Abstract
Processing of the human immunodeficiency virus type 1 (HIV-1) Gag precursor is highly regulated, with differential rates of cleavage at the five major processing sites to give characteristic processing intermediates. We examined the role of the P1 amino acid in determining the rate of cleavage at each of these five sites by using libraries of mutants generated by site-directed mutagenesis. Between 12 and 17 substitution mutants were tested at each P1 position in Gag, using recombinant HIV-1 protease (PR) in an in vitro processing reaction of radiolabeled Gag substrate. There were three sites in Gag (MA/CA, CA/p2, NC/p1) where one or more substitutions mediated enhanced rates of cleavage, with an enhancement greater than 60-fold in the case of NC/p1. For the other two sites (p2/NC, p1/p6), the wild-type amino acid conferred optimal cleavage. The order of the relative rates of cleavage with the P1 amino acids Tyr, Met, and Leu suggests that processing sites can be placed into two groups and that the two groups are defined by the size of the P1′ amino acid. These results point to a trans effect between the P1 and P1′ amino acids that is likely to be a major determinant of the rate of cleavage at the individual sites and therefore also a determinant of the ordered cleavage of the Gag precursor.
Assembly of virus particles is highly regulated to ensure the formation of a complex structure able to leave the cell, bind to and enter a new cell, and initiate a new round of infection. Most retroviruses use a strategy of assembly in which the viral Gag precursor oligomerizes at the plasma membrane of the cell and buds from the cell. The Gag and Gag-Pro-Pol polyprotein precursors that make up the bulk of the virion proteins within the virus envelope are cleaved during virion maturation by the viral protease, which is encoded in the pro gene and initially embedded in the Gag-Pro-Pol precursor (50). The timing of the initiation of these cleavages is regulated such that the cleavage products remain in the virus particle, even though processing appears to initiate while these polyprotein precursors are still cell associated (20, 21, 23, 31).
The human immunodeficiency virus type 1 (HIV-1) Gag precursor is cleaved at least five times to generate a series of mature protein products (16, 17). There is an absolute requirement for cleavage at each of these sites, since substitutions at the cleavage site that block processing invariably lead to the production of aberrant and noninfectious virus particles (36, 58). Cleavage of the Gag precursor at these multiple sites also appears to be highly regulated. Peptide substrates of these different sites are cleaved by the viral protease at different rates, and cleavage of the intact Gag precursor in vitro goes through a distinct series of intermediates that are generated due to different rates of cleavage at the various sites (4, 7, 9, 24, 30, 36, 38, 43, 52). The pattern of intermediates seen in infected cells and in recently budded virus particles is consistent with the ordered cleavage of Gag that has been observed in vitro (11, 12, 29, 36, 56, 58). In the in vitro studies the initial site of cleavage occurs at the site that defines the p2/NC boundary to generate N-terminal and C-terminal intermediates of Gag (9, 24, 36). These intermediates are cleaved at an approximately 10-fold-lower rate, at the MA/CA site in the N-terminal intermediate and at the p1/p6 site in the C-terminal intermediate (36). Finally, C-terminal spacer peptides are trimmed from both the CA and NC proteins at the CA/p2 site and the NC/p1 site in cleavage reactions that are up to several hundred-fold slower than the initial cleavage (36). While the order of these cleavages can be reproduced with peptide and Gag substrates in vitro and a similar order can be inferred in vivo, the absolute rates of cleavage from each of these systems can vary or is not known.
Investigation of the characteristics of the sequences at protease cleavage sites has been an area of interest for understanding the nature of protease-substrate interaction and the regulation of Gag processing and as a starting point for the design of inhibitors of the HIV-1 protease. The amino acids flanking the target scissile bond are generally hydrophobic (16, 33, 37). A number of studies have used peptide or Gag substrates in an attempt to define the role of specific amino acids at different positions flanking the scissile bond as they impact the rate of cleavage by the viral protease (3, 5, 13, 14, 19, 30, 34, 36, 38, 44, 45, 51, 53-55; recently reviewed in reference 27). In the present study we have examined the ability of approximately 15 different amino acids in the P1 position of the five different Gag cleavage sites to support cleavage by the HIV-1 protease (the P1 position is the amino acid immediately upstream of the scissile bond, and the P1′ position is the amino acid immediately downstream of the scissile bond). We used full-length Gag as a substrate for cleavage with recombinant protease. This system allowed us to examine the relative rates of cleavage under conditions that were more physiological than those typically used to detect the cleavage of peptides (which include the use of low pH and high salt concentrations). We confirmed the requirement for a hydrophobic amino acid at the P1 position and found that certain of the substituted amino acids are able to accelerate the rate of cleavage at certain sites, indicating that the presence of suboptimal amino acids is responsible for some of the differences observed in the rates of processing of the various Gag sites. In addition, we were able to classify processing sites into two groups, defined by the size of the amino acid in the P1′ position, which varied in the order of cleavage efficiency of Met, Leu, and Tyr to function as a P1 amino acid.
MATERIALS AND METHODS
Plasmid construction and mutagenesis, and transfection.
The phagemid pGagS was described previously (36) and contains the 5′ noncoding region and gag gene from HIV-1 molecular clone HXB (accession no. NC001802) (42), with the inserted sequences placed between the XbaI and SalI sites of pIBI20 (US Biochemicals). HXB sequences extend from the NarI site at base 182 to the BclI site at base 1974.
The infectious molecular clone, pCadN, is a recombinant between the HXB infectious clone (42) and the NL4-3 infectious clone (1) (accession no. M19921). To create pCadN, a translationally silent unique XbaI site was created in the MA domain of both clones by a single T-to-C transition (HXB base 683; NL4-3 base 1029) using site-directed mutagenesis (2, 25, 36). In pCadN, HXB sequences replaced NL4-3 sequences from the unique XbaI site to the EcoRV site in pol (HXB base 2522; NL4-3 base 2977). The remainder of the viral sequences are derived from NL4-3.
Site-directed mutagenesis, and in vitro transcription and translation.
