Abstract
To elucidate the mechanisms by which Epstein-Barr virus (EBV) latency III gene expression transforms primary B lymphocytes to lymphoblastoid cell lines (LCLs), the associated alterations in cell gene expression were assessed by using 4,146 cellular cDNAs arrayed on nitrocellulose filters and real-time reverse transcription-PCR (RT-PCR). A total of 1,405 of the 4,146 cDNAs were detected using cDNA probes from poly(A)+ RNA of IB4 LCLs, a non-EBV-infected Burkitt's lymphoma (BL) cell line, BL41, or EBV latency III-converted BL41 cells (BL41EBV). Thirty-eight RNAs were consistently twofold more abundant in the IB4 LCL and BL41EBV than in BL41 by microarray analysis. Ten of these are known to be EBV induced. A total of 23 of 28 newly identified EBV-induced genes were confirmed by real-time RT-PCR. In addition, nine newly identified genes and CD10 were EBV repressed. These EBV-regulated genes encode proteins involved in signal transduction, transcription, protein biosynthesis and degradation, and cell motility, shape, or adhesion. Seven of seven newly identified EBV-induced RNAs were more abundant in newly established LCLs than in resting B lymphocytes. Surveys of eight promoters of newly identified genes implicate NF-κB or PU.1 as potentially important mediators of EBV-induced effects through LMP1 or EBNA2, respectively. Thus, examination of the transcriptional effects of EBV infection can elucidate the molecular mechanisms by which EBV latency III alters B lymphocytes.
In primary Epstein-Barr virus (EBV) infection, a substantial fraction of peripheral blood B lymphocytes are latently infected and express virus-encoded proteins that are capable of causing continuous B-cell proliferation. The EBV-encoded proteins engender an unusually vigorous and large T-cell immune response which eliminates most of the virus-infected cells. In severely T-cell immune-compromised or otherwise susceptible people, the EBV-infected B lymphocytes can malignantly proliferate (reviewed in reference 90).
The EBV gene products that are expressed in primary lymphocyte infection and that stimulate cell proliferation include six nuclear antigen proteins (EBNAs), two latent infection integral membrane proteins (LMPs), two small RNAs (EBERs), and BamA rightward transcripts (BARTs) of uncertain function (reviewed in reference 59). This complex infection, termed latency III, is characteristic of EBV gene expression in EBV-transformed lymphoblastoid cell lines (LCLs) and many lymphoproliferations that occur in immune-deficient humans (23, 95).
Many of the EBV effects on cell growth and survival are likely to be mediated by EBV-induced changes in cell gene transcription. Five of the EBNAs can alter cell gene transcription through their direct or indirect interaction with the cellular DNA sequence-specific transcription factors RBP-jκ/CBF1, PU.1, and AUF1 (35, 42, 52, 53, 65, 72, 91, 113, 119, 123). Furthermore, EBV LMP1 is similar to a constitutively activated CD40 receptor and activates NF-κB, Jun/Fos, and AP1 cellular transcription factors (27, 28, 30, 33, 36, 40, 45, 50, 60, 66, 81, 83). Moreover, EBV LMP2 is similar to a constitutively activated B-cell antigen receptor (BCR) in causing a stable small increase in BCR tyrosine kinase signaling and in desensitizing the cell to BCR-type signal transduction (12, 80). Four of the EBNAs that interact with cellular transcription factors and LMP1 are critical for EBV effects on cell growth and survival, while EBNA3B, LMP2, EBERs, and BARTs are not (19, 37, 49, 56, 68, 73, 76, 92, 102, 108, 109).
In the experiments reported here, cDNA arrays were used to investigate the effects of EBV latency III infection on cell gene transcription. cDNA arrays are particularly useful in characterizing changes in expression of large numbers of cellular genes (3, 4, 48, 51, 57, 74, 97, 100). We compared gene expression in BL41, an EBV-negative Burkitt's lymphoma (BL) cell line, with gene expression in two latency III-expressing cell lines—BL41 infected in vitro with EBV (BL41EBV) and an LCL (IB4) (18, 62). In contrast to a comparison of LCLs with resting B lymphocytes, which would identify a large number of cell genes whose expression is up-regulated at various points in the cell cycle, comparison with a non-EBV-infected BL cell line will identify genes that are specifically up-regulated by EBV latency III proteins. However, EBV-regulated genes whose transcription is inherent to the BL phenotype, such as c-myc, may escape detection.
MATERIALS AND METHODS
Cell lines.
BL41 and BL30 are EBV-negative B-lymphoma cell lines with c-myc translocations and p53 point mutations (18, 28a). BL41EBV and BL30EBV are the respective BL lines infected in vitro with the prototypic EBV strain B95-8 (13, 29). The LCL used, IB4, is an EBV-transformed B-lymphoblastoid cell line with four integrated copies of the genome (41, 46). All cell lines were maintained in RPMI (Gibco) plus 10% fetal calf serum (HyClone).
Preparation of RNA.
RNAs were extracted from cells maintained in log-phase growth. Cells were seeded at 200,000 cells per ml, 24 h prior to RNA preparation. RNA was prepared using RNAzol (Tel-test) according to the manufacturer's instructions. RNA was resuspended in diethyl pyrocarbonate-treated water, aliquoted, and stored at −80°C. Poly(A)+ RNA was purified using Oligotex (Qiagen) according to the manufacturer's directions.
Filter hybridizations.
Probes were generated by labeling first-strand cDNA from 5 μg of poly(A)+ RNA. Probes were primed with oligo(dT) and extended at 42°C with 1 mM concentrations of deoxynucleoside triphosphates minus dCTP (Gibco), 60 μCi of [33P]dCTP (NEN), 10 mM dithiothreitol (DTT), 1 U of RNasin (Promega), and first-strand buffer (50 mM Tris-HCl [pH 8.3], 75 mM KCl, 3 mM MgCl2 [Gibco]), and 800 U of Superscript II reverse transcriptase (Gibco). RNA was digested from the probes with RNase H (Gibco) followed by 250 mM NaOH (Sigma) at 65°C. Probes were separated from unincorporated nucleotides by using ProbeQuant G-50 Micro columns (Amersham Pharmacia). All probes were analyzed by polyacrylamide gel electrophoresis and autoradiography to ensure that probes of greater than 300 bp had been generated. Probes were heated to 95°C for 5 min and then hybridized with Human Named GeneFilters (Research Genetics) in Microhyb hybridization solution (Research Genetics) plus 25 μg of poly(dA) (Research Genetics) and 5 μg of heat-denatured Cot-1 DNA (Gibco). Hybridizations were carried out at 42°C for 16 to 24 h. Filters were washed twice with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 1% sodium dodecyl sulfate (SDS) and once with 0.5× SSC and 1% SDS at 55°C. Filters were exposed to phosphorimager cassettes and analyzed with a Phosphorimager SI (Molecular Dynamics).
Analysis of filter data.
Scanned phosphorimages were analyzed using P-scan 1.1 software from the National Institutes of Health (http://abs.cit.nih.gov/pscan). Cluster analysis and profile images were generated using the software Cluster and Treeview from the Eisen lab (http://rana.lbl.gov/EisenSoftware.htm) (26).
Real-time RT-PCR.
