Abstract
Epstein-Barr virus (EBV) strains can be distinguished by specific sequence variations in the LMP1 gene. In this study, a heteroduplex tracking assay (HTA) specific for LMP1 was developed to precisely identify the prototypic undeleted strain B958, other undeleted strains (Ch2, AL, NC, and Med−), and strains with the 30-bp deletion (Med+ and Ch1). This technique also provides an estimate of the relative abundance of strains in patient samples. In this study, EBV strains were identified in 25 hairy leukoplakia (HLP) biopsies and six matched peripheral blood samples and throat washes with the LMP1-HTA. To investigate the relationship of the virus found in the peripheral blood to that in the HLP lesion, the strain variants in the peripheral blood B lymphocytes and those present within the epithelial cells in the HLP lesion and in throat washes were identified. In many of the subjects, compartmental differences in the EBV strain profiles in the oral cavity and peripheral blood were readily apparent. The throat wash specimens usually had a strain profile similar to that within the corresponding HLP sample, which was distinct from the strain profile detected in the peripheral blood. These analyses reveal that the nature of EBV infection can be very dynamic, with changes in relative strain abundance over time as well as the appearance of new strains. The patterns of abundance in the blood and oral cavity provide evidence for compartmentalization and for the transmission of strains between the blood and oropharynx.
Epstein-Barr virus (EBV) is an important human pathogen that persistently infects greater than 90% of the world's population (34). Transmission occurs via infected saliva, which results in the establishment of a persistent latent infection in memory B lymphocytes for the lifetime of the host and a chronic asymptomatic shedding of virions in the oral cavity (22). Latently infected cells express a restricted pattern of viral genes and therefore escape immune surveillance (3). However, reactivated cells have increased viral gene expression that can activate EBV-specific cytotoxic T lymphocytes, which limit the proliferation of EBV-infected lymphocytes (19, 25).
As a consequence of decreased immune response, EBV-associated diseases may appear (43). EBV may cause AIDS-associated lymphomas and an unusual lesion of the tongue, hairy leukoplakia (HLP) (15, 42). HLP is an early indicator of immune suppression and is the only example of pathology caused by permissive EBV infection (15). An unusual characteristic of HLP is the frequent presence of multiple types and strains of EBV (45, 46).
There are two distinct types of EBV, types 1 and 2, marked by considerable sequence divergence in the coding sequences for the EBV nuclear antigen 2 (EBNA2) and EBNA3 genes (7, 36). Type 1 was initially thought to be more prevalent than type 2; however, these studies were based on B-cell lines that were established directly from an individual's peripheral blood or from naïve B cells that were infected with virus from throat washes. Subsequent studies have revealed that type 2 is less efficient at lymphocyte transformation and therefore would be underrepresented in some studies of EBV prevalence in populations (35).
Other studies of transmission have been based on restriction enzyme polymorphisms or changes in protein size (4, 13). EBV strains have also been distinguished by sequence changes in the coding sequences for EBNA1, BZLF1, and LMP1 (5, 24, 29). Multiple forms of LMP1 have been identified that are marked by specific base pair changes, a 30-bp deletion, variable numbers of a 33-bp repeat region, and a 15-bp insertion within one of the 33-bp repeats in the LMP1 coding sequence (24). These LMP1 variants can be found in both types of EBV, although some variants may be found preferentially in either type 1 or 2 (24, 31). It has been shown that variation in the number of repeats may result from recombination during replication within a single strain; therefore, the number of repeats does not distinguish between LMP1 variants (47).
The presence or absence of the 30-bp deletion has been widely used to distinguish EBV strains as deleted or undeleted; however, there are also sequence variations that distinguish strains in addition to the deletion. Seven phylogenetically distinct forms of LMP1 have been identified by their sequence differences and signature amino acid changes (8). An eighth strain was also described, but it was concluded to possibly be a recombinant strain (8). The ability to precisely differentiate between the five undeleted and three deleted strains would enable studies of viral persistence and transmission as well as analysis of the profile of strains during infection.
In this study, seven of the distinct LMP1 variants identified previously were prepared as clones in a 264-bp region within exon 3 of LMP1 that contained representative sequence changes for each of the variants (8). This region of LMP1 was amplified by PCR and tested to develop a heteroduplex tracking assay (HTA) for LMP1. The HTA is a sensitive technique that can distinguish sequences that have small numbers of differences and can reveal the relative abundance of viral variants in infections with multiple strains or changing strains. In particular, the HTA has been used to analyze the evolution of human immunodeficiency virus (HIV) strains in patients who develop resistance to antiviral therapy (33). The LMP1-HTA presented here can distinguish seven LMP1 strains and also identify subspecies of strains that were shown to have additional base pair changes from the known strain controls.
In this study, EBV strains were identified in 25 HLP biopsies and six matched peripheral blood samples and throat wash samples with the LMP1-HTA. The strains present and their relative abundance in the peripheral blood, throat washes, and HLP lesion were determined for six patients. The profile of strains and their patterns of abundance in the blood and oral cavity provide evidence for compartmentalization and for the transmission of strains between the blood and oropharynx.
MATERIALS AND METHODS
Patient samples.
Biopsy specimens were collected at the Infectious Disease Clinic or Dental Clinic at the University of North Carolina Hospitals from five HIV-positive subjects and one HIV-negative medically compromised subject who displayed HLP. One subject also exhibited an oral lymphoma, and a biopsy was obtained. Peripheral blood was collected by routine venipuncture, and lymphocytes were separated by a Ficoll step gradient. Throat wash samples were obtained by having subjects gargle with 5 ml of sterile phosphate-buffered saline. Tongue scrapings were collected and placed in 5 ml of sterile phosphate-buffered saline. Two subjects gave samples at more than one visit.
Nucleic acid isolation.
DNA was isolated from pulverized tissue biopsies with a cesium chloride step gradient as previously described (32). DNA was purified from lymphocytes with the DNeasy tissue kit (Qiagen, Valencia, Calif.). DNA was harvested from 200 μl of throat wash or tongue scraping with the High Pure viral nucleic acid isolation kit (Roche Molecular Biochemicals, Indianapolis, Ind.).
Amplification of LMP1.
LMP1 was amplified from patient samples by nested PCR with 200 ng of DNA template in duplicate independent reactions. The reaction mixture contained 1 U of Taq DNA polymerase (Promega, Madison, Wis.); a 1:10 dilution of reaction buffer; a final concentration of 2.0 mM MgCl2; a final concentration of 10 mM of a mix containing dATP, dGTP, dCTP, and dTTP (Promega); and 25 pmol of each primer in a 50-μl reaction volume. The first round of amplification with primers LMP-3UT (EBV genome coordinates 168017 to 168036; ATCACGAGGAATTCAATGTGGCTTTTCAGCCTAG) (8) and FUC-HIND3 (EBV genome coordinates 168427 to 168408; ATCAGAGAGCTTTGACAATGGCCCACATGACC) yielded a 411- or 381-bp product, depending on the presence or absence of a 30-bp deletion.
The second round of PCR with 6 μl of template from the first reaction was performed with primers FUE-ECO (EBV genome coordinates 168163 to 168183; ATCACGAGGAATTCGTCATAGTAGCTTAGCTGAAC) (24) and FUC-HIND3, which yielded a 264- or 234-bp product. The strain controls for the HTA were amplified from 200 ng of cloned DNA in one round of PCR with primers FUE-ECO and FUC-HIND3. Amplification was performed in a Robocycler Gradient 96 (Stratagene, La Jolla, Calif.) for 30 to 35 cycles for each round of PCR (1 min each of denaturation at 95°C, annealing at 55°C, and extension at 72°C). PCR products were analyzed by 1.5% agarose gel electrophoresis and visualized by ethidium bromide staining.
