Abstract
To define the human immunodeficiency virus type 1 (HIV-1) RNA maturation pathways, we analyzed the intracellular distribution of HIV-1 RNA and the viral regulatory proteins Rev and Tat in transfected COS cells and HIV-1-infected lymphoid C8166 cells by means of ultrastructural in situ hybridization using antisense RNA probes and immunoelectron microscopy. The intranuclear viral RNA occurs in ribonucleoprotein fibrils in the perichromatin and interchromatin regions. The simultaneous demonstration of Rev, Tat, Br-labeled RNA, and cellular proteins SC35 and CRM1 in such fibrils reveals the potential of Rev to associate with nascent HIV pre-mRNA and its splicing complex and transport machinery. In a rev-minus system, the env intron-containing, incompletely spliced viral RNAs are revealed only in the nucleus, indicating that Rev is required to initiate the transport to the cytoplasm. Moreover, env intron sequences frequently occur in the periphery of interchromatin granule clusters, while the probe containing the rev exon sequence does not associate with this nucleoplasmic domain. When cells were treated with the CRM1 inhibitor leptomycin B in the presence of Rev protein, the env intron containing HIV RNAs formed clusters throughout the nucleoplasm and accumulated at the nuclear pores. This suggests that Rev is necessary and probably also sufficient for the accumulation of incompletely spliced HIV RNAs at the nuclear pores while CRM1 is needed for translocation across the nuclear pore complex.
During lentivirus replication, the nuclear export of transcribed viral mRNAs from the nucleus to the cytoplasm via the nuclear pore complex is regulated by factors encoded in the viral genome (reviewed in reference 57). The 116-amino-acid Rev protein of human immunodeficiency virus type 1 (HIV-1), a lentivirus, interacts with the viral RNA and mediates its nuclear export. A short arginine-rich domain of Rev binds directly to a 234-nucleotide RNA sequence termed the Rev responsive element (RRE), which is localized in intron RNA (33; for reviews, see references 9 and 40). Completely spliced RNAs appear in the cytoplasm independently of Rev. From these fully spliced RNAs, Rev, together with the transcriptional elongation factor Tat, is translated early in infection. Rev enters the nucleus to induce transport of the intron-containing, Rev-dependent unspliced and partially spliced RNAs, from which the late structural HIV-1 proteins are translated (28, 40). In addition to the intron-RNA binding capacity, the basic domain of the Rev protein possesses a nuclear localization sequence. Another domain, a short leucine-rich region located at the carboxy terminus of Rev and called a nuclear export signal (NES), is required for interaction with the cellular export receptors (18, 19). Both these domains confer on the Rev protein the possibility of shuttling between the nucleus and the cytoplasm (26, 35, 52, 58).
Molecular arrangements and mechanisms by which Rev controls posttranscriptional export of RNA during the viral replication cycle are partly known. Immunofluorescence-labeling assays suggest that the Rev protein interacts with HIV-1 RNAs at putative sites of mRNA transcription and splicing (4). The following step is an interaction between the NES and an essential nuclear export factor, CRM1/exportin1. The complex RNA-Rev-CRM1 associates with Ran-GTP molecules and is thus recruited to nuclear pores via the direct interaction of CRM1 with nucleoporins. The exact mechanism of translocation to the cytoplasm is not understood. Also unclear is a role of other proposed Rev cofactors, such as the human nucleoporin-like protein hRIP/Rab, eukaryotic initiation factor 5A, or nucleoporin-like protein NLP-1 (15, 45, 51; for reviews, see references 23 and 28). The Rev-dependent nuclear export of HIV-1 RNAs is inhibited by leptomycin B (LMB), which binds directly to CRM1 and thus prevents the translocation of the intron-containing HIV-1 RNA to the cytoplasm (29, 59; for a review, see reference 28).
The application of in situ hybridization (ISH) of HIV-1 RNAs at an ultrastructural level in both HIV-transfected and HIV-infected cells allowed us to obtain novel observations about in situ molecular organization of HIV-1 RNAs. Postembedding immunoelectron microscopy was used for visualizing digoxigenin-labeled probes as well as for localizing viral and cellular proteins involved in posttranscriptional events including export of viral RNAs. This high-resolution microscopic approach allowed us to show that viral RNA occurs in the form of ribonucleoprotein (RNP)-containing structural fibrillar constituents distributed in the nucleus within the perichromatin regions and within the interchromatin space. This RNA was also localized in the cytoplasm and in the viral particles at the same time. Immunolocalization of viral Rev and Tat proteins and of the cellular SC35 splicing factor revealed these factors also on fibrillar RNP constituents. After using LMB in the rev-plus system, viral RNA occurred in a clustered pattern very often observed close to the inner or central portion of nuclear pores.
Our observations demonstrate that Rev is required for transport of incompletely spliced HIV RNAs from its origin to the nuclear pore. The association with CRM1 is needed to complete the translocation of the Rev-RRE-RNA complex across the nuclear pore.
MATERIALS AND METHODS
Cells, transfection, and electron microscopic processing.
COS cells at 80% confluency were transiently transfected with plasmid DNAs as previously described (4). At 24 h after transfection, the cells were seeded onto 10- or 12-mm glass coverslips in 24-well plates. The next day, the cells were fixed with 4% paraformaldehyde in 0.1 M Sörensen phosphate buffer (pH 7.4) for 60 min at 4°C. To visualize nascent RNA transcripts, some cells were labeled with 5-bromo-UTP (BrUTP) using the FuGENE reagent (Roche Molecular Biochemicals, Mannheim, Germany) to mediate the uptake of BrUTP into nonpermeable cells (21), incubated for 10 min, and fixed. To inhibit viral biogenesis, LMB (a kind gift from Barbara Wolff) was added at 5 nM to the incubation medium of some cultures for 3 or 12 h before fixation.
Human T-lymphoblastoid cell line C8166 was grown and infected with HIV-1 as previously described (26). In brief, the C8166 cells were infected by supernatant which was harvested at 48 h posttranfection from COS cells transfected with pSVC21 (see below). When syncytium formation was clearly visible in the infected C8166 cells 3 days later, a new culture of uninfected C8166 cells was inoculated with 1:100 volume of the infected C8166 culture. At 24 h later, when minute syncytia started to appear, the cells were pelleted, resuspended in phosphate-buffered saline (PBS), and then fixed in cold 4% paraformaldehyde in PBS. After centrifugation (200 × g for 5 min at room temperature), the pellets of the fixed C8166 cell were pre-embedded in 2% low-viscosity agarose.
All samples were then dehydrated in ethanol at room temperature, embedded in LRWhite resin, and polymerized at 60°C for 24 h. Some COS and C8166 cells were also dehydrated at progressively lower temperature by using an EM AFS cryoapparatus (Leica, Vienna, Austria), embedded into Lowicryl K4M, and polymerized with UV for 48 h at −35°C (7). After polymerization, blocs with COS cells were separated from glass coverslips and cut parallel to the substrate with a diamond knife using a Leica Ultracut UCT ultramicrotome, while the pelleted C8166 cells were cut directly without any orientation. Ultrathin sections were placed on nickel grids coated with a Formvar-carbon layer and further processed for postembedding ISH and/or for immunogold labeling.
Plasmids and probe preparation.
Preparation and origin of our plasmids are described in detail in previous papers (4, 53). Briefly, plasmid pSVc21 contains the infectious HIV-1 HXBc2 proviral DNA clone and the simian virus 40 origin of replication. The rev-minus pSVc21B has a frameshift mutation in the second exon of the rev gene. The pgTAT subgenomic construct contains the part of the HIV-1 genome, which encodes Tat and Env proteins, while the pRev construct contains cDNA of rev driven by the cytomegalovirus CMV promoter (33).
Two digoxigenin-labeled RNA probes, generated using an in vitro transcription kit (Boehringer Mannheim), were used. (i) The env intron sequence was PCR amplified from a plasmid containing the HIVHXB2 proviral DNA by using the oligonucleotide primers tgtggtccatagtaatc (sense, corresponding to nucleotides 6125 to 6141 of GenBank accession number K03455) and ttgttaatttctctgtcc (antisense, corresponding to nucleotides 8133 to 8116). The resulting 2,006-bp fragment was purified and blunt end ligated into the SmaI site of the polylinker of transcription vector pGEM7Zf (+) (Promega Biotec). Antisense digoxigenin-labeled RNA was transcribed from the EcoRI-linearized version of this construct by using the SP6 RNA polymerase. This HIV env intron probe therefore contains HIV intron sequences located between the major splice donor site at position 6045 and the major splice acceptor site at position 8376 in the env region of HIV-1 RNAs. (ii) The rev exon (nucleotides 5969 to 6044 plus nucleotides 8378 to 8652 of GenBank accession number K03455, corresponding to the first and second rev exon, respectively) was PCR amplified and blunt-end ligated into the EcoRV site of pBluescript II KS (+/−) (Stratagene). Digoxigenin-labeled antisense RNA corresponding to the 351-bp HIVHXB2 Rev cDNA was transcribed using T3 RNA polymerase.
