Abstract
Domains required late in the virus budding process (L domains) have been identified in the Gag proteins of a number of retroviruses. Here we show that the human T-cell leukemia virus type 1 candidate L domain motif PPPY is indeed required for virus production. Strikingly, however, mutation of this motif arrested virus particles at an earlier stage in the budding process than was seen for mutation of the L domain motifs thus far described for retroviruses. In view of the exchangeability of such domains, we propose that the retrovirus budding process may involve a continuum from bud formation to membrane fission.
Retroviruses are enveloped viruses that acquire their lipid membranes upon budding from the plasma membrane of the infected cell (26, 31). Buds are formed by the products of the gag gene that push on the cytoplasmic face of the plasma membrane. The gag gene products are translated as polyprotein precursors. While their size and sequence vary among retroviruses, all Gag precursors contain at least three domains, termed matrix (MA), capsid (CA), and nucleocapsid (NC). In a maturation process that is believed to begin during budding, the Gag precursors are cleaved by the viral protease to yield the separate MA, CA, and NC proteins that compose the capsid shell of the mature virion.
It has been clearly established that expression of the gag gene alone allows production of membrane-enveloped particles in the extracellular medium (2, 6, 10, 36). Thus, although maturation by the viral protease is necessary for the retrovirus particles to gain infectivity (5, 16, 18, 33), the Gag polyprotein precursor has information that is sufficient to mediate its intracellular transport to the cytoplasmic face of the plasma membrane, to direct assembly of the capsid shell, and to drive all steps of the budding process up to virion release. Functionally, three types of domains, the M (membrane binding), I (interaction between Gag proteins), and L (late budding) domains (4, 8, 31), have indeed been identified in the Gag sequences of different retroviruses. The positions and sequences of L domains vary among retroviruses. The L domains of Rous sarcoma virus (RSV), Mason-Pfizer monkey virus (M-PMV), and Moloney murine leukemia virus (MMLV) are located between the MA and CA domains and contain a highly conserved PPPY motif as the core sequence (35, 37, 39-41). In human immunodeficiency virus type 1 (HIV-1), the L domain is located at the C terminus of the Gag sequence and contains a distinct core motif, PTAP (11, 14). While the L domain of equine infectious anemia virus likewise maps to the C terminus of Gag, its critical residues are YPDL (24). However, the L domain sequences can be functionally interchanged among different retroviruses, showing that they provide the same function despite the lack of sequence homology (1, 22, 24, 40). Although the underlying mechanism remains to be fully elucidated, L domains are apparently responsible for a very late step in retrovirus egress. Indeed, mutations affecting the core motif have been shown to cause a significant reduction in virus production despite competence of Gag molecules for membrane binding and interaction (11, 14, 24, 35, 37, 39-41). Moreover, the electron microscopy data available so far on L domain mutants have all revealed an accumulation of assembled particles resembling virions but still tethered to the cell surface and/or to each other via a thin stalk (11, 39, 40). These observations have led to the suggestion that the L domains of retroviruses are required specifically for the ultimate step of the budding process, i.e., the fission event that separates the assembled particle from the plasma membrane of the infected cell (8, 9, 34).
As is the case in a wide variety of retroviruses with the exception of lentiviruses (35), a PPPY motif is present at the C terminus of the MA sequence in human T-cell leukemia virus type 1 (HTLV-1), HTLV-2, and bovine leukemia virus (BLV) (Fig. 1A). While a PTAP sequence is found close to the PPPY motif in the HTLV-1 Gag primary structure, only PPPY is conserved in the retroviruses of the HTLV/BLV genus. Here we have investigated the role of this candidate L domain motif in the context of a full-length, infectious HTLV-1 molecular clone.
FIG. 1.
Sequence alignment for Gag polyproteins of HTLV/BLV retroviruses and description of HTLV-1 PPPY mutants. The PPPY motifs are shown in bold. (A) Sequence alignment for the MA-CA junction of HTLV-1, HTLV-2, and BLV. The HTLV-1 sequence is taken from the work of Seiki et al. (28), the HTLV-2 sequence from Shimotohno et al. (29), and the BLV sequence from Coulston et al. (3). The arrow indicates the putative cleavage site between MA and CA. (B) Description of the mutations introduced into the PPPY motif of HTLV-1.
