Abstract
The major and minor capsid proteins of polyomavirus are preassembled in the cytoplasm and translocated to the nucleus only as a VP1-VP2/VP3 complex. In this study, we describe independent nuclear translocation of the L1 major protein and the L2 minor capsid protein of human papillomavirus type 33 by several approaches. First, we observed that expression and nuclear translocation of L2 in natural lesions precede expression of L1. Second, using a cell culture system for coexpression, we found that accumulation of L2 in nuclear domain 10 (ND10) subnuclear structures precedes L1 by several hours. In contrast, complexes of L2 and mutants of L1 forced to assemble in the cytoplasm are translocated directly to ND10, like L2 expressed alone. Interestingly, accumulation of wild-type L1 is observed only after L2-induced release of the ND10-associated protein Sp100. Third, nuclear translocation of L2 but not of L1 was blocked by the proteasome inhibitor MG132. Our data suggest that L1 and L2 interaction occurs after L2-induced reorganization of ND10 subnuclear domains.
Papillomaviruses are nonenveloped DNA viruses of higher vertebrates that specifically infect epithelia and replicate in terminally differentiating keratinocytes. The papillomavirus capsid has icosahedral symmetry and consists of 72 capsomeres, pentamers of the major capsid protein L1, and several copies of a minor capsid protein, L2, probably 12 per virion (7). The L1 protein self-assembles into virus-like particles (VLPs) in the absence of L2, but the minor capsid protein is integrated into VLPs when it is coexpressed with L1. The L2 protein seems to be essential for the infectivity of virions (9, 19, 21), but its exact role in papillomavirus infection is still unclear. Recent studies using expression of viral genes in cultured cells have revealed that L2 localizes to nuclear substructures, called PML oncogenic domains or nuclear domains 10 (ND10) (1), and recruits L1 into these domains (3). This indicates an important function of L2 in the morphogenesis of papillomaviruses. Okun et al. have identified interaction domains of L2 of bovine papillomavirus type 1 necessary for encapsidation of the viral genome (13), but it is not known at what stage of viral assembly L1 and L2 first interact. Studies on the expression of the polyomavirus major and minor capsid proteins, VP1 and VP2/3, respectively, support the idea that the capsid proteins associate in the cytoplasm and are translocated to the nucleus as a VP1-VP2/3 complex (2, 4, 6, 8).
In this study, we have investigated the interactions of the major and minor capsid proteins of human papillomavirus type 33 (HPV33). More specifically, we have addressed the question of whether L1 and L2 are translocated to the nucleus separately or as a preformed complex. In order to address this issue, we have analyzed expression and cellular distribution of wild-type (wt) and mutant L1 and L2 both in natural lesions and cultured cells, respectively.
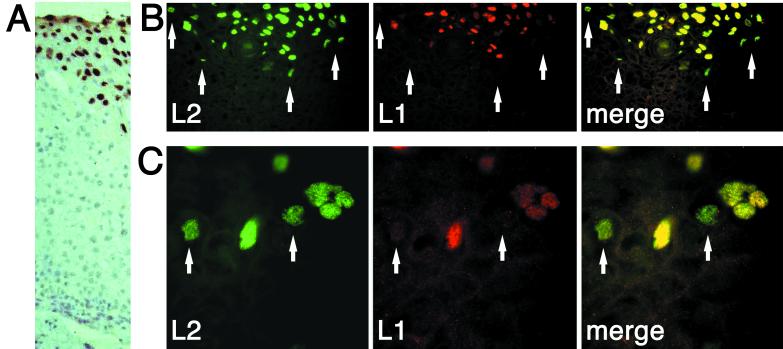
Initially, expression of L1 and L2 was studied in a cervical intraepithelial neoplasia (CIN II) induced by HPV33. Paraffin-embedded thin sections were deparaffinized with xylol (three times for 5 min each time), rehydrated using 90, 70, and 50% ethanol, boiled for 15 min in citrate buffer (pH 6.0), washed for 15 min with phosphate-buffered saline, and used for immunostaining. Figure 1B and C show two examples from different sections. A cross-section of a different lesion stained for L1 by dye precipitation using 33L1-7, horseradish peroxidase-conjugated secondary antibody, and diaminobenzidine (the section was counterstained with hematoxylin as described previously [15]) is also shown in Fig. 1A to indicate the orientation of areas shown in Fig. 1B and C. Possibly due to the previous paraffin and xylol treatments, immunostaining produced some nonspecific background. Nuclear dots resembling ND10 structures could still be detected in these structures, as previously described (5). In the upper layers of keratinocytes, coexpression of L1 and L2 was observed in most cells. In the lower layer of cells, however, where synthesis of the capsid proteins starts, about 5 to 10% of nuclei contained only L2 (Fig. 1B and C). Cells containing L1 only could not be found. This suggests that synthesis and nuclear translocation of L2 precede synthesis of L1 in these lesions. Association of L1 and L2 is clearly not an obligate intermediate for translocation to the nucleus.