The two antisense RNA probes used in the present work are therefore denoted the rev exon probe and env intron probe. While the rev exon probe is expected to hybridize to all HIV-1 RNA species, the env intron probe has the potential to hybridize only to unspliced and partially spliced viral RNAs.
Primary antibodies.
The 8E7F2 and 1G7B11 monoclonal antibodies directed against the Rev protein (26) and the 1D9D5 monoclonal antibody recognizing the Tat protein (54) were used to detect HIV-1 proteins. Moreover, we have used a monoclonal antibody specifically recognizing the cellular non-small nuclear RNP splicing factor SC35 (20) or a monoclonal anti-bromodeoxyuridine antibody that also allowed a specific and efficient visualization of Br-labeled nascent RNA (Partec, Münster, Germany). To localize cellular CRM1 protein, a rabbit antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.) was applied.
Fluorescent microscopy.
The fluorescent ISH protocol, as well as simultaneous detection of the proteins, was described previously (4). Briefly, the transfected cells grown on glass coverslips were hybridized for 4 h at 60°C over 25 μl of hybridization mixture containing 0.5 ng of digoxigenin-labeled probe per ml. After washing and blocking of nonspecific binding of antibodies, the cells were incubated with primary monoclonal antibody (anti-SC35), together with anti-digoxigenin antibody (Boehringer), at room temperature for 40 min. Both antibodies were visualized with fluorescein isothiocyanate-conjugated antibody (Sigma, St. Louis, Mo.) and Texas red-conjugated streptavidin (Pierce, Rockford, Ill.). Confocal microscopy was carried out using the Bio-Rad MRC 1000 system and a Zeiss Axiovert 100 microscope. The fluorochromes were independently recorded following argon ion laser excitation at wavelengths of 488 and 529 nm, respectively.
ISH and immunoelectron microscopic labeling.
ISH and immunolabeling at the ultrastructural level were performed directly on grids with the sections, in a humid chamber, over drops of either the hybridization or antibody solutions.
ISH was performed by a previously reported procedure (17) with some modifications. The grids were incubated for 10 min at room temperature on the surface of a drop of prehybridization buffer (HB) containing yeast competitor transfer RNA (tRNA; Sigma) in saline sodium citrate buffer (1 × SSC [0.15 M NaCl plus 0.015 M sodium citrate; pH 7.0]) to reduce nonspecific adsorption between the probe and the tissue section. The grids were then transferred to hybridization mixture, in which the HIV env intron or rev exon digoxigenin-labeled probes, both diluted 1:300 in the HB, annealed to the single-stranded HIV RNA at 55°C for 4 h. After hybridization, several washes followed by floating the grids over successive drops of 5 × SSC, 2 × SSC, Tris saline buffer containing 0.05% Tween 20 (TBS-T; Sigma), and TRIS saline buffer containing 0.5% blocking reagent (Boehringer) (TBS-BR). Hybrids were detected following incubation of the grids with a drop of mouse monoclonal anti-digoxigenin antibody (Boehringer Mannheim), diluted 1:10 in TBS-BR for 30 min at room temperature, and washed in 3 drops of TBS-T. The signal was finally revealed by using goat anti-mouse antibody conjugated with 12-nm-diameter colloidal gold (GAM 12; Jackson Immunoresearch Laboratories, West Grove, Pa.) diluted 1:10 in TBS-BR for 30 min at room temperature.
For protein immunolabeling, the grids with sections were pretreated with 10% normal goat serum in PBS for 10 min and then reacted, for 17 h at 4°C, with primary antibodies diluted in PBS containing 0.05% Tween 20 (Sigma) and 1% bovine serum albumin (Fluka, Buchs, Switzerland). Then the sections were washed with PBS-Tween and PBS alone, followed by a repeated treatment with normal goat serum for 10 min, and reacted, for 30 min at room temperature, with GAM 12 secondary antibody (Jackson) diluted in PBS. Finally, all grids were thoroughly rinsed with PBS and ultrapure water and air dried.
Double detection of HIV-1 RNA and viral or cellular proteins was performed by two different protocols as follows. (i) After the last washes following ISH, the sections were incubated for 2 h at room temperature with antibodies (diluted in TBS-BR) recognizing various protein factors. These antibodies, together with digoxigenin-labeled probes, were then visualized by means of specific antibodies coupled with colloidal gold particles of different sizes (Aurion, Wageningen, The Netherlands). (ii) The ISH signal was revealed by rabbit anti-digoxigenin antibody (Sigma) followed by colloidal gold-conjugated goat anti-rabbit antibody (Jackson). The grids were then air dried and subsequently treated for protein immunolabeling as mentioned above.
The sections of cells embedded in LRWhite resin were stained by the regressive technique preferential for RNP-containing nuclear structural domains (2) as follows: 4.7% aqueous uranyl acetate for 45 s, 0.02 M EDTA for 4 min, and lead citrate for 45 s. Since chromatin in the material embedded in Lowicryl K4M tends to exhibit a bleached aspect and RNP constituents are very easily distinguished without EDTA treatment (55), such sections were contrasted only with 2.5% uranyl acetate for 5 min followed by lead citrate for 1 min.
As controls, nontransfected cells were processed as above. Moreover, the sections of transfected cells were incubated in the absence of primary antibodies. To confirm the RNA nature of the hybridization signal, some sections were treated, before hybridization, with 0.2% RNase (type IA; Sigma) in 1 mM triethanolamine-acetic acid buffer (pH 7.3) for 18 h at 37°C.
Evaluation of the ISH signal.
To evaluate the extent of the ISH signal, the intensity of labeling was determined in four subnuclear constituents (RNP-containing fibrils, interchromatin granules, condensed chromatin, and nucleolus) and in the cytoplasm. The signal was revealed using the anti-digoxigenin antibody followed by a secondary antibody coupled with 12-nm-diameter colloidal gold particles. We have evaluated separately the signal for env intron or rev exon probe over cells transfected with both rev-minus (pSVc21B) and rev-plus (pSVc21) plasmids. To clearly distinguish the labeled subnuclear constituents, 12 micrographs of each of four chosen labeling experiments were printed at a final magnification of ×52,500. The micrographs were scanned, and the surface of the measured area was morphometrically determined by using the Openlab measurement module (Improvision, Coventry, United Kingdom). The mean measured surface area of analyzed nuclei was 7.8 μm 2. Since it is virtually impossible to measure the surface of areas occupied by dispersed RNP-containing fibrils in the perichromatin and interchromatin region, a labeling density for each structural constituent was expressed as the number of gold particles in a given nuclear constituent per square micrometer of the total nuclear surface. Background labeling was evaluated on neighbouring nontransfected cells or over the resin outside the cells.
For quantitative evaluation of the env intron-ISH signal in the nuclei of the cells transfected with pSVc21 followed by LMB treatment, three nuclear regions were considered for quantification: the interior of the nuclear pores, the region close to the nuclear pore, and the nucleoplasm. Twelve entire nuclei from 3- and 12-h LMB incubations were scanned with a wide-angle slow-scan charge-coupled device camera (Gatan, Pleasanton, Calif.), and the number of gold particles and/or of gold grain clusters in each region was manually counted. Moreover, to obtain quantitative information about the intracellular transport of CRM1-independent RNA under LMB treatment conditions, the total number of gold particles in the nucleus and in the cytoplasm was evaluated in LMB-treated and untreated cells using the rev exon probe.
Statistical analysis of the signal differences between different constituents in the same experimental group was performed by using the Wilcoxon signed ranks test, while labeling differences of the same cell compartment were evaluated in two independent samples or treatments by using the Wilcoxon-Mann-Whitney test, as reported by Siegel and Castellan (47). Statistical significance was set at P < 0.05.
RESULTS
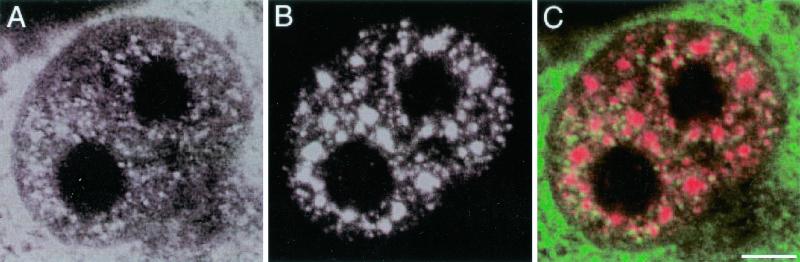
Immunofluorescent ISH images of cells transfected with pSVc21 show the HIV-1 RNA diffusely distributed in the nucleoplasm and cytoplasm, with relative exclusion of the nucleolus (Fig. 1A), while SC35 labeling occurs in the nucleoplasm both diffusely and as strongly labeled speckles (Fig. 1B). An overlay of these two images (Fig. 1C) shows that most of the HIV RNA signal does not colocalize with the SC35-containing speckles.
FIG. 1.
