Abstract
There are several forms of human immunodeficiency virus type 1 (HIV-1) DNA in peripheral blood T cells and lymph nodes in untreated HIV-1-infected individuals and in patients whose plasma HIV-1 RNA levels are suppressed by long-term combination antiretroviral therapy. However, it remains to be established whether the concentration of HIV-1 DNA in cells predicts the clinical outcome of HIV-1 infection. In this report, we measured the concentration of HIV-1 DNA forms which has undergone the second template switch (STS DNA) and 2-long-terminal-repeat DNA circles in peripheral blood mononuclear cell (PBMC) samples. To do this, we used molecular-beacon-based real-time PCR assays and studied 130 patients with hemophilia in the Multicenter Hemophilia Cohort Study. We assessed the influence of baseline HIV-1 STS DNA levels on the progression of HIV-1 disease in the absence of combination antiretroviral therapy by Kaplan-Meier and Cox regression analysis. Among the patients who progressed to AIDS, the median levels (interquartile ranges) of STS HIV-1 DNA in PBMC were significantly higher than those of patients who remained AIDS free during the 16 years of follow-up (1,017 [235 to 6,059] and 286 [31 to 732] copies per 106 PBMC, respectively; P < 0.0001). Rates of progression to death and development of AIDS varied significantly (log rank P < 0.001) by quartile distribution of HIV-1 STS DNA levels. After adjustment for age at seroconversion, baseline CD4+ T-cell counts, plasma viral load, and T-cell-receptor excision circles, the relative hazards (RH) of death and AIDS were significantly increased with higher HIV-1 STS DNA levels (adjusted RH, 1.84 [95% confidence interval {CI}, 1.30 to 2.59] and 2.62 [95% CI, 1.75 to 3.93] per 10-fold increase per 106 PBMC, respectively). HIV-1 STS DNA levels in each individual remained steady in longitudinal PBMC samples during 16 years of follow-up. Our findings show that the concentration of HIV-1 STS DNA in PBMC complements the HIV-1 RNA load in plasma in predicting the clinical outcome of HIV-1 disease. This parameter may have important implications for understanding the virological response to combination antiretroviral therapy.
The rate of progression of human immunodeficiency virus type 1 (HIV-1) disease is highly variable among patients not receiving potent antiretroviral therapy (34, 48). Previous epidemiological studies on untreated HIV-1-infected individuals have revealed that viral load (37, 38, 45, 52), host factors (17, 22, 27, 43, 44), and age at HIV-1 antibody seroroconversion (13) are associated with the variation in disease progression, with plasma HIV-1 RNA load being the most effective independent predictor of the rate of progression of HIV-1 disease (37, 38). The plasma HIV-1 RNA load is now widely considered a direct indicator of the overall level of HIV-1 expression in infected individuals. Commercially available assays for measuring the plasma viral load (12, 41, 61) are currently being used to monitor disease progression in HIV-1-infected patients receiving potent antiretroviral therapy and to assess the efficacy of new antiretroviral regimens to treat HIV-1 infection (18, 21). Most current antiretroviral protocols utilizing drugs that inhibit the HIV-1 reverse transcriptase and protease proteins suppress the replicative ability of HIV-1 to such an extent that circulating HIV-1 in plasma becomes undetectable by the most sensitive viral RNA detection assays (18, 21). Several studies have revealed that there is a direct relationship between reduction in HIV-1 replication as evidenced by plasma HIV-1 RNA concentrations and inhibition of progression of HIV-1 disease (25, 29, 30, 46, 47, 49-51). However, in HIV-1-infected individuals who have prolonged suppression of HIV-1 replication by antiretroviral therapy, peripheral blood mononuclear cells (PMBC), lymphoid tissues, and semen may still contain HIV-1 DNA (9, 14, 63, 64, 66), replication-competent virus (14, 64, 66), and viral products indicative of persistent HIV-1 replication (54) and transcription (15).
Over the past decade, numerous PCR-based methodologies for quantifying cell-associated HIV-1 DNA have been described in various formats (1, 2, 5, 11, 16, 19, 28, 35, 36, 39, 40, 53, 62, 65), some of which have been used to measure the decay characteristics of HIV-1 DNA in individuals whose plasma virus levels are suppressed by antiretroviral therapy (3, 42, 55). Several PCR-based assays to quantify integrated HIV-1 DNA levels in HIV-1-infected cell cultures and patients have been described recently (4, 6, 9, 59). Some of these methodologies have been utilized in studies aimed at elucidating whether long-term suppression of HIV-1 replication can reduce levels of integrated HIV-1 (6-9, 14, 15, 26). Other applications include studies to establish the kinetics of HIV-1 DNA integration (4, 59) and to evaluate the ability of newly developed inhibitors of HIV-1 integrase to obstruct integration in cell culture (60). Despite the growing number of studies involving quantification of cellular HIV-1 DNA, some of the methodologies used either are semiquantitative, have a restricted dynamic range, require laborious post-PCR analytical procedures, or utilize genomic equivalent markers present at an imprecise number of copies per cell to determine the exact number of cells in a sample. Although it is generally recognized that accurate quantification of HIV-1 DNA in peripheral blood cells and tissues is important for monitoring disease progression in patients receiving antiretroviral therapy, it remains to be determined whether the prognosis of HIV-1 infection is associated with the quantity of HIV-1 DNA in cells.
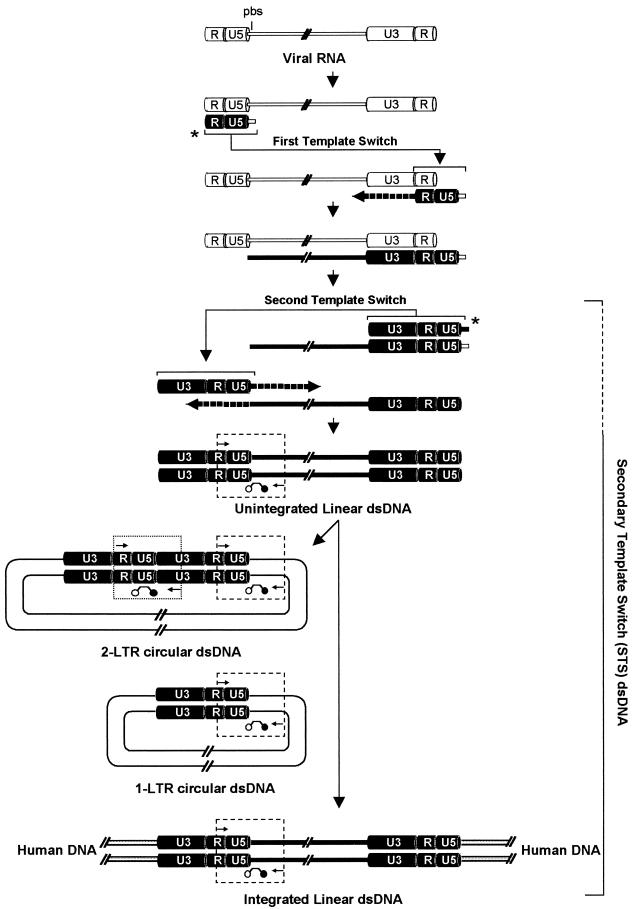
Following reverse transcription of the viral RNA genome, the HIV-1 RNA must be reverse transcribed to linear double-stranded DNA (dsDNA) prior to integration of the proviral HIV-1 DNA genome into the human chromosome. To accomplish a successful reverse transcription, HIV-1 undergoes two template switches by using the R and U5 long terminal repeat (LTR) regions and the primer-binding site (PBS) of its genome. In this study, we report an assay, which is based on real-time PCR and the molecular-beacon detection system, that quantifies PBMC-associated HIV-1 DNA forms that have undergone the second template switch (HIV-1 STS DNA). The HIV-1 STS DNA assay mainly detects a pool of HIV-1 forms that includes unintegrated and integrated linear dsDNA viral genomes and 1- and 2-LTR circles (LTRC and 2LTRC). In contrast, the existing 2LTRC-specific assay quantifies only 2LTRC, as previously described (17, 54, 58).
By using the STS DNA- and 2LTRC-specific assays coupled with a CCR5-specific real-time PCR assay that quantifies genomic equivalent markers with known numbers of copies in human cells, we quantified HIV-1 STS and 2LTRC DNAs in PBMC for a cohort of longitudinally studied HIV-1-infected hemophiliacs. The STS DNA and 2LTRC measurements were used to determine the relationships between STS DNA and 2LTRC in PBMC and HIV-1 RNA in plasma, to study the kinetics of cellular STS DNA throughout the natural history of HIV-1 infection, and to establish the relationship between the PBMC-associated HIV-1 STS DNA load and clinical outcome in untreated HIV-1 infection.
MATERIALS AND METHODS
Study patients.
All clinical samples were obtained from HIV-1-infected Greek study participants enrolled in the Multicenter Hemophilia Cohort Study (MHCS). The Greek component of MHCS consists of 158 white HIV-1-infected hemophiliac men with known HIV-1 antibody seroconversion dates; they have been prospectively monitored for more than 18 years after seroconversion. Clinical and laboratory data were collected for each patient approximately every 6 months (56). For 131 subjects of this cohort, plasma HIV-1 RNA loads and levels of T-cell-receptor excision DNA circles (TREC) taken during the early chronic infection period were shown to be predictive of the progression of HIV-1 disease (22). STS and 2LTRC HIV-1 DNA concentrations were measured in cryopreserved PBMC samples isolated closest to the HIV-1 antibody seroconversion date. In longitudinal studies, STS DNA levels were determined in 655 available cryopreserved PBMC samples isolated from all participants before the initiation of highly effective antiretroviral therapy. Long-term nonprogressors were selected based on the absence of clinical AIDS and high CD4 T-cell counts (more than 500 cells/μl for at least 10 years after HIV-1 antibody seroconversion), and progressors, who developed AIDS during follow-up, were randomly selected. Plasma HIV-1 RNA levels were measured with the ultrasensitive HIV-1 Amplicor Monitor assay (Roche Diagnostics, Alameda, Calif.), which has a detection limit of 50 HIV-1 RNA copies/ml. CD4 T-cell counts were measured by flow cytometry with standard procedures.
Quantitation of PBMC-associated HIV-1 STS and 2LTRC DNAs.
The schematic outline in Fig. 1 illustrates the major HIV-1 DNA structures formed during reverse transcription and the strategy for STS- and 2LTRC-specific assays. To uniquely detect DNA structures that have completed the two template switches, we designed a real-time PCR assay with PCR primers that direct the amplification of viral sequences between the 5′ R-LTR region and the 5′ gag gene (4). This assay measures only HIV-1 DNA structures that have undergone both single-stranded DNA (ssDNA) template switches, including unintegrated and integrated linear viral genomes as well as LTRC and 2LTRC. The 2LTRC-specific assay is based on amplification of a region spanning the 5′- and 3′-end LTR ligation as previously described (54, 58). To count the number of cells in the input DNA, we used a molecular-beacon- and real-time PCR-based assay to quantify a region of the human CCR5 gene adjacent to the Δ32 deletion, which exists at 2 copies per cell (data not shown).
FIG. 1.
Schematic representation summarizes the cellular HIV-1 intermediates formed during reverse transcription. U3, R, and U5 LTR regions are indicated on the 5′ and 3′ ends of the viral genome; solid line, viral gag, pol, and env genomic regions; pbs, primer-binding site; slashed lines, gaps in the HIV-1 genome. Arrowed lines marked by asterisks indicate the primary transfer of the R-U5 negative ssDNA from the 5′ end to the 3′ end of the RNA genome by complementarity between the R regions and the secondary transfer of the U3-R-U5-PBS positive ssDNA from the 3′ end to the 5′ end of the positive ssDNA viral genome by complementarity of the PBS region. Dashed arrows indicate the synthesis of positive and negative ssDNA to form a full-length dsDNA. In the nucleus, some linear dsDNA molecules, by following the integration pathway, are successfully integrated into the human genome, and some are circularized to LTRC and 2LTRC. Viral sequences bound by PCR primers and molecular beacons represent the amplicons used in STS DNA- and 2LTRC-specific molecular-beacon-based real-time PCR assays (enclosed by dashed and dotted boxes, respectively). The pool of HIV-1 DNA forms identified by the STS DNA-specific assay is indicated by the bracket on the right (dashed line indicates uncertainty).
To quantify the concentrations of HIV-1 STS DNA and 2LTRC per cell, we designed two molecular-beacon real-time PCR assays by using the general method of quantifying single sequences with nucleotide-specific molecular beacons (57) and real-time PCR (23) as we have previously described (22). Genomic DNA was isolated from uncultured PBMC by standard procedures. HIV-1 STS DNA was quantified by amplifying a region spanning the LTR-U5 and gag regions. The sequence of the gag-specific molecular beacon was fluorescein-5′-CCGGTCTCCCCCGCTTAATACTGACGCTCTCGACCGG-3′-dabcyl, where dabcyl is the quencher 4-(4′-dimethylamino phenylazo)benzoic acid (underlining indicates the complementary sequences forming the hairpin structure). The target recognition sequence for the molecular beacon was 5′-CGAGAGCGTCAGTATTAAGCGGGGGAGA-3′ (at positions 797 to 824 of the gag region of the HXB2 sequence). Primers used in the real-time PCR were 5′-GCCTCAATAAAGCTTGCCTTGAGTG-3′ (at positions 522 to 546 of the LTR-R region) and 5′-GTTCTTCTGATCCTGTCTGAAGGG-3′ (at positions 989 to 1012 of the gag region). HIV-1 DNAs of circularized HIV-1 genomes containing 2 LTRs (2LTRC) were quantified by amplifying a region spanning the 5′- and 3′-end LTR ligation as previously described (54). The sequence of the LTR-U5-specific molecular beacon was fluorescein-5′-GCGGGTTCTGAGGGATCTCTAGTTACCAGACCCGC-3′-dabcyl. The target recognition sequence for the molecular beacon was 5′-TCTGGTAACTAGAGATCCCTCAGA-3′ (at positions 580 to 603 of the LTR-U5 region of the HXB2 sequence). Primers used in the real-time PCR were 5′-GGTACTAGCTTGAAGCACCATCC-3′ (at positions 129 to 151 of the LTR-U3 region) and LK164 5′-GCCTCAATAAAGCTTGCCTTGAGTG-3′ (at positions 522 to 546 of the LTR-R region). The PCR primers and the target recognition sequence of the molecular beacon were designed to hybridize on conserved regions from all the genetic subtypes within the M group based on a comprehensive DNA sequence alignment from published HIV-1 sequences. HIV-1 STS and 2LTRC amplicons were sequenced and found to contain the correct U5-gag and R-U5-U3 regions, respectively. Each 50-μl PCR mixture contained 0.5 to 5.0 μg of genomic DNA, 0.25 μM each molecular beacon, 0.5 μM each primer, 0.25 mM dATP, 0.25 mM dCTP, 0.25 mM dGTP, 0.25 mM dTTP, 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer), 50 mM KCl, 3.5 mM MgCl2, and 10 mM Tris-HCl (pH 8.3). One cycle of denaturation (94°C for 10 min), followed by 40 cycles of amplification (denaturation at 94°C for 15 s, annealing and data collection at 60°C for 30 s, and polymerization at 72°C for 30 s), was performed in a spectrofluorometric thermal cycler (ABI 7700; Applied Biosystems).
To quantify the cell equivalents in the input genomic DNA, we used a molecular-beacon-based real-time PCR assay with primers and probe within the CCR5 coding region, because it is known that this gene is present at only 2 copies per cell (L. G. Kostrikis, unpublished data). The genomic equivalent number was determined in each cell aliquot by real-time PCR using the CCR5 molecular beacon tetrachloro-fluorescein-5′-GCGCCTATGACAAGCAGCGGCAGGAGGCGC-3′-dabcyl. Primers used in the real-time PCR were 5′-GCTGTGTTTGCGTCTCTCCCAGGA-3′ and 5′-CTCACAGCCCTGTGCCTCTTCTTC-3′. The target recognition sequence for the molecular beacon (5′-TCCTGCCGCT GCTTGTCAT-3′) is adjacent to the Δ32 deletion of CCR5 and therefore recognizes both the wild-type CCR5 and mutant CCR5Δ32 alleles. Each 50-μl PCR mixture contained 0.5 to 5.0 μg of genomic DNA, 0.25 μM each molecular beacon, 0.5 μM each primer, 0.25 mM dATP, 0.25 mM dCTP, 0.25 mM dGTP, 0.25 mM dTTP, 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer), 3.5 mM MgCl2, and 10 mM Tris-HCl (pH 8.3). One cycle of denaturation (94°C for 10 min), followed by 40 cycles of amplification (denaturation at 94°C for 15 s, annealing and data collection at 60°C for 30 s, and polymerization at 72°C for 30 s), was performed. Real-time PCR amplifications were performed in a spectrofluorometric thermal cycler (ABI 7700; Applied Biosystems).
DNA standards.
The DNA external standards used in the three real-time PCR assays were STS DNA, 2LTRC, and CCR5 amplicons with known nucleotide lengths and base compositions, which contain within them the amplicons utilized in the real-time PCR assays. Each amplicon was purified by gel filtration twice to remove excess PCR primers, deoxynucleoside triphosphates, and polymerase, and the amplicon length was compared to dsDNA standards with known lengths by agarose gel electrophoresis. The concentrations of the purified STS DNA, 2LTRC, and CCR5 amplicons were quantified by UV absorbance spectrophotometry using a Cary 219 spectrophotometer. Wavelength accuracy was ±0.2 nm, and wavelength repeatability was within ±0.05 nm. Spectra of DNA in neutral aqueous buffer (10 mM Tris-HCl-0.1 mM EDTA [pH 7.0]) were recorded from 340 to 250 nm at scan rates of 0.5 to 1.0 nm/s, at a period of 0.5 s, and at a slit width of 1.0 nm. Concentrations of purified DNA with negligible (optical density cm−1, <10−4) light scattering for a λ of >320 nm were determined with a scatter-corrected absorbance coefficient of 19.98 mg−1 cm2 at 260 nm. Tenfold serial dilutions of the purified dsDNA transcripts with known molar concentrations were used as standard templates to generate the standard curves in the selective amplification assays using molecular-beacon-based real-time PCR. The slope and correlation coefficient of each standard curve were calculated based on the average threshold cycle (CT) values measured in eight replicates for each dilution point ranging from 106 to 101 DNA templates. The PCR efficiency, E, corresponding to the experimentally derived dynamic range was computed as (10−1/s − 1) × 100, where s is the slope of the standard curve generated.
During the molecular-beacon annealing stage of each PCR cycle, the fluorescence emission spectrum in each sample was automatically recorded from 500 to 650 nm. After completion of the PCR, each spectrum generated was decomposed into prerecorded fluorescein (specific for HIV-1 STS DNA or 2LTRC) and tetrachloro-fluorescein (specific for CCR5) reference spectra. In each experiment, standard curves for HIV-1 STS or 2LTRC and human CCR5 templates were run in duplicate by using six serial dilutions, ranging from 106 to 101 copies, for each DNA template and a no-template negative control. Changes in the emission spectra during amplification were compared with those from control STS DNA, 2LTRC, or CCR5 samples with known concentrations of initial DNA templates. In each genomic DNA sample, the number of cells was quantified as 1 cell per 2 CCR5 copies, because there is experimental evidence that the CCR5 amplicon used in the assay is present at 2 copies per human cell (data not shown), and the HIV-1 STS DNA and 2LTRC levels were calculated per 106 PBMC.
Statistical analysis.
Median levels and interquartile ranges (IQR) of HIV-1 STS DNA and 2LTRC were determined. Spearman correlations were used to evaluate the relationships between HIV-1 STS DNA levels, plasma HIV-1 RNA levels, TREC levels, and CD4 T-cell counts. The relative prognostic values of baseline HIV-1 STS DNA and 2LTRC levels with regard to death and development of clinical AIDS were investigated. Kaplan-Meier survival curves were used to estimate the cumulative incidence of an end point, and the log rank test was used to compare survival curves among different groups. Cox proportional-hazard models for death and AIDS risk were constructed using continuous measures of log10-transformed HIV-1 STS DNA, TREC, and plasma RNA levels and untransformed CD4 T-cell counts. The cohort was analyzed as seroincident (the HIV-1 seroconversion date was used as time zero) by using Cox proportional-hazard models with allowance for late entry; that is, participants entered into the risk set at the time they were first tested for HIV-1 STS DNA and 2LTRC. Temporal trends in STS DNA values were described by fitting random-effects models. These models provide estimates of average marker trends while accounting for correlation of repeated measurements within each individual.
RESULTS
Characteristics of real-time PCR assays for quantifying HIV-1 STS and 2LTRC DNAs in PBMC.
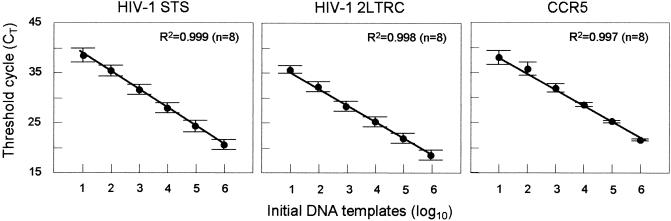
We have established that the STS DNA, 2LTRC, and CCR5 assays are specific and can accurately detect 10 DNA copies with a 6-log10 linear dynamic range. The coefficients of variation were below 20% for 106 copies and below 30% for 101 copies. The slopes of the standard curves (Fig. 2) were between −3.6 and −3.4 cycles/log10 DNA templates, corresponding to PCR efficiencies between 90 and 97%. The capability of the STS DNA assay to detect HIV-1 strains within the M group was evaluated by using DNA extracted from primary PBMC samples isolated from patients infected with HIV-1 strains from genetic subtypes A, B, C, D, and E and two recombinant strains. As expected, based on the designs of the PCR primers and molecular beacon, the 5′-R-U5-gag regions could be amplified from all strains (data not shown). The specificities of the STS DNA and 2LTRC assays against human genomic DNA were examined by using DNA extracted from individuals who were not infected with HIV-1; the results were negative, with no detectable signals after 50 cycles (data not shown).
FIG. 2.
Standard curves for HIV-1 STS DNA and 2LTRC templates and for human CCR5 templates utilized in the real-time PCR assays for quantifying HIV-1 STS and 2LTRC DNAs in human PBMC. Six serial dilutions ranging from 106 to 101 DNA templates were made for each DNA standard, and all standard dilutions were measured by real-time PCR using nucleotide sequence-specific molecular beacons. Average CT values (± standard deviations) were measured for eight replicates for each dilution point of the standard curve. The correlation coefficients (R2) of the three standard curves were >0.995, and the PCR efficiencies were >90%.
The analytical sensitivities of STS DNA- and 2LTRC-specific assays were assessed overall in 743 PBMC samples isolated from Greek individuals in the MHCS and 208 samples taken from the entire MHCS during early chronic infection and prior to effective antiretroviral therapy. Eighty-seven (87%) of the 951 PBMC samples taken prior to effective antiretroviral therapy had positive values for STS DNA (>10 copies/106 PBMC), and 31 (31%) of the samples had positive values for 2LTRC (>10 copies/106 PBMC). Median values were 451 copies/106 PBMC for STS DNA and <10 copies/106 PBMC for 2LTRC.
Baseline values of HIV-1 STS and 2LTRC DNAs.
Quantities of STS DNA and 2LTRC in PBMC were assessed for 130 individuals in the Greek MHCS. The median time between HIV-1 antibody seroconversion and the first PBMC isolation (baseline) was 6.7 years (range, 3.4 to 12.9 years). The mean age (± standard deviation) of the subjects at baseline was 30.4 (±13.7) years (range, 7 to 66 years). Three subjects who had developed AIDS by the time of the first PBMC isolation, with corresponding STS DNA levels of 916, 572, and 47 copies/106 PBMC, were excluded from further analysis. Of the remaining 127 participants, 14 had fewer than 10 STS DNA copies/106 PBMC, with a median CD4 T-cell count of 679 (IQR, 360 to 702) cells/μl.
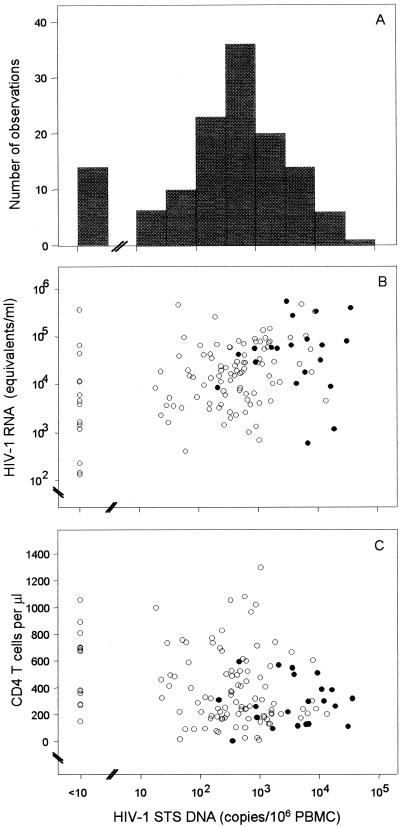
At baseline, the median HIV-1 STS DNA level for subjects without clinical AIDS was 443 copies/106 PBMC (IQR, 123 to 1,505 copies/106 PBMC) (Fig. 3A). We have found that there was no significant association between STS DNA levels and age at time of PBMC isolation. The corresponding median CD4 T-cell count was 320 cells/μl (IQR, 204 to 563 cells/μl), and the median HIV RNA level was 17,850 copies/ml (IQR, 4,580 to 59,860 copies/ml). Of the 127 subjects, only 20 (16.5%) had detectable 2LTRC levels. The median 2LTRC level was <10 copies/106 PBMC (range, <10 to 98 copies/106 PBMC). STS DNA levels were moderately correlated with plasma HIV-1 RNA levels (Spearman's r = 0.38; P < 0.001) (Fig. 3B) and weakly correlated with CD4 T-cell counts (Spearman's r = −0.24; P = 0.007) (Fig. 3C). The majority of subjects with detectable 2LTRC levels (>10 copies/106 PBMC) had high STS DNA levels (>103 copies/106 PBMC) and tended to cluster at higher plasma HIV-1 RNA levels and lower CD4 T-cell counts (Fig. 3B and C).
FIG. 3.
Distribution of baseline HIV-1 STS DNA levels (A), plasma RNA levels (B), and CD4+ T-cell counts (C) among 127 AIDS-free participants from the MHCS. Solid circles indicate subjects with detectable 2LTRC levels (>10 copies/106 PBMC). The lower limits of detection were 10 copies/106 PBMC (1.0 log10 copies/106 PBMC) for HIV-1 STS and 2LTRC DNAs in PBMC and 50 copies/ml (1.7 log10 copies/ml) for plasma HIV-1 RNA load.
Effects of HIV-1 STS DNA and 2LTRC levels on the progression of untreated HIV-1 disease.
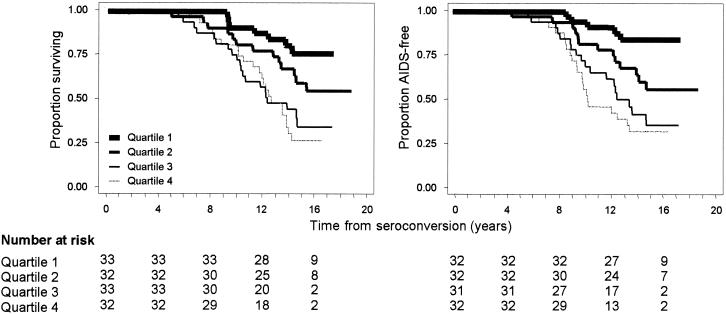
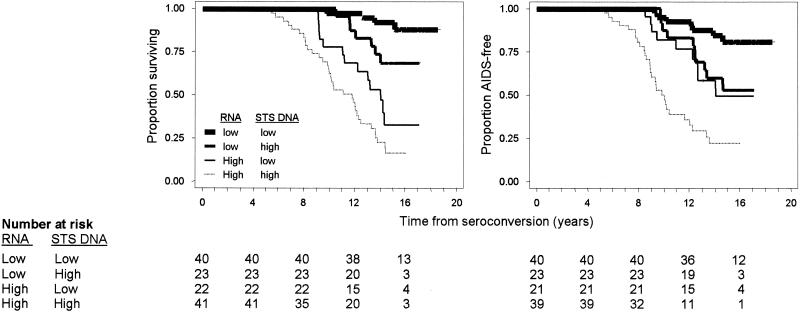
Among the 127 participants without clinical AIDS at baseline, STS DNA levels were higher in the 54 subjects who progressed to AIDS than in those who remained AIDS free. The corresponding median levels were 1,017 (IQR, 235 to 6,059) and 286 (IQR, 31 to 732) copies/106 PBMC, respectively (P < 0.0001). For quartiles of the STS DNA distribution, ranging from lowest to highest values, the cumulative rates of death by 16 years after seroconversion were 23.6% (95% confidence interval [CI], 11.8 to 43.7%), 44.8% (95% CI, 28.5 to 64.9%), 65.0% (95% CI, 47.4 to 82.0%), and 72.6% (95% CI, 55.8 to 87.1%), respectively. The corresponding rates for progression to clinical AIDS were 14.0% (95% CI, 5.5 to 33.2%), 46.0% (95% CI, 29.5 to 66.3%), 64.9% (95% CI, 44.9 to 84.1%), and 70.7% (95% CI, 53.8 to 85.8%) (Fig. 4). The progression rates differed significantly by HIV-1 STS DNA levels (log rank P < 0.001 for both end points).
FIG. 4.
Kaplan-Meier survival curves for participants from the MHCS were examined to assess the effect of baseline HIV-1 STS DNA levels in PBMC on progression to death (left) and clinical AIDS (right). Quartiles 1 through 4 correspond to fewer than 123, 124 to 443, 444 to 1,431, and more than 1,431 STS HIV-1 DNA copies/106 PBMC, respectively. The numbers of patients corresponding to each quartile at HIV-1 antibody seroconversion and every 4 years thereafter are given.
Following adjustment for age at HIV-1 antibody seroconversion, STS DNA levels, concurrent CD4 T-cell counts, plasma viral load, and TREC levels were significantly associated with the hazards of death and of developing clinical AIDS (Table 1). The predictive value of STS DNA levels remained significant and of almost the same magnitude with multivariate adjustment for concurrent CD4 T-cell counts, HIV-1 RNA levels, and TREC levels. In this multivariate model, there was greater attenuation of the prognostic values of concurrent CD4 cell counts and TREC levels, such that they were not statistically significant for time to AIDS (Table 1). Figure 5 shows the cumulative incidence rates of progression to death and to clinical AIDS after stratification of patients by their baseline STS DNA and plasma RNA levels. Higher baseline STS DNA levels were associated with increased risks of AIDS and death for patients with baseline RNA levels either above or below the median (log rank P < 0.05 in both cases). In univariate analysis, detectable versus undetectable 2LTRC levels (more versus fewer than 10 copies/106 PBMC, respectively) were significantly associated with time to death and development of clinical AIDS. However, after adjustment for STS DNA levels, 2LTRC levels did not significantly predict the progression of HIV-1 disease. The frequency of patients with detectable 2LTRC levels in this study is lower than those in previously published studies using similar methodologies (4, 54). This discrepancy may be attributed to differences in real-time-PCR methodologies in quantifying both 2LTRC templates and cell equivalents and to genetic diversity among the HIV-1 strains.
TABLE 1.
Relative risk of HIV-1-induced death and AIDS associated with changes in PBMC HIV-1 STS DNA levels, plasma HIV-1 RNA levels, CD4 T-cell counts, and TREC levelsa
| Outcome and risk factor (increase) | Risk adjusted for age at seroconversion
|
Risk adjusted for all other factors as well
|
||
|---|---|---|---|---|
| RH (95% CI) | P | RH (95% CI) | P | |
| Death | ||||
| HIV-1 STS DNA (10-fold/106 PBMC) | 2.00 (1.48-2.72) | <0.001 | 1.84 (1.30-2.59) | 0.001 |
| HIV-1 RNA (10-fold/ml of plasma) | 3.47 (2.33-5.17) | <0.001 | 3.37 (2.15-5.30) | <0.001 |
| CD4 T-cell count (100 cells/μl) | 0.79 (0.70-0.90) | <0.001 | 0.89 (0.79-1.01) | 0.061 |
| TREC (10-fold/106 PBMC) | 0.48 (0.36-0.65) | <0.001 | 0.55 (0.38-0.82) | 0.002 |
| AIDS | ||||
| HIV-1 STS DNA (10-fold/106 PBMC) | 2.59 (1.82-3.67) | <0.001 | 2.62 (1.75-3.93) | <0.001 |
| HIV-1 RNA (10-fold/ml of plasma) | 3.00 (1.98-4.54) | <0.001 | 3.04 (1.86-4.97) | <0.001 |
| CD4 T-cell count (100 cells/μl) | 0.80 (0.70-0.92) | 0.001 | 0.90 (0.80-1.03) | 0.121 |
| TREC (10-fold/106 PBMC) | 0.51 (0.36-0.71) | 0.001 | 0.65 (0.41-1.03) | 0.070 |
Cox proportional-hazard models were used to evaluate the relative risks of death and AIDS per unit of change in the covariates, as indicated.
FIG. 5.
Kaplan-Meier survival curves for time to death (left) and time to clinical AIDS (right) from combined effects of baseline HIV-1 STS DNA and plasma RNA levels.
Longitudinal HIV-1 STS DNA values.
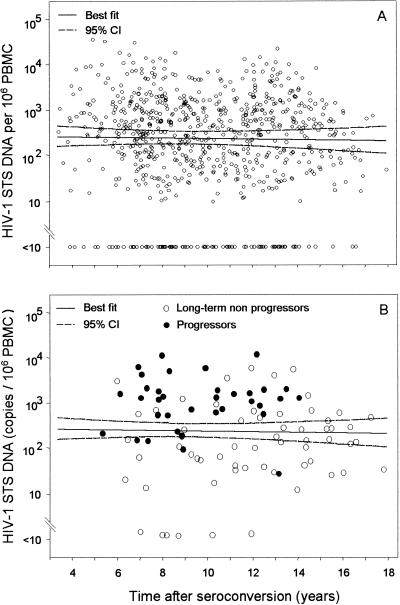
HIV-1 STS DNA levels were measured in all available samples from all patients prior to initiation of effective antiretroviral therapy. From each patient, six longitudinal STS DNA values were determined on average during the 16 years of follow-up, with a median interval of 1 year between successive measurements. The results indicated that during the natural history of HIV-1 infection, there was no evidence of increasing or decreasing levels of STS DNA over time. The overall mean slope of STS levels (change in log10 unit) was −0.001 per year (95% CI, −0.031 to 0.028), corresponding to a 0.3% rate of drop in the original scale (95% CI, 6.8% decrease to 6.6% increase) (Fig. 6A). As shown in Fig. 6B, the majority of the longitudinal STS values from seven randomly selected progressors were above the median values estimated from all study participants, while the majority of the STS values derived from seven selected long-term nonprogressors were below the estimated median values.
FIG. 6.
(A) Distribution of HIV-1 STS DNA levels obtained from longitudinal PBMC samples isolated from all participants. STS DNA levels were measured in all available stored samples isolated after HIV-1 antibody seroconversion and before initiation of effective antiretroviral therapy. The solid line represents the linear regression of the log10-transformed STS DNA levels and time of HIV-1 infection, and the dashed lines represent the corresponding 95% CI. (B) Distribution of STS DNA levels in longitudinal samples from seven participants who were long-term nonprogressors (open circles) and from seven randomly selected progressors (solid circles). HIV-1 STS DNA levels are shown against time of HIV-1 infection and relative to the best-fit curve and its 95% CI, derived from all study participants.
DISCUSSION
Our interest in developing a quantitative real-time PCR assay to measure the PBMC-associated levels of HIV-1 DNA in HIV-1-infected patients in a well-characterized AIDS study cohort originates from our ongoing effort to identify new clinically important prognostic markers of HIV-1 disease. We also want to use this assay to study persistent viral reservoirs and to monitor patients receiving antiretroviral treatment who have undetectable plasma RNA loads. We therefore identified biologically significant pools of HIV-1 DNA structures formed during reverse transcription and DNA integration, and we evaluated whether they correlated with progression of HIV-1 disease independently of the well-known effect of plasma viral load. During the early steps of cellular HIV-1 infection, the viral RNA is reverse transcribed into linear dsDNA and then integrates into the human cell chromosome. The resulting provirus carries out the production of progeny HIV-1 virions. A few linear dsDNA molecules undergo recombination, forming LTRC and 2LTRC (24, 54).
Considering the complex mechanism of HIV-1 reverse transcription and integration, we deemed it prudent to design a molecular-beacon-based real-time PCR assay to quantify the pool of HIV-1 DNA forms that had undergone both template switches. These DNA forms, termed STS DNA, are a prerequisite for integration of the viral genome. Real-time PCR assays that quantitate 2LTRC have been described previously (17, 24, 54, 58). Thus, a real-time PCR assay using a TaqMan-based detection system has been used to quantify HIV-1 DNA in human cells (4, 11). The real-time assay developed by Butler et al. (4), termed “late reverse transcript,” quantitates HIV-1 DNA forms that have undergone two template switches, whereas the assay by Desire et al. (11) detects DNA forms with one template switch. To the best of our knowledge, real-time PCR assays that quantify HIV-1 DNA with one or two template switches have not been used to estimate the prognosis of HIV-1 infection. Two previous studies of the relationship between cell-associated HIV-1 DNA load and clinical outcome by use of non-real-time PCR technologies produced contradictory results (10, 20).
To establish whether the cellular HIV-1 DNA load influences the rate of disease progression, and to monitor the progression of DNA throughout the natural history of HIV-1 infection, we measured HIV-1 STS and 2LTRC DNA levels in PBMC samples isolated from patients in the Greek MHCS. To do this, we used a real-time PCR technology with a molecular-beacon detection system (57). Similar assays based on the same technologies were previously developed to investigate other markers of progression of HIV-1 disease, such as natural polymorphisms on the CCR5 and CCR2 HIV-1 coreceptor genes (31-33) and the concentration of TREC (22). Our results demonstrate that the HIV-1 STS DNA concentration is an important independent predictor of HIV-1 disease. We showed that patients who progressed to AIDS had significantly higher levels of HIV-1 STS DNA than those who remained AIDS free. Importantly, we further showed that STS DNA levels were rather weakly correlated with plasma RNA loads, suggesting a biologically significant and independent role of cellular HIV-1 STS DNA in the pathogenesis of HIV-1 disease. The simplest interpretation of the results obtained in this study suggests that the HIV-1 cellular DNA load may be an indicator of the spread of infection, whereas the plasma RNA load is an indicator of active infection. Furthermore, the results from our longitudinal study showed that the concentration of HIV-1 STS DNA remained constant during the natural course of HIV-1 infection, as previously described (10).
In summary, we have introduced a new real-time PCR assay to measure the level of HIV-1 STS DNA forms, which are viral DNA forms that have undergone two template switches in PBMC. By using the cellular HIV-1 STS DNA assay, we established that the quantity of STS DNA has a significant and independent influence on the rate of disease progression throughout the natural course of HIV-1 infection. Furthermore, we reconfirmed previously published findings that, unlike the plasma RNA load, HIV-1 DNA levels remain constant during the natural course of HIV-1 infection. Similarly, as the term “plasma viral load” has been introduced to denote the concentration of HIV-1 RNA in plasma, we propose the term “cellular viral load” to refer to the concentration of HIV-1 STS DNA in PBMC. The observations we have made about the implications of cellular DNA load for the progression of HIV-1 disease suggest further studies to evaluate the clinical significance of cellular DNA load in patients receiving effective antiretroviral therapy, whose plasma RNA loads are undetectable by currently available assays. Although the biological explanation of the independent role of HIV-1 STS DNA in the progression of HIV-1 disease is not immediately apparent, new lines of experimental studies might emerge from such studies. In time, cellular DNA load might have important implications for therapeutic research and the clinical management of patients.
Acknowledgments
We are grateful to L. A. Day, S. Tyagi and F. R. Kramer for useful discussions and for the use of spectrophotometric facilities. We thank E. Delwart, J. P. Moore, M. Stevenson, and S. M. Wolinsky for critical comments on the manuscript. We are thankful to Zissis Moschidis for technical assistance, to V. Milona for administrative assistance, and to A. Leonditsi for secretarial help. We are indebted to the patients enrolled in the Greek component of the Multicenter Hemophilia Cohort Study.
The Elizabeth Glazer Pediatric AIDS Foundation provided grant support (51086-25-PG, awarded to L.G.K.). C.A. was funded by the Hellenic Center of Infectious Disease Control. The Greek Hemophilia Study Cohort is funded by the National Cancer Institute under contract N01-CP-33002. Additional support was provided by the Hellenic Society for the Study of AIDS and Other Sexually Transmitted Diseases and by the University of Athens Medical School, where L.G.K. is the Athanasios Mantecos Scholar.
REFERENCES
- 1.Aoki, S., R. Yarchoan, R. V. Thomas, J. M. Pluda, K. Marczyk, S. Broder, and H. Mitsuya. 1990. Quantitative analysis of HIV-1 proviral DNA in peripheral blood mononuclear cells from patients with AIDS or ARC: decrease of proviral DNA content following treatment with 2′,3′-dideoxyinosine (ddI). AIDS Res. Hum. Retrovir. 6:1331-1339. [DOI] [PubMed] [Google Scholar]
- 2.Bieniasz, P. D., K. Ariyoshi, M. A. Bourelly, S. Bloor, R. B. Foxall, E. C. Harwood, and J. N. Weber. 1993. Variable relationship between proviral DNA load and infectious virus titre in the peripheral blood mononuclear cells of HIV-1-infected individuals. AIDS 7:803-806. [DOI] [PubMed] [Google Scholar]
- 3.Bruisten, S. M., P. Reiss, A. E. Loeliger, P. van Swieten, R. Schuurman, C. A. Boucher, G. J. Weverling, and J. G. Huisman. 1998. Cellular proviral HIV type 1 DNA load persists after long-term RT-inhibitor therapy in HIV type 1 infected persons. AIDS Res. Hum. Retrovir. 14:1053-1058. [DOI] [PubMed] [Google Scholar]
- 4.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 5.Christopherson, C., Y. Kidane, B. Conway, J. Krowka, H. Sheppard, and S. Kwok. 2000. PCR-based assay to quantify human immunodeficiency virus type 1 DNA in peripheral blood mononuclear cells. J. Clin. Microbiol. 38:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183-188. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., D. Engel, M. M. Berrey, T. Shea, L. