection. Previous studies in our laboratory have shown that HDAg mRNA synthesis is performed by pol II (23); however, a high level of HDAg was also shown to inhibit pol II-mediated, DNA-dependent transcription (15). If HDAg has a similar effect on pol II-mediated HDV RNA-dependent transcription, this could provide a feedback mechanism that could regulate the level of HDAg production and thus the level of HDV RNA synthesis. Thus, following natural HDV infection, we would predict an unregulated, exponential increase in HDV-associated markers until sufficient HDAg is produced to inhibit pol II-mediated transcription and consequently lead to the establishment of steady-state HDV replication, where synthesis and degradation are in balance. In any case, such a regulation is not likely due to the production of L-HDAg. In sum, the observations made in this study necessitates the revision of one aspect of the long-held view of HDV RNA replication, namely, that L-HDAg inhibits HDV replication. Our findings indicate that L-HDAg does not play such a role in the natural HDV replication cycle. Rather, its main function is for virus particle assembly.
Acknowledgments
We thank Stanley Lemon (Department of Microbiology and Immunology, The University of Texas Medical Branch, Galveston) for provision of the L-HDAg-specific rabbit polyclonal antibody, LP3.
M.M.C.L. is an Investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Bergmann, K. F., and J. L. Gerin. 1986. Antigens of hepatitis delta virus in the liver and serum of humans and animals. J. Infect. Dis. 154:702-706. [DOI] [PubMed] [Google Scholar]
- 2.Bonino, F., K. H. Heermann, M. Rizzetto, and W. H. Gerlich. 1986. Hepatitis delta virus: protein composition of delta antigen and its hepatitis B virus-derived envelope. J. Virol. 58:945-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Branch, A. D., and H. D. Robertson. 1984. A replication cycle for viroids and other small infectious RNAs. Science 223:450-455. [DOI] [PubMed] [Google Scholar]
- 4.Chang, F.-L., P.-J. Chen, S.-J. Tu, C.-J. Wang, and D.-S. Chen. 1991. The large form of hepatitis delta antigen is crucial for the assembly of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 88:8490-8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, M., S.-Y. Hsieh, and J. Taylor. 1990. Role of two forms of hepatitis delta virus antigen: evidence for a mechanism of self-limiting genome replication. J. Virol. 64:5066-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, P.-J., F.-L. Chang, C.-J. Wang, C.-J. Lin, S.-Y. Sung, and D.-S. Chen. 1992. Functional studies of hepatitis delta virus large antigen in packaging and replication inhibition: role of the amino-terminal leucine zipper. J. Virol. 66:2853-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P.-J., G. Kalpana, J. Goldberg, W. Mason, B. Werner, J. L. Gerin, and J. Taylor. 1986. Structure and replication of the genome of hepatitis delta virus. Proc. Natl. Acad. Sci. USA 83:8774-8778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glenn, J. S., and J. M. White. 1991. trans-dominant inhibition of human hepatitis delta virus genome replication. J. Virol. 65:2357-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowans, E. J., B. M. Baroudy, F. Negro, A. Ponzetto, R. H. Purcell, and J. L. Gerin. 1988. Evidence for replication of hepatitis delta virus RNA in hepatocyte nuclei after in vivo infection. Virology 167:274-278. [DOI] [PubMed] [Google Scholar]
- 10.Harpold, M. M., R. M. Evans, M. Salditt-Georgieff, and J. E. Darnell. 1979. Production of mRNA in Chinese hamster cells: relationship of the rate of synthesis to the cytoplasmic concentration of nine specific mRNA sequences. Cell 17:1025-1035. [DOI] [PubMed] [Google Scholar]
- 11.Hwang, S. B., and M. M. C. Lai. 1993. Isoprenylation mediates direct protein-protein interactions between hepatitis large delta antigen and hepatitis B virus surface antigen. J. Virol. 67:7659-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeng, K. S., P. Y. Su, and M. M. C. Lai. 1996. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J. Virol. 70:4205-4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo, M. Y.-P., M. Chao, and J. Taylor. 1989. Initiation of replication of the human hepatitis delta virus genome from cloned DNA: role of delta antigen. J. Virol. 63:1945-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lazinski, D. W., and J. M. Taylor. 1994. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J. Virol. 68:2879-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo, K., G. T. Sheu, and M. M. C. Lai. 1998. Inhibition of cellular RNA polymerase II transcription by delta antigen of hepatitis delta virus. Virology 247:178-188. [DOI] [PubMed] [Google Scholar]