Confocal optical sections of a COS cell transfected with plasmid pSVc21 containing the complete HIV-1 provirus, showing the fluorescent ISH signal for HIV-1 RNA (env intron probe) and indirect immunofluorescent signal for SC35 splicing factor. (A) The typical nucleoplasmic and cytoplasmic diffuse distribution of HIV-1 RNA is demonstrated. Large unlabeled areas correspond to nucleoli. (B) Splicing factor SC35 occurs diffusely distributed throughout the nucleoplasm as well as in several speckled regions. (C) Merged image of the signal for viral RNA (in green) and SC35 (in red) demonstrates colocalization in the nucleoplasm, while SC35-labeled speckles are mostly devoid of env intron-containing RNA. Bar, 3.0 μm.
To obtain novel observations on regulation of one of the essential HIV biogenesis stages, we took advantage of ultrastructural ISH and immunoelectron microscopy to investigate the intracellular distribution of HIV-1 RNA and of the viral regulatory proteins Rev and Tat in transfected COS cells.
For transfection of the cells, we have used four distinct plasmids. The pSVc21 proviral plasmid expressed all viral RNAs and proteins, while its rev-minus frameshift derivative pSVc21B did not express functional Rev protein. The subgenomic construct pgTat produced only two species of mRNAs: the incompletely spliced, env intron-containing and Rev/CRM1-dependent mRNA encoding Env protein, and the completely spliced, Rev-independent mRNA encoding Tat protein. To specifically analyze the export of the Rev-dependent Env mRNA, the pgTat plasmid was also used for cotransfection together with the pRev plasmid encoding only Rev protein. Moreover, to avoid potential problems associated with overexpression in transfected cells, HIV-infected lymphoid C8166 cells were also used for experiments.
Of the two probes used for our ISH assays, the rev exon probe was able to reveal all HIV-1 pre-mRNA and other HIV-1 RNA species, while the env intron probe recognized only unspliced or partially spliced viral RNAs.
All control assays carried out for ISH as well as for immunolabeling were negative and gave rise to a signal corresponding to background level. Moreover, RNase treatment of sections prior to ISH completely prevented the hybridization signal. This verified the RNA nature of the hybrids.
Plasmid pSVc21 and expression of progeny virions.
Plasmid pSVc21 transfected into COS cells was selected as the system of choice for the examination of Rev-dependent and CRM-dependent ultrastructural localization of HIV mRNA. The functionality of pSVc21 was demonstrated by the visualization of abundant progeny virions in transfected cells. Although the efficiency of transfection was lower (approximately 25%) than that of infection (approximately 40%), the transfected COS cells produced approximately 50% more virions than did the HIV-1-infected C8166 cells. The morphological appearance of progeny virions was similar in the two systems.
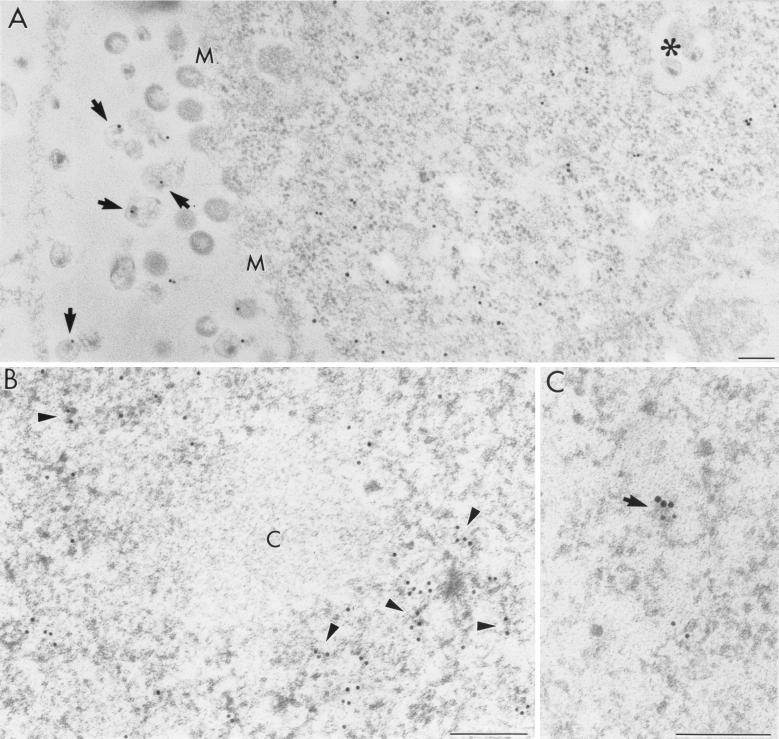
The viral particles were observed outside the cells, budding from the cell surface, as well as within internal cytoplasmic membrane vesicles. They varied in morphological features. Some particles displayed a typical appearance of wild-type HIV having two distinctive parts: a centric or an eccentric electron-dense cone-shaped core and a bilayer envelope. However, many particles displayed abnormal features without the electron-dense core and/or without a clearly recognizable viral envelope (Fig. 2A).
FIG. 2.
Immunoelectron microscopic localization of in situ hybridization signal for env intron-containing HIV-1 RNAs in pSVc21-transfected COS cells. (A) A 15-nm gold marker labels the cytoplasm and mature virions (arrows). M, cell membrane; asterisk, virions in a cytoplasmic vesicle. (B) Nuclear distribution of HIV-1 RNAs (6-nm colloidal-gold-conjugated antibody) occurs on electron-dense RNP-containing fibrils which are distributed in the nucleoplasm (some fibrils are indicated by arrowheads). Condensed chromatin (c) is virtually devoid of ISH signal. (C) A high-power micrograph shows an RNP fibril (arrow) revealing colocalization of the ISH signal (large grains, 15 nm) with brominated RNA (small grains, 10 nm). Bars, 0.2 μm.
Localization of the env intron sequence of HIV-1 mRNAs in the presence of Rev.
Rev-dependent incompletely spliced HIV RNAs were detected by ultrastructural ISH with the digoxigenin-labeled env intron probe, which is complementary to the entire env intron sequence. ISH was carried out on ultrathin sections of pSVc21 plasmid-transfected COS cells as well as on sections of infected C8166 cells, and the ISH signal was revealed by anti-digoxigenin antibodies conjugated with colloidal gold.
Using the env intron probe, we showed that Rev-dependent HIV mRNAs localize in both the cell nucleus and the cytoplasm (Table 1) as well as in progeny virions. The ISH signal occurred on the virions, regardless of their localization or morphological appearance (Fig. 2A). Moreover, a rather strong diffuse signal was observed in the cytoplasm of transfected cells (Fig. 2A). Unfortunately, it is difficult to precisely localize the ISH signal with regard to the cytoplasmic compartments or organelles because, after paraformaldehyde fixation, the ultrastructural morphology of the cytoplasmic constituents is not very well preserved. Nuclear labeling mostly occurred regularly distributed on RNP-containing fibrils located within both the perichromatin regions and the interchromatin space without any apparent local accumulation (Fig. 2B). Double-labeling experiments after short BrUTP incorporation demonstrated that many fibrils represent nascent RNA and that some of them showed colocalization with the ISH probe signal (Fig. 2C). Furthermore, the ISH signal sometimes occurred close to both sides of the nuclear pores or inside them. Condensed chromatin areas, interchromatin granules (IG), and nucleoli were devoid of the ISH label (Table 1).
TABLE 1.
Quantitative analysis of ISH labeling in rev-plus and rev-minus cells, comparing the signal density over nuclear compartments and cytoplasm of transfected cells with background label as described in Materials and Methods
| Labeling | No. of grains/μm 2 (mean ± SE) ina:
|
|||||
|---|---|---|---|---|---|---|
| RNP | IGs | CHR | NU | CY | BG | |
| rev plus/env intron | 12.31 ± 1.76 (P < 0.01) | 0.28 ± 0.08 (NSb) | 0.74 ± 0.18 (NS) | 0.38 ± 0.07 (NS) | 13.11 ± 2.37 (P < 0.01) | 0.64 ± 0.41 |
| rev minus/env intron | 14.07 ± 1.42 (P < 0.001) | 2.81 ± 0.12 (P < 0.05) | 0.71 ± 0.13 (NS) | 0.54 ± 0.06 (NS) | 0.88 ± 0.20 (NS) | 0.45 ± 0.09 |
| rev plus/rev exon | 11.91 ± 1.76 (P < 0.01) | 0.20 ± 0.03 (NS) | 0.79 ± 0.15 (NS) | 0.42 ± 0.08 (NS) | 12.73 ± 1.33 (P < 0.001) | 0.41 ± 0.08 |
| rev minus/rev exon | 12.42 ± 1.85 (P < 0.01) | 0.43 ± 0.11 (NS) | 0.48 ± 0.1 (NS) | 0.66 ± 0.1 (NS) | 10.12 ± 1.51 (P < 0.01) | 0.48 ± 0.09 |
RNP, RNP fibrils; IGs, interchromatin granule clusters; CHR, condensed chromatin; NU, nucleolus; CY, cytoplasm; BG, background.
NS, not significant.
Comparable ISH results were obtained for both COS cells transfected with pSVc21 and C8166 cells infected with HIV-1.
Localization of HIV mRNA using the rev exon probe in the rev-plus system.