FIG. 1.
Cellular localization of L1 and L2 proteins in CIN II lesions induced by HPV33. Thin sections were deparaffinized and immunostained for L1 and L2. (A) Cross-section of a lesion stained for L1 using dye precipitation and hematoxylin counterstaining as described previously (15) to indicate the orientation of the sections shown in panels B and C. The pictures in panels B and C were taken of keratinocyte layers at the onset of capsid protein expression. Two magnifications from different areas are shown. Arrows indicate nuclei exclusively displaying L2 staining.
To extend and confirm these data, we then analyzed expression and nuclear translocation of L1 and L2 in COS-7 or HuTK−143 B cells, using the T7/vaccina virus system (12), as previously described (21). Cells were grown at 37°C in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and antibiotics (Life Technologies) and infected at a multiplicity of infection of 2 for each virus. Mouse monoclonal (15, 16) and rabbit polyclonal (22) antibodies were used to visualize the capsid proteins by indirect immunofluorescence. Cells grown on coverslips were fixed with methanol-0.02 M EGTA (−20°C for at least 10 min), washed with phosphate-buffered saline, and blocked with 5% goat serum. Incubation with the specific antibodies for 1 h at 37°C was followed by incubation with Cy3-conjugated Affinipure goat anti-rabbit immunoglobulin G (IgG) and DTAF-conjugated goat anti-mouse IgG (Jackson Immunoresearch Products). Coverslips were washed and mounted onto slides using Vectashield mounting medium (Camon). Pictures were taken with a Leica DM RBE fluorescence microscope and a KOHU digital camera at an instrumental magnification of ×630. Software for merging pictures was obtained from Applied Imaging Corp.
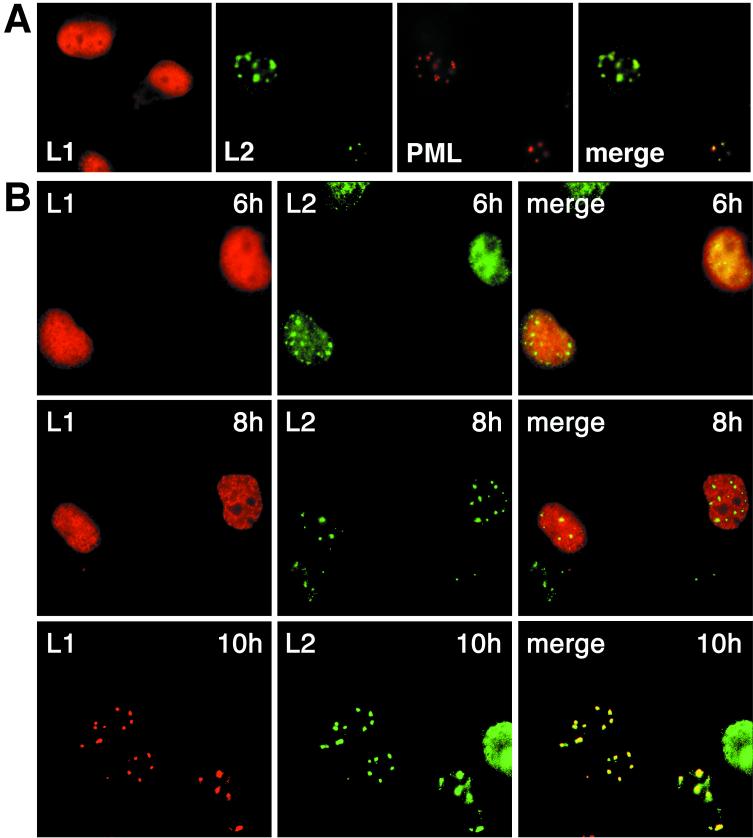
Using this system for expression and visualization of the capsid proteins, L1 was detected exclusively in the nucleus about 4 to 5 h after infection of cells with the recombinant virus. Its nuclear distribution was essentially homogeneous and did not change over time when expressed in the absence of L2 (Fig. 2A). In contrast, L2 assembled in nuclear dots, the ND10, colocalizing with promyelocytic leukemia protein (Fig. 2A) (3, 5). The time after infection when L2 localized exclusively in ND10 varied slightly between experiments, possibly depending on the condition of cells or virus. Usually 6 to 7 h after infection, it was mainly detected in ND10.
