Abstract
Nuclear magnetic resonance spectroscopy demonstrates that the rhesus rotavirus hemagglutinin specifically binds α-anomeric N-acetylneuraminic acid with a Kd of 1.2 mM. The hemagglutinin requires no additional carbohydrate moieties for binding, does not distinguish 3′ from 6′ sialyllactose, and has approximately tenfold lower affinity for N-glycolylneuraminic than for N-acetylneuraminic acid. The broad specificity and low affinity of sialic acid binding by the rotavirus hemagglutinin are consistent with this interaction mediating initial cell attachment prior to the interactions that determine host range and cell type specificity.
Rotavirus causes approximately 600,000 childhood deaths through dehydrating diarrhea annually (43) and is also an important veterinary pathogen. Understanding early events in cell entry by rotavirus could aid in the development of therapeutic and preventive measures against rotavirus and could help explain the host range and cell type specificity of the virus. A number of rotavirus strains agglutinate human type O red blood cells (31, 39). Hemagglutination and infection of cultured cells by these strains is sensitive to the treatment of cells by neuraminidase (8, 9, 11, 25), indicating that these strains bind sialic acid (N-acetylneuraminic acid or NeuNAc) on the cell surface. In contrast, a majority of rotavirus strains, including most of those that cause disease in humans, do not hemagglutinate, nor do they require cell surface sialic acid for entry (9, 31).
The specificity of viral receptors for specific linkages of sialic acid is a significant determinant of phenotype in other viruses. For influenza viruses, sialic acid specificity influences host range: avian and equine strains of influenza virus recognize sialic acid in an α2,3 linkage to galactose, while human strains prefer sialic acid in an α2,6 linkage (10, 36). Changes in sialic acid binding specificity have been linked to the spread of influenza virus strains from animal to human populations (28). For polyomaviruses, sialic acid specificity correlates with cell tropism and pathogenicity: while all polyomavirus strains recognize straight-chain oligosaccharides ending in an α2,3-linked sialic acid, strains that also recognize branched oligosaccharides containing an additional α2,6-linked sialic acid are less tumorigenic and spread less extensively in mice (5, 14, 17).
The sialic acid dependence of rotavirus strains is associated with one key difference in entry. While sialic acid-independent strains can infect cells through either the apical or basolateral membrane, sialic acid-dependent strains infect cells only through the apical membrane (7). Sialic acid binding by rotavirus has not, however, been clearly linked to any other host range restriction, cell tropism, or pathogenicity.
The specificity of rotavirus for sialic acid-containing oligosaccharides and sialylmimetics has been examined by using small molecules as competitors of hemagglutination, infection, or binding to cells (15, 22, 24, 27, 37, 44). The low affinity of rotavirus for monovalent sialosides complicates this approach. Nevertheless, the results of two studies have suggested that rotavirus inhibition is not strictly dependent on the glycosidic linkage of the sialic acid moiety: 2.7 mM 3′ sialyllactose inhibited the binding of rotavirus strain OSU to cultured cells by 50%, as did 1.9 mM 6′ sialyllactose (37), and a synthetic thiosialoside at 6.25 mM in either α2,3 or α2,4 linkage to galactose inhibited rotavirus strain NCDV infection of monolayers by 50% (22).
Studies of rotavirus binding have suggested that either glycoproteins or glycolipids may be the initial sialic acid-containing cellular receptors for some rotavirus strains (11, 16, 37, 41, 44). One intriguing result from such studies is the finding that some rotavirus strains (SA11, OSU, and NCDV) preferentially bind immobilized gangliosides containing the N-glycolyl variant of sialic acid, which is hydroxylated on C11 (Fig. 1) (11, 37). Since their divergence from chimpanzees, humans have sustained a mutation that prevents the synthesis of N-glycolylneuraminic acid from an N-acetylneuraminic acid precursor (6). As a result, little if any N-glycolylneuraminic acid is present on the surface of human cells (32). A widespread preference of sialic acid-dependent rotavirus strains for N-glycolylneuraminic acid would offer an explanation for the sialic acid independence of most human rotavirus strains (15 out of 16 tested [9]).