The rev exon probe was expected to hybridize to all HIV mRNA species. However, it does not contain nucleotide sequences complementary to the rev/tat intron, which is excised from HIV pre-mRNA by splicing. The overall distribution pattern of the rev exon probe ISH signal was comparable to that shown by means of the env intron probe: in the nucleus, viral RNAs mainly occurred on nucleoplasmic fibrils visualized by preferential staining for RNP in the perichromatin regions and in the interchromatin space. Furthermore, we have observed significant labeling inside or close to the nuclear pores, in the cytoplasm, and in the progeny virions (Table 1). Interestingly, we found comparable levels of labeling in the nucleus and in the cytoplasm with both the rev exon and the env intron probe in rev-plus cells, similar to earlier FISH observations (4). Whether this is due to a minimum length of target sequence exposed in the specimen in order to obtain an ISH signal or to another reason remains unclear.
Are the HIV-1 Rev and HIV-1 Tat proteins and the cellular factors SC35 and CRM1 detected in the same nuclear domains as HIV-1 RNAs?
Viral and cellular proteins were detected by postembedding immunolabeling with corresponding primary antibodies followed by secondary gold-conjugated probes. All anti-Rev, anti-Tat, anti-SC35, and anti-CRM1 antibodies used in our immunoelectron microscopic assays revealed significant signal both in the COS cells transfected with the complete proviral DNA and in the infected C8166 cells. No detectable differences in the overall distribution of proteins between the two cell lines were found.
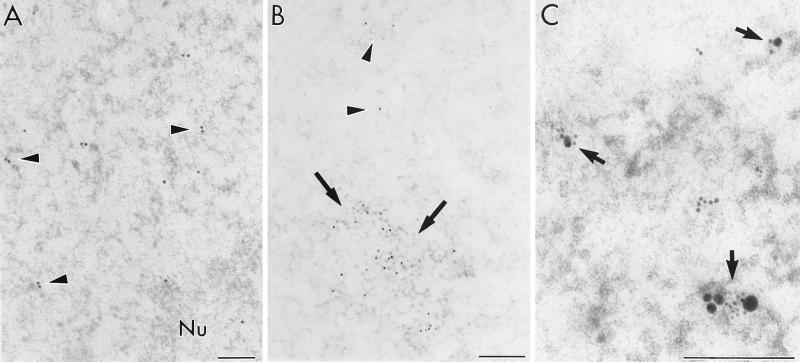
The anti-Tat antibody showed that the distribution of this viral transcription factor was restricted mainly to the nucleoplasmic RNP fibrils in the perichromatin regions and in the interchromatin space, similar to the ISH signal (Fig. 3A). Moreover, anti-SC35 antibody recognized this splicing factor within clusters of IG (seen as “speckles” by immunofluorescence), in addition to nucleoplasmic RNP fibrils (Fig. 3B). Finally, when double labeling of the Rev protein and HIV RNAs was performed, the RNP-fibrillar structures were the major nuclear constituent in which protein and ISH signal are colocalized (Fig. 3C). A similar situation was observed in such double-labeling experiments using anti-CRM1 antibodies (data not shown).
FIG. 3.
Immunocytochemical labeling of protein factors. (A) RNP fibrils in C8166-infected cells exhibit anti-Tat labeling visualized with 15-nm colloidal gold (arrowheads). Moreover, the Tat protein also occurs in the nucleolus (Nu). (B) Splicing factor SC35 occurs within clusters of interchromatin granules (arrows) and on nucleoplasmic RNP fibrils diffusely distributed in the nucleoplasm (arrowheads). (C) Colocalization of Rev protein (1G7B11 monoclonal antibody with 6-nm colloidal-gold particles) with rev exon probe (15-nm grains) is visible on the RNP-containing fibrils in the nucleoplasm of transfected cells (arrows). Bars, 0.2 μm.
However, in contrast to nuclear HIV RNA label, both the viral proteins also occurred in the nucleolus, associated with its dense fibrillar and granular components (Fig. 3A). Some gold grains representing viral proteins were found in the cytoplasm. However, neither Rev nor Tat was revealed in association with the progeny virions.
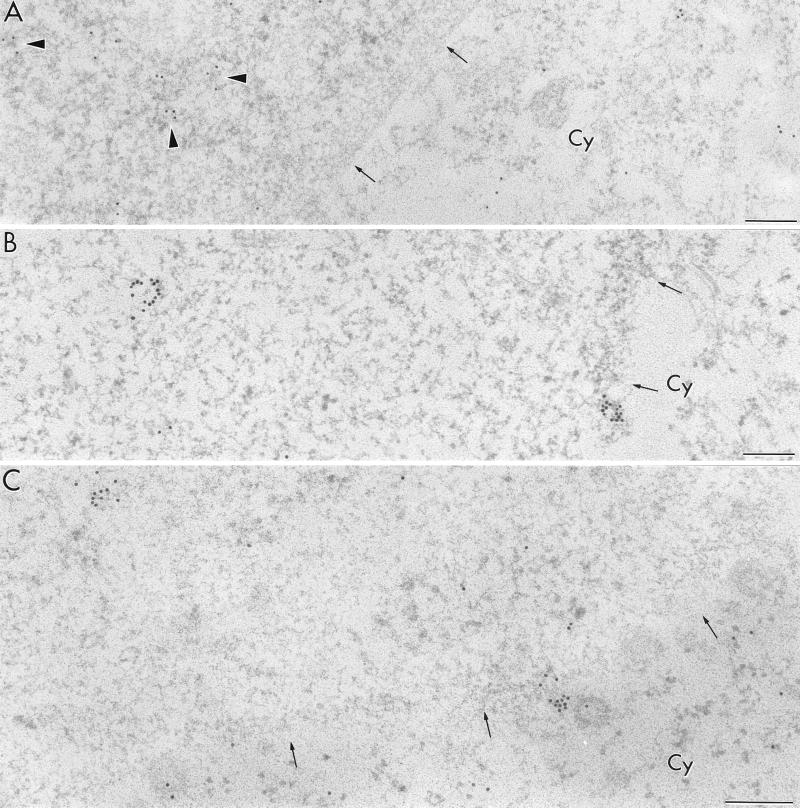
Transfection of COS cells with rev-minus plasmid pSVc21B.
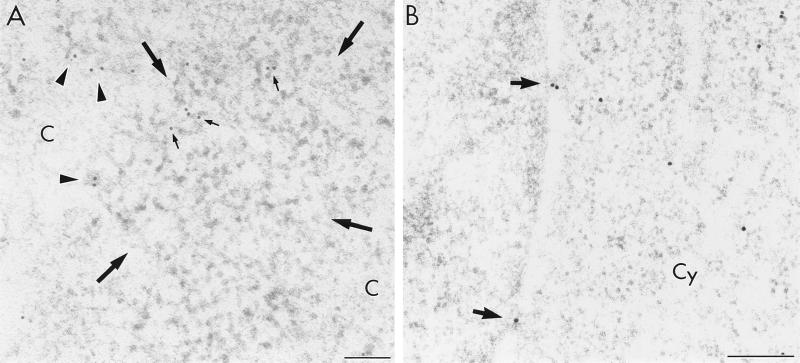
When plasmid pSVc21B, which is a rev-minus derivative of pSVc21, was used for cell transfection, no viral particles were observed. Moreover, no env intron hybridization signal was found in the cytoplasm (Table 1). However, the signal of the env intron probe was abundant in the nucleus of rev-minus cells, where it was localized mostly in perichromatin or interchromatin regions. Distinct from the rev-plus system, env intron-containing RNA also occurred close to the clusters of IG, while the interior of such clusters remained devoid of label (Fig. 4A). Finally, no qualitative difference of rev exon-labeling pattern between the rev-plus and the rev-minus systems occurred in the nucleus or in the cytoplasm. We also observed migration of the viral RNAs containing complementary sequence to the rev exon probe from the nucleus to the cytoplasm in cells lacking the Rev protein (Table 1; Fig. 4B). The condensed chromatin and the nucleolus were devoid of any labeling after hybridization with either the env intron or the rev exon probe (Table 1). Finally, in the cells transfected with plasmid pSVc21B, the immunolocalization patterns of viral Tat and cellular SC35 protein did not change compared with those in the rev-plus cells (not shown).
FIG. 4.
Electron microscopic localization of HIV-1 RNA in a COS cell transfected with rev-minus plasmid (pSVc21B). (A) The env intron probe visualized with 12-nm colloidal-gold particles is localized on the RNP-containing fibrils (arrowheads) as well as on the periphery of a cluster of interchromatin granules (small arrows); large arrows delimit the IG cluster; c, condensed chromatin. (B) The rev exon containing RNA (15-nm gold particles) is shown to migrate across the nuclear pores (arrows) to the cytoplasm (Cy). Bars, 0.2 μm.
Effect of LMB on nuclear export of HIV-1 RNAs.