Reverse transcription (RT) was carried out using 400 ng of total RNA and the TaqMan reverse transcription reagents (PE Applied Biosystems) in a 50-μl reaction mixture. Four microliters of the RT reaction mixture was used for real-time PCR using the SYBR green PCR core reagents (PE Applied Biosystems) and sequence-specific primers. The primers used were interferon-inducible 9-27 forward 5′TCAACATCCACAGCGAGACC3′ and reverse 5′GAAGAGGGTGTTGAACAGGGAC3′, cystatin B forward 5′GAGCGTGCACTTGTGATCCTAA3′ and reverse 5′GCCCCTTCCACCCCAA3′, enolase 2 forward 5′TAGTTCACCCCCTGAGATCCC3′ and reverse 5′CAGCGGAGCAGGTCAATCA3′, YMP forward 5′CTATGCCACCGGCCTCTG3′ and reverse 5′AGATCAAGGCGCCAGTAAACA3′, BASP1 (NAP-22) forward 5′CCCAAGCCGGTGGAGG3′ and reverse 5′TGTCCTTGTCACTCTTTCACGG3′, NK4 forward 5′AGAAAGAGATGGATTACGGTGCC3′ and reverse 5′TCGGTTGCGGGATCCTC3′, PAC-1 forward 5′AGGCTTCATTGACTGGGTGAA3′ and reverse 5′AGATACCCGCCTGGCAGTG3′, phosphoglycerate kinase forward 5′GCTGGCTGGATGGGCTT3′ and reverse 5′TTAGCCCGAGTGACAGCCTC3′, TRAP delta forward 5′ATTTCCATCATCCCGCCTCT3′ and reverse 5′AGGGCCCGTTCCAAGTG3′, interferon-inducible IP30 forward 5′CGGAGTGTTTGCTTCGAGTGT3′ and reverse 5′AGTTCCCACTCGCCTTCCAT3′, ATPase subunit M9.2 forward 5′TACCATGTTGGTGACCTGTTCAG3′ and reverse 5′GGGTTGAGTTGGGCCAGAA3′, BRF1 forward 5′TCCGGCCCATGTCCG3′ and reverse 5′CGAGAGAGAATCCTGAGGGCT3′, DNAS1L3 forward 5′CCCCAAGAAGGCCTGGAA3′ and reverse 5′TTGGTCCCCGATCAGCC3′, HCK forward 5′AGCGCCAACTGCTGGCT3′ and reverse 5′TCTCGCTATCCCGGATCATG3′, HPK1 forward 5′GGGATCCCAGATGCAGACTG3′ and reverse 5′TCTCCATTCCTCGGAGCTTC3′, heat shock factor 1 forward 5′AACAGAAAGTCGTCAACAAGCTCAT3′ and reverse 5′CTTTCTCTTCACCCCCAGGAT3′, human transcription factor forward 5′GGAGGGCAACTTATCAGGCATG3′ and reverse 5′TCGGACACTTCCCTTTCTGC3′, initiation factor 4B forward 5′CCAATTGACCGTTCCATCCT3′ and reverse 5′GGTCGATATTGGGTTCCCG3′, interferon-inducible 1-8U forward 5′ATCCACATCCGCAGCGAG3′ and reverse 5′AGGGTGTTGAACAGGGACCA3′, galectin 9 forward 5′TTACCCAGACAGTCATCCACACA3′ and reverse 5′GGGATGGCGGGAGTAGAGAA3′, interferon-inducible 1-8D forward 5′ACCGCCAAGTGCCTGAAC3′ and reverse 5′GGATGATGACGAGCAGAATGG3′, aldolase A forward 5′CACATCTACCTGGAAGGCACC3′ and reverse 5′CTTCTGAGTGCAAGCATGGC3′, cathepsin C forward 5′TTTCTATGGAGGCTGCAATGAA3′ and reverse 5′AGCAACTGCCATGGGCC3′, FYN forward 5′ACAAGGTGCAAAGTTCCCCA3′ and reverse 5′TGAACCTCCCGTACAGGGC3′, interferon-induced microtubule aggregating protein forward 5′CGTTACAGCCCTGCATTTGA3′ and reverse 5′ATTGCGGCACACACCAGTACAG3′, 40S ribosomal protein S8 forward 5′CATCTCTCGGGACAACTGGC3′ and reverse 5′TCTTGTGGTAGGGCTTTCTCTTG3′, hepatocyte nuclear factor dimerization factor forward 5′TTGCTGTGGGATGTGCCA3′ and reverse 5′ GAAGAGAGTGGTGCAGGGAAAA3′, alpha 1 interferon forward 5′CCGAACTCTACCAGCAGCTGA3′ and reverse 5′GGAGTTTCTCCCACCCTCTCC3′, beta interferon forward 5′CTCCGAAACTGAAGATCTCCTAGC3′ and reverse 5′TGCTGGTTGAAGAATGCTTGA3′, and gamma interferon forward 5′GCAGCTAAAACAGGGAAGCG3′ and reverse 5′GGACAACCATTACTGGGATGCT3′. PCRs were cycled in a GeneAmp 5700 sequence detection system and analyzed with GeneAmp 5700 SDS software (version 1.1; PE Applied Biosystems). Cycle times were analyzed at a reading of 0.2 fluorescence units. All reactions were done in duplicate. Cycle times that varied by more than 1.0 between the duplicates were discarded. The duplicate cycle times were then averaged and the cycle time of p100 was subtracted from them for a normalized value. The normalized values for BL41EBV or IB4 were subtracted from values for BL to determine a normalized cycle time difference for the appearance of the transcript. For the comparisons of resting B lymphocytes to LCLs and BL30 to BL30EBV, no normalization to p100 was performed because p100 mRNA levels varied substantially in these comparisons, whereas it does not vary in BL41, BL41EBV, and IB4. Rather, equal masses of total RNA from multiple RNA preparations were used, yielding consistent results.
Promoter analysis.
Five hundred base pairs 5′ of the transcriptional start site were identified using TRASER (http://genome-www6.stanford.edu/cgi-bin/Traser/traser) and analyzed using AliBaba2.1 and the Transfac database (http://www.gene-regulation.com).
Establishment of LCLs and purification of resting B cells.
Peripheral blood mononuclear cells (PBMC) from normal donors were infected with the B95-8 strain of EBV in the presence of 0.5 μg of cyclosporine A/ml. Total RNA was extracted from bulk cultures at 3 months (LCL1) and 2 months (LCL2) postinfection. Resting B cells were purified from 108 PBMC by negative selection using a human B-lymphocyte prep column (Caltag) according to the manufacturer's instructions.
RESULTS
Effect of EBV latency III infection on cell gene expression.
Research Genetics gf211 filters that have 4,146 partially redundant IMAGE cDNA array elements, 178 total genomic control spots, and 1,436 no-DNA control spots were successively hybridized to poly(A)+ RNA-directed first-strand cDNA probes from EBV-negative BL41 cells, from BL41 stably converted to latency III by EBV infection (BL41EBV), and from IB4 LCLs. BL41EBV and IB4 cells express EBNAs and LMP1 that are characteristic of latency III EBV infection. Poly (A)+ RNA was used in these experiments because on average 1,352 cDNAs were detectable at twofold over background with poly(A)+ RNA-directed probes, whereas only 930 cDNAs were detected with total RNA-directed probes (data not shown). Signal intensities were analyzed by P-SCAN 1.1 software, which normalizes each cDNA signal to the average signal intensity for all array elements on the filter. Normalized intensities were then used to compare hybridizations of different probes to the same filter.
Array elements were considered to have detected RNA if their hybridized radioactivity was at least twice the mean background with probes from BL41, BL41EBV, or IB4 RNAs. Of the 1,436 no-DNA control spots, only 2 to 3% were greater than two times the mean on any filter, whereas of 4,146 cDNA array elements, 34% (1,405) gave a signal that was at least twice the background with repeat hybridizations using probes from BL41, BL41EBV, or IB4 poly(A)+ RNA.
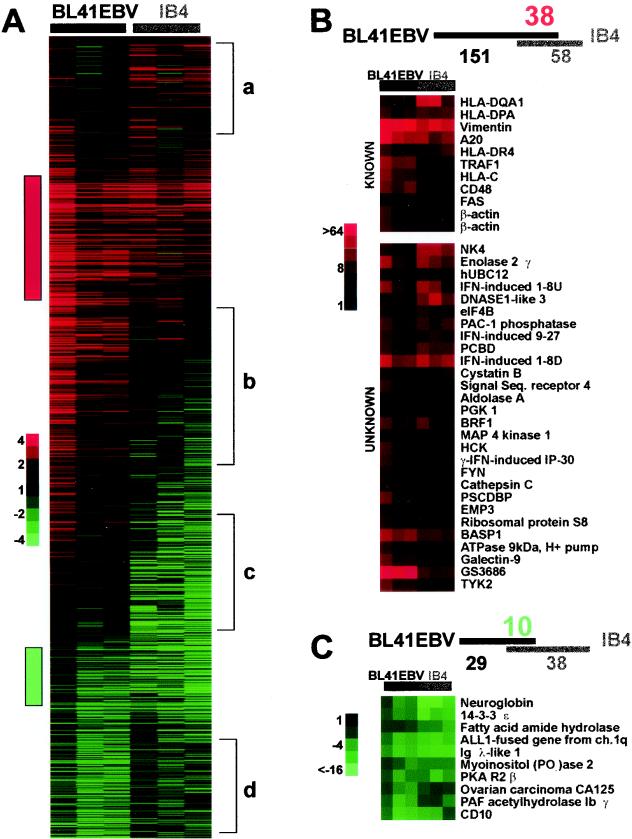
In Fig. 1A, each column represents an individual filter and each row represents a particular array element. The data are presented as the ratio of normalized intensities for each gene with BL41EBV or IB4 cells divided by the normalized intensities with non-EBV-infected BL41 cells on the identical filter. The ratios for BL41EBV to BL41 and IB4 to BL41 were ordered simultaneously using the Cluster self-organizing map (SOM) function (26). A strong red or green signal indicates fourfold higher or lower abundance, respectively, than in BL41. Black is indicative of no difference from the level in uninfected BL41 cells. Many genes were affected by EBV infection, as evidenced by the red or green.
FIG. 1.
(A) Effect of latency III EBV infection on cellular gene expression. Vertical columns represent hybridization to a single filter; a row represents a single cDNA array element. Values are displayed as fold changes (normalized counts detected from 33P-labeled first-strand cDNAs generated from BL41EBV or IB4 divided by those from BL41). Red bar, genes highly expressed in both BL41EBV and IB4. Green bar, genes suppressed by EBV in BL41EBV and IB4. Group a genes are highly expressed in IB4 but unaffected by EBV in BL41; group b genes are highly induced by EBV in BL41EBV but not highly expressed in IB4; and group c genes are unaffected by EBV in BL41 but expressed at a low level in IB4; group d genes are repressed by EBV in BL41EBV and are unaffected in IB4. (B) Intersection of genes induced by EBV twofold or greater in both BL41EBV and IB4, including known and newly identified genes. (C) Intersection of genes repressed by EBV twofold or greater in both BL41EBV and IB4. Scales are indicated.
The bars labeled a or b at the right of the SOM in Fig. 1A demarcate RNAs that are expressed at a higher level in IB4 (group a) or BL41EBV (group b) relative to BL41 but not in both BL41EBV and IB4, whereas the bars labeled c or d demarcate RNAs that are expressed at a lower level in IB4 (group c) or BL41EBV (group d) relative to BL41 but not in both BL41EBV and IB4. These seeming inconsistencies may be due to constitutive c-myc expression or some other dominant transcription factor effect in the BL cells that may obscure the effect of EBV on these genes in the BL41 background or may alter expression in BL41 relative to IB4. Less likely, some RNAs may be differentially expressed in BL41EBV or IB4 because BL41EBV lacks EBER expression and IB4 lacks EBNA3B expression. However, EBERS and EBNA3B are not critical for B-lymphocyte conversion to LCLs (102, 108). Interestingly, a number of genes in group a have related functions, such as cyclophilin C, cyclophilin B, and FKBP-associated protein 48 or coronin-1 and p40phox.