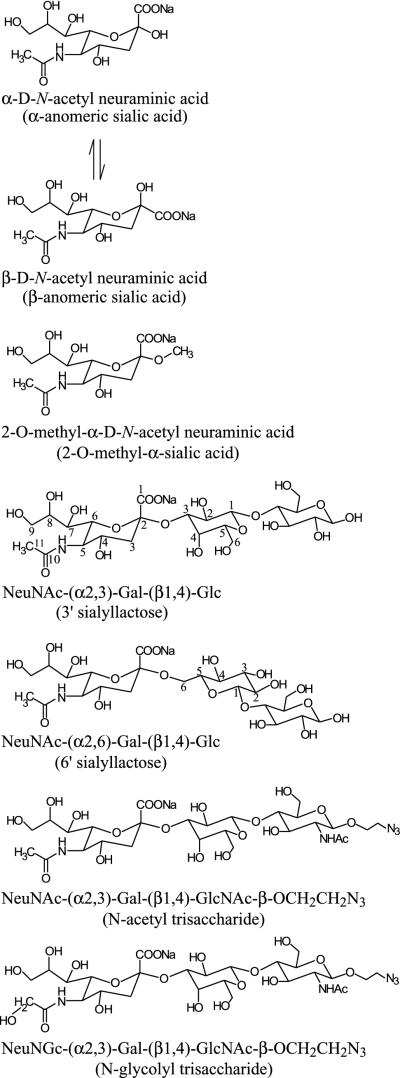
FIG. 1.
Structures of the sialosides used in the binding analysis. A short form of the name for each compound is given in parentheses below the complete name. The numbering scheme for the carbons of sialic acid is shown on 3′ sialyllactose, and the numbering scheme for the carbons of galactose is shown on 3′ and 6′ sialyllactose. The N-glycolyl and N-acetyl trisaccharides differ only at C-11 of the sialic acid moiety, where a hydrogen on the N-acetyl trisaccharide is replaced by a hydroxyl on the N-glycolyl trisaccharide.
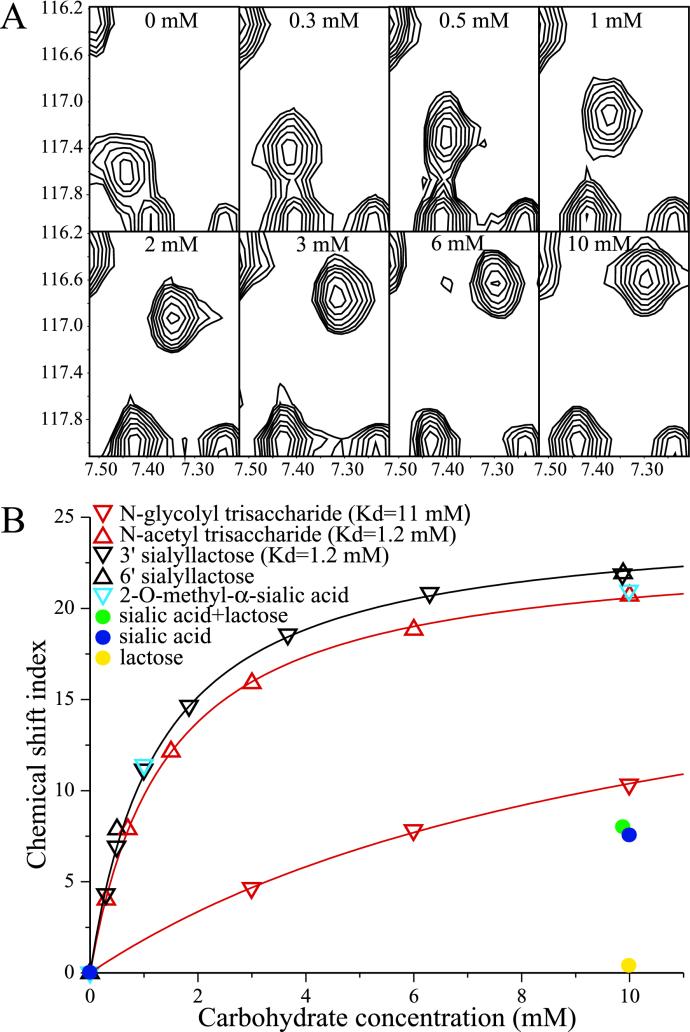
We have measured the specificity and affinity of the rhesus rotavirus (RRV) hemagglutination domain for sialosides by nuclear magnetic resonance (NMR) spectroscopy. NMR can be used to determine dissociation constants in equilibrium mixtures of receptors and ligands in solution; it has been used previously to study sialoside binding by influenza virus hemagglutinin (38). In the NMR experiments reported here, we titrated the rotavirus hemagglutination domain with sialosides and detected binding by monitoring the changes in the protein backbone amide nitrogen and hydrogen chemical shifts. The characteristic pattern of affected chemical shifts ensured the specificity of the observed changes. Figure 2A shows the migration of the R101 backbone amide peak induced by the binding of 3′ sialyllactose.