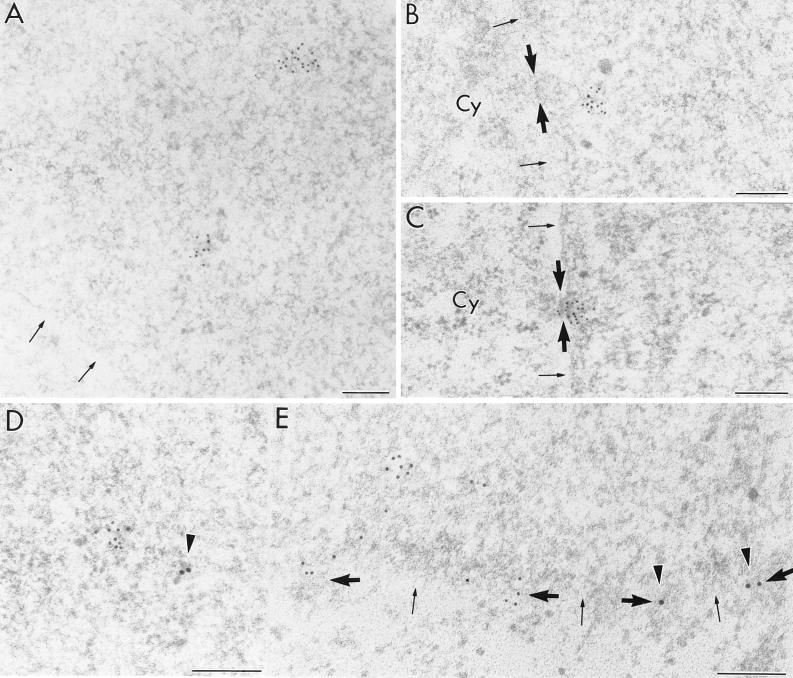
LMB specifically inhibits NES-dependent nuclear export by virtue of alkylation of a cysteine residue in CRM1 (30). Although LMB is known to be a cytotoxin, we have not been able to detect any effect on cellular morphology or behavior in culture at the concentration and incubation periods used in our assays. After 3 h of incubation with LMB, the localization pattern of the env intron probe in the COS cells transfected with the rev-plus plasmid pSVc21 was dramatically changed. The more or less diffuse pattern of label observed over the nucleoplasm in the absence of the drug changed into a clustered labeling pattern consisting of focally accumulated gold grains (Fig. 5A; Table 2). Moreover, aggregates of gold grains corresponding to the env intron probe were also found close to the nuclear side of the nuclear pores (Fig. 5B). Occasionally such clusters were identified within the pores (Fig. 5C). Approximately 10% of all visible nuclear pores were in contact with aggregates of env intron-containing RNAs. After 12 h of incubation with LMB, no evident variation of the above labeling pattern could be observed, except that the gold grain aggregates occurring in the proximity of the nuclear pores sometimes appeared larger. The statistical significance was demonstrated for the difference between the number of gold grains in clusters in the nucleoplasm and in clusters that are in contact with the nuclear pores (Table 2, P < 0.05) in transfected COS cells after LMB incubation for 12 h.
FIG. 5.
Transfected rev-plus COS cells treated with LMB for 3 h (A, B, C, and D) or 12 h (E). (A) env intron containing RNA occurs in clusters throughout the nucleoplasm (arrows). (B and C) Aggregates of env intron-containing RNA are often observed at the proximity of the nuclear pores (B) or sometimes directly inside the nuclear pores (C) (thick arrows). (D and E) Following double labeling with the env intron probe (small grains) and anti-CRM1 (large grains, arrowheads), no close colocalization is observed either in the nucleoplasm (D) or in the vicinity of the nuclear pores (thick arrows) (E). Thin arrows point to the nuclear periphery. Cy, cytoplasm. Bars, 0.2 μm.
TABLE 2.
Quantitative evaluation of clustered env intron ISH signal in the nucleus of rev-plus cells treated with LMB
| LMB treatment | Nuclear areaa | No. of clusters (mean ± SE) | No. of gold grains in clusters (mean ± SE) |
|---|---|---|---|
| 3 h | NPC-in | 3.92 ± 0.32 | 16.43 ± 1.85 |
| NPC-close | 2.78 ± 0.26 | 14.13 ± 2.08 | |
| Nucleoplasm | 3.52 ± 0.55 | 11.67 ± 1.47 | |
| 12 h | NPC-in | 4.28 ± 0.52 | 20.12 ± 2.51b |
| NPC-close | 2.45 ± 0.43 | 19.73 ± 3.27b | |
| Nucleoplasm | 3.76 ± 0.81 | 13.85 ± 1.91b |
NPC-in, labeling within the nuclear pore complex; NPC-close, labeling occurring in the immediate vicinity of the nuclear pore complex but not directly inside.
Statistically significant difference (P < 0.05) between the number of gold grains occurring in clusters in the nucleoplasm and in clusters that are observed either in contact with or within the nuclear pores (NPC-close and NPC-in), after 12 h of LMB treatment. No cluster formation was found in cells incubated in the absence of LMB.
No gold particle aggregates were found on the cytoplasmic side of the nuclear pores or in the cytoplasm, where some diffuse labeling occurred regardless of the duration of the LMB incubation period.
When the transfected and LMB-treated cells were hybridized with the rev exon probe, the clustered pattern of viral RNA labeling in the nucleus was also found. However, a considerable part of the rev exon signal remained in diffuse form, and a strong signal without clustering of colloidal gold particles was found in the cytoplasm of transfected cells (Table 3). No clustered signal was observed when LMB was applied to cells transfected with rev-minus pSVc21B plasmid. These results, together with findings that labeling patterns for the Rev, Tat, SC35, and CRM1 proteins also were not evidently altered after the LMB treatment compared with control cells, led to the conclusion that LMB treatment did not have an influence on the fully spliced Rev-independent HIV-1 mRNAs. When the double-labeling assay was performed with anti-CRM1 antibody and RNA probes, no close colocalization was found between the LMB-induced clusters of HIV-1 RNAs and CRM1 protein (Fig. 5D and E).
TABLE 3.
Quantitative evaluation of rev exon ISH signal in the nucleus of rev-plus cells treated with LMB, showing the labeling density over the nucleus and the cytoplasm of pSVc21 transfected cells
| COS cell treatment | ISH signala (no. of gold grains/μm2, mean ± SE) in:
|
|
|---|---|---|
| Cytoplasm | Nucleus | |
| None | 12.73 ± 1.33 | 13.32 ± 2.02 |
| LMB 3 h | 11.34 ± 1.16 | 12.94 ± 0.64 |
| LMB 12 h | 11.82 ± 1.09 | 13.45 ± 1.04 |
No significant difference between LMB-treated and untreated cells was found. Statistical significance was set at P ≤ 0.05.
Localization of viral mRNA in COS cells transfected with subgenomic constructs.
pSVc21 and the derived rev-minus pSVc21B were chosen as the preferred experimental system following a careful initial comparison with several subgenomic constructs. The main reasons were that pSVc21 contains the complete HIV-1 provirus and expresses all viral RNAs and proteins and that the distributions of RNA between different transfected single cells and between experiments were most consistent. The subgenomic pgTat, which can be supplemented with Rev in trans when the pRev is cotransfected, has been previously used in many studies of Rev (see, e.g., reference 33). Since only the env pre-mRNA is made from pgTat, the use of this system may look like a cleaner experimental design. In the pSVc21 model, the env intron probe detects both unspliced gag/pol mRNA and singly spliced env mRNA. The theoretical disadvantage of the pgTat system, however, is that the lack of important cis elements in the env pre-mRNA which are present in the full-length HIV pre-mRNA (splice signal, CRS/IN elements) may compromise the initial presentation of the env pre-mRNA to the host cell RNA-processing apparatus. Thus, a proportion of pgTat- and pgTat/pRev-transfected cells exhibited some viral RNA nuclear accumulation (25).
Although the proviral construct pSVc21 is more representative of the natural situation of Rev and incompletely spliced HIV RNA, we were able to find a similar viral RNA distribution using the pgTat model. Viral RNAs occurred regularly distributed on RNP-containing fibrils located within both the perichromatin and the interchromatin regions (Fig. 6A). Moreover, ISH with the env intron probe on cells treated with LMB also revealed clustered pattern of colloidal gold particles in this subgenomic model (Fig. 6B). Finally, cytoplasmic localization of the rev exon probe in such LMB-treated cells again showed that LMB gives rise to relocalization and clustering of only Rev/CRM1-dependent viral mRNAs and not all mRNAs (Fig. 6C). In addition, in LMB-treated cells transfected only with pgTat, no clustered gold particles were observed (data not shown).
FIG. 6.
In situ hybridization in cells transfected with subgenomic pgTat/pRev constructs. (A) Signal for env intron probe (12-nm colloidal gold) is diffusely distributed on the RNP-containing nuclear fibrils (arrowheads) and in the cytoplasm. (B) LMB treatment gives rise to viral RNA clustering, revealed here by the env intron probe. The gold particle clusters occur in the nucleoplasm and at the nuclear pores, while no signal is detected in the cytoplasm. (C) In LMB-treated cells, viral RNA detection by the rev exon probe gives rise to both clustered and diffuse labeling patterns in the nucleoplasm and also reveals RNAs diffusely distributed in the cytoplasm (arrowheads). Thin arrows show the nuclear periphery. Cy, cytoplasm. Bars, 0.2 μm.