HTA.
The design of the LMP1-HTA and the construction of the probes were modeled after studies on the hypervariable V3 region of gp120 in HIV (26). Clones were established for each of seven LMP1 variants (B958, Ch2, AL, NC, Med−, Med+, and Ch1), which correspond to EBV coordinates 168163 to 168427 (8). The 264- or 234-bp fragments, depending on the presence or absence of the 30-bp deletion, were ligated into the PGEM3Z vector (Promega). The probes were made by digesting 5 μg of plasmid with EcoRI (New England Biolabs, Beverly, Mass.) to linearize the vector. The plasmid was end labeled for 30 min at room temperature in a reaction volume of 30 μl containing 25 μCi of [35S]dATP (Amersham Pharmacia Biotech, Arlington Heights, Ill.) and 1 U of Klenow enzyme (New England Biolabs); following the incubation, the enzyme was heat inactivated. The linearized vector was digested with HindIII (New England Biolabs) to release the radiolabeled fragment from the vector. Unincorporated nucleotides were removed, and the probe was purified with a Quick Spin Sephadex G-25 fine column (Boehringer Mannheim, Indianapolis, Ind.).
The heteroduplex formation reaction was performed with 8 μl of PCR product of a strain control or a patient sample, 1 μl of annealing buffer (1 M NaCl, 100 mM Tris-HCl [pH 7.5], 20 mM EDTA), and 1 μl of radiolabeled probe. The mixtures were denatured for 5 min at 100°C and allowed to reanneal for 4 min at 4°C. The reactions were separated in nondenaturing 10% polyacrylamide gels (Hoefer apparatus; Pharmacia Biotech, San Francisco, Calif.) and subsequently dried in a gel drier (Bio-Rad, Hercules, Calif.) and exposed to a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
Sequence analysis.
Sequence analysis was performed to confirm the results of the HTA. PCR samples were sequenced directly with the primer FUE-ECO or the undeleted specific primer LMP-FUD (EBV genome coordinates 168267 to 168287; GCCGGAATCATGACTATGAC). The samples were sequenced at the University of North Carolina-Chapel Hill Automated DNA Sequencing Facility on a model 377 DNA sequencer (Applied Biosystems Division, Perkin-Elmer Cetus, Norwalk, Conn.) with the ABI Prism dye terminator cycle sequencing ready reaction kit with AmpliTaq DNA polymerase FS (Applied Biosystems Division, Perkin-Elmer Cetus).
RESULTS
Development of LMP1-HTA.
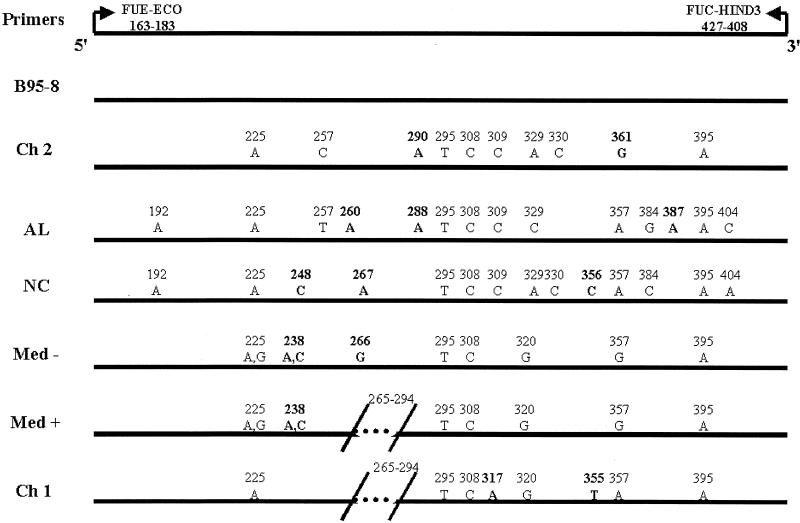
To more finely characterize EBV strain variation, an HTA specific for LMP1 was developed. The phylogenetically distinct LMP1 variants that have been identified include the prototypic undeleted strain B958, other undeleted strains (Ch2, AL, NC, and Med−), and strains with the 30-bp deletion (Med+ and Ch1) (Fig. 1) (8). There are multiple strain-specific sequence changes throughout the molecule; however, changes within the carboxyl terminus are the most informative (8, 24). A 264-bp region in exon 3 of LMP1 (EBV gene coordinates 168163 to 168427) that has multiple signature changes that might enable discrimination with an HTA for EBV genotyping was identified. Clones of this region from each of the seven strains were produced and differed by the presence or absence of a 30-bp deletion and 7 to 14 base pair changes compared to the B958 strain, with each strain containing at least one signature base pair change within this region (Fig. 1). There are several common positions that have changes in this region compared to the B958 strain, which may indicate that the strains experienced a similar evolutionary pressure.
FIG. 1.
EBV gene coordinates 168163 to 168427 used in this study to identify LMP1 strain variants. The locations of the primers that were used for PCR amplification are indicated. Sequence changes relative to the prototype B958 sequence are labeled for each strain. Signature changes for each strain are in bold. The slash marks at positions 265 to 294 denote the 30-bp deletion.
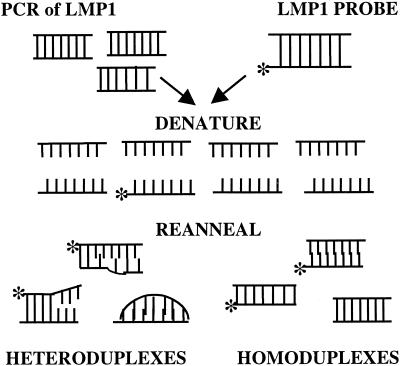
HTA analysis is based on the principle that completely homologous DNA molecules, termed homoduplexes, will migrate according to size on a polyacrylamide gel, while heteroduplexes will migrate more slowly because of malformations in the DNA helix corresponding to regions of mismatch (6). The greater the mismatch of two sequences, the slower the migration of the heteroduplex through a gel matrix. The heteroduplex formation reaction was performed by mixing partially homologous PCR-amplified LMP1 sequences with a radiolabeled probe of the same region, denaturing combined DNAs and subsequently reannealing them (Fig. 2). The resulting product contained heteroduplexes and homoduplexes, which were then subjected to electrophoresis through a nondenaturing gel.
FIG. 2.
Schematic of the LMP1-HTA. The first step included mixing of the PCR-amplified LMP1 and the radiolabeled LMP1 probe of the same region. The mixture was denatured and allowed to reanneal. The resulting product contained heteroduplexes and homoduplexes. The asterisk denotes the radiolabeled strand of DNA.