FIG. 2.
(A) Change in the HSQC amide peak position for R101 in the presence of various concentrations of 3′ sialyllactose (indicated at the top of each frame). The 15N-labeled Ec-VP846-231 concentration is 0.306 mM. The backbone amide 15N chemical shift (in parts per million) is given on the y axis, and the proton chemical shift (in parts per million) is given on the x axis. The R101 peak is the central peak seen migrating from the lower left to the upper right of the frames. (B) Plot of chemical shift index (ΔI) versus concentration of carbohydrate. The Ec-VP846-231 concentrations are between 0.282 and 0.306 mM, except for the samples mixed with lactose, in which the protein concentration is 0.228 mM. Measurements of chemical shifts in the presence of the N-glycolyl trisaccharide at 15 and 20 mM are not shown in the figure but were included in the determination of Kd. The determined Kds and hyperbolic fit curves used to determine ΔIholo are shown for the N-glycolyl trisaccharide, the N-acetyl trisaccharide, and 3′ sialyllactose.
Both the protein and the carbohydrates used in this binding study have been extensively characterized. The protein is an Escherichia coli-expressed construct (EcVP846-231) that corresponds to a protease-resistant core of the VP8* region of the RRV outer capsid spike protein, VP4 (12). The construct contains the rotavirus hemagglutination domain, which consists of residues L65 to L224. The construction, purification, and NMR solution structure of this recombinant protein and the crystal structure of a similar recombinant protein complexed with a sialoside are described elsewhere (13). The recombinant VP8* core is monomeric (12), so multivalent binding does not complicate the affinity measurements.
The monovalent sialoside ligands used in this study are synthetic products with the structures shown in Fig. 1. N-Acetylneuraminic acid (sialic acid), 2-O-methyl α-d-N-acetylneuraminic acid (2-O-methyl-α-sialic acid), and lactose were obtained from Sigma. NeuNAc-(α2,3)-Gal-(β1,4)-Glc (3′ sialyllactose) and NeuNAc-(α2,6)-Gal-(β1,4)-Glc (6′ sialyllactose) were obtained from Glyko, Inc. NeuNAc-(α2,3)-Gal-(β1,4)-GlcNAc-β-OCH2CH2N3 (N-acetyl trisaccharide) and NeuNGc-(α2,3)-Gal-(β1,4)-GlcNAc-β-OCH2CH2N3 (N-glycolyl trisaccharide) were synthesized by using a combined chemical and enzymatic synthesis procedure (3, 4). All charged carbohydrates were prepared as sodium salts at pH 7.0.