The red bar to the left of the SOM in Fig. 1A demarcates 195 RNAs that are expressed at least twofold higher in at least two of three hybridizations with BL41EBV and IB4 cDNAs, whereas the green bar indicates 344 RNAs that are expressed at least twofold lower in at least two of three hybridizations with BL41EBV and IB4 cDNAs. The former are likely to be EBV latency III-induced genes, whereas the latter are likely to be EBV latency III-repressed genes. These genes were further investigated.
EBV-induced genes.
Of the 195 putative RNAs indicated with a red bar, 151 in BL41EBV and 58 in IB4 were at least twofold more abundant than in BL41 in three of three hybridizations. Thirty-eight unique genes encoded the RNAs that were twofold more abundant in all experiments with both BL41EBV and IB4 cDNAs (Fig. 1B). Included in the 38 are 10 previously known EBV-induced genes, such as major histocompatibility antigens, vimentin, A20, TRAF1, CD48, and FAS, all of which are LMP1 induced and NF-κB up-regulated (6, 8, 24, 63, 66, 120). Neither A1/Bfl1 nor IκBα were among the 38 because they were more than twofold induced in only five of six filters. ICAM1 was not among the 38 because it was not detected. Of the 28 new candidate EBV-induced genes, NK4 and PAC-1 may have been anticipated to be EBV induced because they were known to be NF-κB regulated (70, 99), and EBV LMP1 strongly up-regulates NF-κB.
Of the genes newly identified as EBV induced, four are known to be interferon (IFN) induced and one, Tyk2, is important in IFN signaling (5, 69, 112). Since IFNs are not among the arrayed cDNAs, real-time RT-PCR was used to quantify steady-state IFN mRNA levels. Although IFN-α1, -β, and -γ were two- to fourfold more abundant in LCLs than in BL41 cells, IFN-γ was unaffected by EBV infection in BL41EBV, and IFN-α and IFN-β were suppressed two- to fourfold. Thus, increased expression of IFN-regulated genes in BL41EBV is not likely to be due to IFN induction (data not shown).
EBV-repressed genes.
Although 344 array elements clustered as EBV repressed (Fig. 1A), most changed less than twofold. Only 29 array elements were less abundant in all three BL41EBV RNA hybridizations, 38 were less abundant in all three IB4, and 10 genes were less abundant in both BL41EBV and IB4 cells (Fig. 1C). The 10 less abundant genes included the gene for CD10; CD10 is known to be repressed by latency III EBV infection in BL cells (25, 115). For the remaining genes, there is no indication of a common pathway that may impact their promoters. However, 14-3-3 epsilon and protein kinase A (PKA) regulatory subunit 2 beta may be functionally related since 14-3-3 proteins can affect the subcellular localization of PKA-phosphorylated proteins (84).
RT-PCR validation of EBV-induced genes.
Expression of EBV-induced genes was validated by real-time RT-PCR with p100 RNA as an internal control; p100 RNA abundance was the same in BL41, BL41EBV, and IB4 by Northern blotting (data not shown). Titration of either first-strand cDNA or plasmid DNA for p100 showed that one cycle change correlates with an approximate twofold difference in RNA abundance (data not shown). For most genes the fold change on profiling correlated with the cycle time change on real-time RT-PCR, but the magnitude of change by real-time RT-PCR was generally greater. GS3686, for example, was 68-fold more abundant in BL41EBV profiling, and the RT-PCR signal appeared 10 cycles earlier with BL41EBV RNA, indicative of a 1,000-fold change. Of 26 genes tested, 13 had replicated changes of at least one cycle for both BL41EBV and IB4 compared to BL, indicating a more than twofold induction (Table 1). Eleven others had positive changes in most experiments, confirming EBV induction. ENO2 failed to confirm in BL41EBV but was 12-fold more abundant in IB4 than in BL cells. IFI30 and ALDOA were four- and threefold increased by array analysis but were minimally increased by RT-PCR. One gene, hUBC12, was incorrectly annotated as HSP1. Real time RT-PCR failed to show EBV induction of HSP1 (data not shown), and hUBC12 has not yet been tested.
TABLE 1.
Assay of EBV-induced genes by real-time RT-PCR
| Gene symbol/name | Proposed function | BL41EBV
|
IB4
|
||
|---|---|---|---|---|---|
| Profiling | RTb | Profiling | RTb | ||
| Interferon induced | |||||
| GS3686/mRNA expressed in osteoblasts | Homology to an interferon-induced, microtubule aggregating protein | 68 | 10/10 | 6.2 | 7.6/6.8 |
| IFITM2/interferon-inducible protein (1-8D) | Transmembrane protein | 13 | 3.7/4.9 | 15 | 5.9/5.5 |
| IFITM3/interferon-inducible protein (1-8U) | Transmembrane protein | 7.3 | 3.2/5.2 | 9.9 | 3.8/4.5 |
| IFITM1/interferon-inducible protein (9-27) | Growth suppression, homotypic adhesion | 5.0 | 2.2/0.72 | 5.4 | 2.3/2.4 |
| Adhesion and cytoskeleton | |||||
| NK4/natural killer cell protein 4 | Induced by antigen receptors, RGD seq. suggest role in ECM/adhesion | 3.4 | 6.7/5.6 | 13 | 9/10 |
| PSCDBP/pleckstrin homology, Sec7 and coiled/coil domains, binding protein | Binds to cytohesin 1 | 6.5 | 0.96/1.5 | 3.5 | 0.56/4.0 |
| BASP1/brain-abundant, membrane-attached signal protein 1 (NAP-22) | Found in rafts, Ca+/CaM binding protein, actin binding protein | 13 | 3.5/3.2 | 6.7 | 10/2.3 |
| LGALS9/lectin, galactoside-binding, soluble 9 (galectin 9, ecalectin) | Eosinophil chemoattractant, causes apoptosis of thymocytes | 6.9 | 0.6/5.3 | 3.0 | 1.47/4.2 |
| Metabolism | |||||
| PCBD/6-pyruvoyl-tetrahydropterin synthase (TCF1) | Synthesis of (BH(4), a cofactor for amino acid hydroxylases | 4.2 | 7.5/7 | 7.8 | 9/3.5 |
| ATP6H/ATPase, H+ transporting, 9 kDa | Vacuolar proton pump | 4.8 | 1.2/2.3 | 2.7 | 4.4/1.2 |
| PGK1/phosphoglycerate kinase 1 | Glycolysis, PGK | 3.0 | 0.89/0.71 | 3.2 | 8.63/.89 |
| DNASE1L3/DNase I-like 3 | DNA metabolism | 4.1 | 5.1/5.7 | 13 | 7.7/8.3 |
| Signaling | |||||
| UBE2M/ubiquitin-conjugating enzyme E2M, homologous to yeast UBC12 | NEDD8-specific conjugating enzyme | 3.0 | NDa | 4.3 | NDa |
| TYK2/TYK2 tyrosine kinase | Cytokine-JAK/STAT signaling | 5.7 | 3.05/1.4 | 2.13 | ND |
| HCK/hemopoietic cell kinase | Src-family kinase, FCγR signaling | 4.4 | 3.0/2.3 | 3.7 | 3.3/2.5 |
| FYN/FYN | Src-family kinase, TCR signaling | 3.6 | 2.3/6.1 | 3.4 | 3.1/2.8 |
| MAP4K1/mitogen-activated protein kinase kinase kinase kinase 1 (HPK1) | Activator of the stress-activated protein kinase pathway | 3.0 | 0.52/1.0 | 3.2 | 1.92/−0.6 |
| DUSP2/dual-specificity (PO4)ase PAC-1 | MAP kinase (ERK1/ERK2) phosphatase | 5.6 | 2.6/2.6 | 6.7 | 3.8/7.8 |
| BRF1/butyrate response factor 1 | Immediate-early response transcription factor (Tis11 D family) | 5.7 | 3.3/3.6 | 6.0 | 2.3/3.1 |
| EMP3/epithelial membrane protein 3 | MP20/PMP22 tetraspanin superfamily | 3.2 | 0.34/1.6 | 2.9 | 0.67/5.8 |
| Protein biosynthesis and degradation | |||||
| EIF4B/elongation initiation factor 4B | Aids unwinding of 5′ UTR | 3.2 | 0.22/3.6 | 4.0 | 1.67/−0.08 |
| RPS8/ribosomal protein S8 | Component of the ribosome | 4.8 | 0.52/1.4 | 3.1 | 1/0.48 |
| SSR4/signal sequence receptor 4 | Cotranslation membrane targeting | 3.8 | 1.21/0.82 | 3.1 | 1.33/0.79 |
| CTSC/cathepsin C | Papain family thiol-protease | 3.6 | 1.6/5.6 | 3.5 | 3.2/2.5 |
| Confirmed in IB4 and not BL41EBV | |||||
| CSTB/cystatin B | Cathepsin L, H, and B inhibitor | 3.9 | 1.8/−1.9 | 4.6 | 5.12/1.5 |
| ENO2/enolase 2 γ | Glycolysis, phosphopyruvate hydrate | 8.7 | 0.22/−0.37 | 10 | 4.6/4.7 |
| Less than twofold by real-time PCR | |||||
| IFI30/IFN-γ-inducible protein IP-30 | Lysosomal thiol reductase | 4.5 | 0.89/−0.51 | 3.4 | 0.43/0.68 |
| ALDOA/aldolase A | Glycolysis, fructose-biphosphate aldolase | 2.7 | 0.47/−0.59 | 2.8 | 0.75/0.7 |
ND, not done due to errors in annotation of the RG filters.
Profiling results are fold changes as in Fig. 1. RT results are the change in cycle number at which a given gene reaches the threshold fluorescence in BL41EBV or IB4 minus that for BL in two independent experiments. One cycle change is approximately twofold.
EBV-induced genes in LCLs versus primary B lymphocytes.
To investigate the relevance of the newly identified EBV-induced gene set to EBV latency III effects in primary B-lymphocyte conversion to LCLs, the abundances of seven induced RNAs were compared by real time RT-PCR using RNAs from recently established LCLs and from primary B lymphocytes. GS3686, ENO2, DNASE1L3, TYK2, HCK, FYN, and MAP4K1(HPK) RNAs were greater than sixfold more abundant in LCLs than in resting B lymphocytes (Table 2). Thus, most of the newly identified EBV-induced genes are likely to be relevant to the effects of latency III EBV infection of primary B lymphocytes.
TABLE 2.
Quantitation of EBV-induced genes in resting B lymphocytes, LCLs, BL30, and BL30EBV by real time RT-PCR
| Gene symbol | Ct in resting Ba | Ct in LCLa | Δb | Avg Δ (BL30EBV − BL30)c |
|---|---|---|---|---|
| GS3686 | 28.59, 28.18 | 21.89, 22.75 | 6.06 | 8.80 |
| ENO2 | 29.69, 28.58 | 24.28, 24.62 | 4.69 | 0.83 |
| DNASE1L3 | 27.51, 28.19 | 25.05, 25.14 | 2.75 | 1.82 |
| TYK2 | 32.30, 32.36 | 28.49, 28.50 | 3.83 | 0.73 |
| HCK | 31.16, 29.74 | 26.94, 27.13 | 3.42 | 0.22 |
| FYN | 29.48, 29.48 | 27.02, 26.14 | 2.90 | −0.815 |
| MAP4K1 | 33.14, 31.47 | 28.55, 28.36 | 3.85 | −0.17 |
Results (mean cycle at threshold [Ct]) from two experiments, performed in triplicate, with two different B-lymphocyte preparations and LCLs, respectively.
LCL average − resting B average.
Average of three experiments performed in duplicate.
EBV-induced genes in BL30EBV versus BL30.
Various BL cell lines differ in their response to EBV latency III infection; similarities and some differences have been previously noted between BL41 and BL30 (13, 14). In real-time RT-PCR assays, GS3686, ENO2, DNASE1L3, and TYK2 were at least 1.7-fold more abundant in BL30EBV than in BL30, whereas HCK, FYN, or MAP4K1 were not more abundant in BL30EBV versus BL30 (Table 2). BL30EBV and BL41EBV cell lines are similar in EBV latency III protein expression; however, the EBER status of BL30EBV is unknown (data not shown). Likely, differences between BL41 and BL30 cells result in disparate effects of EBV latency III on cellular gene expression.
DISCUSSION
EBV infects resting B lymphocytes and growth transforms them by altering cellular gene expression through the expression of the essential viral proteins EBNA2, EBNALP, EBNA3A, EBNA3C, EBNA1, and LMP1 (reviewed in reference 59). Since most of the latency III proteins that are required for LCL outgrowth impact transcriptional pathways, changes in cell gene transcription are likely to be critical to EBV effects on cell growth and survival. Indeed, EBV reverse genetic analyses identify the transcriptional effector domains of EBNA2 and LMP1 as critical domains for B-lymphocyte conversion to LCLs (49, 50, 119). EBNA2, EBNA3A, EBNA3B, and EBNA3C regulate transcription at least in part through interactions with RBP-Jκ/CBF1, a transcription factor that mediates Notch receptor effects (35, 42, 52, 53, 65, 72, 91, 113, 119, 123). LMP1 is a tumor necrosis factor receptor analogue which constitutively activates NF-κB, JNK, and p38 signaling pathways, thereby altering cell gene transcription (27, 28, 30, 33, 36, 40, 45, 50, 60, 66, 81, 83).
These experiments are the first broad survey of the potential effects of latency III EBV infection on 4,146 cDNAs of known or imputed function. Thus, we were able to identify EBV-regulated genes and compare the transcriptional profile of latency III-infected cells to those profiles established for other B-cell malignancies.
Of the 38 RNAs that are up-regulated by latency III EBV infection in BL41EBV, 11 were known to be up-regulated by EBV. Except for β-actin, all are induced by LMP1 through NF-κB activation (6, 8, 24, 63, 66, 120). Based on the key role of LMP1 activation of NF-κB in 10 of the 11 previously known EBV-induced genes, many of the 24 newly identified EBV-induced genes are likely to be up-regulated by LMP1. Indeed, NK4 and PAC-1 are known to be NF-κB responsive and are therefore likely to be LMP1 induced (70, 99). Aside from NF-κB, LMP1 can effect transcription through AP-1 and SP1 activation (60, 104). EBNA2 is also a strong activator of cell gene expression, and its effects are potentiated by EBNALP and EBNA3C (38, 72, 85). EBNA2 can up-regulate CD21 and CD23 with LMP1, and it also up-regulates c-myc and c-fgr (2, 20, 54, 64, 115, 116).
HCK and FYN, two other Src family tyrosine kinases (89, 98, 125), have now been found to be induced by latency III EBV infection (Table 1) and may also be EBNA2 up-regulated. HCK and FYN are up-regulated fourfold in BL41EBV and IB4 cells compared to BL41 cells and were induced approximately eightfold in newly established LCLs relative to resting B cells. Up-regulation of specific Src family kinases may affect signal transduction from receptor tyrosine kinases, from receptors which bind Src family tyrosine kinases through their SH2 or SH3 domains, and from GPCRs that stimulate Gαs and GαI (1, 9, 96). The Src family kinases can further alter cell transcription, growth, or survival through multiple signal cascades, including the mitogen-activated protein (MAP) kinase pathway. We also identified two regulators of the MAP kinase cascade that are EBV induced. MAP4K1 is expressed at threefold higher levels in BL41EBV and in IB4 cells relative to BL41, and PAC-1 is expressed at sixfold higher levels. MAP4K1 interacts with BLNK and a novel SLP-76-related molecule, CLNK/MIST, to couple BCR signaling to NF-κB activation, interleukin-2 promoter activity, and MAP kinase activation (44, 58, 118, 121). PAC-1 dephosphorylates ERK1 and ERK2, and this may prevent desensitization as a consequence of continuous signaling from latency III EBV infection (94, 117).
Four of the identified EBV-induced genes are also IFN inducible. Latency III EBV infection has been associated with direct IFN-β induction as well as with cytokine-mediated IFN induction (5, 55, 105). However, IFN-α1, -β1, and -γ1 RNAs were not up-regulated in BL41EBV. IFN-γ alpha receptor and Tyk2 were up-regulated in BL41EBV and IB4 and could increase IFN signaling. Latency III EBV infection may up-regulate an IFN that would not have been detected with the probes for IFN-α1, -β1, or -γ1, or the IFN effects may be augmented by signaling from another EBV-inducible cytokine and receptor pathway. Cytomegalovirus, another human herpesvirus, is known to induce IFN response genes immediately following infection of cells in the absence of IFN induction (124).
Five EBV-induced genes, including the IFN-induced gene GS3686, encode proteins that are involved in cell adhesion or structure. LMP1 up-regulates expression of vimentin intermediate filaments, a p55 actin bundling protein, MARCKS, LFA-1, LFA-3, and ICAM-1 (10, 13, 47, 101, 106, 107, 114-116). GS3686, the most highly EBV-induced mRNA, encodes a putative microtubule aggregating protein (43, 103). PSCDBP binds to cytoadhesin 1 (61), and BASP1 is an N-terminally myristolated, Ca+ and calmodulin binding protein found in plasma membrane microdomains. BASP1 colocalizes with MARCKS, another EBV-induced protein (7), and is also a PKC substrate (82, 88). Natural killer cell protein 4 may have a role in cell adhesion (21), whereas galectin 9 has been implicated in eosinophil chemotaxis (77).
Ten other EBV-induced genes are constituents of metabolic pathways, including protein biosynthesis and degradation, PCBD/6-pyruvoly-tetrahydropterin synthase, vacuolar ATPase subunit H, phosphoglycerate kinase 1 (PGK1), enolase 2, DNase I-like elongation and initiation factor 4B, ribosomal protein S8, signal sequence receptor 4, cystatin B, and cathepsin C (11, 17, 22, 31, 39, 67, 75, 78, 79, 86, 93, 111, 122). Up-regulation of these RNAs may enhance the ability of latency III-infected cells to rapidly alter location, metabolism, or proliferation.
Analysis of potential transcriptional regulatory sites near promoters of these new EBV-induced genes may identify new signaling pathways impacted by latency III EBV infection. Programs associated with the Transcription Factor database and the completed human genomic sequences were used to identify putative transcription factor binding sites 500 bases upstream of the Tyk2, PGK1, PCBD, MAP4K1, hUBC12, GS3686, and ENO2 promoters. All except for Tyk2 have predicted NF-κB sites and are therefore likely to be LMP1 responsive. The Tyk2, PGK1, GS3686, and ENO2 promoters have predicted PU.1 sites that may enable EBNA2 responsiveness. These promoters also have predicted AP-1, AP-2, and SP1 binding sites that can be affected by LMP1 (28, 104). Thus, most of the genes identified in this study are likely to be impacted by pathways already known to be EBV induced.
Of the 10 RNAs that were down-regulated twofold or more in BL41EBV and IB4, CD10 is known to be down-regulated by LMP1 (34, 71, 115). CD10 is a neutral endopeptidase that is down-regulated in Alzheimer's disease patients. Potentially of interest is the recent observation that B cells of Alzheimer's disease patients transform more rapidly to LCLs following in vitro infection with EBV than do cells from age-matched controls (87).
BL41EBV and IB4 are good model cell lines for the identification of EBV-induced genes, since EBV has robust effects in BL41. Furthermore, IB4 cell growth is still dependent on EBNA2 interactions with RBP-Jκ (A. Cooper, submitted for publication). In fact, of the genes tested, all showed EBV induction in the newly established LCLs compared to resting B cells, validating our model. HCK, FYN, and MAP4K1, was not confirmed as being EBV induced in the comparison of BL30EBV to BL30, whereas GS3686, ENO2, DNASE1L3, and TYK2 were. This likely reflects a difference in the basal gene expression between BL41 and BL30, possibly due to variations in accumulated mutations. BL41 and BL30 have c-myc translocations and p53 mutations; their BCL-6 status is unknown.
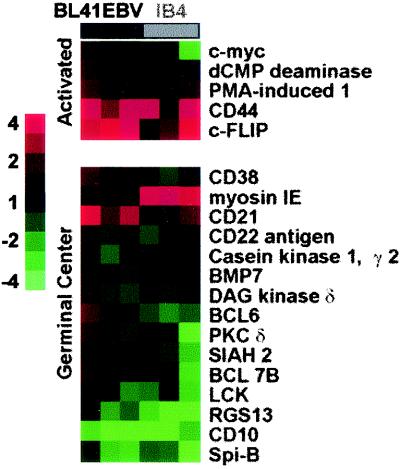
In studies of diffuse large B-cell lymphomas, some genes are abundant in activated antigen receptor-stimulated B cells and scarce in germinal center (tonsil) B cells. These genes constitute an activated B-cell signature. In contrast, genes that are expressed at a high level in tonsils and at a low level in activated B cells constitute a germinal center signature (4). Five activated and 15 germinal center B-cell signature cDNAs were detected on the Research Genetics gf211 filters (Fig. 2). Two of five activation signature genes, CD44 and c-FLIP, were up-regulated in at least five of six hybridizations with BL41EBV and IB4 RNAs. Three of five activated signature genes, c-myc, phorbol myristate acetate (PMA)-induced 1, and dCMP deaminase, were unaffected by EBV infection. c-myc is constitutively expressed at a high level in BL cells and would not be expected to change in these studies (110). PMA-induced 1 and dCMP deaminase are transiently induced by antigen receptor signaling, with expression returning to resting levels at 48 h (4). Thus, the effect of EBV infection on activated and germinal center genes is difficult to predict.
FIG. 2.
Effect of latency III EBV infection on activated and germinal center signature genes. Fold changes are displayed as described in the legend for Fig. 1.
EBV did not consistently up-regulate any germinal center signature gene. Rather, the only significant effect of EBV infection on germinal center signature genes was down-regulation. RGS13, Spi-B, and CD10 were expressed at a low level in BL41EBV and in IB4 cells, consistent with active inhibition by latency III EBV infection. BCL-6 was less abundant in LCLs compared to BL, as was expected since LMP1 expression can down-regulate BCL-6 (15, 16). However, BCL-6 levels were unaffected in BL41EBV, probably due to BL-associated mutations in the BCL-6 promoter (32). Overall, latency III EBV-infected BL41EBV and IB4 cells have increased expression of the activation markers CD44 and c-FLIP and decreased expression of the germinal center markers RGS13, Spi-B, and CD10 (Fig. 2).
In this study, we have focused on genes that are robustly induced and highly expressed in latency III EBV-infected cells. Relaxation of these criteria to include genes that are up-regulated twofold in more than half of the repeated hybridizations expands the set of EBV-induced genes to 80 cDNAs, which include almost all previously identified EBV-induced genes. Further studies are necessary to characterize gene expression in EBV-associated malignancies that express latency I or latency II. These future studies, and our present one, may enable the identification of EBV signatures associated with different types of latency that can lead to therapeutic targets for the treatment of EBV-associated diseases.
Acknowledgments
K. L. Carter and E. Cahir-McFarland contributed equally to this work.
We thank Peter Munson for providing the PSCAN 1.1 software and for assisting us in its use; Emily Klotz for providing guidance with protocols; Danielle Rizzo for technical help; and Jena Giltnane, Louis Staudt, and the Kieff lab members for helpful discussions.
K.L.C., E.C.-M, and E.K. were supported by Public Health Service grants CA47006, CA85180, CA87661, CA75646, and CA76727. E.C.-M is a Special Fellow of the Leukemia and Lymphoma Society.
K. L. Carter and E. Cahir-McFarland contributed equally to this work.
REFERENCES
- 1.Abu-Ghazaleh, R., J. Kabir, H. Jia, M. Lobo, and I. Zachary. 2001. Src mediates stimulation by vascular endothelial growth factor of the phosphorylation of focal adhesion kinase at tyrosine 861, and migration and anti-apoptosis in endothelial cells. Biochem. J. 360:255-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alfieri, C., M. Birkenbach, and E. Kieff. 1991. Early events in Epstein-Barr virus infection of human B lymphocytes. Virology 181:595-608. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh, A., M. Eisen, R. E. Davis, C. Ma, H. Sabet, T. Tran, J. I. Powell, L. Yang, G. E. Marti, D. T. Moore, J. R. Hudson, Jr., W. C. Chan, T. Greiner, D. Weisenburger, J. O. Armitage, I. Lossos, R. Levy, D. Botstein, P. O. Brown, and L. M. Staudt. 1999. The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harbor Symp. Quant. Biol. 64:71-78. [DOI] [PubMed] [Google Scholar]
- 4.Alizadeh, A. A., M. B. Eisen, R. E. Davis, C. Ma, I. S. Lossos, A. Rosenwald, J. C. Boldrick, H. Sabet, T. Tran, X. Yu, J. I. Powell, L. Yang, G. E. Marti, T. Moore, J. Hudson, Jr., L. Lu, D. B. Lewis, R. Tibshirani, G. Sherlock, W. C. Chan, T. C. Greiner, D. D. Weisenburger, J. O. Armitage, R. Warnke, L. M. Staudt, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503-511. [DOI] [PubMed] [Google Scholar]
- 5.Andersson, U., O. Martinez-Maza, J. Andersson, S. Britton, H. Gadler, M. De Ley, and S. Modrow. 1984. Secretion of gamma-interferon at the cellular level. Induction by Epstein-Barr virus. Scand. J. Immunol. 20:425-432. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin, A. S., Jr., and P. A. Sharp. 1987. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2Kb class I major histocompatibility gene. Mol. Cell. Biol. 7:305-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birkenbach, M., K. Josefsen, R. Yalamanchili, G. Lenoir, and E. Kieff. 1993. Epstein-Barr virus-induced genes: first lymphocyte-specific G protein-coupled peptide receptors. J. Virol. 67:2209-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenbach, M., D. Liebowitz, F. Wang, J. Sample, and E. Kieff. 1989. Epstein-Barr virus latent infection membrane protein increases vimentin expression in human B-cell lines. J. Virol. 63:4079-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisotto, S., and E. D. Fixman. 2001. Src-family tyrosine kinases, phosphoinositide 3-kinase and Gab1 regulate extracellular signal-regulated kinase 1 activation induced by the type A endothelin-1 G-protein-coupled receptor. Biochem. J. 360:77-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnefoy, J. Y., T. Defrance, C. Peronne, C. Menetrier, F. Rousset, J. Pene, J. E. De Vries, and J. Banchereau. 1988. Human recombinant interleukin 4 induces normal B cells to produce soluble CD23/IgE-binding factor analogous to that spontaneously released by lymphoblastoid B cell lines. Eur. J. Immunol. 18:117-122. [DOI] [PubMed] [Google Scholar]
- 11.Brenner, V., G. Nyakatura, A. Rosenthal, and M. Platzer. 1997. Genomic organization of two novel genes on human Xq28: compact head to head arrangement of IDH gamma and TRAP delta is conserved in rat and mouse. Genomics 44:8-14. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell, R. G., J. B. Wilson, S. J. Anderson, and R. Longnecker. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405-411. [DOI] [PubMed] [Google Scholar]
- 13.Calender, A., M. Billaud, J. P. Aubry, J. Banchereau, M. Vuillaume, and G. M. Lenoir. 1987. Epstein-Barr virus (EBV) induces expression of B-cell activation markers on in vitro infection of EBV-negative B-lymphoma cells. Proc. Natl. Acad. Sci. USA 84:8060-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calender, A., M. Cordier, M. Billaud, and G. M. Lenoir. 1990. Modulation of cellular gene expression in B lymphoma cells following in vitro infection by Epstein-Barr virus (EBV). Int. J. Cancer 46:658-663. [DOI] [PubMed] [Google Scholar]
- 15.Carbone, A., G. Gaidano, A. Gloghini, L. M. Larocca, D. Capello, V. Canzonieri, A. Antinori, U. Tirelli, B. Falini, and R. Dalla-Favera. 1998. Differential expression of BCL-6, CD138/syndecan-1, and Epstein-Barr virus-encoded latent membrane protein-1 identifies distinct histogenetic subsets of acquired immunodeficiency syndrome-related non-Hodgkin's lymphomas. Blood 91:747-755. [PubMed] [Google Scholar]
- 16.Carbone, A., G. Gaidano, A. Gloghini, C. Pastore, G. Saglio, U. Tirelli, R. Dalla-Favera, and B. Falini. 1997. BCL-6 protein expression in AIDS-related non-Hodgkin's lymphomas: inverse relationship with Epstein-Barr virus-encoded latent membrane protein-1 expression. Am. J. Pathol. 150:155-165. [PMC free article] [PubMed] [Google Scholar]
- 17.Citron, B. A., S. Kaufman, S. Milstien, E. W. Naylor, C. L. Greene, and M. D. Davis. 1993. Mutation in the 4a-carbinolamine dehydratase gene leads to mild hyperphenylalaninemia with defective cofactor metabolism. Am. J. Hum. Genet. 53:768-774. [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen, J. H., E. Fischer, M. D. Kazatchkine, G. M. Lenoir, C. Lefevre-Delvincourt, and J. P. Revillard. 1987. Expression of CR1 and CR2 complement receptors following Epstein-Barr virus infection of Burkitt's lymphoma cell lines. Scand. J. Immunol. 25:587-598. [DOI] [PubMed] [Google Scholar]
- 19.Cohen, J. I., F. Wang, J. Mannick, and E. Kieff. 1989. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc. Natl. Acad. Sci. USA 86:9558-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cordier, M., A. Calender, M. Billaud, U. Zimber, G. Rousselet, O. Pavlish, J. Banchereau, T. Tursz, G. Bornkamm, and G. M. Lenoir. 1990. Stable transfection of Epstein-Barr virus (EBV) nuclear antigen 2 in lymphoma cells containing the EBV P3HR1 genome induces expression of B-cell activation molecules CD21 and CD23. J. Virol. 64:1002-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dahl, C. A., R. P. Schall, H. L. He, and J. S. Cairns. 1992. Identification of a novel gene expressed in activated natural killer cells and T cells. J. Immunol. 148:597-603. [PubMed] [Google Scholar]
- 22.Davies, B., and M. Fried. 1993. The structure of the human intron-containing S8 ribosomal protein gene and determination of its chromosomal location at 1p32-p34.1. Genomics 15:68-75. [DOI] [PubMed] [Google Scholar]
- 23.Delecluse, H. J., E. Kremmer, J. P. Rouault, C. Cour, G. Bornkamm, and F. Berger. 1995. The expression of Epstein-Barr virus latent proteins is related to the pathological features of post-transplant lymphoproliferative disorders. Am. J. Pathol. 146:1113-1120. [PMC free article] [PubMed] [Google Scholar]
- 24.Devergne, O., E. Cahir-McFarland, G. Mosialos, K. M. Izumi, C. F. Ware, and E. Kieff. 1998. Role of the TRAF binding site and NF-κB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J. Virol. 72:7900-7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehlin-Henriksson, B., A. Manneborg-Sandlund, and G. Klein. 1987. Expression of B-cell-specific markers in different Burkitt lymphoma subgroups. Int. J. Cancer 39:211-218. [DOI] [PubMed] [Google Scholar]
- 26.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eliopoulos, A. G., N. J. Gallagher, S. M. Blake, C. W. Dawson, and L. S. Young. 1999. Activation of the p38 mitogen-activated protein kinase pathway by Epstein-Barr virus-encoded latent membrane protein 1 coregulates interleukin-6 and interleukin-8 production. J. Biol. Chem. 274:16085-16096. [DOI] [PubMed] [Google Scholar]
- 28.Eliopoulos, A. G., and L. S. Young. 1998. Activation of the cJun N-terminal kinase (JNK) pathway by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1). Oncogene 16:1731-1742. [DOI] [PubMed] [Google Scholar]
- 28a.Farrell, P. J., G. J. Allan, F. Shanahan, K. H. Vousden, and T. Crook. 1991. p53 is frequently mutated in Burkitt's lymphoma cell lines. EMBO J. 10:2879-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Favrot, M. C., O. Maritaz, T. Suzuki, M. Cooper, I. Philip, T. Philip, and G. Lenoir. 1986. EBV-negative and -positive Burkitt cell lines variably express receptors for B-cell activation and differentiation. Int. J. Cancer 38:901-906. [DOI] [PubMed] [Google Scholar]
- 30.Floettmann, J. E., and M. Rowe. 1997. Epstein-Barr virus latent membrane protein-1 (LMP1) C-terminus activation region 2 (CTAR2) maps to the far C terminus and requires oligomerisation for NF-κB activation. Oncogene 15:1851-1858. [DOI] [PubMed] [Google Scholar]
- 31.Freemont, P. S., B. Dunbar, and L. A. Fothergill-Gilmore. 1988. The complete amino acid sequence of human skeletal-muscle fructose-bisphosphate aldolase. Biochem. J. 249:779-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaidano, G., A. Carbone, and R. Dalla-Favera. 1998. Genetic basis of acquired immunodeficiency syndrome-related lymphomagenesis. J. Natl. Cancer Inst. Monogr. 23:95-100. [DOI] [PubMed] [Google Scholar]
- 33.Gires, O., U. Zimber-Strobl, R. Gonnella, M. Ueffing, G. Marschall, R. Zeidler, D. Pich, and W. Hammerschmidt. 1997. Latent membrane protein 1 of Epstein-Barr virus mimics a constitutively active receptor molecule. EMBO J. 16:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gregory, C. D., C. F. Edwards, A. Milner, J. Wiels, M. Lipinski, M. Rowe, T. Tursz, and A. B. Rickinson. 1988. Isolation of a normal B cell subset with a Burkitt-like phenotype and transformation in vitro with Epstein-Barr virus. Int. J. Cancer 42:213-220. [DOI] [PubMed] [Google Scholar]
- 35.Grossman, S. R., E. Johannsen, X. Tong, R. Yalamanchili, and E. Kieff. 1994. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc. Natl. Acad. Sci. USA 91:7568-7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hammarskjold, M. L., and M. C. Simurda. 1992. Epstein-Barr virus latent membrane protein transactivates the human immunodeficiency virus type 1 long terminal repeat through induction of NF-κB activity. J. Virol. 66:6496-6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 38.Harada, S., and E. Kieff. 1997. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J. Virol. 71:6611-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hart, T. C., P. S. Hart, D. W. Bowden, M. D. Michalec, S. A. Callison, S. J. Walker, Y. Zhang, and E. Firatli. 1999. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. J. Med. Genet. 36:881-887. [PMC free article] [PubMed] [Google Scholar]
- 40.Hatzivassiliou, E., W. E. Miller, N. Raab-Traub, E. Kieff, and G. Mosialos. 1998. A fusion of the EBV latent membrane protein-1 (LMP1) transmembrane domains to the CD40 cytoplasmic domain is similar to LMP1 in constitutive activation of epidermal growth factor receptor expression, nuclear factor-kappa B, and stress-activated protein kinase. J. Immunol. 160:1116-1121. [PubMed] [Google Scholar]
- 41.Henderson, A., S. Ripley, M. Heller, and E. Kieff. 1983. Chromosome site for Epstein-Barr virus DNA in a Burkitt tumor cell line and in lymphocytes growth-transformed in vitro. Proc. Natl. Acad. Sci. USA 80:1987-1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henkel, T., P. D. Ling, S. D. Hayward, and M. G. Peterson. 1994. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein Jκ. Science 265:92-95. [DOI] [PubMed] [Google Scholar]
- 43.Honda, Y., J. Kondo, T. Maeda, Y. Yoshiyama, E. Yamada, Y. K. Shimizu, T. Shikata, and Y. Ono. 1990. Isolation and purification of a non-A, non-B hepatitis-associated microtubular aggregates protein. J. Gen. Virol. 71:1999-2004. [DOI] [PubMed] [Google Scholar]
- 44.Hu, M. C., W. R. Qiu, X. Wang, C. F. Meyer, and T. H. Tan. 1996. Human HPK1, a novel human hematopoietic progenitor kinase that activates the JNK/SAPK kinase cascade. Genes Dev. 10:2251-2264. [DOI] [PubMed] [Google Scholar]
- 45.Huen, D. S., S. A. Henderson, D. Croom-Carter, and M. Rowe. 1995. The Epstein-Barr virus latent membrane protein-1 (LMP1) mediates activation of NF-κB and cell surface phenotype via two effector regions in its carboxy-terminal cytoplasmic domain. Oncogene 10:549-560. [PubMed] [Google Scholar]
- 46.Hurley, E. A., L. D. Klaman, S. Agger, J. B. Lawrence, and D. A. Thorley-Lawson. 1991. The prototypical Epstein-Barr virus-transformed lymphoblastoid cell line IB4 is an unusual variant containing integrated but no episomal viral DNA. J. Virol. 65:3958-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hurley, E. A., and D. A. Thorley-Lawson. 1988. B cell activation and the establishment of Epstein-Barr virus latency. J. Exp. Med. 168:2059-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. Lee, J. M. Trent, L. M. Staudt, J. Hudson, Jr., M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283:83-87. [DOI] [PubMed] [Google Scholar]
- 49.Izumi, K. M., K. M. Kaye, and E. D. Kieff. 1997. The Epstein-Barr virus LMP1 amino acid sequence that engages tumor necrosis factor receptor associated factors is critical for primary B lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 94:1447-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Izumi, K. M., and E. D. Kieff. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc. Natl. Acad. Sci. USA 94:12592-12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 96:13118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johannsen, E., E. Koh, G. Mosialos, X. Tong, E. Kieff, and S. R. Grossman. 1995. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J. Virol. 69:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johannsen, E., C. L. Miller, S. R. Grossman, and E. Kieff. 1996. EBNA-2 and EBNA-3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J. Virol. 70:4179-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaiser, C., G. Laux, D. Eick, N. Jochner, G. W. Bornkamm, and B. Kempkes. 1999. The proto-oncogene c-myc is a direct target gene of Epstein-Barr virus nuclear antigen 2. J. Virol. 73:4481-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kanda, K., B. Kempkes, G. W. Bornkamm, A. von Gabain, and T. Decker. 1999. The Epstein-Barr virus nuclear antigen 2 (EBNA2), a protein required for B lymphocyte immortalization, induces the synthesis of type I interferon in Burkitt's lymphoma cell lines. Biol. Chem. 380:213-221. [DOI] [PubMed] [Google Scholar]
- 56.Kaye, K. M., K. M. Izumi, and E. Kieff. 1993. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc. Natl. Acad. Sci. USA 90:9150-9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khan, J., M. L. Bittner, L. H. Saal, U. Teichmann, D. O. Azorsa, G. C. Gooden, W. J. Pavan, J. M. Trent, and P. S. Meltzer. 1999. cDNA microarrays detect activation of a myogenic transcription program by the PAX3-FKHR fusion oncogene. Proc. Natl. Acad. Sci. USA 96:13264-13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kiefer, F., L. A. Tibbles, M. Anafi, A. Janssen, B. W. Zanke, N. Lassam, T. Pawson, J. R. Woodgett, and N. N. Iscove. 1996. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 15:7013-7025. [PMC free article] [PubMed] [Google Scholar]
- 59.Kieff, E., and A. B. Rickinson. 2001. Epstein-Barr virus and its replication, p. 2511-2574. In D. Knipe, P. Howley, D. Griffin, M. Martin, R. Lamb, B. Roizman, and S. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott-Raven, Philadelphia, Pa.