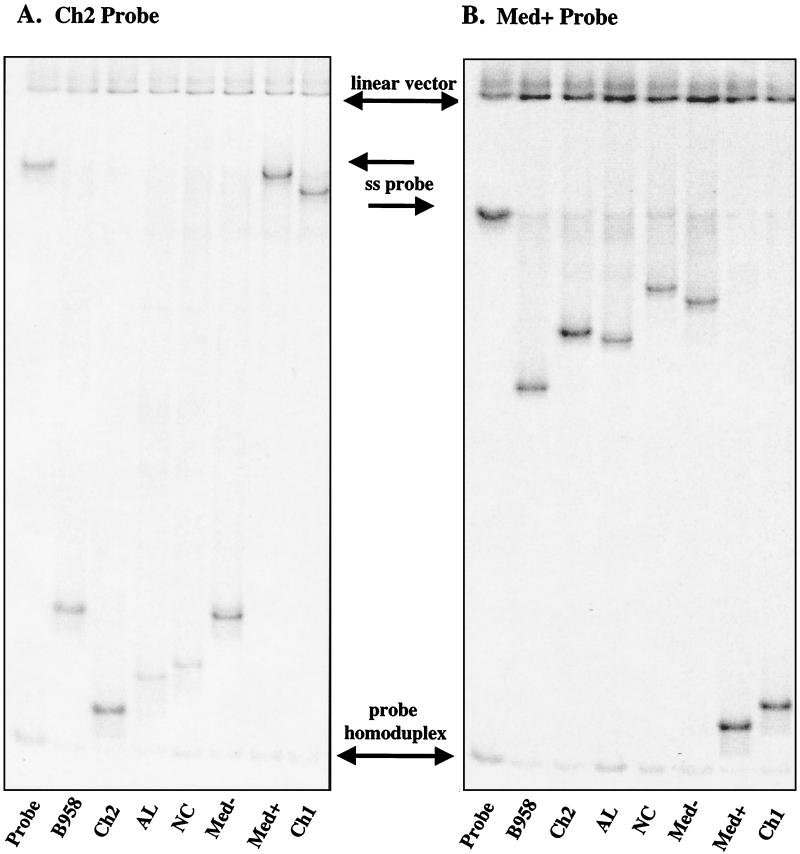
Distinct migration patterns of heteroduplexes resulted, depending on the strain that was used as the probe. In particular, when a deleted or undeleted strain probe was used, the location of the heteroduplexes in the gel was quite different. When an undeleted probe such as Ch2 was used, the heteroduplexes formed with undeleted strains migrated to the bottom of the gel above the probe homoduplex, while heteroduplexes with deleted strains were restricted in their migration due to the 30-bp mismatch bubble and were detected higher in the gel near the single-stranded probe (Fig. 3). When a deleted probe such as Med+ was used, the heteroduplexes with undeleted strains were located high in the gel below the single-stranded probe, while deleted strains bound to deleted probes migrated more according to size just above the probe homoduplex.
FIG. 3.
Comparison of migration of heteroduplexes with undeleted and deleted probes. PCR-amplified control strains B958, Ch2, AL, NC, Med−, and Med+ were hybridized to (A) the undeleted Ch2 probe and (B) the deleted Med+ probe. Each lane contained the linearized vector, excess single-stranded (ss) probe, and probe homoduplex. The lanes labeled probe contained only probe in the hybridization reaction.
Regardless of which strain probe was used, each lane consistently contained three bands that represent probe components (Fig. 3). The linearized vector migrated at the top of the gel, representing the radiolabeled vector cleaved from the LMP1 fragment. The single-stranded probe migrated in the upper middle portion of the gel and was detected as a result of excess probe. The probe homoduplex was formed when the labeled strand of the probe reannealed to the complementary unlabeled probe and migrated according to size. All strains bound to the probe, including the homoduplex strain, migrated slightly higher than the probe homoduplex because of short extensions on the ends of the PCR products derived from the primers. The intensity of the probe homoduplex and single-stranded probe varied depending on the specific activity of the radiolabeled probe and the relative ratio of PCR product to probe.
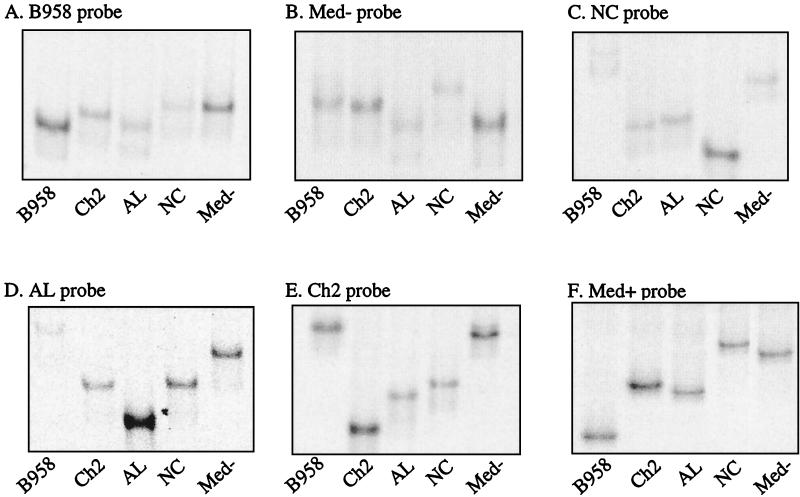
In order to determine which strains would serve as the most informative probes and produce the most discriminating pattern of heteroduplexes, probes of each of the five undeleted strains and the two deleted strains were prepared. Each probe was heteroduplexed to each PCR-amplified strain, and the migration patterns were determined (Fig. 4 and 5). Comparing the patterns of migration of heteroduplexes formed with undeleted strains indicated that the B958 probe did not result in dramatic migration shifts when heteroduplexed to other undeleted strains (Fig. 4A). This result was not surprising because each strain is unlike the B958 sequence to a similar extent. When bound to the B958 probe, Med− and NC had the most restricted migration, followed by Ch2 and AL.
FIG. 4.
Migration patterns of undeleted control strain heteroduplexes bound to each strain probe. (A) B958 probe, (B) Med− probe, (C) NC probe, (D) AL probe, (E) Ch2 probe, and (F) Med+ probe. The Ch1 probe yielded a pattern identical to that with the Med+ probe.
FIG. 5.
Migration patterns of deleted strain heteroduplexes. The strains used as the probe for each panel were (A) B958, (B) Med+, and (C) Ch1. The Ch2, NC, and Med− probes produced a heteroduplex pattern for the deleted strains similar to that represented in panel A. The AL probe produced a heteroduplex pattern for the deleted strains similar to that represented in panel B.
The Med− probe also did not produce a dramatic pattern of migrating heteroduplexes (Fig. 4B). In five of the eight positions where there were sequence changes in Med−, there were also changes in the other strains; therefore, the heteroduplexes were only slightly impeded in their migration due to relatively little mismatch. Compared to Med−, NC had two additional changes at positions 330 and 356 that increased the mismatch in that region, so that the NC/Med− heteroduplex had the slowest migration, followed by B958, Ch2, and AL (Fig. 4B).
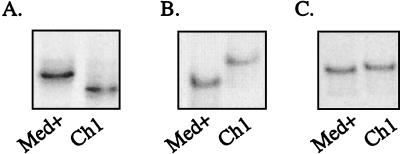
The remaining undeleted probes, Ch2, AL, and NC, provided more-useful heteroduplex migration profiles that more clearly distinguished other undeleted LMP1 variants. The sequences of Ch2, AL, and NC are most unlike the sequences of B958 and Med−; therefore, the heteroduplexes of these probes to B958 and Med− were the most restricted in their migration for each of these probes. When NC was used as the probe, the migrations of the Ch2/NC and AL/NC heteroduplexes were quite similar (Fig. 4C). When AL was used as the probe, the migrations of the Ch2/AL and NC/AL heteroduplexes were identical (Fig. 4D). When Ch2 was used as the probe, NC was slightly more impeded than AL (Fig. 4E and 3A). The clustering of similar sequence changes within Ch2, AL, and NC was responsible for the similar migration patterns. When either of the deleted strains, Med+ and Ch1, was used as the probe, the heteroduplexes that formed with undeleted strains had identical patterns and migrated considerably higher in the gel (Fig. 4F and 3B). Med− and NC exhibited the most restricted migration, followed by Ch2, AL, and B958 when a deleted probe was used (Fig. 4F). Three possible migration patterns resulted for deleted strains, depending on the probe used. The migration of Med+ was slightly more impeded than that of Ch1 when B958, Ch2, NC, or Med− was the probe (Fig. 5A and 3A). In contrast, when Med+ or AL was the probe, Ch1 migrated slightly higher than Med+ (Fig. 5B and 3B). The two deleted strains were indistinguishable when Ch1 was the probe (Fig. 5C).