Table 1 summarizes the results of investigations of the intracellular distribution of viral RNA revealed by a postembedding in situ RNA-RNA hybridization method. The quantitative evaluation of gold grain ISH signal in the nuclei of transfected and subsequently LMB-treated cells is summarized in Tables 2 and 3. Our results demonstrate that RNP fibrils are the major nucleoplasmic structural domains labeled with different antiviral probes. Furthermore, in the absence of Rev protein, the env intron-containing viral RNAs accumulate in the nucleoplasm, frequently in the periphery of IGs, indicating a possible role of this nucleoplasmic compartment in RNA degradation or sequestering. Finally, inhibition of Rev-dependent RNA export by LMB does not arrest the intranuclear RNA migration; Rev-dependent RNAs occur in an aggregated form and move throughout the nucleoplasm up to the nuclear pores but are unable to translocate across the nuclear envelope.
DISCUSSION
In the present work we have been able to demonstrate, using high-resolution ISH, the intracellular distribution of incompletely spliced HIV-1 mRNA and the viral (and cellular) proteins. This has been achieved by directly hybridizing ultrathin sections of transfected or HIV-1-infected cells with digoxigenin-labeled RNA probes, with the signal being revealed by colloidal-gold-conjugated antibodies. This method, avoiding denaturation of nucleic acid in the specimen, allowed us to keep the ultrastructural details of the cell nucleus well preserved as well as to see nuclear RNP-containing structural constituents under high contrast. Moreover, the method was suitable for simultaneous detection of viral RNA and proteins.
Observations carried out in our work on fine-structure localization of HIV-1 RNA and of Rev, Tat, and SC35 in COS cells transfected with proviral as well as subgenomic constructs and in HIV-1-infected CD4-positive lymphoid cells demonstrate the following points. (i) RNP fibrils observed in both perichromatin and interchromatin regions of transfected cell nuclei represent the major nucleoplasmic constituent labeled with all viral RNA molecular probes or antiprotein antibodies as well as with the anti-bromodeoxyuridine antibody revealing nascent RNA after BrUTP incorporation. No accumulation or track-like distribution of the labeled fibrils was observed. (ii) The extranuclear ISH signal is detected within the viral particles as well as in the ground cytoplasm. (iii) Following rev-minus transfection of the cells, the viral particles are not observed, the env intron-containing RNA accumulates in the nucleus, and only the RNA detected by the rev exon probe is revealed in the cytoplasm. Moreover, env intron-containing RNA also tends to occur at the periphery of interchromatin granule clusters. (iv) When LMB is used to inhibit Rev-dependent RNA export in the rev-plus system, the env intron probe reveals aggregation of viral RNA throughout the nucleoplasm and in the vicinity of the nuclear pores.
It has been demonstrated that perichromatin regions of the nucleoplasm are the major sites of RNA polymerase II transcription and that perichromatin fibrils (PF) represent the in situ forms of pre-mRNA transcripts. Moreover, numerous data indicate that this nucleoplasmic constituent is also the site of major pre-mRNA-processing events (for more details, see references 8, 12, 42, and 48). Furthermore, a migration of a fraction of PFs from perichromatin regions toward the nuclear interior was previously reported (13, 41). Our observations demonstrate that all viral RNA occurs in the nucleus mainly in the form of perichromatin fibril-like constituents and consequently strongly suggest that splicing of viral pre-mRNA on the one hand and binding of Rev protein to the Rev responsive element on the other hand take place on these structural constituents. In addition, occasional env intron and rev exon labeling was found close to or on the inside of the nuclear pores. In contrast, the condensed chromatin, the IG clusters, and the nucleolus did not accumulated HIV-1 RNA signal. It is known that the molecular mechanisms for the export of incompletely spliced HIV RNAs, being Rev and CRM1 dependent, differ from the mechanisms for the export of spliced HIV mRNA and host cell mRNA. Our observations show that Rev is present at the sites where the HIV-1 RNA occurs and is probably processed in the interchromatin and perichromatin regions. Pairwise, our colocalization studies demonstrate the occurrence of the viral RNA and of Rev protein or BrUTP-label on RNP fibrils in the above regions. Furthermore, both anti-Rev and anti-Tat antibodies labeled the nucleolar dense fibrillar and granular components, in agreement with previously reported data (10, 34), while the HIV RNA and the anti-SC35 probes never localized to the nucleolus. Therefore, we cannot confirm previous data suggesting nucleolar trafficking of HIV-1 RNA (36).
The present conclusion, therefore, is that the Rev-dependent RNA is not found in any special nuclear structural constituent different from other RNA polymerase II transcripts. However, while the cellular nucleoplasmic RNA is transcribed essentially in the perichromatin regions (see, e.g., reference 8), the transfected viral genome, which does not integrate into chromosomes, can obviously be transcribed and processed within both the perichromatin and interchromatin regions of nucleoplasm. Moreover, similarly to cellular pre-mRNA, the viral RNA does not occur in IGs, which accumulate the SC35 protein (reference 49 and this study) as well as other splicing factors (14, 43, 50). Furthermore, our observations do not support the idea that Rev-dependent HIV pre-mRNA and Rev-independent pre-mRNA accumulate in different nuclear domains (31, 32, 37).
Both the env intron and rev exon probes, as well as Rev protein, were also visualized in the cytoplasm without an evident accumulation, sometimes close to the cytoplasmic side of the nuclear pores. However, the paraformaldehyde fixation used in our assays is not optimal for the preservation of cytoplasmic details. The hybridization probes also detected the viral RNA in the viral particles, especially on the electron-dense core of the virions. The present method therefore has the potential for a simultaneous study of HIV-1 structure, assembly, and budding in the context of HIV-1 RNA and protein colocalization. We were unable to detect any Rev or Tat signal associated with the viral particles. The presence of the particles shows that our transfection conditions are consistent with the completion of the HIV-1 life cycle. Since different forms of defective particles have been reported in addition to the typical viral particles in infected lymphocytes (39), the occurrence of particles exhibiting abnormal features in our experiments is not surprising.
We were unable to observe any track-like features of the HIV RNA or Rev protein, and consequently we cannot favor the idea of a tracking pattern of the Rev-mediated export pathway (44). In addition, we have never seen any extended filamentous structures in the cell nucleus after either aldehydic fixation or cryofixation, so that, in contrast to a recent study reporting an association of viral RNA or Rev with nuclear fibrillar cable-like actin-containing structures (27), we have no evidence for such an association.
Hypothetically, along the route of RNA export there could exist transit regions, which would be detected only following treatment with inhibitors at those stages. As a first approach, the localization of the HIV pre-mRNA was determined when the Rev function was abrogated by a mutation introduced into pSVc21 to derive pSVc21B. The specificity of the system was verified by the disappearance of HIV virions and by the lack of cytoplasmic env intron labeling in the rev-minus system. This confirmed the results of previous studies of Rev function (reviewed, e.g., in reference 22). While the rev exon probe labeled both the nucleoplasm and the cytoplasm, the env intron signal was strictly confined to the nucleus in a pattern rather similar to that in the rev-plus system.
One notable difference was, however, an association of the env intron signal with the periphery of the IGs in rev-minus cells. This suggests that partially spliced or unspliced RNA could, in the absence of Rev protein, follow unusual intranuclear pathways and be attracted to domains known as splicing factor accumulation sites in the nucleus (12, 42, 48). The previous literature on the relationship between HIV RNA and speckles (IGs) is inconclusive (3, 4, 16, 46, 60). In investigations using more complete primary HIV transcripts, no enrichment in the interior of the SC35-containing speckles was found (3, 4) but deletion or addition of specific sequences in such HIV RNAs redirected the RNAs to the speckles (3). In the latter study, the fully spliced HIV mRNA was also found to accumulate in SC35-containing speckles (3), in contrast to results reported by Bøe et al. (4) and to our present observation. In studies of subgenomic HIV env RNA transcripts, no association of the env intron sequence with SC35-containing speckles (46, 60) was reported. Furthermore, we were not able to observe any association of the rev exon probe with the IGs in rev-minus cells. Whether this association of env intronic HIV RNA sequences with IGs reflects a dead-end pathway of Rev-dependent incompletely spliced HIV RNAs unable to move to the cytoplasm in the absence of Rev (24) or a possible accumulation of env introns resulting from increased splicing of Rev-unstabilized HIV pre-mRNA still remains to be elucidated. Both possibilities suggest an involvement of IGs in sequestering and/or degradation of some RNA sequences in the nucleus under conditions of altered processing.
Concerning the association of HIV-1 RNA with the nuclear pore complex, we have not been able to detect this when the env intron probe was assayed on the rev-minus system, suggesting a necessary role of Rev during the migration of incompletely spliced HIV RNA from the transcription site to the nuclear pores.