We replaced each of the four residues of the PPPY motif separately with a nonconservative amino acid via oligonucleotide-directed mutagenesis of the sequence encoding the HTLV-1 MA protein by the Kunkel method, as described previously (20). The mutated gag fragments were then inserted into the complete infectious HTLV-1 provirus XMT (7) and sequenced. The mutants were named according to the pattern “X amino acid position-Y,” where X and Y are the wild-type and replacement amino acids, respectively, and amino acid position 1 corresponds to the initiator methionine of the HTLV-1 MA protein (Fig. 1B).
To determine the effect of each mutation of the PPPY motif on the intracellular synthesis and processing of the Gag precursor, 293-TSA cells (13) were transfected with wild-type or PPPY-mutated XMT proviruses using the FUNGENE protocol (Boehringer), and cell lysates obtained 24 h later were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis. Western blotting was then performed with a pool of sera from HTLV-1-infected individuals (a gift from J. Coste, CRTS, Montpellier, France) as the primary antibody and I125-labeled protein A (Amersham) as the second-step reagent (20) (Fig. 2A). The wild-type HTLV-1 Gag precursor was processed into MA and CA mature proteins (the NC protein is not revealed in this assay) (Fig. 2A, lane 1). All four mutations of the PPPY motif also allowed precursor cleavage (Fig. 2A, lanes 2 to 5). For the Tyr121-Asp mutant, the MA protein migrated more rapidly in an SDS-polyacrylamide gel electrophoresis gel than did that of the wild type or the other mutants (Fig. 2A, lane 5). However, replacement of the tyrosine by an alanine or a phenylalanine resulted in an MA protein with the same apparent size as that of the wild-type MA protein (data not shown). Thus, the shift observed for the Tyr121-Asp mutant could not be attributed to a putative phosphorylation of the tyrosine. Instead, the shift might be due to cleavage of the MA protein at an additional site that is normally inaccessible to the viral protease.
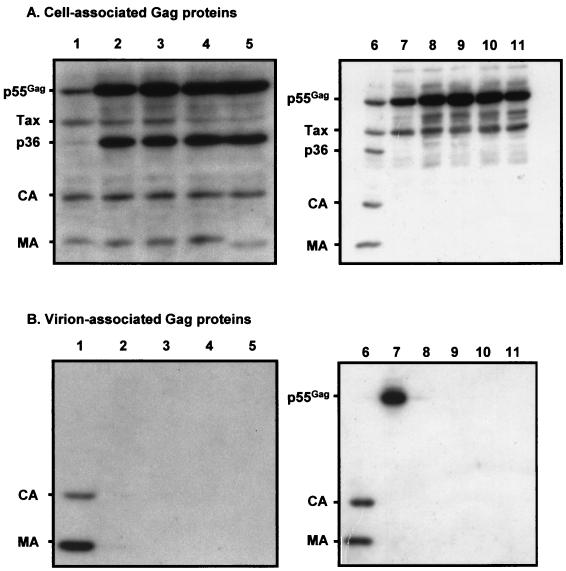
FIG. 2.
Western blots of the HTLV-1 Gag proteins from transfected-cell lysates (A) and virus pellets (B). Lanes 1 and 6, wild-type provirus; lanes 2, Pro118-Leu; lanes 3, Pro119-Leu; lanes 4, Pro120-Leu; lanes 5, Tyr121-Asp; lanes 7, wild-type/PR(−); lanes 8, Pro118-Leu/PR(−); lanes 9, Pro119-Leu/PR(−); lanes 10, Pro120-Leu/PR(−); lanes 11, Tyr121-Asp/PR(−). p55Gag, Gag precursor; Tax, transactivator protein; p36, Gag cleavage intermediate.