Based on the migration patterns of the heteroduplexes that were produced, combined analysis with the Ch2 and Med+ probes was determined to be the most effective approach to precisely identify the strains present in a patient sample. If an additional base pair change is present within the defined region of LMP1, the heteroduplex may migrate to a different position because of the increased mismatch. For example, B958 with a base pair change at position 266 (see subject 4, Fig. 8A) migrated higher than the known B958 sequence when bound to the Med+ probe. Ch1 strains that lack the characteristic change at position 317 migrated similarly to Med+ when heteroduplexed to the B958 probe (data not shown). However, when bound to the Med+ probe, the Ch1 substrain migrated to the same position as the control Ch1 strain. This effect also suggests the importance of using two strain probes to analyze a patient sample.
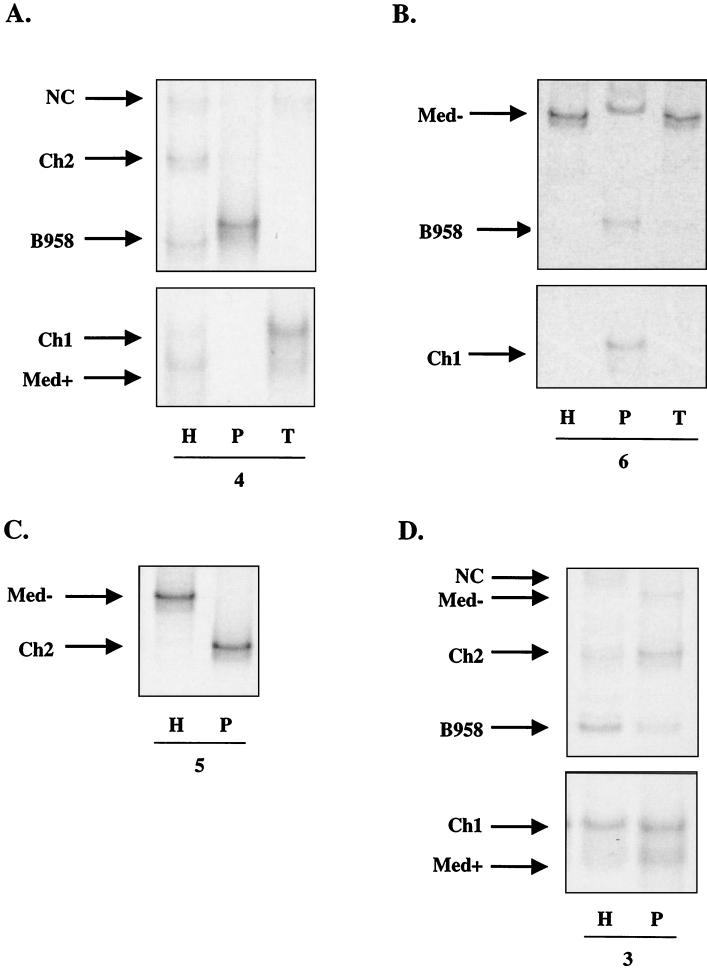
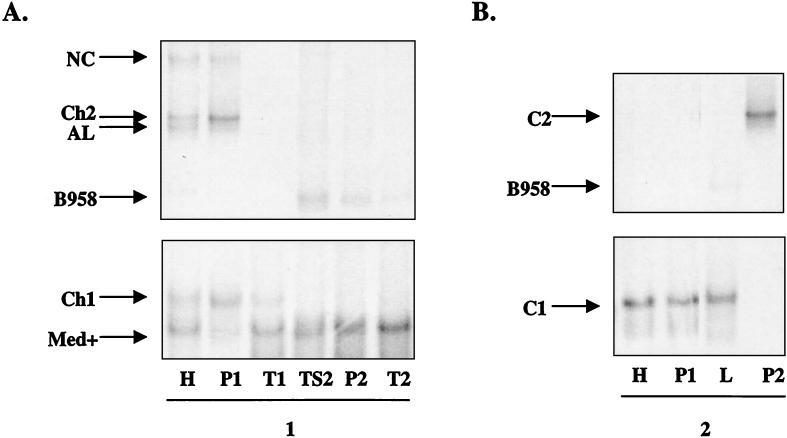
FIG. 8.
(A and B) LMP1-HTA (Med+ probe) of EBV strain profiles from the HLP lesion (H), the peripheral blood (P), and throat wash (T) samples of subjects 4 (A) and 6 (B). (C and D) EBV strain profiles from the HLP lesion (H) and the peripheral blood (P) samples of subjects 5 (C) and 3 (D).
Determination of detection limits of Med+ and Ch2 probes.
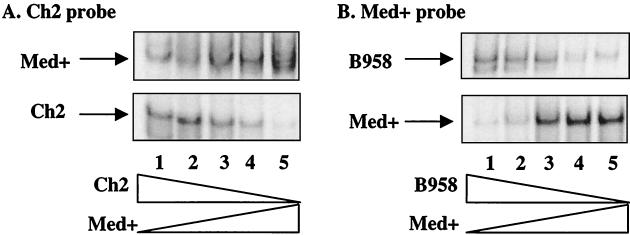
The comparisons of migration patterns for each probe indicated that a probe might bind to its homoduplex strain with more affinity than other strains, as detected by the intensity of the homoduplex band compared to the heteroduplex bands (Fig. 4). To evaluate the potential bias in relative abundance, mixing experiments were performed to determine the binding preference of a probe for its homoduplex strain versus other strains present in the population (Fig. 6). These experiments analyzed the binding competition that may occur between the probe and mixed strains in a PCR sample. The Ch2 and Med+ probes were mixed with increasing or decreasing concentrations of strains representing 1, 10, 50, 90, and 99% of the total population. The Ch2 and Med+ probes could detect Med+ and Ch2, and B958 and Med+, respectively, representing 1% of the mixture (Fig. 6).
FIG. 6.
Detection limitations for the Med+ and Ch2 probes were determined by mixing various strain DNAs at 1, 10, 50, 90, and 99% of the total template, which were then subjected to PCR and HTA. (A) The Ch2 probe hybridized to decreasing amounts of Ch2 and increasing amounts of Med+. (B) The Med+ probe hybridized to decreasing amounts of B958 and increasing amounts of Med+.
Other strain mixtures that were tested included B958 and Med− and Med− and Med+ (data not shown). Each probe was able to detect the presence of the strains at their lowest concentration; however, the Med+ homoduplex strain was detected with a greater intensity than the B958 heteroduplex strain at the higher concentrations (Fig. 6B). These results indicate that the HTA can depict the relative abundance of strains in a mixed population with sufficient sensitivity to detect strains at very low abundance. However, to eliminate any potential bias that may be inherent in this assay, each patient sample was characterized with two distinct probes to determine the relative abundance of the strains.
LMP1 strain variants in HLP patient samples.
The presence of HLP is an indicator of immune system status in HIV-positive subjects; however, HLP is not restricted to HIV-infected persons (45). HLP is a keratinized hyperproliferative lesion on the lateral borders of the tongue that is caused by permissive EBV infection (15). In HLP, EBV replicates in the upper differentiated epithelial layers of the tongue, but it has not been detected in the basal layers in a latent or lytic form (28). It is thought that HLP may develop from virus released from mucosal lymphoid or epithelial cells (12, 46). Coinfection of the lesion with multiple EBV types has been shown, which permits the recombination of EBV genomes within the lesion (46). The presence of multiple strains in HLP could be due to infection and superinfection from endogenous and exogenous sources.