Mutagenesis of the codon encoding the P1 amino acid of the five major Gag processing sites in pGagS was performed as previously described using uracil-substituted single-stranded templates (2, 25, 36). Individual oligonucleotides 30 nucleotides in length and degenerate for the P1 codon were used during site-directed mutagenesis to generate a library of random substitution mutations for each site. Mutations were identified by DNA sequencing with modified T7 DNA polymerase (U.S. Biotechnologies).
pGagS, linearized with SalI, was used in in vitro transcription with T7 RNA polymerase (Promega) to produce capped mRNA transcripts of gag as described previously (36). Translation of the synthetic mRNA in a rabbit reticulocyte lysate (Promega) to generate radiolabeled Gag protein was performed as described previously (36) with 50-μl reaction mixtures containing 50 μCi of [35S]cysteine (1,000 Ci/mM) (DuPont NEN). The completed translations were either used immediately or stored briefly at −20°C prior to use.
In vitro assay for the proteolytic processing of Gag.
The Gag precursor was processed in 50-μl reaction mixtures containing 5 μl of the Gag-containing rabbit reticulocyte lysate and 0.2 to 5 μg of purified recombinant protease in phosphate-buffered saline at pH 7.0 (0.8% [wt/vol] NaCl, 0.02% KCl, 0.012% KH2PO4, 0.091% Na2HPO4). Purified protease was prepared as described previously (36). The amount of added protease was adjusted for the particular site examined so that nearly complete cleavage of the wild-type sequence at that site occurred roughly 1 h into the time course of the reaction. Reactions were performed for 6 h at 30°C. At various times, 5-μl aliquots were removed and the reaction was stopped by the addition of an equal volume of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (26). Products of the processing reaction were heated to 100°C prior to separation by electrophoresis in a 17% polyacrylamide gel with a Tris-glycine buffer (46). After electrophoresis, gels were fixed in 10% acetic acid and dried followed by autoradiography and phosphorimaging.
Densitometic analysis of Gag processing reactions and calculation of the relative rates of cleavage.
Relative quantitation of Gag precursor and products was performed by densitometric analysis using a Molecular Dynamics 840 Storm PhosphorImager. The relative rates of cleavage between P1-mutated Gag substrates for a particular site were estimated as described previously (36, 49). The estimate of the relative amount of each protein species was first corrected for the number of cysteine residues available to label in that species. The amount of uncleaved substrate for a particular mutation was determined as the total sum of precursors and intermediates containing the uncleaved site; reduced processing at some sites resulted in the appearance of new intermediates, whose identities were inferred based on size (36). In some instances the amounts of some intermediates could not be determined because of unincorporated radioactivity in the range of 7 to 10 kDa present in gels or because of secondary cleavage at minor sites within the precursor. In these cases, the indiscernible intermediates were not included in the calculation of the total uncleaved substrate. In all cases, total inhibition of cleavage at a site was judged by the absence of the final processed product. The calculations of the relative rates of cleavage were performed as described previously (36) from semilog (first-order) plots. The slope of the line derived from points in the midcourse of cleavage (20 to 80% cleavage) was used as a numerical value of relative rate.
Construction of infectious HIV-1 DNA clones, transfection, and Western blot analysis of released virions.
DNA fragments containing mutations at the NC/p1 site were removed from pGagS by digestion with ApaI and BclI and used to replace the same fragment in pCadN. A 20-μg aliquot of purified pCadN was transfected into an 80% confluent monolayer of HeLa cells in a 100-mm-diameter plate by the CaCl2 procedure described by Chen and Okayama (6). At 48 h posttransfection, the medium was collected and clarified by low-speed centrifugation (5,000 × g for 10 min). The relative amount of released virus in the medium was determined using a reverse transcriptase assay (32). Similar amounts of virus (from approximately 10 ml of medium) were pelleted by centrifugation at 100,000 × g for 90 min. After removal of the supernatant, the pellet was resuspended in 100 μl of SDS-PAGE buffer and heated to 100°C prior to Western blot analysis.
A 10-μl portion of the resuspended virion protein solution was electrophoresed in a 12% polyacrylamide-Tris-tricine gel (46) and then subjected to western blot analysis (36). After transfer to a polyvinylidene difluoride membrane (Schleicher & Schuell), blots were blocked with 1% bovine serum albumin in Tris-buffered saline-0.1% Tween 20 and then exposed to a 1:1,000 dilution of anti-nucleocapsid polyclonal primary antibody from rabbit in phosphate-buffered saline (48, 49). The colormetric signal was developed as described previously (35, 36).
RESULTS
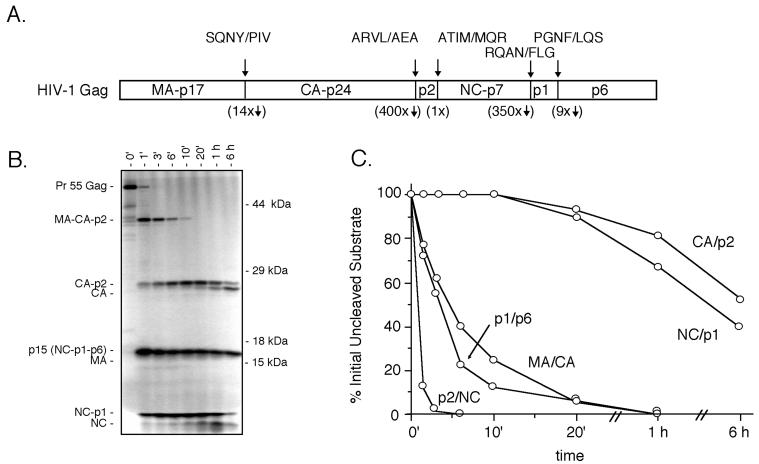
There are five major cleavage sites in the HIV-1 Gag precursor (Fig. 1A). Cleavage of the Gag precursor in vitro with the viral protease gives rise to a series of distinct intermediates (Fig. 1B). These intermediates arise because the various cleavage sites are cleaved at roughly three distinct rates (Fig. 1C), with the initial cleavage occurring at the p2/NC site to generate a 41-kDa intermediate (MA-CA-p2) and the p15 intermediate (NC-p1-p6). Cleavage of these two intermediates follows, at the MA/CA site and the p1/p6 site and at a rate approximately 10-fold lower than the initial cleavage. Finally, the two C-terminal spacer peptides, at the CA/p2 boundary and the NC/p1 boundary, are cleaved at a rate approximately 400-fold slower than the initial cleavage at the p2/NC site. We explored the potential role of the P1 amino acid (i.e., the amino acid immediately upstream of the scissile bond and generating the C-terminal carboxylate after cleavage of the peptide bond) in regulating the rate of cleavage at these sites. The position of the codon encoding the P1 amino acid was randomly mutagenized in the gag expression vector pGagS (36), and individual clones were screened by sequence analysis to identify a majority of the possible substitution mutations at each site. Between 12 and 17 of the 19 possible substitution mutants at each of the P1 positions at the five major cleavage sites were identified. In this system it is possible to determine if a substitution enhances the rate of cleavage, and the sensitivity for detecting reduced rates of cleavage extends to the range of a 20- to 30-fold reduction in rate relative to the wild-type site. Cleavage that occurs at a rate lower than 20- to 30-fold that of the wild-type site would not be detected and is reported as not cleaved.