- 16.Luo, G., M. Chao, S. Y. Hsieh, C. Sureau, K. Nishikura, and J. Taylor. 1990. A specific base transition occurs on replicating hepatitis delta virus RNA. J. Virol. 64:1021-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macnaughton, T. B., E. J. Gowans, A. R. Jilbert, and C. J. Burrell. 1990. Hepatitis delta virus RNA, protein synthesis and associated cytotoxicity in a stably transfected cell line. Virology 177:692-698. [DOI] [PubMed] [Google Scholar]
- 18.Macnaughton, T. B., E. J. Gowans, B. Reinboth, A. R. Jilbert, and C. J. Burrell. 1990. Stable expression of hepatitis delta virus antigen in a eukaryotic cell line. J. Gen. Virol. 71:1339-1345. [DOI] [PubMed] [Google Scholar]
- 19.Macnaughton, T. B., and M. M. C. Lai. 2002. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J. Virol. 76:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Macnaughton, T. B., S. T. Shi, L. E. Modahl, and M. M. C. Lai. 2002. Rolling-circle replication of hepatitis delta virus RNA is carried out by two different cellular RNA polymerases. J. Virol. 76:3920-3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Modahl, L. E., and M. M. C. Lai. 2000. The large delta antigen of hepatitis delta virus potently inhibits genomic but not antigenomic RNA synthesis: a mechanism enabling initiation of viral replication. J. Virol. 74:7375-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modahl, L. E., and M. M. C. Lai. 1998. Transcription of hepatitis delta antigen mRNA continues throughout hepatitis delta virus (HDV) replication: a new model of HDV RNA transcription and replication. J. Virol. 72:5449-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Modahl, L. E., T. B. Macnaughton, N. Zhu, D. L. Johnson, and M. M. C. Lai. 2000. RNA-dependent replication and transcription of hepatitis delta virus RNA involve distinct cellular RNA polymerases. Mol. Cell. Biol. 20:6030-6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moraleda, G., and J. Taylor. 2001. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J. Virol. 75:10161-10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakabayashi, H., K. Taketa, K. Miyano, T. Yamane, and J. Sato. 1982. Growth of human hepatoma cell lines with differentiated function in chemically defined medium. Cancer Res. 42:3858-3863. [PubMed] [Google Scholar]
- 26.Netter, H. J., T. B. Macnaughton, W. P. Woo, R. Tindle, and E. J. Gowans. 2001. Antigenicity and immunogenicity of novel chimeric hepatitis B surface antigen particles with exposed hepatitis C virus epitopes. J. Virol. 75:2130-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rizzetto, M., M. G. Canese, S. Arico, O. Crivelli, C. Trepo, F. Bonino, and G. Verme. 1977. Immunofluorescence detection of a new antigen-antibody system (delta/anti-delta) associated with hepatitis B virus in liver and serum of HBsAg carrier. Gut 18:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryu, W.-S., H. Netter, J., M. Bayer, and J. Taylor. 1993. Ribonucleoprotein complexes of hepatitis delta virus. J. Virol. 67:3281-3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, J. G., J. Cullen, and S. M. Lemon. 1992. Immunoblot analysis demonstrates that the large and small forms of hepatitis delta virus antigen have different C-terminal amino acid sequences. J. Gen. Virol. 73:183-188. [DOI] [PubMed] [Google Scholar]
- 30.Wang, K.-S., Q.-L. Choo, A. J. Weiner, J.-H. Ou, R. C. Najerian, R. M. Thayer, G. T. Mullenbach, K. J. Denniston, J. L. Gerin, and M. Houghton. 1986. Structure, sequence and expression of the hepatitis δ viral genome. Nature 323:508-514. (Author's correction, 328:456, 1987.) [DOI] [PubMed] [Google Scholar]
- 31.Wu, J. C., T. Z. Chen, Y. S. Huang, F. S. Yen, L. T. Ting, W. Y. Sheng, S. H. Tsay, and S. D. Lee. 1995. Natural history of hepatitis D viral superinfection: significance of viremia detected by polymerase chain reaction. Gastroenterology 108:796-802. [DOI] [PubMed] [Google Scholar]