To more thoroughly characterize EBV strain variation in HLP, strains were identified by LMP1-HTA in the HLP lesion of 25 patients, 6 of whom also contributed blood samples and 3 of whom contributed a throat wash sample. In addition to HLP, one subject exhibited an EBV-positive oral lymphoma that was also biopsied and analyzed. Multiple samples were available from two subjects. The strains present in the samples were determined by HTA analysis, and in some cases the PCR products were sequenced to confirm the HTA (Fig. 7; Table 1). These analyses indicated that a single strain was present in 3 of 25 specimens, two strains were detected in 11 of 25 specimens, and three or more strains were detected in 11 of 25 specimens (Table 1). The number of strains detected in a subject did not correlate with their CD4+ cell count or HIV RNA load. In fact, five strains were detected in the HLP of the HIV-negative medically compromised subject (subject 4), indicating that the presence of multiple strains is not restricted to HIV-infected subjects.
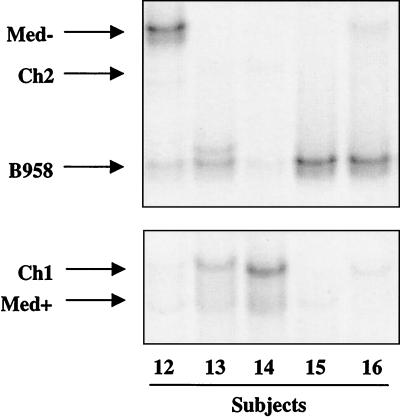
FIG. 7.
Examples of LMP1-HTA patterns for HLP biopsies from five subjects (12 to 16) analyzed with the Med+ probe. Only the regions of the gel that contained heteroduplexes are shown.
TABLE 1.
EBV strain profiles determined by HTA analysis
| Patient no. | Samplea | Strain(s) detectedb |
|---|---|---|
| 1 | HLP | B958, AL, NC, Ch2, Med+, Ch1 |
| Blood | AL, NC, Ch2, Med+, Ch1 | |
| TW | AL, Med+, Ch1 | |
| TS (27 mo) | B958, Med+ | |
| Blood (27 mo) | B958, Med+ | |
| TW (27 mo) | B958, Med+ | |
| 2 | HLP | Ch1 (317G)* |
| Blood | Ch1 (317G)* | |
| Lymphoma | Ch1 (317G)*, B958 | |
| Blood (2 mo) | Ch2 | |
| 3 | HLP | B958*, NC, Ch2, Med+, Ch1* |
| Blood | B958, Ch2, Med−, Med+, Ch1 | |
| 4 | HLP | B958, NC, Ch2, Med+, Ch1 |
| Blood | B958 (266T)* | |
| TW | NC, Med+, Ch1* | |
| 5 | HLP | Med− |
| PBL | Ch2 | |
| 6 | HLP | B958, Med−* |
| PBL | B958*, Med−, Ch1 | |
| TW | B958, Med−* | |
| 7 | HLP | B958*, Ch1 (357C)* |
| 8 | HLP | Med−* |
| 9 | HLP | B958*, Ch1 |
| 10 | HLP | Med−, Ch1 |
| 11 | HLP | B958*, Ch1 (317G)* |
| 12 | HLP | B958, Ch2, Med− Ch1, Med+ |
| 13 | HLP | B958, Ch2, Ch1, Med+ |
| 14 | HLP | B958, Ch2, Ch1, Med+ |
| 15 | HLP | B958, Med+ |
| 16 | HLP | B958, Med−, Ch1 |
| 17 | HLP | B958, Ch2, Med−, Ch1, Med+ |
| 18 | HLP | B958, Med− |
| 19 | HLP | B958, Med−, Med+, Ch1 |
| 20 | HLP | B958, Ch1 |
| 21 | HLP | B958, Ch1 |
| 22 | HLP | B958, Med+ |
| 23 | HLP | B958, Med+, Ch1 |
| 24 | HLP | B958, Ch1 |
| 25 | HLP | B958, Med− Med+ |
TW, throat wash; TS, tongue scraping; PBL, peripheral blood lymphocytes.
An asterisk indicates that the strain was sequenced. The predominant strain is shown in boldface.
Within the HLP specimen, the relative abundance of strains was variable, with one predominant strain in 20 of 22 samples. In some samples, multiple deleted and undeleted strains were present that would not have been identified in studies that distinguished strains by the deletion alone. In addition, some strains were detected at such low abundance that their presence could easily be missed in studies that detected strains by cloning and sequencing.
To investigate the relationship of the virus found in the peripheral blood to that in the HLP lesion, the strain variants in the peripheral blood B lymphocytes and those present within the epithelial cells in the HLP lesion (Fig. 8) and in throat washes were identified (Fig. 8A, 8B, and 9A). In most cases, the throat wash specimens had a strain profile similar to that within the corresponding HLP sample, which was distinct from the strain profile detected in the peripheral blood. The HLP sample of subject 4 contained five strains that were present in similar amounts (B958, NC, Ch2, Med+, and Ch1), with a subset of these (NC, Med+, and Ch1) detected in the throat washes (Fig. 8A). In contrast, the pattern in the peripheral blood sample of subject 4 was quite distinct, with only a single strain present that was a variant of the B958 strain with an additional base pair change (266 A→Τ). In this subject, the source of strains for the HLP lesion does not appear to be the blood.
FIG. 9.
LMP1-HTA (Med+ probe) of EBV strain profiles from two time points for the same subjects. (A) Subject 1. HLP (H), peripheral blood (P1), and throat wash (T1) samples from the first collection, and normal tongue scraping (TS2), peripheral blood (P2), and throat wash (T2) samples from the second collection 27 months later. (B) Subject 2. HLP (H), peripheral blood (P1), and oral lymphoma (L) samples from the first collection and peripheral blood (P2) sample 2 months later.
Similarly, the HLP sample from subject 6 primarily contained the Med− strain, with very low levels of the B958 strain, and the throat wash sample contained only Med− (Fig. 8B). The peripheral blood contained equal levels of B958, Ch1, and a variant of Med− that displayed a unique migration pattern. Subject 5 also had striking compartmental differences, with a single Ch2 strain in the blood and Med− predominantly in the HLP sample, with a trace of Ch2 (Fig. 8C). Subject 3 had a complex pattern, with at least five strains in the HLP and five in the blood. However, one strain in each compartment was not detected in the other, and the relative abundance of the strains was also distinct (Fig. 8D).
These similarities in strain patterns and abundance in HLP and throat washes suggest that the HLP lesion may be the source of strains in the oral cavity in these patients, although every strain that was detected in the corresponding HLP sample was not present in the throat washes. The striking differences in some subjects between the prevalent strains in the blood and those in the HLP and throat wash samples suggest that the virus in the oropharynx may not be due to reactivation of virus from peripheral blood lymphocytes and that there may be distinct compartments of infection.
To identify possible transmission of virus between blood and oropharynx, strains were identified in the two compartments at two time points. In subject 1, Med+, Ch1, and a very low level of AL were detected in the throat washes, while the HLP specimen had the same strains with the additional presence of NC, Ch2, and trace amounts of B958 (Fig. 9A). The peripheral blood was similar to the HLP, with detection of NC, Ch2, AL, Ch1, and Med+. The B958 strain was only detected in the HLP at this time. At the second time point, subject 1 had an undetectable HIV load and was on highly active antiretroviral therapy; therefore, the subject did not have HLP. In a scraping of the subject's pathologically normal tongue and also in the throat wash and peripheral blood samples, the pattern of strain abundance was dramatically different. B958 and Med+ were now the most abundant strains in all three samples, with trace levels of NC and AL. The detection of B958, which was previously only detected in the HLP, may indicate that the B958 strain was transmitted from the oropharynx to the blood. Alternatively, B958 may have been present in the blood but was below the level of detection or was sequestered in lymphoid tissue, possibly in the spleen or bone marrow.