FIG. 2.
Cellular localization of wt L1 and L2 proteins. (A) COS-7 cells were infected with vaccinia viruses recombinant for HPV33 L1 or HPV33 L2 and immunostained after 8 h. Colocalization of L2 with promyelocytic leukemia protein (PML) is shown. (B) COS-7 cells were coinfected with both viruses; at the indicated times after infection the cells were fixed, and the capsid proteins were visualized by immunofluorescence.
We then studied coexpression and nuclear distribution of L1 and L2 as a function of time. Six hours after infection with the recombinant viruses, L1 was homogeneously distributed throughout the nucleus, whereas L2 was accumulating in ND10 (Fig. 2B). This was the same distribution as observed on separate expression of the two proteins (Fig. 2A). Two hours later, when L2 was exclusively detected in ND10, L1 was still largely dispersed throughout the nucleus (Fig. 2B). Colocalization with L2 yielding a punctate L1 pattern characteristic of ND10 was observed only 10 h after infection (Fig. 2B). This indicates that L1 and L2 colocalize only after separate nuclear translocation.
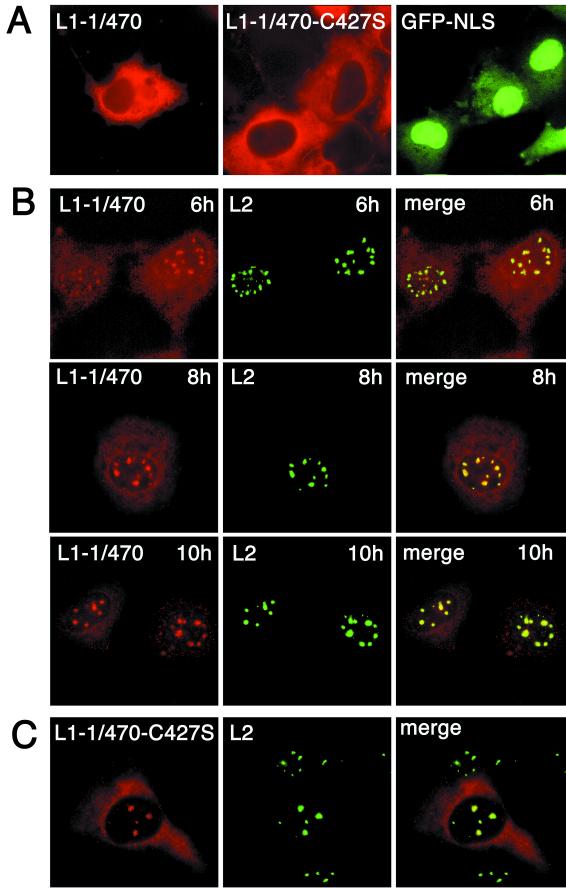
To verify this conclusion, we wondered if we could, for a control, enforce cytoplasmic assembly and nuclear cotranslocation of L1 and L2. This was achieved by deleting the nuclear localization signal (NLS) of L1. The only NLS of HPV33 L1 corresponds to the seven most C-terminal amino acids (amino acids 493 to 499, KRKKVKK). This is a complete and functional NLS, as shown by fusion to green fluorescent protein (GFP) (GFP-NLS [Fig. 3A]). L1 mutants lacking this sequence were arrested in the cytoplasm (Fig. 3A). Deletion of up to 29 amino acids from the C terminus did not prevent assembly into VLPs (14, 18). One of these deletion mutants, L1-1/470, was selected for further study.
FIG. 3.