Two-dimensional 15N-1H heteronuclear single quantum correlation (HSQC) spectra of mixtures of the VP8* core and carbohydrates were obtained at 25°C by using a 500-MHz Varian spectrometer or a 500-MHz Bruker spectrometer equipped with a cryoprobe. Because only the protein was isotopically labeled with 15N, as previously described (13), only protein resonances were measured. The NMR buffer consisted of 18 mM Na2HPO4/NaH2PO4 (pH 7.0), 9 mM NaCl, 0.018% sodium azide, and 10% D2O in water. Data were processed with PROSA software (19), spectra were analyzed with the XEASY program (1), binding curves were fit with Origin 5.0 (Microcal Software, Inc.), and figures were prepared with XEASY, Origin 5.0, Illustrator 8.0 (Adobe Systems, Inc.), and ISIS/DRAW 2.4 (MDL Information Systems, Inc.).
To determine binding affinities, we followed the sialoside-induced changes in the backbone amide chemical shifts of 10 residues. R101, D142, Y155, G156, A166, V167, N178, G179, K187, and T192 were selected for the test set on the basis of their large binding-induced changes in chemical shift. K145, Y175, Y189, T191, and Y194 showed equally large sialoside-induced changes, but their peak positions could not be determined unambiguously at all sialoside concentrations due to chemical shift overlap and conformational averaging. The NMR and X-ray crystallographic structures of the VP8* core (13) show that most of the residues listed above either bind the sialoside directly or participate in interactions that stabilize sialoside-binding residues. A166 and V167 do not appear to stabilize the binding site, but their backbone amides are positioned close to several aromatic rings so that small binding-induced movements may significantly change their electronic environment. N178 and G179 also do not appear to participate directly in sialoside binding but are adjacent to an area of multiple conformations in the bound structure near the sialoside-binding site.
To determine the binding affinities, the binding-induced changes in chemical shifts were quantified by the following index (40): ΔI = Σ{[(N0 − N)/0.5]2 + [(H0 − H)/0.1]2}1/2, where N0 and N are the amide 15N chemical shifts in the absence and presence of ligand, respectively; H0 and H are the backbone amide hydrogen chemical shifts in the absence and presence of ligand, respectively; chemical shifts are expressed in parts per million; and the summation is over the 10 residues listed above. The total concentration of protein, [Ptot], was determined by absorbance using an extinction coefficient at 280 nm of 39.6 × 103 M−1 cm−1, based on prediction from the primary amino acid sequence by Protean (DNAStar, Inc.). The total concentration of ligand, [Ltot], was determined by weight. Under conditions of rapid chemical exchange, the equilibrium chemical shifts of VP8* core atoms in the presence of ligand are the average of the shifts of the VP8* core in an unbound and a ligand-bound state, weighted by the proportion of the VP8* core in each state. The chemical shifts in the unbound state were measured directly. The chemical shifts in the fully bound state were extrapolated from the chemical shifts measured during the titration (Fig. 2B) by using a hyperbolic function, consistent with a simple adsorption isotherm. These extrapolated chemical shifts were used to estimate ΔIholo, the chemical shift index of the VP8* core saturated with ligand. The portion of the VP8* core in the bound state (v) is given by v = ΔIobs/ΔIholo, where ΔIobs is the observed chemical shift index. The concentration of free ligand, [Lfree], is given by [Lfree] = [Ltot] − v[Ptot]. The Kd was determined by fitting the binding data to the following formula: 1/v = 1 + Kd/[Lfree], with [Lfree] in millimolar.
The chief sources of error in the calculations of Kd are the errors of predicting an extinction coefficient from sequence data (to determine [Ptot]), of weighing ligands (to determine [Ltot]), and of extrapolating ΔIholo (to determine v). Due to these uncertainties, we estimate that the final error of the Kd calculations is approximately ±20%.