- 60.Kieser, A., E. Kilger, O. Gires, M. Ueffing, W. Kolch, and W. Hammerschmidt. 1997. Epstein-Barr virus latent membrane protein-1 triggers AP-1 activity via the c-Jun N-terminal kinase cascade. EMBO J. 16:6478-6485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim, H. S. 1999. Assignment of the human B3-1 gene (PSCDBP) to chromosome 2 band q11.2 by radiation hybrid mapping. Cytogenet. Cell Genet. 84:95.. [DOI] [PubMed] [Google Scholar]
- 62.King, W., A. L. Thomas-Powell, N. Raab-Traub, M. Hawke, and E. Kieff. 1980. Epstein-Barr virus RNA. V. Viral RNA in a restringently infected, growth-transformed cell line. J. Virol. 36:506-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klaman, L. D., and D. A. Thorley-Lawson. 1995. Characterization of the CD48 gene demonstrates a positive element that is specific to Epstein-Barr virus-immortalized B-cell lines and contains an essential NF-κB site. J. Virol. 69:871-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knutson, J. C. 1990. The level of c-fgr RNA is increased by EBNA-2, an Epstein-Barr virus gene required for B-cell immortalization. J. Virol. 64:2530-2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krauer, K. G., N. Kienzle, D. B. Young, and T. B. Sculley. 1996. Epstein-Barr nuclear antigen-3 and -4 interact with RBP-2N, a major isoform of RBP-Jκ in B lymphocytes. Virology 226:346-353. [DOI] [PubMed] [Google Scholar]
- 66.Laherty, C. D., H. M. Hu, A. W. Opipari, F. Wang, and V. M. Dixit. 1992. The Epstein-Barr virus LMP1 gene product induces A20 zinc finger protein expression by activating nuclear factor kappa B. J. Biol. Chem. 267:24157-24160. [PubMed] [Google Scholar]
- 67.Lay, A. J., X. M. Jiang, O. Kisker, E. Flynn, A. Underwood, R. Condron, and P. J. Hogg. 2000. Phosphoglycerate kinase acts in tumour angiogenesis as a disulphide reductase. Nature 408:869-873. [DOI] [PubMed] [Google Scholar]
- 68.Lee, M. A., M. E. Diamond, and J. L. Yates. 1999. Genetic evidence that EBNA-1 is needed for efficient, stable latent infection by Epstein-Barr virus. J. Virol. 73:2974-2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lewin, A. R., L. E. Reid, M. McMahon, G. R. Stark, and I. M. Kerr. 1991. Molecular analysis of a human interferon-inducible gene family. Eur. J. Biochem. 199:417-423. [DOI] [PubMed] [Google Scholar]
- 70.Li, J., G. W. Peet, D. Balzarano, X. Li, P. Massa, R. W. Barton, and K. B. Marcu. 2001. Novel NEMO/IκB kinase and NF-κB target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579-18590. [DOI] [PubMed] [Google Scholar]
- 71.Liebowitz, D., J. Mannick, K. Takada, and E. Kieff. 1992. Phenotypes of Epstein-Barr virus LMP1 deletion mutants indicate transmembrane and amino-terminal cytoplasmic domains necessary for effects in B-lymphoma cells. J. Virol. 66:4612-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin, J., E. Johannsen, E. Robertson, and E. Kieff. 2002. Epstein-Barr virus nuclear antigen 3C putative repression domain mediates coactivation of the LMP1 promoter with EBNA-2. J. Virol. 76:232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Longnecker, R., C. L. Miller, X. Q. Miao, B. Tomkinson, and E. Kieff. 1993. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J. Virol. 67:2006-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu, Y. J., D. Williamson, J. Clark, R. Wang, N. Tiffin, L. Skelton, T. Gordon, R. Williams, B. Allan, A. Jackman, C. Cooper, K. Pritchard-Jones, and J. Shipley. 2001. Comparative expressed sequence hybridization to chromosomes for tumor classification and identification of genomic regions of differential gene expression. Proc. Natl. Acad. Sci. USA 98:9197-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ludwig, J., S. Kerscher, U. Brandt, K. Pfeiffer, F. Getlawi, D. K. Apps, and H. Schagger. 1998. Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J. Biol. Chem. 273:10939-10947. [DOI] [PubMed] [Google Scholar]
- 76.Mannick, J. B., J. I. Cohen, M. Birkenbach, A. Marchini, and E. Kieff. 1991. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J. Virol. 65:6826-6837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matsumoto, R., H. Matsumoto, M. Seki, M. Hata, Y. Asano, S. Kanegasaki, R. L. Stevens, and M. Hirashima. 1998. Human ecalectin, a variant of human galectin-9, is a novel eosinophil chemoattractant produced by T lymphocytes. J. Biol. Chem. 273:16976-16984. [DOI] [PubMed] [Google Scholar]
- 78.Mendel, D. B., P. A. Khavari, P. B. Conley, M. K. Graves, L. P. Hansen, A. Admon, and G. R. Crabtree. 1991. Characterization of a cofactor that regulates dimerization of a mammalian homeodomain protein. Science 254:1762-1767. [DOI] [PubMed] [Google Scholar]
- 79.Milburn, S. C., J. W. Hershey, M. V. Davies, K. Kelleher, and R. J. Kaufman. 1990. Cloning and expression of eukaryotic initiation factor 4B cDNA: sequence determination identifies a common RNA recognition motif. EMBO J. 9:2783-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller, C. L., A. L. Burkhardt, J. H. Lee, B. Stealey, R. Longnecker, J. B. Bolen, and E. Kieff. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155-166. [DOI] [PubMed] [Google Scholar]
- 81.Mitchell, T., and B. Sugden. 1995. Stimulation of NF-κB-mediated transcription by mutant derivatives of the latent membrane protein of Epstein-Barr virus. J. Virol. 69:2968-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mosevitsky, M. I., J. P. Capony, G. Skladchikova, V. A. Novitskaya, A. Plekhanov, and V. V. Zakharov. 1997. The BASP1 family of myristoylated proteins abundant in axonal termini. Primary structure analysis and physico-chemical properties. Biochimie 79:373-384. [DOI] [PubMed] [Google Scholar]
- 83.Mosialos, G., M. Birkenbach, R. Yalamanchili, T. VanArsdale, C. Ware, and E. Kieff. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389-399. [DOI] [PubMed] [Google Scholar]
- 84.Muslin, A. J., J. W. Tanner, P. M. Allen, and A. S. Shaw. 1996. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell 84:889-897. [DOI] [PubMed] [Google Scholar]
- 85.Nitsche, F., A. Bell, and A. Rickinson. 1997. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression: a role for the W1W2 repeat domain. J. Virol. 71:6619-6628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Oliva, D., L. Cali, S. Feo, and A. Giallongo. 1991. Complete structure of the human gene encoding neuron-specific enolase. Genomics 10:157-165. [DOI] [PubMed] [Google Scholar]
- 87.Ounanian, A., B. Guilbert, and J. M. Seigneurin. 1992. Characteristics of Epstein-Barr virus transformed B cell lines from patients with Alzheimer's disease and age-matched controls. Mech. Ageing Dev. 63:105-116. [DOI] [PubMed] [Google Scholar]
- 88.Park, S., Y. I. Kim, B. Kim, C. Seong, Y. Oh, K. Baek, and J. Yoon. 1998. Characterization of bovine and human cDNAs encoding NAP-22 (22 kDa neuronal tissue-enriched acidic protein) homologs. Mol. Cells 8:471-477. [PubMed] [Google Scholar]
- 89.Quintrell, N., R. Lebo, H. Varmus, J. M. Bishop, M. J. Pettenati, M. M. Le Beau, M. O. Diaz, and J. D. Rowley. 1987. Identification of a human gene (HCK) that encodes a protein-tyrosine kinase and is expressed in hemopoietic cells. Mol. Cell. Biol. 7:2267-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rickinson, A., and E. Kieff. 2001. Epstein-Barr virus, p. 2575-2628. In D. Knipe, P. Howley, D. Griffin, M. Martin, R. Lamb, B. Roizman, and S. Straus (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 91.Robertson, E. S., S. Grossman, E. Johannsen, C. Miller, J. Lin, B. Tomkinson, and E. Kieff. 1995. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J. Virol. 69:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Robertson, E. S., B. Tomkinson, and E. Kieff. 1994. An Epstein-Barr virus with a 58-kilobase-pair deletion that includes BARF0 transforms B lymphocytes in vitro. J. Virol. 68:1449-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rodriguez, A. M., D. Rodin, H. Nomura, C. C. Morton, S. Weremowicz, and M. C. Schneider. 1997. Identification, localization, and expression of two novel human genes similar to deoxyribonuclease I. Genomics 42:507-513. [DOI] [PubMed] [Google Scholar]