In subject 2, the same strain was detected in the HLP, peripheral blood lymphocytes, and an oral lymphoma, which was a subspecies of Ch1 (lacking the characteristic change at position 317) (Fig. 9B). Trace levels of B958 were also detected in the oral lymphoma sample, which may represent infected lymphocytes at the biopsy site, although B958 was not detected in the peripheral blood or HLP. It is possible that the prevalent strains in mucosal oral lymphocytes are distinct from the prevalent strains detected in the peripheral blood. A second blood sample from subject 2 was obtained 2 months later, which revealed the presence of the Ch2 strain that was not detected in any sample at the collection 2 months prior. These analyses reveal that EBV infection can be very dynamic, with changes in relative strain abundance over time as well as the appearance of new strains.
DISCUSSION
The identification of EBV strains has been used in many studies to investigate questions concerning the biology of infection and the nature of viral persistence. Many techniques have been used to identify variants of EBV in infected people. The establishment of lymphocyte cell lines in vitro from blood samples or by infecting naïve B lymphocytes with virus in throat washes has often been used to identify the EBV variants present in a sample (4, 14, 37). DNA or protein can be harvested from lymphocyte cell lines and subjected to various methods to identify differences in EBV types or strains. Sequence differences in the EBNA2 and -3 genes distinguish between EBV types 1 and 2, respectively (7, 36). EBV type 2 is less efficient at lymphocyte transformation; consequently, early studies that analyzed the prevalence of type 1 or 2 in lymphocyte cell lines likely underestimated the prevalence of type 2 virus (35, 38).
A process called Ebnotyping refers to the study of EBV nuclear antigens. The EBNAs are expressed during latent infection and can vary slightly in protein size depending on the number of repeated sequences in the gene; thus, EBV strains have also been distinguished by the relative pattern of the size of the EBNA proteins. Ebnotyping has been useful for studies of transmission and persistence (13).
EBV variants that are marked by sequence polymorphisms can be distinguished by restriction enzyme polymorphisms, single-strand conformation polymorphism analysis, PCR, and Southern blotting (1, 10, 18, 21, 40). However, all strains may not be identified by these methods, which rely on specific sequence changes or relatively large sequence differences. Similarly, the repeat region of LMP1 has been used as a marker of EBV strains (31, 37). Because the repeat region is subject to intra- and interstrain recombination, the number of repeats cannot determine the number of strains that are present, nor can this number provide information about transmission (24). Different strains frequently have the same number of repeats, and the same strain can have a different number of repeat units (47). Another method to analyze strain variants involves obtaining viral DNA directly from the human tissue, subjecting it to PCR, and cloning and sequencing it (16, 47). This approach is labor intensive and may not provide a complete representation of less-abundant strain variants.
The HTA was developed and used in this study to identify EBV strains that have signature changes in a defined region of LMP1 as well as provide information about the relative abundance of LMP1 variants within a patient sample. Seven distinct EBV strains have been clearly identified; an eighth strain was reported in 2 of 92 specimens but was concluded to be either a very rare strain or a Ch1/B958 double recombinant strain (8). It is possible that more than the eight described strains exist. The data presented here indicate that a given strain has a consistent migration pattern with each individual probe. The HTA is also sufficiently discriminatory to distinguish substrains, which have a unique heteroduplex migration that represents minor base pair changes that were confirmed by sequence analysis. For example, a Ch1 strain that lacked the 317G→A change was detected in subjects 2 and 10 (Fig. 9B; Table 1), and a Ch1 strain that lacked the 357C→A change was detected in subject 7 (Table 1). Variants of B958 and Med− were also detected in subjects 4 and 6, respectively (Fig. 8A and B). In each case, the additional base pair changes resulted in a migration difference slightly above the position of the parental strain.
In the study presented here, the HTA was used to analyze strain variants in 25 subjects with the HIV-associated pathology HLP, which has been shown to contain multiple types and strains of EBV. The HTA was used to analyze the strain profiles in the HLP and blood specimens from six subjects, the HLP, blood, and throat wash specimens from three subjects, and the HLP, blood, and oral lymphoma specimens from one subject. These analyses revealed a complex pattern of EBV strain variants that demonstrated the persistence of some strains and the appearance and disappearance of other strains.
The strains in the HLP specimens had variable patterns of abundance, with one strain typically being predominant and several minor variants also being present. The differences in strain abundance within the HLP itself and compared to the peripheral blood may reflect differences in the replicative efficiency of the strains represented by LMP1 variants. It is important to note that although differences in biological properties of LMP1 variants have been described in vitro, this study did not identify a correlation between the presence of certain LMP1 variants and the existence of HLP (9, 17, 23).
Intertypic recombinants between the type-specific EBNA2 and EBNA3 genes have been identified, and the LMP1 variants have been shown not to cosegregate with EBV types (48). Therefore, the LMP1 variants may be linked to different allelic forms of multiple EBV genes that would influence replication, tissue tropism, or transformation. A specific viral isolate from an individual may contain a collection of changes in many different regions of the viral genome that may collectively confer a growth advantage, such that it would be difficult to link a particular strain to a disease state.
The life cycle of EBV involves two compartments, the peripheral blood and the oral cavity. The relationship of the viruses found in these two compartments is unresolved. Latently infected memory B lymphocytes circulate in the peripheral blood and traffic through lymphoid tissues and are believed to be the site of EBV latency and persistence. Oropharyngeal and salivary gland epithelial cells have been shown to harbor replicative EBV; however, several studies have suggested that the source of virus in saliva is permissively infected tonsillar lymphocytes (2, 20, 27, 39, 41).
The data presented here revealed completely different profiles of EBV strains in the oropharynx and peripheral blood in some individuals, calling into question the relationship of these two compartments. A recent study used variation in the repeat region of LMP1, the presence or absence of the 30-bp deletion, and variation in the EBNA genes as markers for strain variants to investigate the trafficking of virus between the peripheral blood and the HLP lesion in 16 subjects (30). This study provided the initial evidence for a relationship between strains present in the blood and HLP. In the data presented here, several subjects had a similar strain profile in their blood and HLP lesion, suggesting that there can be compartmental crossover of virus. The subsequent detection in the peripheral blood of strains initially unique to the HLP lesion, as demonstrated in subject 1, provides additional evidence for transmission between the oral and blood compartments.
In many of the subjects analyzed in this study, compartmental differences in the EBV strain profiles in the oral cavity and peripheral blood were readily apparent, with one subject exhibiting a dominant strain in the HLP that was not detected in the blood. The normal tongue epithelium of an HIV-infected person has also been shown to contain replicative EBV (44). Furthermore, the data presented here reveal that the normal tongue epithelium can be infected with multiple strains of EBV. In each subject the throat wash sample contained a subset of strains that were present in the HLP sample. As HLP represents a lytic form of infection that is releasing virus particles into the oral cavity, it is not surprising that the strains detected in the HLP were detected in the throat wash. However, it is interesting that if a strain was not present in the HLP lesion, it was not detected in the throat wash.