This analysis shows that the VP8* core binds 3′ sialyllactose with a Kd of 1.2 mM (Fig. 2B). Chemical shifts in the presence of 0.5 or 9.9 mM sialic acid in an α2,6 linkage to lactose (6′ sialyllactose) match those in the presence of the same quantities of sialic acid in an α2,3 linkage to lactose (3′ sialyllactose) (Fig. 2B). Free sialic acid in aqueous solution is in equilibrium between the α and β anomers (Fig. 1), with 7% in the α-anomeric configuration (20). Free sialic acid at a concentration of 10.0 mM produces the same ΔI value as 0.56 mM 3′ sialyllactose (Fig. 2B), consistent with the binding of approximately 6% of the free sialic acid. In 2-O-methyl α-sialic acid, the lactose moiety of 3′ or 6′ sialyllactose is replaced by a methyl group, which locks the sialoside in the α anomer (Fig. 1). The changes in chemical shift induced by 1.0 or 10.0 mM 2-O-methyl α-neuraminic acid match those induced by 3′ and 6′ sialyllactose (Fig. 2B). Taken together, these data demonstrate that the RRV VP8* core is specific for the α anomer of sialic acid, that no sugar residues other than sialic acid are required for binding, that the presence of linked galactose does not significantly enhance binding, and that the VP8* core binds sialic acid in α2,3 and α2,6 linkages to the galactose moiety of lactose equally well.
The VP8* core binds a trisaccharide containing N-glycolylneuraminic acid with a Kd of 11 mM, while it binds an equivalent trisaccharide containing N-acetylneuraminic acid with a Kd of 1.2 mM (Fig. 2B). This strong preference for an N-acetyl sialoside over an N-glycolyl sialoside indicates that the sialic acid independence of most human rotavirus strains cannot be explained by a general preference of sialic acid-dependent rotavirus strains for N-glycolylneuraminic acid. Rather, there appears to be strain variation in the preference for N-glycolylneuraminic or N-acetylneuraminic acid. The possibility that strain variation in sialic acid binding arises during passage in cell culture cannot be ruled out, as this phenomenon has been observed with passage of influenza virus (35).
The variation in preference for N-acetylneuraminic or N-glycolylneuraminic acid may be linked to a specific genetic variation. The strains documented to preferentially bind N-glycolylneuraminic acid-containing glycosphingolipids (OSU, SA11, and NCDV [11, 37]) have a glycine at position 187, while RRV and strain UK (which gives mixed results in glycosphingolipid binding [11]) have lysine in this position (12, 21, 33, 34). Residue 187 is adjacent to the C-11 methyl group of bound N-acetylneuraminic acid (13), and a lysine in this position might sterically hinder binding of N-glycolylneuraminic acid, which is hydroxylated at C-11 (Fig. 1). Two studies have linked a lysine-to-arginine mutation at residue 187 of RRV VP8* to the acquisition of sialic acid-independent entry despite preserved hemagglutination, suggesting that mutations at this site may affect the specificity, affinity, or conformational effects of sialoside binding (26, 29). Future studies will address the effects of mutations at this site on binding affinity and specificity.
The relatively low affinity of the VP8* core for sialosides parallels NMR measurements of the affinity of influenza virus hemagglutinin for sialosides: influenza virus hemagglutinin (H3, strain X-31) binds 3′ sialyllactose with a Kd of 3.2 mM and 6′ sialyllactose with a Kd of 1.9 mM (38). The multivalency resulting from the 120 molecules of VP4 arrayed on the rotavirus surface and the many sialic acid moieties presented on cell surface glycolipids and glycoproteins should allow for relatively tight binding despite the weak individual interactions. Consistent with this conclusion, sialic acid presented on sialylphospholipid vesicles inhibits rotavirus infection with 1,000-fold greater potency than does soluble sialic acid (23). Tight binding due to multivalency has been quantitated for rosettes of influenza virus hemagglutinin, which bind fetuin with a Kd of 100 nM, as determined by surface plasmon resonance (42). The equivalent affinities of the VP8* core for 3′ sialyllactose, 6′ sialyllactose, and 2-O-methyl α-sialic acid indicate that it has a significantly broader binding specificity than influenza virus hemagglutinin.
The finding that 2-O-methyl α-sialic acid is the minimal sialoside with a defined anomeric state required for binding to the rotavirus hemagglutination domain provided the basis for the crystallization of the liganded VP8* core and the determination of the structure of the complex to a resolution of 1.4 Å (13). The determination of the binding affinity and specificity of the RRV hemagglutinin in a monomeric, monovalent, well-defined, bimolecular system supports the conclusion, based on more complicated systems, that sialic acid binding provides a relatively nonspecific initial interaction between rotavirus and the cell. As the cell type restriction of RRV can be bypassed by the transfection of double-layered subviral particles into cells (2), an entry event is probably responsible for cell specificity. The data presented here support the case that more-specific interactions, subsequent to sialic acid binding, are responsible for rotavirus cell type and host specificity (18, 30).
Acknowledgments
We thank Marina Babyonyshev for her skillful technical assistance; Harry Greenberg for providing scientific advice and materials; Steve Litster and Ulug Unligil for computational expertise; Genfa Zhou, Don Wiley, and Stephen DeWall for helpful and stimulating discussions; and Max Ciarlet and Mary Estes for sharing unpublished data.
This work was supported by National Institutes of Health grants K08 AI 001496 to P.R.D., GM 47467 and P41-RR00995 to G.W., and CA 13202 to S.C.H. S.C.H. is an investigator for the Howard Hughes Medical Institute.
REFERENCES
- 1.Bartels, C., T.-H. Xia, M. Billeter, P. Guntert, and K. Wuthrich. 1995. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 5:1-10. [DOI] [PubMed] [Google Scholar]
- 2.Bass, D. M., M. R. Baylor, C. Chen, E. M. Mackow, M. Bremont, and H. B. Greenberg. 1992. Liposome-mediated transfection of intact viral particles reveals that plasma membrane penetration determines permissivity of tissue culture cells to rotavirus. J. Clin. Investig. 90:2313-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blixt, O., K. Allin, L. Pereira, A. Datta, and J. C. Paulson. 2002. Efficient chemoenzymatic synthesis of O-linked sialyl oligosaccharides. J. Am. Chem. Soc. 124:5739-5746. [DOI] [PubMed] [Google Scholar]
- 4.Blixt, O., J. Brown, M. J. Schur, W. Wakarchuk, and J. C. Paulson. 2001. Efficient preparation of natural and synthetic galactosides with a recombinant β-1,4-galactosyltransferase-/UDP-4′-gal epimerase fusion protein. J. Org. Chem. 66:2442-2448. [DOI] [PubMed] [Google Scholar]
- 5.Cahan, L. D., R. Singh, and J. C. Paulson. 1983. Sialyloligosaccharide receptors of binding variants of polyoma virus. Virology 130:281-289. [DOI] [PubMed] [Google Scholar]
- 6.Chou, H. H., H. Takematsu, S. Diaz, J. Iber, E. Nickerson, K. L. Wright, E. A. Muchmore, D. L. Nelson, S. T. Warren, and A. Varki. 1998. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc. Natl. Acad. Sci. USA 95:11751-11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ciarlet, M., S. E. Crawford, and M. K. Estes. 2001. Differential infection of polarized epithelial cell lines by sialic acid-dependent and sialic acid-independent rotavirus strains. J. Virol. 75:11834-11850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciarlet, M., and M. K. Estes. 1999. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J. Gen. Virol. 80:943-948. [DOI] [PubMed] [Google Scholar]
- 9.Ciarlet, M., J. E. Ludert, M. Iturriza-Gomara, F. Liprandi, J. J. Gray, U. Desselberger, and M. K. Estes. 2002. The initial interaction of rotavirus strains with N-acetylneuraminic (sialic) acid residues on the cell surface correlates with VP4 genotype, not species of origin. J. Virol. 76:4087-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Connor, R. J., Y. Kawaoka, R. G. Webster, and J. C. Paulson. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205:17-23. [DOI] [PubMed] [Google Scholar]
- 11.Delorme, C., H. Brussow, J. Sidoti, N. Roche, K. A. Karlsson, J. R. Neeser, and S. Teneberg. 2001. Glycosphingolipid binding specificities of rotavirus: identification of a sialic acid-binding epitope. J. Virol. 75:2276-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dormitzer, P. R., H. B. Greenberg, and S. C. Harrison. 2001. Proteolysis of monomeric recombinant rotavirus VP4 yields an oligomeric VP5* core. J. Virol. 75:7339-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dormitzer, P. R., Z.