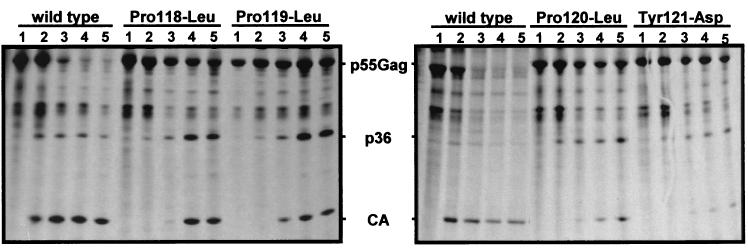
For each of the PPPY mutants, the mature Gag products were present in amounts similar to those obtained with the wild-type provirus. However, the intensities of the bands corresponding to the mutated Gag precursors were greater than that observed with the wild-type provirus (Fig. 2A, compare lanes 2 to 5 to lane 1). The p36 intermediate cleavage product was also more strongly represented in the case of the PPPY mutants than in the wild type. We performed pulse-chase experiments to investigate whether the greater amounts of Gag precursor and p36 could be attributed to a greater stability of these Gag products. Twenty-four hours after transfection of 293-TSA cells with the wild-type or mutated proviral contructs, the cells were starved for 1 h in methionine- and cysteine-free medium and pulse-labeled for 15 min at 37°C with Promix labeling medium (1.5 mCi/ml) containing [35S]cysteine and [35S]methionine (Amersham). After a variable chase period in complete, nonradioactive medium, the cells were lysed and the Gag proteins were immunoprecipitated with a pool of sera from HTLV-1-infected individuals (20) (Fig. 3). Immediately after pulse-labeling, similar amounts of Gag precursor proteins were immunoprecipitated from cells transfected with the wild-type provirus or the four mutants, indicating that similar amounts of these proteins were synthesized (Fig. 3, lanes 1). However, the half-lives of the mutated Gag precursors were longer than that of the wild-type precursor (Fig. 3, compare lanes 5 for the wild type and the mutants). Moreover, while the p36 product appeared after a 1-h chase for the wild type as well as the mutants, the wild-type p36 subsequently diminished, whereas the mutated p36 accumulated. Finally, the CA protein appeared after a 2-h chase in cells transfected with the wild-type provirus, whereas it appeared only after a 4-h chase with the mutated proviruses (Fig. 3, lanes 2 and 3). We conclude that mutation of the HTLV-1 PPPY motif causes a delay in Gag maturation and an accumulation of the Gag precursor within the cells.
FIG. 3.
Accumulation of Gag precursor proteins in cells transfected with PPPY-mutated HTLV-1 proviruses. After a 15-min pulse with radioactive medium, a chase with nonradioactive medium was performed for various amounts of time, and HTLV-1 Gag proteins were immunoprecipitated from transfected-cell lysates. Immunoprecipitates were electrophoresed in SDS-13% polyacrylamide gels and visualized by autoradiography. Lanes 1, no chase; lanes 2, 2-h chase; lanes 3, 4-h chase; lanes 4, 8-h chase; lanes 5, 12-h chase. p55Gag, Gag precursor; p36, Gag cleavage intermediate.
We next tested whether the intracellular accumulation of Gag proteins observed with the PPPY mutants could be related to a defect in virus particle release. Virus pellets were obtained from the transfected-cell supernatants by centrifugation at 3,000 × g for 15 min, filtration of the supernatant through filters with 0.45-μm pores, and centrifugation using an SW41 Beckman rotor at 25,000 rpm for 2 h (20). Western blotting was then performed as described above for cell lysates to compare the amounts of Gag proteins in viral pellets from the supernatants of cells transfected with the wild type and the mutated provirus constructs (Fig. 2B). Virions released from cells transfected with the wild-type provirus were characterized by the presence of mature CA and MA proteins (Fig. 2B, lane 1). In contrast, no viral protein was detected in the supernatants of cells transfected with any of the four PPPY-mutated proviruses (Fig. 2B, lanes 2 to 5). These results indicate that mutation of the HTLV-1 PPPY motif abolishes virus particle production.