FIG. 1.
Processing of the HIV-1 Gag precursor in vitro. (A) Schematic representation of the Pr55 Gag precursor with the five major processing sites shown as vertical lines. The sequence of the individual sites from the P4 through P3′ is shown above. The rates of cleavage of the five sites relative to the initially cleaved p2/NC site is shown below, as determined by cleavage in vitro with recombinant protease (36). (B) Representative SDS-PAGE gel showing ordered processing in vitro of the wild-type Gag precursor as a function of time in the presence of recombinant HIV-1 protease. The positions of the Gag precursor, processing intermediates, and mature products are shown on the left. The positions of molecular mass markers are shown on the right. (C) Plot of the time course of cleavage of the five processing sites as derived from densitometric analysis of gels of the processing reaction. The plot is derived from the summation of the percent initially uncleaved substrate containing each individual processing site over time.
gag mRNA was generated from each of the mutagenized plasmids in the pGagS backbone by using an in vitro transcription system. The mRNA was translated in vitro in the presence of [35S]cysteine to generate a radiolabeled Gag substrate. The translation product was incubated for various periods with recombinant HIV-1 protease, and the cleavage products were resolved by PAGE under reducing and denaturing conditions. The relative rate of cleavage at each of the major Gag cleavage sites was monitored as a function of time, and the different cleavage products were quantified by PhosphorImager analysis. The relative rate of cleavage at each mutated site was determined from the percentage of the total precursors and intermediates containing the uncleaved site under study, as described in the Materials and Methods.
Mutagenesis of the P1 position of the p2/NC cleavage site.
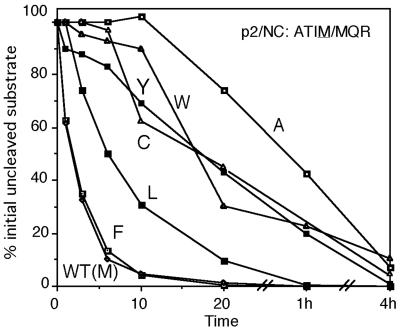
Each of 16 P1 substitution mutants was analyzed in parallel reactions for the relative rate of cleavage at the p2/NC site (ATIM/MQR). These results are shown in Fig. 2 and summarized in Table 1. The mutants fall into five categories with respect to their effect on cleavage at the p2/NC site: Phe was cleaved at a rate equivalent to the wild type (Met); Leu was cleaved at a rate estimated to be approximately three times lower; Cys, Trp, and Tyr were cleaved approximately 10 times slower than the wild type; and Ala was cleaved 22 times slower than the wild type. The remaining amino acids did not support any detectable cleavage (with the exception of His, Lys, and Asn, which were not tested).
FIG. 2.
Effect of substitutions of the P1 amino acid on the cleavage of the p2/NC site in vitro. The plot shows the percentage of initially uncleaved p2/NC site (as precursor and intermediates) over the time course of the assay. Only substitutions that allowed some degree of cleavage are shown in the plot.
TABLE 1.
Activity of P1 substitution mutants for cleavage by the HIV-1 protease
| Cleavage site (wild-type P1) | Relative activity of P1 amino acid mutanta:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | C | D | E | F | G | H | I | K | L | M | N | P | Q | R | S | T | V | W | Y | |
| MA/CA (Y) | (−)b | 0.05 | (−) | (−) | 1.5 | (−) | (−) | (−) | (−) | 0.1 | 0.25 | NDc | (−) | (−) | (−) | (−) | (−) | (−) | ND | 1 |
| CA/p2 (L) | (−) | 0.35 | (−) | ND | 2 | (−) | (−) | (−) | ND | 1 | ND | (−) | (−) | ND | ND | (−) | (−) | (−) | (−) | 2 |
| p2/NC (M) | 0.0 | 0.15 | (−) | (−) | 1 | (−) | ND | (−) | ND | 0.35 | 1 | ND | (−) | (−) | (−) | (−) | (−) | (−) | 0.1 | 0.1 |
| NC/p1 (N) | 0.5 | 3 | ND | ND | >60 | 0.05 | ND | 0.1 | (−) | 30 | >60 | 1 | ND | (−) | ND | (−) | ND | 0.15 | ND | 20 |
| p1/p6 (F) | (−) | (−) | (−) | (−) | 1 | (−) | (−) | (−) | (−) | (−) | 0.15 | (−) | (−) | ND | ND | (−) | ND | ND | (−) | ND |
Rates are estimated from the slope of the line describing the rate of loss of the processing intermediates with that site relative to the rate of cleavage of the wild-type site.
(−), not cleaved at a rate at least 3 to 5% of the wild-type rate.
ND, not done (this mutation was not recovered in the library of random P1 mutations).
Mutagenesis of the P1 position of the MA/CA cleavage site.