Cellular localization of mutant L1 and wt L2 proteins. (A) COS-7 cells were infected for 10 h with recombinant vaccinia virus encoding mutant L1-1/470 or L1-1/470-C427S or were transfected with pEGFPGFP-NLS coding for the HPV33 L1-NLS fused to the carboxy terminus of dimeric GFP (GFP-NLS). Capsid proteins were visualized after 10 h by immunofluorescence. GFP was visualized 40 h after transfection. (B) COS-7 cells were coinfected with recombinant vaccinia viruses encoding wt L2 and mutant L1-1/470, respectively. Capsid proteins were visualized by immunofluorescence at the indicated times after infection. (C) COS-7 cells were infected with recombinant viruses encoding wt L2 and mutant L1-1/470-C427S, respectively. Cells were fixed 8 h after infection, and the capsid proteins were visualized by immunostaining.
Coexpression of L1-1/470 with wt L2 yielded a cellular distribution that was entirely different from the distribution observed with wt L1. At 6 h after infection, the L1-1/470 protein localized partly to ND10 and partly to the cytoplasm (Fig. 3B). L2 and the mutant L1 continued to accumulate in ND10, although some L1 protein remained cytoplasmic 10 h after infection (Fig. 3B). This demonstrates that wt L2 can complement NLS-defective L1 in nuclear translocation. Association with L2 in the cytoplasm and passage of the complex to the nucleus by virtue of the NLS of L2 must explain relocation of L1-1/470 from the cytoplasm to the nucleus. Since L1 and L2 appeared in ND10 simultaneously, they obviously remained associated in the nucleus. This is fundamentally different from the nuclear translocation of wt L1 and L2.
To extend this control, we studied nuclear translocation of capsomeres of L1 by L2. To do so, we used a mutant of L1 in which one of the essential cysteines required for disulfide bonding of capsomeres into capsids was replaced by serine. This mutant, L1-C427S, forms capsomeres but no capsids (17). The double mutant L1-1/470-C427S is therefore blocked in the cytoplasm (Fig. 3A) where it can form no higher-order structure than a capsomere. When this mutant was coexpressed with wt L2, L1 was again found in ND10 colocalizing with L2 as soon as the two proteins could be detected (Fig. 3C). The conclusion that L2 can translocate capsomeres of L1 is in agreement with the previous results of Merle et al. who had shown that capsomeres can pass nuclear pores in vitro (11). More importantly, the enforced translocation of these capsomeres by L2 was again entirely different from the translocation of wt L1 protein.
To account for the different phenotypes of wt and NLS-defective L1 proteins with regard to nuclear cotranslocation with L2, it is conceivable that the NLS-defective L1 floats around in the cytoplasm and is accessible to L2, whereas binding of L1-specific importins to wt L1 and subsequent nuclear trafficking may well prevent effective interaction with L2. This is in line with the observation that wt L1 is hardly detectable in the cytoplasm, suggesting a fast and efficient nuclear translocation of wt L1.
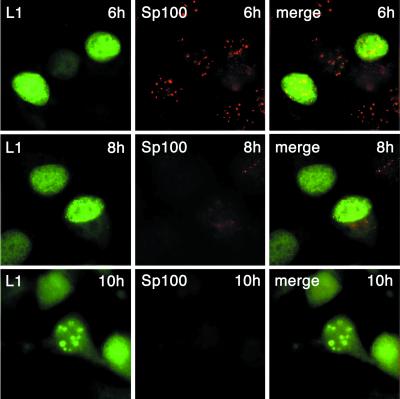
We have previously observed that L2 induces the release of Sp100, a major ND10-associated protein, which is subsequently degraded (5). When L2 was coexpressed together with wt L1, the effect on Sp100 was essentially the same (Fig. 4). Using a monoclonal antibody kindly donated by H. Will, Sp100 was still detected in ND10 6 h after infection with the L1 and L2 recombinant viruses. At 8 h, it started to disappear and was completely missing 10 h after infection. At no time could colocalization of Sp100 and L1 at ND10 be observed. In contrast to L2, wt L1 was detected only in ND10 after the L2-induced release of Sp100 was complete (Fig. 4), demonstrating separate translocation to ND10 of L1 and L2.
FIG. 4.
Cellular localization of L1 and Sp100. HuTK− cells were coinfected with recombinant vaccina viruses encoding wt L1 and L2, respectively. Cells were fixed at the indicated times after infection. L1 and Sp100 were visualized by immunostaining.