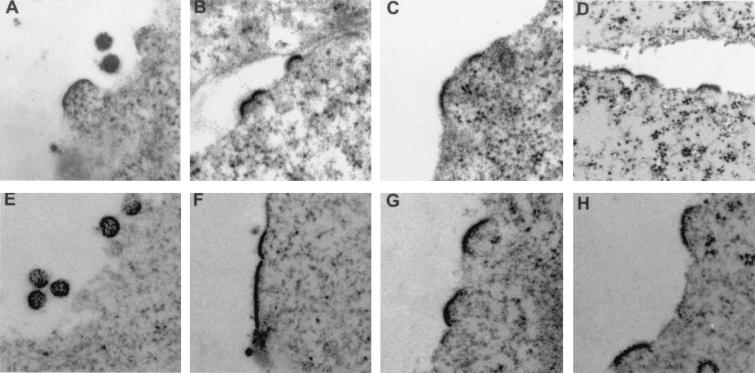
Both the intracellular accumulation of Gag proteins and the abolition of virus release into the supernatant suggested that the PPPY mutants might have a budding defect. We therefore examined their phenotype by electron microscopy. Transfected 293-TSA cells were fixed for 1 h at 4°C in a solution containing 1.6% glutaraldehyde, 1% OsO4, and 0.05 M phosphate buffer, rinsed three times with distilled water, and stained overnight in 0.5% uranyl acetate. The cells were then dehydrated in a graded series of ethanol solutions (from 25 to 100%) before being embedded in epoxy resin at 60°C for 48 h. Ultrathin sections were cut on a Leica Ultratuc UCT microtome and then examined under a JEOL 1200 EX electron microscope (Fig. 4). Virus particles were present in the extracellular medium after transfection of the wild-type provirus construct, and a few buds characterized by curved, electron-dense thickenings underneath the plasma membrane were also visible (Fig. 4A). In contrast, no virus particle could be detected in the extracellular medium of cells transfected with the PPPY mutants. However, electron-dense thickenings of variable size were found underneath the plasma membrane, appearing like budding structures arrested early in the budding process (Fig. 4B to D). Importantly, we never observed any assembled particles attached to the cell surface or to each other via a tether despite careful examination of numerous different cell sections in several independent experiments. This contrasted strongly with what was previously seen for HIV (11), M-PMV (39), or MMLV (40) L domain mutants. Thus, mutation of the HTLV-1 PPPY motif appeared to arrest virus particles at an earlier stage in the budding process than that reported for the mutations of the L domains thus far described in retroviruses.
FIG. 4.
Electron micrographs of 293-TSA cells transfected with wild-type or PPPY mutant HTLV-1 proviruses. (A) Wild-type provirus. (B) Pro119-Leu. (C) Pro120-Leu. (D) Tyr121-Asp. (E) Wild type/PR(−). (F) Pro118-Leu/PR(−). (G) Pro120-Leu/PR(−). (H) Tyr121-Asp/PR(−).
Under some experimental conditions, the budding defect caused by mutation of the HIV-1 PTAP motif can be suppressed by inactivation of the viral protease (14). This prompted us to study the effects of HTLV-1 PPPY mutations in the context of a defective protease. Residues of the putative active site of the HTLV-1 protease (DTG) (21) were replaced by nonconservative amino acids (VLV). Mutagenesis was accomplished by the Kunkel method with the PGEM-CA-PR plasmid, which contains a 1,529-bp NcoI-BglII fragment from the XMT proviral clone [positions 1254 to 2783 in Seiki's sequence (28) inserted into the PGEM-5Zf(+) vector (Promega)]. The mutated pro fragment was then excised and inserted into the wild-type or PPPY mutant XMT plasmids. The mutants were named wild type/PR(−) and X-amino acid position-Y/PR(−), respectively. For simplicity the latter were collectively designated PPPY/PR(−).