In a further attempt to reveal the transit sites of Rev-dependent HIV RNA transport, COS cells transfected with pSVC21 were treated with LMB for 3 or 12 h. While the rev exon probe signal distribution pattern was only moderately influenced by this treatment, the env intron probe distribution changed markedly. Similarly to pSVc21 plasmid, this alternation was also found in cells transfected with subgenomic pgTat/pRev plasmid. Much evidence indicates that the LMB interference with Rev activity is very specific (59). Presumably, the LMB-mediated alkylation of Cys529 of CRM1 prevents the functional association between Rev and CRM1 (29, 30). However, all known types of nuclear import and non-NES-dependent nuclear export can continue (6).
Our quantitative evaluation of the rev exon probe distribution reveals significant signal in the cytoplasm of rev-plus cells after LMB treatment. This shows that completely spliced and Rev-independent HIV-1 RNAs, not recognized by the env intron probe, are transported from the nucleus to the cytoplasm. Moreover, the clustered signal pattern identified by both probes in the nuclei of LMB-treated cells is similar after proviral or subgenomic transfection. Furthermore, LMB treatment does not give rise to viral RNA nuclear clustering in the rev-minus system (plasmids pSVc21B or pgTat) and the rev exon probe demonstrates the presence of spliced RNAs in the cytoplasm (Table 3). Finally, immunolabeling distribution patterns for the viral Rev and Tat, as well as for the cellular SC35 and CRM1 proteins, also do not seem to be modified after the LMB treatment compared with those in untreated cells. Taken together, these control observations strongly suggest that the LMB treatment does not influence the intracellular distribution of the fully spliced, Rev-independent HIV-1 mRNAs and that the viral RNA clustering and inhibition of HIV-1 transport by this drug is not likely to be a consequence of LMB cytotoxicity.
It is not clear whether the LMB-induced modification of CRM1 prevents its physical association with the NES sequences of Rev. Some previous data suggest that in in vitro assays only the association of Rev/CRM1 with RanGTP is inhibited by LMB (1) and, moreover, that LMB can even increase the interaction of CRM1 with some NES-containing proteins (5). Furthermore, a recent report indicates that a single amino acid mutation outside the NES region can restore the translocation of the complex through the nuclear pores independently of LMB action in a herpes simplex virus type 1 model (38). Our colocalization experiments with LMB-treated cells did not show a close association between the clusters of env intronic HIV RNA and the protein CRM1. This tends to support the former hypothesis about LMB preventing CRM1 association with Rev. However, the question regarding interactions of CRM1 with the HIV-1-Rev complex under LMB treatment conditions obviously still needs further investigations.
The increased accumulation of HIV env intron RNA at the nuclear pore in LMB-treated cells is therefore likely to demonstrate the critical requirement of CRM1 for the transit of Rev and its cargo RNA through the nuclear pore. It is probable that Rev protein is capable of transporting the Rev-dependent mRNA, although in an unusual clustered form, throughout the nucleoplasm toward the nuclear pores, while CRM1 is inactivated by LMB. Several reports have addressed the affinity between Rev and phenylalanine-glycine (FG)-containing nuclear pore proteins, but it is still controversial whether this affinity is mediated entirely by interposition of the CRM1 or whether some additional direct binding between Rev and the FG-containing nuclear pore proteins occurs (reference 15 and references therein).
Previous immunofluorescence studies showed that LMB treatment retained influenza virus RNPs in the nuclei of infected cells and that the viral RNA binding nucleoprotein redistributed mainly to the nuclear periphery (11, 31, 56). Unfortunately, RNA was not directly visualized in those studies and the resolution of fluorescence microscopy precludes a detailed localization with regards to nuclear structural domains.
Since nucleoplasmic clusters of incompletely spliced HIV RNA occur only in rev-plus cells following LMB treatment, it is likely that the association of the Rev-RNA complex and CRM1 takes place in the nucleoplasm early after formation of the Rev-RNA complex. Whether such clustering reflects some kind of channeled or directed movement different from free diffusion remains to be elucidated.
Furthermore, we observed, an accumulation of gold grains in clusters at the nuclear pores after 12 h of LMB incubation, compared with 3 h of incubation, while the overall level of labeling in the nucleus remained similar (Table 2).
Our findings demonstrate that Rev itself, in the absence of its functional association with CRM1, is necessary and probably also sufficient for accumulation of unspliced HIV RNA at the nuclear pore complexes, while CRM1 is needed for further translocation across the pore. Considering the lack of evidence for a direct interaction of Rev with nucleoporins, it is not clear whether Rev is involved in directing its cargo RNA through the nucleus toward the pores or in docking it at the nuclear pore complexes in the case of diffusion migration within the nucleus. Whether the labeled clusters, occurring when the CRM1-Rev interaction is not functional, reflect an RNA accumulation through some unusual Rev association or further multimerization remains a matter for speculation.
Acknowledgments
We thank J. Fakan, V. Mamin, F. Voinesco, and B. Johannesen for excellent technical assistance and W. Blanchard for photographic work. We also thank J. Cmarko for assistance with the preparation of the manuscript, B. Wolff for the kind gift of leptomycin B, and D. E. Helland for the use of the anti-Tat antibody 1D9D5.
C.S. benefited from a fellowship from the University of Perugia and from the Fondation du 450e Anniversiare de l'Université de Lausanne. This work was supported by the Swiss National Science Foundation (grant 31-53944.98 to S.F.). K.-H.K. was supported by the Research Council of Norway, The Norwegian Cancer Society, and the Family Blix' Legacy; and G.H. was supported by the Sonneborn Charitable Trust.
REFERENCES
- 1.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273:33414-33422. [DOI] [PubMed] [Google Scholar]
- 2.Bernhard, W. 1969. A new staining procedure for electron microscopical cytology. J. Ultrastruct. Res. 27:250-265. [DOI] [PubMed] [Google Scholar]
- 3.Berthold, E., and F. Maldarelli. 1996. cis-Acting elements in human immunodeficiency virus type 1 RNAs direct viral transcription to distinct intranuclear location. J. Virol. 70:4667-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bøe, S.-O., B. Bjorndal, B. Rosok, A. M. Szilvay, and K.-H. Kalland. 1998. Subcellular localization of human immunodeficiency virus type 1 RNAs, Rev, and the splicing factor SC-35. Virology 244:473-482. [DOI] [PubMed] [Google Scholar]
- 5.Brennan, C. M., I.-E. Gallouzi., and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brokstad, K. A., K.-H. Kalland, W. C. Russell, and D. A. Matthews. 2001. Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 281:1161-1169. [DOI] [PubMed] [Google Scholar]
- 7.Carlemalm, E., R. M. Garavito, and W. Villiger. 1982. Resin development for electron microscopy and an analysis of embedding at low temperature. J. Microsc. 126:123-143. [DOI] [PubMed] [Google Scholar]
- 8.Cmarko, D., P. J. Verschure, T. E. Martin, M. E. Dahmus, S. Krause, X.-D. Fu, R. van Driel, and S. Fakan. 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol. Biol. Cell 10:211-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen, B. R. 1998. Retroviruses as model systems for the study of nuclear RNA export pathways. Virology 249:203-210. [DOI] [PubMed] [Google Scholar]
- 10.Dundr, M., G. H. Leno, M.-L. Hammarskjöld, D. Rekosh, C. Helga-Maria, and M. O. J. Olson. 1995. The role of nuclear structure and function in the subcellular location of the HIV-1 Rev protein. J. Cell Sci. 108:2811-2823. [DOI] [PubMed] [Google Scholar]
- 11.Elton, D., M. Simpson-Holley, K. Archer, L. Medcalf, R. Hallam, J. McCauley, and P. Digard. 2001. Ineraction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fakan, S. 1994. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 4:86-90. [DOI] [PubMed] [Google Scholar]
- 13.Fakan, S., E. Puvion, and G. Spohr. 1976. Localization and characterization of newly synthesized nuclear RNA in isolated rat hepatocytes. Exp. Cell Res. 99:155-164. [DOI] [PubMed] [Google Scholar]
- 14.Fakan, S., G. Leser, and T. E. Martin. 1984. Ultrastructural distribution of nuclear ribonucleoproteins as visualized by immunocytochemistry on thin sections. J. Cell Biol. 98:358-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farjot, G., A. Sergeant, and I. Mikaélian. 1999. A new nucleoporin-like protein interacts with both HIV-1 Rev nuclear export signal and CRM-1. J. Biol. Chem. 274:17309-17317. [DOI] [PubMed] [Google Scholar]
- 16.Favaro, J. P., K. T. Borg, S. J. Arrigo, and M. G. Schmidt. 1998. Effect of Rev on the intranuclear localization of HIV-1 unspliced RNA. Virology 249:286-296. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, D., D. Weisenberger, and U. Scheer. 1996. In situ hybridization of DIG-labeled rRNA probes to mouse liver ultrathin sections, p. 148-151. In Nonradioactive in situ hybridization. Application manual, 2nd ed. Boehringer, Mannheim, Germany.