In several examples, strains in the HLP lesion were not detectable in the blood. This finding may indicate exogenous infection with additional EBV strains or reflect virus reactivation and replication in salivary epithelial cells or mucosal lymphocytes that harbor distinct strains from peripheral lymphocytes. These striking differences in the relative abundance of the EBV strains would not have been revealed with other methods used to distinguish EBV variants. Interestingly, a similar study of HIV compartmentalization by HTA analysis revealed complete correlation between the strains detected and their relative abundance in the peripheral blood and those present in saliva (11). This finding confirmed that the blood was the source of HIV in saliva. The differences in EBV strain abundance between the throat wash and blood samples detected here may reflect the ability of EBV to infect and replicate in epithelial cells and possibly indicate that epithelial cells are an additional distinct reservoir of persistent chronic infection.
The HTA is a powerful technique for characterizing strain variation and may provide insight into the pathogenesis of EBV infection. The true utility of this assay will be demonstrated in long-term studies that will provide information about the persistence of strains and how the strain profiles change over time. Understanding how EBV infection changes throughout the lifetime of the host may reveal key elements about the development of pathology or the maintenance of the asymptomatic state. The complete identification of EBV strains that infect an individual is important to elucidate the elements of EBV infection, including virus transmission, compartmentalization, persistence, and the development of pathology, all of which are poorly understood.
Acknowledgments
We thank Julie Nelson and Ron Swanstrom for assistance with the development of the HTA.
This study was supported by U.S. Public Health Service grants DE11644 and CA32979 to N.R.-T. D.S.-G. is supported by NIH training grant DE07310.
REFERENCES
- 1.Abdel-Hamid, M., J. J. Chen, N. Constantine, M. Massoud, and N. Raab-Traub. 1992. EBV strain variation: geographical distribution and relation to disease state. Virology 190:168-175. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, I., M. Hummel, C. Kreschel, and H. Stein. 1995. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood 85:744-750. [PubMed] [Google Scholar]
- 3.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 4.Berger, C., D. van Baarle, M. J. Kersten, M. R. Klein, A. S. Al-Homsi, B. Dunn, C. McQuain, R. van Oers, and H. Knecht. 1999. Carboxy terminal variants of Epstein-Barr virus-encoded latent membrane protein 1 during long-term human immunodeficiency virus infection: reliable markers for individual strain identification. J. Infect. Dis. 179:240-244. [DOI] [PubMed] [Google Scholar]
- 5.Bhatia, K., A. Raj, M. I. Guitierrez, J. G. Judde, G. Spangler, H. Venkatesh, and I. T. Magrath. 1996. Variation in the sequence of Epstein Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt's lymphomas. Oncogene 13:177-181. [PubMed] [Google Scholar]
- 6.Bhattacharyya, A., and D. M. Lilley. 1989. The contrasting structures of mismatched DNA sequences containing looped-out bases (bulges) and multiple mismatches (bubbles). Nucleic Acids Res. 17:6821-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dambaugh, T., K. Hennessy, L. Chamnankit, and E. Kieff. 1984. U2 region of Epstein-Barr virus DNA may encode Epstein-Barr nuclear antigen 2. Proc. Natl. Acad. Sci. USA 81:7632-7636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards, R. H., F. Seillier-Moiseiwitsch, and N. Raab-Traub. 1999. Signature amino acid changes in latent membrane protein 1 distinguish Epstein-Barr virus strains. Virology 261:79-95. [DOI] [PubMed] [Google Scholar]
- 9.Fielding, C., K. Sandvej, A. Mehl, P. Brennan, M. Jones, and M. Rowe. 2001. Epstein-Barr virus LMP-1 natural sequence variants differ in their potential to activate cellular signaling pathways. J. Virol. 75:9129-9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frank, D., E. Cesarman, Y. F. Liu, R. E. Michler, and D. M. Knowles. 1995. Posttransplantation lymphoproliferative disorders frequently contain type A and not type B Epstein-Barr virus. Blood 85:1396-1403. [PubMed] [Google Scholar]
- 11.Freel, S., J. Williams, J. Nelson, L. Patton, S. Fiscus, R. Swanstrom, and D. Shugars. 2001. Characterization of human immunodeficiency virus type 1 in saliva and blood plasma by V3-specific heteroduplex tracking assay and genotype analyses. J. Virol. 75:4936-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilligan, K., P. Rajadurai, L. Resnick, and N. Raab-Traub. 1990. Epstein-Barr virus small nuclear RNAs are not expressed in permissively infected cells in AIDS-associated leukoplakia. Proc. Natl. Acad. Sci. USA 87:8790-8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gratama, J. W., M. A. Oosterveer, G. Klein, and I. Ernberg. 1990. EBNA size polymorphism can be used to trace Epstein-Barr virus spread within families. J. Virol. 64:4703-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratama, J. W., M. A. Oosterveer, W. Weimar, K. Sintnicolaas, W. Sizoo, R. L. Bolhuis, and I. Ernberg. 1994. Detection of multiple ′Ebnotypes' in individual Epstein-Barr virus carriers following lymphocyte transformation by virus derived from peripheral blood and oropharynx. J. Gen. Virol. 75:85-94. [DOI] [PubMed] [Google Scholar]
- 15.Greenspan, J. S., D. Greenspan, E. T. Lennette, D. I. Abrams, M. A. Conant, V. Petersen, and U. K. Freese. 1985. Replication of Epstein-Barr virus within the epithelial cells of oral “hairy” leukoplakia, an AIDS-associated lesion. N. Engl. J. Med. 313:1564-1571. [DOI] [PubMed] [Google Scholar]
- 16.Henry, S., C. Sacaze, L. Berrajah, H. Karray, M. Drira, A. Hammami, J. Icart, and B. Mariame. 2001. In nasopharyngeal carcinoma-bearing patients, tumors and lymphocytes are infected by different Epstein-Barr virus strains. Int. J. Cancer 91:698-704. [DOI] [PubMed] [Google Scholar]
- 17.Hu, L. F., F. Chen, X. Zheng, I. Ernberg, S. L. Cao, B. Christensson, G. Klein, and G. Winberg. 1993. Clonability and tumorigenicity of human epithelial cells expressing the EBV encoded membrane protein LMP1. Oncogene 8:1575-1583. [PubMed] [Google Scholar]
- 18.Hu, L. F., E. R. Zabarovsky, F. Chen, S. L. Cao, I. Ernberg, G. Klein, and G. Winberg. 1991. Isolation and sequencing of the Epstein-Barr virus BNLF-1 gene (LMP1) from a Chinese nasopharyngeal carcinoma. J. Gen Virol. 72:2399-2409. [DOI] [PubMed] [Google Scholar]
- 19.Khanna, R., and S. Burrows. 2000. Role of cytotoxic T lymphocytes in Epstein-Barr virus-associated diseases. Annu. Rev. Microbiol. 54:19-48. [DOI] [PubMed] [Google Scholar]
- 20.Lemon, S., L. Hutt, J. Shaw, J. Li, and J. Pagano. 1977. Replication of EBV in epithelial cells during infectious mononucleosis. Nature 268:268-270. [DOI] [PubMed] [Google Scholar]
- 21.Lung, M. L., R. S. Chang, M. L. Huang, H. Y. Guo, D. Choy, J. Sham, S. Y. Tsao, P. Cheng, and M. H. Ng. 1990. Epstein-Barr virus genotypes associated with nasopharyngeal carcinoma in southern China. Virology 177:44-53. [DOI] [PubMed] [Google Scholar]
- 22.Miller, G. 1990. Epstein-Barr virus: biology, pathogenesis and medical aspects, p. 1921-1958. In B. Fields and D. Knipe (ed.), Fields virology, 2nd ed. Raven Press, New York, N.Y.