-Y. J. Sun, G. Wagner, and S. C. Harrison. 2002. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 21:885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dubensky, T. W., R. Freund, C. J. Dawe, and T. L. Benjamin. 1991. Polyomavirus replication in mice: influences of VP1 type and route of inoculation. J. Virol. 65:342-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fazli, A., S. J. Bradley, M. J. Kiefel, C. Jolly, I. H. Holmes, and M. von Itzstein. 2001. Synthesis and biological evaluation of sialylmimetics as rotavirus inhibitors. J. Med. Chem. 44:3292-3301. [DOI] [PubMed] [Google Scholar]
- 16.Fiore, L., H. B. Greenberg, and E. R. Mackow. 1991. The VP8 fragment of VP4 is the rhesus rotavirus hemagglutinin. Virology 181:553-563. [DOI] [PubMed] [Google Scholar]
- 17.Freund, R., A. Calderone, C. J. Dawe, and T. L. Benjamin. 1991. Polyomavirus tumor induction in mice: effects of polymorphisms of VP1 and large T antigen. J. Virol. 65:335-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guerrero, C. A., E. Mendez, S. Zarate, P. Isa, S. Lopez, and C. F. Arias. 2000. Integrin αvβ3 mediates rotavirus cell entry. Proc. Natl. Acad. Sci. USA 97:14644-14649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guntert, P., V. Dotsch, G. Wider, and K. Wuthrich. 1992. Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR 2:619-629. [Google Scholar]
- 20.Haverkamp, J., H. van Halbeek, L. Dorland, J. F. Vliegenthart, R. Pfeil, and R. Schauer. 1982. High-resolution 1H-NMR spectroscopy of free and glycosidically linked O-acetylated sialic acids. Eur. J. Biochem. 122:305-311. [DOI] [PubMed] [Google Scholar]
- 21.Kantharidis, P., M. L. Dyall-Smith, G. W. Tregear, and I. H. Holmes. 1988. Nucleotide sequence of UK bovine rotavirus segment 4: possible host restriction of VP3 genes. Virology 166:308-315. [DOI] [PubMed] [Google Scholar]
- 22.Kiefel, M. J., B. Beisner, S. Bennett, I. D. Holmes, and M. von Itzstein. 1996. Synthesis and biological evaluation of N-acetylneuraminic acid-based rotavirus inhibitors. J. Med. Chem. 39:1314-1320. [DOI] [PubMed] [Google Scholar]
- 23.Koketsu, M., T. Nitoda, H. Sugino, L. R. Juneja, M. Kim, T. Yamamoto, N. Abe, T. Kajimoto, and C. H. Wong. 1997. Synthesis of a novel sialic acid derivative (sialylphospholipid) as an antirotaviral agent. J. Med. Chem. 40:3332-3335. [DOI] [PubMed] [Google Scholar]
- 24.Kuhlenschmidt, T. B., W. P. Hanafin, H. B. Gelberg, and M. S. Kuhlenschmidt. 1999. Sialic acid dependence and independence of group A rotaviruses. Adv. Exp. Med. Biol. 473:309-317. [DOI] [PubMed] [Google Scholar]
- 25.Ludert, J. E., N. Feng, J. H. Yu, R. L. Broome, Y. Hoshino, and H. B. Greenberg. 1996. Genetic mapping indicates that VP4 is the rotavirus cell attachment protein in vitro and in vivo. J. Virol. 70:487-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludert, J. E., B. B. Mason, J. Angel, B. Tang, Y. Hoshino, N. Feng, P. T. Vo, E. M. Mackow, F. M. Ruggeri, and H. B. Greenberg. 1998. Identification of mutations in the rotavirus protein VP4 that alter sialic-acid-dependent infection. J. Gen. Virol. 79:725-729. [DOI] [PubMed] [Google Scholar]
- 27.Mackow, E. R., J. W. Barnett, H. Chan, and H. B. Greenberg. 1989. The rhesus rotavirus outer capsid protein VP4 functions as a hemagglutinin and is antigenically conserved when expressed by a baculovirus recombinant. J. Virol. 63:1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matrosovich, M. N., S. Krauss, and R. G. Webster. 2001. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology 281:156-162. [DOI] [PubMed] [Google Scholar]