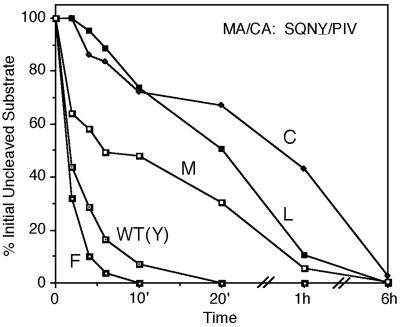
A total of 17 substitution mutations of the wild-type Tyr were tested at the P1 position of the MA/CA cleavage site (SQNY/PIV; Asn and Trp were not tested). Only four amino acids other than the wild type supported cleavage (Fig. 3; Table 1). A Phe substitution accelerated cleavage at this site nearly 2-fold, while Met, Leu, and Cys substitutions were cleaved between 4- and 14-fold slower.
FIG. 3.
Effect of substitutions of the P1 amino acid on the cleavage of the MA/CA site in vitro. The plot shows the amount of initially uncleaved MA/CA processing site over the time course of the assay. Only substitutions that allowed some degree of cleavage are shown in the plot.
Mutagenesis of the P1 position of the p1/p6 cleavage site.
Only one of the 14 substitutions tested to replace the wild-type Phe supported cleavage at this site (PGNF/LQS); a Met substitution supported cleavage at a rate approximately sixfold lower than the wild type (Table 1). The other 13 amino acid substitutions tested did not support measurable cleavage (Gln, Arg, Thr, Val, and Tyr were not tested).
Mutagenesis of the P1 position of the CA/p2 cleavage site.
Of the 14 amino acid substitutions tested 2 supported cleavage of the CA/p2 site (ARVL/AEA) faster than the wild-type Leu, with both Phe and Tyr showing accelerated cleavage at approximately a twofold higher rate (Table 1). Cys was the only other substitution that supported cleavage but at a rate threefold lower than the wild type (Glu, Lys, Met, Gln, and Arg, were not tested). Using peptide substrates, Ridky et al. (45) reported an increase in the rate of cleavage with a Phe but a slight decrease with a P1 Tyr at this site.
Mutagenesis of the P1 position of the NC/p1 cleavage site.
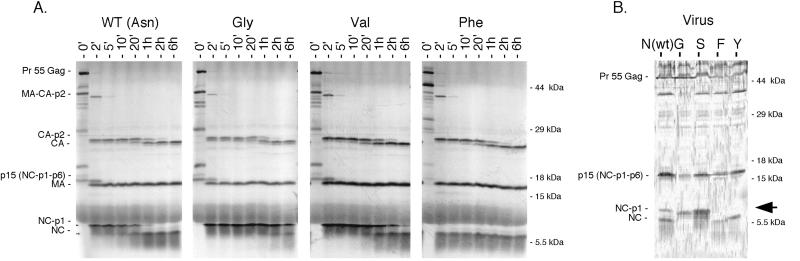
A total of 12 substitutions were tested in place of the wild-type Asn at the P1 position of the NC/p1 cleavage site (RQAN/FLG; Asp, Glu, His, Pro, Arg, Thr, and Trp were not tested). Surprisingly, only 3 amino acid substitutions failed to permit cleavage, while 9 of the 12 substitutions tested gave detectable cleavage. Furthermore, the wild-type Asn supported cleavage poorly relative to five of the substitution mutants, with Phe and Met accelerating cleavage by at least 60-fold (Table 1) and Leu, Tyr, and Cys all accelerating cleavage 30- to 3-fold. Finally, even amino acids with beta-branched side chains (Val and Ile) were able to support a low level of cleavage (Table 1). Examples of the patterns of Gag cleavage are shown in Fig. 4A for a subset of NC/p1 mutants, with examples of a Gly substitution blocking cleavage, a Phe substitution accelerating cleavage, and a Val substitution slowing cleavage. Note that these different rates of NC/p1 processing were seen while the rates of cleavage of other sites in Gag were similar (e.g., compare CA/p2 cleavage). The wide range and rates of cleavage of the substitution mutants with mutations at the NC/p1 cleavage site contrasts with the restricted range of allowed substitutions at CA/p2, even though these two sites have similar rates of cleavage for the wild-type sequences.
FIG. 4.
Cleavage of the NC/p1 site in vitro and in vivo. (A) Time courses of cleavage in vitro of the wild type (WT) and P1 mutants with mutations of the NC/p1 site. Gag processing intermediates (including NC-p1) are shown on the left, as well as the mature products (including NC). The P1 substitution mutant is noted above the relevant gel. (B) Extent of processing of NC-containing Gag products in released virus with P1 substitutions at the NC/p1 site. Virion-associated precursors, intermediates, and products containing NC were detected with a specific antibody reagent in a Western analysis as described in Material and Methods. The compositions of NC-containing precursors and products are shown on the left, and the positions of molecular mass markers are shown on the right. The P1 substitution mutation of the NC/p1 cleavage site is shown at the top of the relevant lane.
More complete cleavage of mutant NC/p1 cleavage sites in the virus.
We examined the potential of selected mutants of the NC/p1 cleavage site to alter the extent of cleavage of the NC/p1 intermediate in the assembled virus. HeLa cells were transfected with a full-length HIV-1 genome or with a version of this genome encoding an NC/p1 cleavage site with a substitution at the P1 position. The Gly, Ser, Phe, and Tyr substitution mutants were tested in this system as examples of substitutions that mediated cleavage over a wide range of rates. Supernatant virus that was produced after the transfection was collected, concentrated by centrifugation, and analyzed by SDS-PAGE followed by Western analysis with an antibody specific for NC. In this system the wild-type virions displayed the expected processing products (including mature NC) as well as several processing intermediates including the p15 (NC-p1-p6) and NC-p1 intermediates (Fig. 4B). The Ser substitution blocked the processing of NC/p1 since no mature NC protein was present in virus particles; similarly, this Ser substitution blocked cleavage at this site in vitro (Table 1). Gly, which supported only a low level of processing in vitro (Fig. 4A), blocked the processing of NC/p1 as seen by the absence of mature NC in virus particles (Fig. 4B). In contrast, both Phe and Tyr, each of which accelerated the rate of processing in vitro (Table 1, Fig. 4A), supported more extensive cleavage of NC/p1 in vivo compared to the wild type (Fig. 4B). Thus, for this site the in vitro results were consistent with the effect on processing as inferred from the protein composition of virus particles.
In an initial analysis of the effect of these mutations on virus infectivity, we have placed several of these mutations into a full-length infectious clone. Using a single-cycle infectivity assay, we found that the Tyr and Phe substitutions were compatible with a high level of infectivity while the Ser substitution supported only a very low level of infectivity (data not shown). Thus, while limiting cleavage at this site severly affected infectivity, mutations that strongly accelerated the rate of cleavage in vitro were not significantly deleterious to infectivity.
DISCUSSION
In these studies we have characterized the role of the P1 amino acid in determining the rate of cleavage by the HIV-1 protease at the five predominant processing sites in the HIV-1 Gag precursor. Approximately 15 different amino acids were tested at each of these sites for their ability to support cleavage by the viral protease. These analyses were carried out using full-length Gag precursor as substrate with more physiological reaction conditions of neutral pH and low salt concentration rather than the conditions of low pH and high salt concentration that are typically used in the analysis of cleavage of peptide substrates. Our results confirmed previous observations that the P1 position prefers a hydrophobic and non-beta-branched amino acid. In addition, we were able to add sufficient quantitation to these studies to be able to draw several unexpected conclusions.
One surprising outcome of these studies is the observation that for three of the five sites the rate of cleavage could be improved with the introduction of specific alternative amino acids at the P1 position (summarized in Fig. 5). Only the p2/NC site and the p1/p6 site could not be improved. This observation provides clear evidence that the rate of cleavage at these sites, and perhaps their order of cleavage, is regulated by the use of suboptimal amino acids in the cleavage sites. Using peptide substrates, Ridky et al. (45) observed a 42% increase in the rate of cleavage at the CA/p2 site with a P1 Phe in place of the wild-type Leu, although they observed a decrease in the rate of cleavage with a P1 Tyr at this site. Tritch et al. (55) observed an increase in the rate of cleavage of the MA/CA site with a P1 Phe in place of Tyr, similar to our observation (Fig. 3), and Partin et al. (34) observed that Phe readily substituted for Tyr at this site. Both increased and decreased rates of cleavage for a P1 Phe have been reported for equivalent peptide substrates of the MA/CA site (3, 54).
FIG. 5.
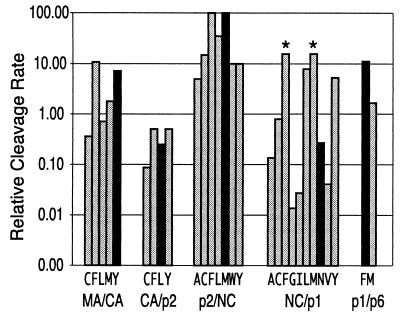
Summary of the activity of Gag P1 processing site mutations. The values presented in Table 1 were corrected for the rate of cleavage of the individual sites relative to each other, as described in the legend to Fig. 1 and reference 36. After correcting for the relative rates, all rates were multiplied by 100 so that the rate of the first site cleaved (p2/NC) has a value of 100. For each site, the rate of cleavage of the wild-type sequence is shown by the black bar. The individual sites and the substitutions that supported detectable cleavage are shown at the bottom of the graph. The asterisks indicate that the actual rate was greater than the rate shown but by an unknown amount.
There are constraints that may dictate the use of some suboptimal amino acids. The P1′ Pro in the MA/CA site is required to participate in a specific interaction that is unique to the processed form of CA and an integral part of virion maturation (10, 57). Pro is the P1′ amino acid in several processing sites, although this distinctive requirement for Pro after cleavage is known only for CA, and this is a conserved feature of the upstream cleavage site of CA among retroviruses (37, 57). It may be that the requirement for Pro in this postcleavage interaction provides the selective pressure to be able to accommodate Pro in the P1′ site of the protease target sequence, defining a maximum rate of cleavage even with an optimal P1 amino acid. The P1 amino acid Asn of the NC/p1 site contributes its third codon position U to the slippery sequence that directs the −1 frameshifting event that allows expression of the Gag-Pro-Pol precursor (18, 59). The potential importance of this P1 amino acid (or its underlying coding sequence) can be seen in the fact that a compensatory mutation to increase the rate of cleavage at this site in the presence of a protease inhibitor-resistant protease occurs through a change in the P2 position (Ala to Val) (8, 28, 61). Introduction into a virus (with a wild-type protease) of the P2 position Val at the NC/p1 site has a small negative effect on infectivity (8, 28). However, the NC/p1 site does not appear to be overly sensitive to up regulation since a Phe or Tyr substitution at the P1 site retains significant infectivity (data not shown).
The other surprising conclusion derived from these experiments is that the processing sites appear to fall into two groups, based on the pattern of activity associated with a specific subset of P1 amino acids: Phe, Met, Tyr, and Leu. In group 1 sites (p2/NC and NC/p1), the aliphatic amino acids Met and Leu confer activity that is more similar to Phe, while Tyr is the least active of this group of amino acids. In contrast, the group 2 sites (MA/CA and CA/p2) display activity with Tyr similar to that with Phe, while Met and Leu show lesser activity. For the p1/p6 site, Met is significantly less active than Phe, with Leu being inactive; Tyr was not tested at this site, but overall the site appears to follow the group 2 pattern (Fig. 5).
An examination of the amino acids flanking the P1 position in the cleavage sites suggests an explanation for the existence of these two groups of sites. In group 1 sites (P1 Phe/Met/Leu), the P1′ amino acid is large, either Phe or Met. In the group 2 sites (P1 Phe/Tyr), the P1′ amino acid is smaller (Pro, Ala, and Leu). Given that the substrate cleavage site assumes a beta-sheet-like conformation in the active site of the protease (60), this linkage suggests a significant trans interaction between the P1 and P1′ amino acids flanking the scissile bond. The trans interaction between P1 and P1′ appears to provide a better explanation of the data than the trans interaction between P1 and P2 or the cis interactions between P1 and P3 or P1 and P2′. In some HIV-1 isolates, the P1′ Leu is replaced with Pro in the p1/p6 site, maintaining an amino acid in the small group (group 2). In contrast, increases in the rate of processing of the p1/p6 site in the presence of a protease inhibitor-resistant protease are effected by a P1′ change of Leu to Phe (8, 61), introducing a P1′ amino acid from the large group (group 1).