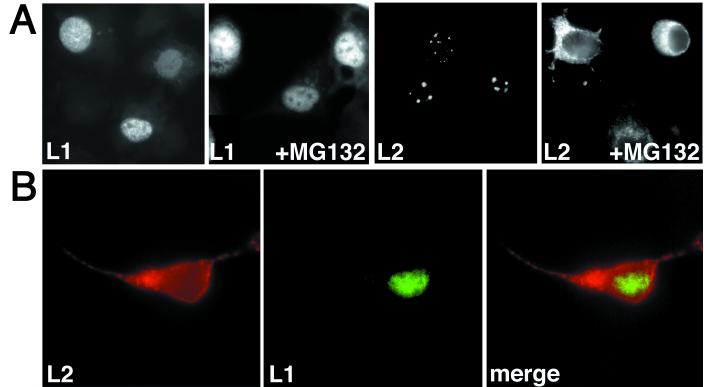
A final clue in the analysis of nuclear translocation of L1 and L2 was provided by an unexpected observation. When we studied whether the L2-induced degradation of Sp100 could be inhibited by peptidyl aldehydes such as MG132 (10), which have recently been discovered to act as inhibitors of the proteasome, we found that these inhibitors blocked the nuclear translocation of L2 (5). This prompted us to examine whether L1 was similarly affected. L1 and L2 were expressed using recombinant vaccina viruses, as described above, and 5 μM MG132 (carbobenzyloxy-ll-al; Sigma) was added 3 h after infection. As shown in Fig. 5, nuclear translocation of L2, but not of wt L1, was impaired when the two proteins were expressed either separately (Fig. 5A) or together (Fig. 5B). If a cytoplasmic L1/L2 complex were an intermediate in nuclear translocation, one would expect both capsid proteins to be affected in the same way. At the present time, the molecular mechanism of the inhibition of L2 translocation is not known. This will be the subject of further study.
FIG. 5.
Localization of wt L1 and L2 in cells treated with MG132. HuTK− cells were coinfected with vaccinia viruses encoding wt L1 and L2, respectively. MG132 was added 3 h after infection, and the capsid proteins were visualized 6 h later using immunofluorescence. The cells were infected with one virus (A) or two viruses (B). (A) The nuclear dots lacking L1, as seen in the L1 +MG132 panel, represent nucleoli.
To conclude, we have presented evidence that the assembly of the papillomavirus capsid differs from polyomavirus assembly. Whereas a polyomavirus VP1-VP2/3 complex is assembled in the cytoplasm and acts as the building block of the capsid after translocation to the nucleus (2), the papillomavirus L1 and L2 proteins are separately translocated, as shown by the following observations. (i) L2 synthesis and nuclear translocation precede L1 synthesis and nuclear translocation in HPV-infected lesions. (ii) L2 accumulation in ND10 precedes L1 accumulation by several hours. (iii) The proteasome inhibitor MG132 blocks L2 nuclear translocation, whereas L1 is unaffected. In addition, a mutant L1/wt L2 complex, which is forced to assemble in the cytoplasm, is directly translocated to ND10, resulting in the concomitant appearance of L1 and L2 in these subnuclear structures.
It is tempting to speculate that the differences between polyomaviruses and papillomaviruses with regard to assembly and nuclear translocation of the major and minor capsid proteins is reflected in the stoichiometry of the capsid proteins in the viral capsids. Whereas each polyomavirus VP1 pentamer is associated with one molecule of either VP2 or VP3 (2), only a small subset of papillomavirus L1 capsomeres, probably 12 (20), are associated with a molecule of the minor capsid protein L2.
Taken together with previous observations (5), our results suggest the following steps in the assembly of the papillomavirus capsid. Synthesis of L2 initiates prior to synthesis of L1 in the terminally differentiating keratinocytes. L2 is translocated into the nucleus independently of L1 and induces the reorganization of ND10. L1 is assembled into capsomeres in the cytoplasm. Capsomeres are translocated into the nucleus and recruited to ND10 after L2-induced release of Sp100. L2 and capsomeres then further assemble into capsids.
Acknowledgments
We gratefully acknowledge H. Will for monoclonal antibodies and K. Sotlar for CIN sections.
This work was supported in part by the Deutsche Forschungsgemeinschaft and Stiftung Rheinland-Pfalz. L.F. is a fellow of Graduiertenkolleg 194.