As expected for the wild type/PR(−) mutant, processing of the Gag precursor into mature Gag proteins was abolished (Fig. 2A, lane 7). We also verified that inactivation of the HTLV-1 protease did not prevent the release of virus particles into the extracellular medium. Indeed, both the wild type and the wild type/PR(−) mutant produced virus particles, as shown by the presence of Gag proteins in virus pellets from transfected cells; the virus particles contained mature MA and CA proteins (Fig. 2B, lane 6) and uncleaved Gag precursor (Fig. 2B, lane 7), respectively. Moreover, pulse-chase experiments indicated that the kinetics of virus particle release were comparable for the wild type and the wild type/PR(−) mutant (data not shown). We conclude that inactivation of the HTLV-1 protease has no effect on the production of virus particles.
In the context of PPPY mutations the protease mutation resulted in the absence of Gag precursor cleavage, as was seen for wild type/PR(−) (Fig. 2A, lanes 8 to 11). However, an intracellular accumulation of the PPPY/PR(−) Gag precursors was observed, compared to the wild type/PR(−) Gag precursor (Fig. 2A, compare lanes 8 to 11 to lane 7). Pulse-chase experiments indicated that the half-lives of the PPPY/PR(−) Gag precursors were indeed longer than that of the wild type/PR(−) Gag precursor (data not shown). Finally, no viral protein could be detected in the supernatants of cells transfected with the PPPY/PR(−) proviruses (Fig. 2B, lanes 8 to 11). These results demonstrate that the defect in virus production caused by mutation of the HTLV-1 PPPY motif cannot be suppressed by inactivation of the viral protease.
We finally asked whether the morphology of the buds observed in electron microscopy for the PPPY mutants could be related to defects in the Gag maturation process. We therefore examined the phenotype of the PR(−) mutants by electron microscopy. As expected, viral particles were observed in the extracellular medium of cells transfected with the wild type/PR(−) provirus (Fig. 4E). No particle was detected in the case of cells transfected with PPPY/PR(−) proviruses. Again, we observed an accumulation of buds stopped at an early step of budding, characterized by slightly curved, electron-dense thickenings underneath the cell surface (Fig. 4F to H). As was the case for PPPY mutants with an active protease, no particles tethered to the plasma membrane or to each other could ever be visualized. Thus, the stage at which the virus budding arrest caused by HTLV-1 PPPY mutations occurs seems to be independent of Gag maturation by the viral protease.
In this study, we have assessed the effects of individual substitutions of each of the amino acids of the PPPY motif located at the C terminus of the HTLV-1 MA sequence on the production of viral particles. The results indicate that the integrity of the HTLV-1 PPPY motif is absolutely required for virus production, whether the viral protease is active or not. The phenotype of the HTLV-1 PPPY mutants is reminiscent of that conferred by mutations of the L domain in other retroviruses, which have also been shown to impair virus budding (11, 14, 24, 35, 37, 39-41), but it appears to be more drastic, both quantitatively and qualitatively. Indeed, each of the mutations of the HTLV-1 PPPY motif completely abolished the release of virus particles into the extracellular medium. Moreover, the budding of virions off the plasma membrane was blocked at an early stage, characterized by the presence of slightly curved, electron-dense thickenings beneath the cell surface.