The issue of cis and trans interactions has been addressed with the HIV-1 protease by Ridky et al. (44), who examined the effects of single and double substitutions in peptide substrates representing the Rous sarcoma virus NC/PR cleavage site (PAVS/LAM). They noted that a P1 Trp or a P1 Leu improved the activity of a substrate with a P1′ Ala beyond what was expected and suggested that the large P1 amino acid positioned the small Ala side chain deeper in the S1′ subsite. We considered a similar structural model to explain the observed trans effect inferred from our studies. However, in comparing the crystal structures of the protease with six peptide substrates (representing the cleavage sites at MA/CA, CA/p2, p2/NC, p1/p6, RT/RH, and RH/IN) (40, 41), we found no evidence for the physical displacement of either the P1 or the P1′ amino acid side chain due to a trans effect. As an alternative model, we have considered the possibility that the sizes of the P1 and P1′ amino acids are somehow additive and can be too large for optimal cleavage. Thus, a P1 Tyr would be most active with small P1′ amino acids and less active with Tyr or Phe in the P1′ position. We previously observed that aliphatic amino acids at the P1′ position of the MA/CA cleavage site (with a P1 Tyr) were more active than Phe or Tyr (22), although other studies using similar peptide substrates have not observed this rank order (3, 54). Ridky et al. (45) reported that aliphatic amino acids and Phe, when used instead of the P1′ Ala in the CA/p2 site, enhanced the rate of cleavage. In the substrate-PR crystal structures, Tyr makes a significant number of contacts in either the P1 or P1′ position, and it is also able, through its side chain hydroxyl, to make additional contacts with solvent, perhaps enhancing its binding energy. The additive effects of the P1 and P1′ amino acids in defining the rate of cleavage and in passing through an optimum are consistent with the data obtained in this study (Table 1).
Hydrophobic beta-branched amino acids are never found in naturally occurring protease cleavage sites (37, 39), and a structural reason for their exclusion has been proposed (54). In contrast, a site with a P1 Ile substitution of the RSV NC/PR is cleaved by the Rous sarcoma virus protease (47). We have now observed a low level of cleavage by the HIV-1 protease at the NC/p1 site with either Val or Ile at the P1 position (Table 1; Fig. 5). We did not see cleavage at the CA/p2 site with either Val or Ile, even though the rates of cleavage at the NC/p1 and CA/p2 wild-type sequences are similar. This difference may be due to the large P1′ amino acid in the NC/p1 site contributing to an enhanced rate of cleavage with suboptimal P1 amino acids. Thus, we confirm that the protease can cleave a peptide bond with a beta-branched P1 amino acid, although in our case we did not analyze the position of the cleaved site by protein sequencing.
In summary, our results point to significant trans interactions between the P1 and P1′ side chains of HIV-1 protease substrates in determining the rate of cleavage. Phe appears best suited to productively interacting with the S1 subsite and perhaps the S1′ subsite. Met and Leu are more active when an amino acid with a larger P1′ side chain occupies the S1′ subsite, and Tyr is more active with amino acids with smaller P1′ side chains occupying the S1′ subsite. These results suggest a basis for two groupings of the diverse cleavage site sequences as an alternative to older classification schemes that had focused largely on the presence or absence of a P1′ Pro (13, 15, 37).
Acknowledgments
This work was supported by NIH grant R01-Al25321 to R.S. and American Cancer Society grant RPG-99-213-01-MBC to C.A.S. In addition, this work was supported by the UNC Center for AIDS Research (NIH grant P30-Al50410). G.J.H. was supported by NIH training grant T32 Al07419.
We thank Moses M. Prabu-Jeyablan for assistance with the structural analysis and Noah Hoffman for assistance with the figures.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bebenek, K., and T. A. Kunkel. 1989. The use of native T7 DNA polymerase for site-directed mutagenesis. Nucleic Acids Res. 17:5408.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Billich, A., and G. Winkler. 1991. Analysis of subsite preferences of HIV-1 proteinase using MA/CA junction peptides substituted at the P3-P1′ positions. Arch. Biochem. Biophys. 290:186-190. [DOI] [PubMed] [Google Scholar]
- 4.Billich, S., M. T. Knoop, J. Hansen, P. Strop, J. Sedlacek, R. Mertz, and K. Moelling. 1988. Synthetic peptides as substrates and inhibitors of human immune deficiency virus-1 protease. J. Biol. Chem. 263:17905-17908. [PubMed] [Google Scholar]
- 5.Cameron, C. E., B. Grinde, P. Jacques, J. Jentoft, J. Leis, A. Wlodawer, and I. T. Weber. 1993. Comparison of the substrate-binding pockets of the Rous sarcoma virus and human immunodeficiency virus type 1 proteases. J. Biol. Chem. 268:11711-11720. [PubMed] [Google Scholar]
- 6.Chen, C., and H. Okayama. 1987. High efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7:2754-2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darke, P. L., R. F. Nutt, S. F. Brady, V. M. Garsky, T. M. Ciccarone, C. T. Leu, P. K. Lumma, R. M. Freidinger, D. F. Veber, and I. S. Sigal. 1988. HIV-1 protease specificity of peptide cleavage is sufficient for processing of gag and pol polyproteins. Biochem. Biophys. Res. Commun. 156:297-303. [DOI] [PubMed] [Google Scholar]