- 23.Miller, W. E., J. L. Cheshire, A. S. Baldwin, Jr., and N. Raab-Traub. 1998. The NPC derived C15 LMP1 protein confers enhanced activation of NF-kappa B and induction of the EGFR in epithelial cells. Oncogene 16:1869-1877. [DOI] [PubMed] [Google Scholar]
- 24.Miller, W. E., R. H. Edwards, D. M. Walling, and N. Raab-Traub. 1994. Sequence variation in the Epstein-Barr virus latent membrane protein 1. J. Gen. Virol. 75:2729-2740. (Erratum, 76:1305.) [DOI] [PubMed] [Google Scholar]
- 25.Murray, R. J., M. G. Kurilla, J. M. Brooks, W. A. Thomas, M. Rowe, E. Kieff, and A. B. Rickinson. 1992. Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J. Exp. Med. 176:157-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson, J. A. E., S. A. Fiscus, and R. Swanstrom. 1997. Evolutionary variants of the human immunodeficiency virus type 1 V3 region characterized by using a heteroduplex tracking assay. J. Virol. 71:8750-8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niedobitek, G., A. Agathanggelou, N. Steven, and L. Young. 2000. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol. Pathol. 53:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niedobitek, G., L. S. Young, R. Lau, L. Brooks, D. Greenspan, J. S. Greenspan, and A. B. Rickinson. 1991. Epstein-Barr virus infection in oral hairy leukoplakia: virus replication in the absence of a detectable latent phase. J. Gen. Virol. 72:3035-3046. [DOI] [PubMed] [Google Scholar]
- 29.Packham, G., M. Brimmell, D. Cook, A. J. Sinclair, and P. J. Farrell. 1993. Strain variation in Epstein-Barr virus immediate-early genes. Virology 192:541-550. [DOI] [PubMed] [Google Scholar]
- 30.Palefsky, J. M., J. Berline, D. Greenspan, and J. Greenspan. 2002. Evidence for trafficking of Epstein-Barr virus strains between hairy leukoplakia and peripheral blood lymphocytes. J. Gen. Virol. 83:317-321. [DOI] [PubMed] [Google Scholar]
- 31.Palefsky, J. M., J. Berline, M. E. Penaranda, E. T. Lennette, D. Greenspan, and J. S. Greenspan. 1996. Sequence variation of latent membrane protein-1 of Epstein-Barr virus strains associated with hairy leukoplakia. J. Infect. Dis. 173:710-714. [DOI] [PubMed] [Google Scholar]
- 32.Raab-Traub, N., and K. Flynn. 1986. The structure of the termini of the Epstein-Barr virus as a marker of clonal cellular proliferation. Cell 47:883-889. [DOI] [PubMed] [Google Scholar]
- 33.Resch, W., N. Parkin, E. Stuelke, T. Watkins, and R. Swanstrom. 2001. A multiple-site-specific heteroduplex tracking assay as a tool for the study of viral population dynamics. Proc. Natl. Acad. Sci. USA 98:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. Fields, D. Knipe, and P. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa. [Google Scholar]
- 35.Rickinson, A. B., L. S. Young, and M. Rowe. 1987. Influence of the Epstein-Barr virus nuclear antigen EBNA 2 on the growth phenotype of virus-transformed B cells. J. Virol. 61:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sample, J., L. Young, B. Martin, T. Chatman, E. Kieff, and A. Rickinson. 1990. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J. Virol. 64:4084-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sandvej, K., J. W. Gratama, M. Munch, X. G. Zhou, R. L. Bolhuis, B. S. Andresen, N. Gregersen, and S. Hamilton-Dutoit. 1997. Sequence analysis of the Epstein-Barr virus (EBV) latent membrane protein-1 gene and promoter region: identification of four variants among wild-type EBV isolates. Blood 90:323-330. [PubMed] [Google Scholar]
- 38.Sculley, T. B., A. Apolloni, L. Hurren, D. J. Moss, and D. A. Cooper. 1990. Coinfection with A- and B-type Epstein-Barr virus in human immunodeficiency virus-positive subjects. J. Infect. Dis. 162:643-648. [DOI] [PubMed] [Google Scholar]
- 39.Sixbey, J. W., J. G. Nedrud, N. Raab-Traub, R. A. Hanes, and J. S. Pagano. 1984. Epstein-Barr virus replication in oropharyngeal epithelial cells. N. Engl. J. Med. 310:1225-1230. [DOI] [PubMed] [Google Scholar]
- 40.Sung, N. S., R. H. Edwards, F. Seillier-Moiseiwitsch, A. G. Perkins, Y. Zeng, and N. Raab-Traub. 1998. Epstein-Barr virus strain variation in nasopharyngeal carcinoma from the endemic and non-endemic regions of China. Int. J. Cancer 76:207-215. [DOI] [PubMed] [Google Scholar]
- 41.Tao, Q., G. Srivastava, A. C. Chan, L. P. Chung, S. L. Loke, and F. C. Ho. 1995. Evidence for lytic infection by Epstein-Barr virus in mucosal lymphocytes instead of nasopharyngeal epithelial cells in normal individuals. J. Med. Virol. 45:71-77. [DOI] [PubMed] [Google Scholar]
- 42.Triantos, D., S. R. Porter, C. Scully, and C. G. Teo. 1997. Oral hairy leukoplakia: clinicopathologic features, pathogenesis, diagnosis, and clinical significance. Clin. Infect. Dis. 25:1392-1396. [DOI] [PubMed] [Google Scholar]
- 43.van Baarle, D., E. Hovenkamp, M. Callan, K. Wolthers, S. Kostense, L. Tan, H. Niesters, A. Osterhaus, A. McMichael, M. van Oers, and F. Miedema. 2001. Dysfunctional Epstein-Barr virus (EBV)-specific CD8+ T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood 98:146-155. [DOI] [PubMed] [Google Scholar]
- 44.Walling, D., C. Flaitz, C. Nichols, S. Hudnall, and K. Adler-Storthz. 2001. Persistent productive Epstein-Barr virus replication in normal epithelial cells in vivo. J. Infect. Dis. 184:1499-1507. [DOI] [PubMed] [Google Scholar]
- 45.Walling, D. M., N. M. Clark, D. M. Markovitz, T. S. Frank, D. K. Braun, E. Eisenberg, D. J. Krutchkoff, D. H. Felix, and N. Raab-Traub. 1995. Epstein-Barr virus coinfection and recombination in non-human immunodeficiency virus-associated oral hairy leukoplakia. J. Infect. Dis. 171:1122-1130. [DOI] [PubMed] [Google Scholar]
- 46.Walling, D. M., S. N. Edmiston, J. W. Sixbey, M. Abdel-Hamid, L. Resnick, and N. Raab-Traub. 1992. Coinfection with multiple strains of the Epstein-Barr virus in human immunodeficiency virus-associated hairy leukoplakia. Proc. Natl. Acad. Sci. USA 89:6560-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walling, D. M., N. Shebib, S. C. Weaver, C. M. Nichols, C. M. Flaitz, and J. Webster-Cyriaque. 1999. The molecular epidemiology and evolution of Epstein-Barr virus: sequence variation and genetic recombination in the latent membrane protein-1 gene. J. Infect. Dis. 179:763-774. [DOI] [PubMed] [Google Scholar]
- 48.Yao, Q. Y., R. J. Tierney, D. Croom-Carter, G. M. Cooper, C. J. Ellis, M. Rowe, and A. B. Rickinson. 1996. Isolation of intertypic recombinants of Epstein-Barr virus from T-cell-immunocompromised individuals. J. Virol. 70:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]