- 94.Rohan, P. J., P. Davis, C. A. Moskaluk, M. Kearns, H. Krutzsch, U. Siebenlist, and K. Kelly. 1993. PAC-1: a mitogen-induced nuclear protein tyrosine phosphatase. Science 259:1763-1766. [DOI] [PubMed] [Google Scholar]
- 95.Rowe, M., A. L. Lear, D. Croom-Carter, A. H. Davies, and A. B. Rickinson. 1992. Three pathways of Epstein-Barr virus gene activation from EBNA1-positive latency in B lymphocytes. J. Virol. 66:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sasaki, A., H. Yasukawa, A. Suzuki, S. Kamizono, T. Syoda, I. Kinjyo, M. Sasaki, J. A. Johnston, and A. Yoshimura. 1999. Cytokine-inducible SH2 protein-3 (CIS3/SOCS3) inhibits Janus tyrosine kinase by binding through the N-terminal kinase inhibitory region as well as SH2 domain. Genes Cells 4:339-351. [DOI] [PubMed] [Google Scholar]
- 97.Schena, M., D. Shalon, R. W. Davis, and P. O. Brown. 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270:467-470. [DOI] [PubMed] [Google Scholar]
- 98.Semba, K., M. Nishizawa, N. Miyajima, M. C. Yoshida, J. Sukegawa, Y. Yamanashi, M. Sasaki, T. Yamamoto, and K. Toyoshima. 1986. yes-related protooncogene, syn, belongs to the protein-tyrosine kinase family. Proc. Natl. Acad. Sci. USA 83:5459-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shaffer, A. L., A. Rosenwald, E. M. Hurt, J. M. Giltnane, L. T. Lam, O. K. Pickeral, and L. M. Staudt. 2001. Signatures of the immune response. Immunity 15:375-385. [DOI] [PubMed] [Google Scholar]
- 100.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sugden, B., and S. Metzenberg. 1983. Characterization of an antigen whose cell surface expression is induced by infection with Epstein-Barr virus. J. Virol. 46:800-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Swaminathan, S., B. Tomkinson, and E. Kieff. 1991. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc. Natl. Acad. Sci. USA 88:1546-1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Takahashi, K., N. Kitamura, T. Shibui, M. Kamizono, R. Matsui, Y. Yoshiyama, T. Maeda, J. Kondo, Y. Honda, E. Yamada, et al. 1990. Cloning, sequencing and expression in Escherichia coli of cDNA for a non-A, non-B hepatitis-associated microtubular aggregates protein. J. Gen. Virol. 71:2005-2011. [DOI] [PubMed] [Google Scholar]
- 104.Takeshita, H., T. Yoshizaki, W. E. Miller, H. Sato, M. Furukawa, J. S. Pagano, and N. Raab-Traub. 1999. Matrix metalloproteinase 9 expression is induced by Epstein-Barr virus latent membrane protein 1 C-terminal activation regions 1 and 2. J. Virol. 73:5548-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tang, H., T. Matthes, N. Carballido-Perrig, R. H. Zubler, and V. Kindler. 1993. Differential induction of T cell cytokine mRNA in Epstein-Barr virus-transformed B cell clones: constitutive and inducible expression of interleukin-4 mRNA. Eur. J. Immunol. 23:899-903. [DOI] [PubMed] [Google Scholar]
- 106.Thorley-Lawson, D. A., L. M. Nadler, A. K. Bhan, and R. T. Schooley. 1985. BLAST-2 [EBVCS], an early cell surface marker of human B cell activation, is superinduced by Epstein Barr virus. J. Immunol. 134:3007-3012. [PubMed] [Google Scholar]
- 107.Thorley-Lawson, D. A., R. T. Schooley, A. K. Bhan, and L. M. Nadler. 1982. Epstein-Barr virus superinduces a new human B cell differentiation antigen (B-LAST 1) expressed on transformed lymphoblasts. Cell 30:415-425. [DOI] [PubMed] [Google Scholar]
- 108.Tomkinson, B., and E. Kieff. 1992. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J. Virol. 66:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tomkinson, B., E. Robertson, and E. Kieff. 1993. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J. Virol. 67:2014-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Torsteinsdottir, S., M. L. Andersson, J. Avila-Carino, B. Ehlin-Henriksson, M. G. Masucci, G. Klein, and E. Klein. 1989. Reversion of tumorigenicity and decreased agarose clonability after EBV conversion of an IgH/myc translocation-carrying BL line. Int. J. Cancer. 43:273-278. [DOI] [PubMed] [Google Scholar]
- 111.Turk, V., and W. Bode. 1991. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 285:213-219. [DOI] [PubMed] [Google Scholar]
- 112.Velazquez, L., M. Fellous, G. R. Stark, and S. Pellegrini. 1992. A protein tyrosine kinase in the interferon alpha/beta signaling pathway. Cell 70:313-322. [DOI] [PubMed] [Google Scholar]
- 113.Waltzer, L., F. Logeat, C. Brou, A. Israel, A. Sergeant, and E. Manet. 1994. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA2 protein to its DNA responsive elements. EMBO J. 13:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang, D., D. Liebowitz, F. Wang, C. Gregory, A. Rickinson, R. Larson, T. Springer, and E. Kieff. 1988. Epstein-Barr virus latent infection membrane protein alters the human B-lymphocyte phenotype: deletion of the amino terminus abolishes activity. J. Virol. 62:4173-4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang, F., C. Gregory, C. Sample, M. Rowe, D. Liebowitz, R. Murray, A. Rickinson, and E. Kieff. 1990. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J. Virol. 64:2309-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang, F., C. D. Gregory, M. Rowe, A. B. Rickinson, D. Wang, M. Birkenbach, H. Kikutani, T. Kishimoto, and E. Kieff. 1987. Epstein-Barr virus nuclear antigen 2 specifically induces expression of the B-cell activation antigen CD23. Proc. Natl. Acad. Sci. USA 84:3452-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ward, Y., S. Gupta, P. Jensen, M. Wartmann, R. J. Davis, and K. Kelly. 1994. Control of MAP kinase activation by the mitogen-induced threonine/tyrosine phosphatase PAC1. Nature 367:651-654. [DOI] [PubMed] [Google Scholar]
- 118.Yablonski, D., and A. Weiss. 2001. Mechanisms of signaling by the hematopoietic-specific adaptor proteins, SLP-76 and LAT and their B cell counterpart, BLNK/SLP-65. Adv. Immunol. 79:93-128. [DOI] [PubMed] [Google Scholar]
- 119.Yalamanchili, R., X. Tong, S. Grossman, E. Johannsen, G. Mosialos, and E. Kieff. 1994. Genetic and biochemical evidence that EBNA 2 interaction with a 63-kDa cellular GTG-binding protein is essential for B lymphocyte growth transformation by EBV. Virology 204:634-641. [DOI] [PubMed] [Google Scholar]
- 120.Yokoyama, S., D. Staunton, R. Fisher, M. Amiot, J. J. Fortin, and L. D. Thorley. 1991. Expression of the Blast-1 activation/adhesion molecule and its identification as CD48. J. Immunol. 146:2192-2200. [PubMed] [Google Scholar]
- 121.Yu, J., C. Riou, D. Davidson, R. Minhas, J. D. Robson, M. Julius, R. Arnold, F. Kiefer, and A. Veillette. 2001. Synergistic regulation of immunoreceptor signaling by SLP-76-related adaptor Clnk and serine/threonine protein kinase HPK-1. Mol. Cell. Biol. 21:6102-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zeng, Z., D. Parmelee, H. Hyaw, T. A. Coleman, K. Su, J. Zhang, R. Gentz, S. Ruben, C. Rosen, and Y. Li. 1997. Cloning and characterization of a novel human DNase. Biochem. Biophys. Res. Commun. 231:499-504. [DOI] [PubMed] [Google Scholar]
- 123.Zhao, B., D. R. Marshall, and C. E. Sample. 1996. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J. Virol. 70:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhu, H., J. P. Cong, and T. Shenk. 1997. Use of differential display analysis to assess the effect of human cytomegalovirus infection on the accumulation of cellular RNAs: induction of interferon-responsive RNAs. Proc. Natl. Acad. Sci. USA 94:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ziegler, S. F., J. D. Marth, D. B. Lewis, and R. M. Perlmutter. 1987. Novel protein-tyrosine kinase gene (hck) preferentially expressed in cells of hematopoietic origin. Mol. Cell. Biol. 7:2276-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]