- 8.Doyon, L., G. Croteau, D. Thibeault, F. Poulin, L. Pilote, and D. Lamarre. 1996. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J. Virol. 70:3763-3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erickson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retroviruses 5:577-591. [DOI] [PubMed] [Google Scholar]
- 10.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 11.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of the human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gowda, S., B. Stein, and E. Engleman. 1989. Identification of protein intermediates in the processing of the p55 HIV-1 Gag precursor in cells infected with recombinant vaccinia virus. J. Biol. Chem. 264:8459-8462. [PubMed] [Google Scholar]
- 13.Griffiths, J. T., L. H. Phylip, J. Konvalinka, P. Strop, A. Gustchina, A. Wlodawer, R. J. Davenport, R. Briggs, B. M. Dunn, and J. Kay. 1992. Different requirements for productive interaction between the active site of HIV-1 proteinase and substrates containing -hydrophobic∗hydrophobic- or -aromatic∗pro- cleavage sites. Biochemistry 31:5193-5200. [DOI] [PubMed] [Google Scholar]
- 14.Grinde, B., C. E. Cameron, J. Leis, I. T. Weber, A. Wlodawer, H. Burstein, and A. M. Skalka. 1992. Analysis of substrate interactions of the Rous sarcoma virus wild type and mutant proteases and human immunodeficiency virus-1 protease using a set of systematically altered peptide substrates. J. Biol. Chem. 267:9491-9498. [PubMed] [Google Scholar]
- 15.Henderson, L. E., R. E. Benveniste, R. Sowder, T. D. Copeland, A. M. Schultz, and S. Oroszlan. 1988. Molecular characterization of gag proteins from simian immunodeficiency virus (SIVMne). J. Virol. 62:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, L. E., M. A. Bowers, R. d. Sowder, S. A. Serabyn, D. G. Johnson, J. J. Bess, L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson, L. E., R. C. Sowder, T. D. Copeland, S. Oroszlan, and R. E. Benveniste. 1990. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 19:411-419. [PubMed] [Google Scholar]
- 18.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 19.Jupp, R. A., L. H. Phylip, J. S. Mills, F. J. Stuart, S. F. Le Grice, and J. Kay. 1991. Mutating P2 and P1 residues at cleavage junctions in the HIV-1 pol polyprotein. Effects on hydrolysis by HIV-1 proteinase. FEBS Lett. 283:180-184. [DOI] [PubMed] [Google Scholar]
- 20.Kaplan, A. H., M. Manchester, and R. Swanstrom. 1994. The activity of the protease of HIV-1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J. Virol. 68:6782-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 22.Kassel, D. B., M. D. Green, R. S. Wehbie, R. Swanstrom, and J. Berman. 1995. HIV-1 protease specificity derived from a complex mixture of synthetic substrates. Anal. Biochem. 228:259-266. [DOI] [PubMed] [Google Scholar]
- 23.Krausslich, H. G. 1992. Specific inhibitor of human immunodeficiency virus proteinase prevents the cytotoxic effects of a single-chain proteinase dimer and restores particle formation. J. Virol. 66:567-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krausslich, H. G., R. H. Ingraham, M. T. Skoog, E. Wimmer, P. V. Pallai, and C. A. Carter. 1989. Activity of purified biosynthetic proteinase of human immunodeficiency virus on natural substrates and synthetic peptides. Proc. Natl. Acad. Sci. USA 86:807-811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kunkel, T. A., K. Bebenek, and J. McClary. 1991. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 204:125-139. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacterophage T4. Nature 277:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Louis, J. M., I. T. Weber, J. Tozser, G. M. Clore, and A. M. Gronenborn. 2000. HIV-1 protease: maturation, enzyme specificity, and drug resistance. Adv. Pharmacol. 49:111-146. [DOI] [PubMed] [Google Scholar]
- 28.Mammano, F., C. Petit, and F. Clavel. 1998. Resistance-associated loss of viral fitness in human immunodeficiency virus type 1: phenotypic analysis of protease and gag coevolution in protease inhibitor-treated patients. J. Virol. 72:7632-7637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mervis, R. J., N. Ahmad, E. P. Lillehoj, M. G. Raum, F. H. Salazar, H. W. Chan, and S. Venkatesan. 1988. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslational modifications, and evidence for alternative gag precursors. J. Virol. 62:3993-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore, M. L., W. M. Bryan, S. A. Fakhoury, V. W. Magaard, W. F. Huffman, B. D. Dayton, T. D. Meek, L. Hyland, G. B. Dreyer, B. W. Metcalf, J. E. Strickler, J. G. Gorniak, and C. Debouck. 1989. Peptide substrates and inhibitors of the HIV-1 protease. Biochem. Biophys. Res. Comm. 159:420-425. [DOI] [PubMed] [Google Scholar]
- 31.Nakai, M., and T. Goto. 1996. Ultrastructure and morphogenesis of human immunodeficiency virus. J. Electron Microsc. 45:247-257. [DOI] [PubMed] [Google Scholar]
- 32.Olsen, J. C., and R. Swanstrom. 1985. A new pathway in the generation of defective retrovirus DNA. J. Virol. 56:779-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oroszlan, S., and T. B. Luftig. 1990. Retroviral proteinases. Curr. Top. Microbiol. Immunol. 157:153-185. [DOI] [PubMed] [Google Scholar]
- 34.Partin, K., H. G. Krausslich, L. Ehrlich, E. Wimmer, and C. Carter. 1990. Mutational analysis of a native substrate of the human immunodeficiency virus type 1 proteinase. J. Virol. 64:3938-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettit, S. C., M. S. Horwitz, and J. A. Engler. 1989. Mutations of the precursor to the terminal protein of adenovirus serotypes 2 and 5. J. Virol. 63:5244-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettit, S. C., J. Simsic, D. D. Loeb, L. Everitt, C. A. Hutchison III, and R. Swanstrom. 1991. Analysis of retroviral protease cleavage sites reveals two types of cleavage sites and the structural requirements of the P1 amino acid. J. Biol. Chem. 266:14539-14547. [PubMed] [Google Scholar]