- 18.Fischer, U., J. Huber, W. C. Bolens, I. W. Mattaj, and R. Lührmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 19.Fischer, U., V. W. Pollard, R. Lührmann, M. Teufel, M. W. Michael, G. Dreyfuss, and M. H. Malim. 1999. Rev mediated nuclear export of RNA is dominant over nuclear retention and is coupled to the Ran-GTPase cycle. Nucleic Acids Res. 27:4128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, X.-D., and T. Maniatis. 1990. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature 343:437-441. [DOI] [PubMed] [Google Scholar]
- 21.Haukenes, G., and K.-H. Kalland. 1998. Visualisation of ribosomal RNA synthesis in eukaryotic cells in culture. Methods Cell Sci. 19:295-302. [Google Scholar]
- 22.Heguy, A. 1997. Inhibition of the HIV Rev transactivator: a new target for therapeutic intervention. Front. Biosci. 2:283-293. [DOI] [PubMed] [Google Scholar]
- 23.Hope, T. J. 1999. The ins and outs of HIV Rev. Arch. Biochem. Biophys. 365:186-191. [DOI] [PubMed] [Google Scholar]
- 24.Iacampo, S., and A. Cochrane. 1996. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lawrence, J. B., A. W. Cochrane, C. V. Johnson, A. Perins, and C. A. Rosen. 1991. The HIV-1 Rev protein: a model system for coupled RNA transport and translation. New Biol. 3:1221-1232. [PubMed] [Google Scholar]
- 26.Kalland, K.-H., A. M. Szilvay, K. A. Brokstad, W. Sætrevik, and G. Haukenes. 1994. The human immunodeficiency virus type 1 (HIV-1) Rev protein shuttles between the cytoplasm and nuclear compartments. Mol. Cell. Biol. 14:7436-7444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kimura, T., I. Hashimoto, A. Yamamoto, M. Nishikawa, and J. Fujisawa. 2000. Rev-dependent association of the intron-containing HIV-1 gag mRNA with the nuclear actin bundles and the inhibition of its nucleoplasmic transport by latrunculin-B. Genes Cells 5:289-307. [DOI] [PubMed] [Google Scholar]
- 28.Kjems, J., and P. Askjaer. 2000. The Rev-protein and its cellular partners. Adv. Pharmacol. 48:251-297. [DOI] [PubMed] [Google Scholar]
- 29.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 30.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cystein residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma, K., A.-M. M. Roy, and G. R. Whittaker. 2001. Nuclear export of influenza virus ribonucleoproteins: identification of an export intermediate at the nuclear periphery. Virology 282:215-220. [DOI] [PubMed] [Google Scholar]
- 32.Malim, M. H., and B. R. Cullen. 1993. Rev and the fate of pre-mRNA in the nucleus: implications for the regulation of RA processing in eukaryotes. Mol. Cell. Biol. 13:6180-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malim, M. H., J. Hauber, S. Y. Le, J. V. Maizel, and B. R. Cullen. 1989. The HIV-1 rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 338:254-257. [DOI] [PubMed] [Google Scholar]
- 34.Marasco, W. A., A. M. Szilvay, K.-H. Kalland, D. G. Helland, H. M. Reyes, and R. J. Walter. 1994. Spatial association of HIV-1 tat protein and the nucleolar transport protein B23 in stably transfected Jurkat T-cells. Arch. Virol. 139:133-154. [DOI] [PubMed] [Google Scholar]
- 35.Meyer, B. E., and M. H. Malim. 1994. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 8:1538-1547. [DOI] [PubMed] [Google Scholar]
- 36.Michienzi, A., L. Cagnon, I. Bahner, and J. J. Rossi. 2000. Ribozyme-mediated inhibition of HIV-1 suggests nucleolar trafficking of HIV-1 RNA. Proc. Natl. Acad. Sci. USA 97:8955-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikaelian, I., M. Krieg, M. J. Gait, and J. Karn. 1996. Interactions of INS (CRS) elements and the splicing machinery regulate the production of Rev-responsive mRNA. J. Mol. Biol. 257:246-264. [DOI] [PubMed] [Google Scholar]
- 38.Murata, T., F. Goshima, T. Koshizuka, H. Takakuwa, and Y. Nishiyama. 2001. A sungle amino acid substitution in the ICP27 protein of herpes simplex virus type 1 is responsible for its resistance to leptomycin B. J. Virol. 75:1039-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakai, M., and T. Goto. 1996. Ultrastructure and morphogenesis of human immunodeficiency virus. J. Electron Microsc. 45:247-157. [DOI] [PubMed] [Google Scholar]
- 40.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 41.Puvion, E., and G. Moyne. 1978. Intranuclear migration of newly synthesized extranucleolar ribonucleoproteins. A high resolution quantitative autoradiographical and cytochemical study. Exp. Cell Res. 115:79-88. [DOI] [PubMed] [Google Scholar]
- 42.Puvion, E., and F. Puvion-Dutilleul. 1996. Ultrastructure of the nucleus in relation to transcription and splicing: roles of perichromatin fibrils and interchromatin granules. Exp. Cell Res. 229:217-225. [DOI] [PubMed] [Google Scholar]
- 43.Puvion, E., A. Viron, C. Assens, E. H. Leduc, and P. Jeanteur. 1984. Immunocytochemical identification of nuclear structures containing snRNPs in isolated rat liver cells. J. Ultrastruct. Res. 87:180-189. [DOI] [PubMed] [Google Scholar]
- 44.Romanov, V. I., A. S. Zolotukhin, N. N. Aleksandroff, P. Pinto da Silva, and B. K. Felber. 1997. Posttranscriptional regulation by Rev protein of human immunodeficiency virus type 1 results in nonrandom nuclear localization of gag mRNA. Virology 228:360-370. [DOI] [PubMed] [Google Scholar]
- 45.Rosorius, O., B. Reichert, F. Krätzer, P. Heger, M.-C. Dabauvalle, and J. Hauber. 1999. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J. Cell Sci. 112:2369-2380. [DOI] [PubMed] [Google Scholar]
- 46.Séguin, B., A. Staffa, and A. Cochrane. 1998. Control of human immunodeficiency virus type 1 RNA metabolism: role of spliced sites and intron sequences in unspliced viral RNA subcellular distribution. J. Virol. 72:9503-9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siegel, S., and N. J. Castellan. 1988. Nonparametric statistics for the behavioral sciences. McGraw-Hill Book Co, New York, N.Y.
- 48.Spector, D. L. 1996. Nuclear organization and gene expression. Exp. Cell Res. 229:189-197. [DOI] [PubMed] [Google Scholar]
- 49.Spector, D. L., X.-D. Fu, and T. Maniatis. 1991. Association between distinct pre-mRNA splicing components and the cell nucleus. EMBO J. 10:3467-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spector, D. L., W. H. Schrier, and H. Busch. 1983. Immunoelectron microscopic localization of snRNPs. Biol. Cell. 49:1-10. [DOI] [PubMed] [Google Scholar]
- 51.Stutz, F., E. Izaurralde, I. W. Mattaj, and M. Rosbash. 1996. A role for nucleoporin FG repeat domains in export of human immunodeficiency virus type 1 Rev protein and RNA from the nucleus. Mol. Cell. Biol. 16:7144-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Szilvay, A. M., K. A. Brokstad, R. Kopperud, G. Haukenes, and K.-H. Kalland. 1995. Nuclear export of the nucleoplasmic shuttle protein HIV-1 Rev is mediated by its activation domain and is blocked by transdominant negative mutants. J. Virol. 69:3315-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Szilvay, A. M., S.-O. Bøe, and K.-H. Kalland. 1999. Co-expression of a trans-dominant negative mutant of the human immunodeficiency virus type 1 (HIV-1) Rev protein affects the Rev-dependent splicing pattern and expression of HIV-1 RNAs. J. Gen. Virol. 80:1965-1974. [DOI] [PubMed] [Google Scholar]
- 54.Valvatne, H., A. M. Szilvay, and D. E. Helland. 1996. A monoclonal antibody defines a novell HIV-1 Tat domain involved in trans-cellular trans-activation. AIDS Res. Hum. Retroviruses 12:611-619. [DOI] [PubMed] [Google Scholar]
- 55.von Schack, M., and S. Fakan. 1993. The study of the cell nucleus using cryofixation and cryosubstitution. Micron 24:507-519. [Google Scholar]
- 56.Watanabe, K., N. Takizawa, M. Katoh, K. Hoshida, N. Kobayashi, and K. Nagata. 2001. Inhibition of nuclear export of ribonucleoprotein complexes of influenza virus by leptomycin B. Virus Res. 77:31-42. [DOI] [PubMed] [Google Scholar]
- 57.Whittaker, G. R., and A. Helenius. 1998. Nuclear import and export of viruses and virus genomes. Virology 246:1-23. [DOI] [PubMed] [Google Scholar]
- 58.Wolff, B., G. Cohen, J. Hauber, D. Meshceryakova, and C. Rabeck. 1995. Nucleocytoplasmic transport of the Rev protein of human immunodeficiency virus type 1 is dependent on the activation domain of the protein. Exp. Cell Res. 217:31-41. [DOI] [PubMed] [Google Scholar]
- 59.Wolff, B., J.-J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, G., M. L. Zapp, G. Yan, and M. R. Green. 1996. Localization of HIV-1 RNA in mammalian nuclei. J. Cell Biol. 135:9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]