REFERENCES
- 1.Ascoli, C. A., and G. G. Maul. 1991. Identification of a novel nuclear domain. J. Cell Biol. 112:785-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barouch, D. H., and S. C. Harrison. 1994. Interactions among the major and minor coat proteins of polyomavirus. J. Virol. 68:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delos, S. E., L. Montross, R. B. Moreland, and R. L. Garcea. 1993. Expression of the polyoma VP2 and VP3 proteins in insect cells: coexpression with the major capsid protein VP1 alters VP2/3 localization. Virology 194:393-398. [DOI] [PubMed] [Google Scholar]
- 5.Florin, L., F. Schäfer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein L2. Virology 295:97-107. [DOI] [PubMed] [Google Scholar]
- 6.Forstova, J., N. Krausewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin. 1993. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J. Virol. 67:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, vol. 2. Lippincott Raven, Philadelphia, Pa. [Google Scholar]
- 8.Ishii, N., A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu. 1994. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J. Virol. 68:8209-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawana, Y., K. Kawana, H. Yoshikawa, Y. Taketani, K. Yoshiike, and T. Kanda. 2001. Human papillomavirus type 16 minor capsid protein L2 N-terminal region containing a common neutralization epitope binds to the cell surface and enters the cytoplasm. J. Virol. 75:2331-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meriin, A. B., V. L. Gabai, J. Yaglom, V. I. Shifrin, and M. Y. Sherman. 1998. Proteasome inhibitors activate stress kinases and induce Hsp 72. J. Biol. Chem. 273:6373-6379. [DOI] [PubMed] [Google Scholar]
- 11.Merle, E., R. C. Rose, L. LeRoux, and J. Moroianu. 1999. Nuclear import of HPV11 L1 capsid protein is mediated by karyopherin α2β1 heterodimers. J. Cell. Biochem. 74:628-637. [PubMed] [Google Scholar]
- 12.Moss, B., O. Elroy-Stein, T. Mizukami, W. A. Alexander, and T. R. Fuerst. 1990. Product review. New mammalian expression vectors. Nature 348:91-92. [DOI] [PubMed] [Google Scholar]
- 13.Okun, M. M., P. M. Day, H. L. Greenstone, F. P. Booy, D. R. Lowy, J. T. Schiller, and R. B. Roden. 2001. L1 interaction domains of papillomavirus L2 necessary for viral genome encapsidation. J. Virol. 75:4332-4342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paintsil, J., M. Muller, M. Picken, L. Gissmann, and J. Zhou. 1996. Carboxyl terminus of bovine papillomavirus type-1 L1 protein is not required for capsid formation. Virology 223:238-244. [DOI] [PubMed] [Google Scholar]
- 15.Sapp, M., U. Kraus, C. Volpers, P. J. Snijders, J. M. Walboomers, and R. E. Streeck. 1994. Analysis of type-restricted and cross-reactive epitopes on virus-like particles of human papillomavirus type 33 and in infected tissues using monoclonal antibodies to the major capsid protein. J. Gen. Virol. 75:3375-3383. [DOI] [PubMed] [Google Scholar]
- 16.Sapp, M., C. Volpers, M. Muller, and R. E. Streeck. 1995. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J. Gen. Virol. 76:2407-2412. [DOI] [PubMed] [Google Scholar]
- 17.Sapp, M., C. Fligge, I. Petzak, J. R. Harris, and R. E. Streeck. 1998. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J. Virol. 72:6186-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schäfer, F., L. Florin, and M. Sapp. 2002. DNA binding of L1 is required for human papillomavirus morphogenesis in vivo. Virology 295:172-181. [DOI] [PubMed] [Google Scholar]
- 19.Stauffer, Y., K. Raj, K. Masternak, and P. Beard. 1998. Infectious human papillomavirus type 18 pseudovirions. J. Mol. Biol. 283:529-536. [DOI] [PubMed] [Google Scholar]
- 20.Trus, B. L., R. B. S. Roden, H. L. Greenstone, M. Vrhel, J. T. Schiller, and F. P. Booy. 1997. Novel structural features of bovine papillomavirus capsid revealed by three-dimensional reconstruction to 9 Å resolution. Nat. Struct. Biol. 4:413-420. [DOI] [PubMed] [Google Scholar]
- 21.Unckell, F., R. E. Streeck, and M. Sapp. 1997. Generation and neutralization of pseudovirions of human papillomavirus type 33. J. Virol. 71:2934-2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volpers, C., M. Sapp, C. A. Komly, P. Richalet-Secordel, and R. E. Streeck. 1993. Development of type-specific and cross-reactive serological probes for the minor capsid protein of human papillomavirus type 33. J. Virol. 67:1927-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]