- 29.Mendez, E., C. F. Arias, and S. Lopez. 1996. Interactions between the two surface proteins of rotavirus may alter the receptor-binding specificity of the virus. J. Virol. 70:1218-1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendez, E., S. Lopez, M. A. Cuadras, P. Romero, and C. F. Arias. 1999. Entry of rotaviruses is a multistep process. Virology 263:450-459. [DOI] [PubMed] [Google Scholar]
- 31.Mochizuki, M., and O. Nakagomi. 1995. Haemagglutination by rotaviruses in relation to VP4 genotypes. Res. Virol. 146:371-374. [DOI] [PubMed] [Google Scholar]
- 32.Muchmore, E. A., S. Diaz, and A. Varki. 1998. A structural difference between the cell surfaces of humans and the great apes. Am. J. Phys. Anthropol. 107:187-198. [DOI] [PubMed] [Google Scholar]
- 33.Nishikawa, K., and M. Gorziglia. 1988. The nucleotide sequence of the VP3 gene of porcine rotavirus OSU. Nucleic Acids Res. 16:11847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishikawa, K., K. Taniguchi, A. Torres, Y. Hoshino, K. Green, A. Z. Kapikian, R. M. Chanock, and M. Gorziglia. 1988. Comparative analysis of the VP3 gene of divergent strains of the rotaviruses simian SA11 and bovine Nebraska calf diarrhea virus. J. Virol. 62:4022-4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rogers, G. N., and B. L. D'Souza. 1989. Receptor binding properties of human and animal H1 influenza virus isolates. Virology 173:317-322. [DOI] [PubMed] [Google Scholar]
- 36.Rogers, G. N., and J. C. Paulson. 1983. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology 127:361-373. [DOI] [PubMed] [Google Scholar]
- 37.Rolsma, M. D., T. B. Kuhlenschmidt, H. B. Gelberg, and M. S. Kuhlenschmidt. 1998. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 72:9079-9091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sauter, N. K., M. D. Bednarski, B. A. Wurzburg, J. E. Hanson, G. M. Whitesides, J. J. Skehel, and D. C. Wiley. 1989. Hemagglutinins from two influenza virus variants bind to sialic acid derivatives with millimolar dissociation constants: a 500-MHz proton nuclear magnetic resonance study. Biochemistry 28:8388-8396. [DOI] [PubMed] [Google Scholar]
- 39.Spence, L., M. Fauvel, R. Petro, and L. A. Babiuk. 1978. Comparison of rotavirus strains by hemagglutination inhibition. Can. J. Microbiol. 24:353-362. [DOI] [PubMed] [Google Scholar]
- 40.Sun, Z.-Y., V. Dotsch, M. Kim, J. Li, E. L. Reinherz, and G. Wagner. 1999. Functional glycan-free adhesion domain of human cell surface receptor CD58: design, production and NMR studies. EMBO J. 18:2941-2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Superti, F., and G. Donelli. 1991. Gangliosides as binding sites in SA-11 rotavirus infection of LLC-MK2 cells. J. Gen. Virol. 72:2467-2474. [DOI] [PubMed] [Google Scholar]
- 42.Takemoto, D. K., J. J. Skehel, and D. C. Wiley. 1996. A surface plasmon resonance assay for the binding of influenza virus hemagglutinin to its sialic acid receptor. Virology 217:452-458. [DOI] [PubMed] [Google Scholar]
- 43.World Health Organization. 1999. Rotavirus vaccines. Wkly. Epidemiol. Rec. 74:33-38.
- 44.Yolken, R. H., R. Willoughby, S. B. Wee, R. Miskuff, and S. Vonderfecht. 1987. Sialic acid glycoproteins inhibit in vitro and in vivo replication of rotaviruses. J. Clin. Investig. 79:148-154. [DOI] [PMC free article] [PubMed] [Google Scholar]