- 38.Phylip, L. H., A. D. Richards, J. Kay, J. Kovalinka, P. Strop, I. Blaha, J. Velek, V. Kostka, A. J. Ritchie, A. V. Broadhurst, W. G. Farmerie, P. E. Scarborough, and B. M. Dunn. 1990. Hydrolysis of synthetic chromogenic substrates by HIV-1 and HIV-2 proteinases. Biochem. Biophys. Res. Commun. 171:439-444. [DOI] [PubMed] [Google Scholar]
- 39.Poorman, R. A., A. G. Tomasselli, R. L. Heinrikson, and F. J. Kezdy. 1991. A cumulative specificity model of proteases from human immunodeficiency virus types 1 and 2, inferred from statistical analysis of an extended substrate data base. J. Biol. Chem. 266:14554-14561. [PubMed] [Google Scholar]
- 40.Prabu-Jeyabalan, M., E. Nalivaika, and C. A. Schiffer. 2000. How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease. J. Mol. Biol. 301:1207-1220. [DOI] [PubMed] [Google Scholar]
- 41.Prabu-Jeybalan, M. M., E. Nalivaika, and C. A. Schiffer. 2001. Substrate shape determines specificity of recognition for HIV-1 protease: analysis of crystal structures of six substrate complexes. Structure 10:369-381. [DOI] [PubMed] [Google Scholar]
- 42.Ratner, L., A. Fisher, L. L. Jagodzinski, H. Mitsuya, R.-S. Liou, R. C. Gallo, and F. Wong-Staal. 1987. Complete nucleotide sequences of functional clones of the AIDS virus. AIDS Res. Hum. Retroviruses 3:57-69. [DOI] [PubMed] [Google Scholar]
- 43.Richards, A. D., L. H. Phylip, W. G. Farmerie, P. E. Scarborough, A. Alvarez, B. M. Dunn, P. H. Hirel, J. Konvalinka, P. Strop, L. Pavlickova, V. Kostka, and J. Kay. 1990. Sensitive, soluble chromogenic substrates for HIV-1 proteinase. J. Biol. Chem. 265:7733-7736. [PubMed] [Google Scholar]
- 44.Ridky, T. W., C. E. Cameron, J. Cameron, J. Leis, T. Copeland, A. Wlodawer, I. T. Weber, and R. W. Harrison. 1996. Human immunodeficiency virus, type 1 protease substrate specificity is limited by interactions between substrate amino acids bound in adjacent enzyme subsites. J. Biol. Chem. 271:4709-4717. [DOI] [PubMed] [Google Scholar]
- 45.Ridky, T. W., A. Kikonyogo, J. Leis, S. Gulnik, T. Copeland, J. Erickson, A. Wlodawer, I. Kurinov, R. W. Harrison, and I. T. Weber. 1998. Drug-resistant HIV-1 proteases identify enzyme residues important for substrate selection and catalytic rate. Biochemistry 37:13835-13845. [DOI] [PubMed] [Google Scholar]
- 46.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 47.Schatz, G., I. Pichova, and V. M. Vogt. 1997. Analysis of cleavage site mutations between the NC and PR Gag domains of Rous sarcoma virus. J. Virol. 71:444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sheng, N., and S. Erickson-Viitanen. 1994. Cleavage of p15 protein in vitro by human immunodeficiency virus type 1 protease is RNA dependent. J. Virol. 68:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng, N., S. C. Pettit, R. J. Tritch, D. H. Ozturk, M. M. Rayner, R. Swanstrom, and V. S. Erickson. 1997. Determinants of the human immunodeficiency virus type 1 p15NC-RNA interaction that affect enhanced cleavage by the viral protease. J. Virol. 71:5723-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanstrom, R., and J. Wills. 1997. Retroviral gene expression: synthesis, processing, and assembly of viral proteins, p. 263-334. In H. E. Varmus, J. Coffin, and S. Hughes (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 51.Tomasselli, A. G., J. O. Hui, T. K. Sawyer, D. J. Staples, C. Bannow, I. M. Reardon, W. J. Howe, D. L. DeCamp, C. S. Craik, and R. L. Heinrikson. 1990. Specificity and inhibition of proteases from human immunodeficiency viruses 1 and 2. J. Biol. Chem. 265:14675-14683. [PubMed] [Google Scholar]
- 52.Tozser, J., I. Blaha, T. D. Copeland, E. M. Wondrak, and S. Oroszlan. 1991. Comparison of the HIV-1 and HIV-2 proteinases using oligopeptide substrates representing cleavage sites in Gag and Gag-Pol polyproteins. FEBS Lett. 281:77-80. [DOI] [PubMed] [Google Scholar]
- 53.Tozser, J., A. Gustchina, I. T. Weber, I. Blaha, E. M. Wondrak, and S. Oroszlan. 1991. Studies on the role of the S4 substrate binding site of HIV proteinases. FEBS Lett. 279:356-360. [DOI] [PubMed] [Google Scholar]
- 54.Tozser, J., I. T. Weber, A. Gustchina, I. Blaha, T. D. Copeland, J. M. Louis, and S. Oroszlan. 1992. Kinetic and modeling studies of S3-S3′ subsites of HIV proteinases. Biochemistry 31:4793-4800. [DOI] [PubMed] [Google Scholar]
- 55.Tritch, R. J., Y. E. Cheng, F. H. Yin, and S. Erickson-Viitanen. 1991. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J. Virol. 65:922-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Veronese, F. D., R. Rahman, T. D. Copeland, S. Oroszlan, R. C. Gallo, and M. G. Sarngadharan. 1987. Immunological and chemical analysis of p6, the carboxyl-terminal fragment of HIV p15. AIDS Res. Hum. Retroviruses 3:253-264. [DOI] [PubMed] [Google Scholar]
- 57.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wilson, W., M. Braddock, S. E. Adams, P. D. Rathjen, S. M. Kingsman, and A. J. Kingsman. 1988. HIV expression strategies: ribosomal frameshifting is directed by a short sequence in both mammalian and yeast systems. Cell 55:1159-1169. [DOI] [PubMed] [Google Scholar]
- 60.Wlodawer, A., and J. W. Erickson. 1993. Structure-based inhibitors of HIV-1 protease. Annu. Rev. Biochem. 62:543-585. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]