The mutations of the HTLV-1 PPPY motif not only abolished virus production but also delayed the process of Gag maturation. It is striking that the mutations introduced so far in the L domains of retroviruses all resulted to various extents in an impairment of Gag processing (11, 14, 35, 37, 39-41). These observations raise the possibility of a causal relationship between the defect in virus budding and the defect in Gag processing, although it is difficult to unravel which is the cause and which is the consequence. On the one hand, the block in virus budding, which causes an abnormal accumulation of Gag proteins beneath the plasma membrane, may indirectly result in a processing defect due to inaccessibility of the protease recognition sites. On the other hand, the delay in Gag processing may disturb the timing of structural rearrangements of the Gag proteins that is likely to be required for driving the budding process in the context of an active viral protease. Interestingly, virus budding was shown to be arrested at an early stage by mutations of the spacer peptide that normally separates the CA and NC domains of the HIV-1 Gag precursor and is liberated during processing (12, 19). Furthermore, mutations that induced premature processing of the RSV Gag precursor also reduced the release of viral particles (38). In the latter case, however, inactivation of the viral protease could rescue virus production, whereas it could not suppress the budding defect caused by mutations of the L domain in RSV (35), M-PMV (39), and, at least under certain experimental conditions, HIV-1 (1). Likewise, our HTLV-1 mutants are incompetent for virus budding whether the viral protease is active or not, suggesting that the PPPY motif is primarily a budding motif. Finally, it is possible that the defects in virus budding and in Gag processing are totally unrelated. Had the mutations not delayed Gag processing, however, one might speculate that the budding defect would have gone unrecognized by electron microscopic analysis. Indeed, the mature Gag proteins, generated prematurely with regard to virus budding, would probably have disassembled, thus preventing visualization of electron-dense budding structures beneath the plasma membrane. With this in mind, future studies designed to define budding motifs in the Gag sequence of retroviruses should initially be conducted in the context of viral protease inactivation.
The mechanism by which the PPPY motif promotes virus budding remains to be established. Recent findings have linked L domain function to components of the ubiquitination pathway and/or the cellular trafficking machinery (9, 17, 23, 25, 27, 30, 32). A plausible hypothesis is that L domains work by usurping the mechanical device that is normally used for endosomal multivesicular body formation, a cellular process that involves membrane bending away from the cytosol and is thus topologically analogous to virus budding (9, 23). While the underlying mechanism needs further characterization, the current model proposes that L domain function is required specifically for the fission event that separates the virion from the cell membrane (8, 9, 34). This assumption originated from the electron microscopy data available on L domain mutants, which all showed assembled particles arrested at the ultimate stage of the budding process (11, 39, 40). It is very likely that, like the L domains described so far, the HTLV-1 PPPY motif mediates Gag interactions with host cell factors. Consistent with this hypothesis, ubiquitination of a chimeric HIV-1 Gag protein was observed when the protein was fused either to the L domain of RSV or to the PPPY-containing domain of HTLV-1 (30). However, the current model of retrovirus budding cannot be directly transposed to HTLV-1. Indeed, our electron microscopy study showed buds arrested at an early stage of the budding process, implying that the HTLV-1 PPPY motif is not, or not only, involved in the fission step.
One explanation for the peculiar phenotype of our HTLV-1 mutants might be that the HTLV-1 PPPY motif drives an earlier step in the budding process than the PPPY motifs of M-PMV and MMLV or the PTAP motif of HIV-1. In other words, retrovirus budding might involve two discrete steps, each requiring a distinct functional domain. Keeping with this hypothesis, it would be interesting to investigate whether the PTAP sequence that is situated just downstream of the PPPY motif in HTLV-1 Gag primary structure mediates the ultimate stage of HTLV-1 budding. However, the exchangeability of all retroviral L domains defined so far, whatever their core motif (1, 22, 24, 40), suggests that the PPPY motif of HTLV-1 is more likely to provide the same function as the L domains. If this is indeed the case, then our electron microscopic observations would suggest that the budding process involves a continuum from early to late stages rather than two discrete steps. As to fission of biological membranes, two principal mechanisms can be distinguished: either the machinery is distinct from that mediating budding or fission is the consequence of the same mechanism that mediates budding (15). The “pinching off” of virus particles might conform to the second model.
Acknowledgments
We thank Frédéric Delebecque for helpful advice and Linda L. Pritchard for critical reading of the manuscript. We also thank Céline Hebras for her technical assistance.
This work was supported by a grant from the Agence Nationale de Recherches sur le SIDA (Paris, France), and I.L.B. received a fellowship from the Fondation pour la Recherche Médicale (Paris, France).
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boulanger, P., and I. Jones. 1996. Use of heterologous expression systems to study retroviral morphogenesis. Curr. Top. Microbiol. Immunol. 214:237-260. [DOI] [PubMed] [Google Scholar]
- 3.Coulston, J., H. Naif, R. Brandon, S. Kumar, S. Khan, R. C. Daniel, and M. F. Lavin. 1990. Molecular cloning and sequencing of an Australian isolate of proviral bovine leukaemia virus DNA: comparison with other isolates. J. Gen. Virol. 71:1737-1746. [DOI] [PubMed] [Google Scholar]
- 4.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, S., and S. P. Goff. 1985. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J. Virol. 53:899-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delchambre, M., D. Gheysen, D. Thines, C. Thiriart, E. Jacobs, E. Verdin, M. Horth, A. Burny, and F. Bex. 1989. The GAG precursor of simian immunodeficiency virus assembles into virus-like particles. EMBO J. 8:2653-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derse, D., J. Mikovits, D. Waters, S. Brining, and F. Ruscetti. 1996. Examining the molecular genetics of HTLV-I with an infectious molecular clone of the virus and permissive cell culture systems. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:1-5. [DOI] [PubMed] [Google Scholar]
- 8.Garnier, L., J. B. Bowzard, and J. W. Wills. 1998. Recent advances and remaining problems in HIV assembly. AIDS 12:S5-S16. [PubMed] [Google Scholar]
- 9.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Côté, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 10.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 11.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harrison, T., F. Graham, and J. Williams. 1977. Host-range mutants of adenovirus type 5 defective for growth in HeLa cells. Virology 77:319-329. [DOI] [PubMed] [Google Scholar]
- 14.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huttner, W. B., and J. Zimmerberg. 2001. Implications of lipid microdomains for membrane curvature, budding and fission. Curr. Opin. Cell Biol. 13:478-484. [DOI] [PubMed] [Google Scholar]
- 16.Katoh, I., Y. Yoshinaka, A. Rein, M. Shibuya, T. Odaka, and S. Oroszlan. 1985. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology 145:280-292. [DOI] [PubMed] [Google Scholar]
- 17.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kräusslich, H.-G., M. Fäcke, A.-M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Blanc, I., A. R. Rosenberg, and M.-C. Dokhélar. 1999. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J. Virol. 73:1860-1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nam, S. H., M. Kidokoro, H. Shida, and M. Hatanaka. 1988. Processing of Gag precursor polyprotein of human T-cell leukemia virus type I by virus-encoded protease. J. Virol. 62:3718-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Puffer, B. A., L. J. Parent, J. W. Wills, and R. C. Montelaro. 1997. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J. Virol. 71:6541-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puffer, B. A., S. C. Watkins, and R. C. Montelaro. 1998. Equine infectious anemia virus Gag polyprotein late domain specifically recruits cellular AP-2 adapter protein complexes during virion assembly. J. Virol. 72:10218-10221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakalian, M., and E. Hunter. 1998. Molecular events in the assembly of retrovirus particles. Adv. Exp. Med. Biol. 440:329-339. [DOI] [PubMed] [Google Scholar]
- 27.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H.-G. Kräusslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with Gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimotohno, K., Y. Takahashi, N. Shimizu, T. Gojobori, D. W. Golde, I. S. Y. Chen, M. Miwa, and T. Sugimura. 1985. Complete nucleotide sequence of an infectious clone of human T-cell leukemia virus type II: an open reading frame for the protease gene. Proc. Natl. Acad. Sci. USA 82:3101-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strack, B., A. Calistri, M. A. Accola, G. Palù, and H. G. Göttlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hugues, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 32.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogt, V. M. 1996. Proteolytic processing and particle maturation. Curr. Top. Microbiol. Immunol. 214:95-131. [DOI] [PubMed] [Google Scholar]
- 34.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wills, J. W., R. C. Craven, and J. A. Achacoso. 1989. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J. Virol. 63:4331-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiang, Y., T. W. Ridky, N. K. Krishna, and J. Leis. 1997. Altered Rous sarcoma virus Gag polyprotein processing and its effects on particle formation. J. Virol. 71:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]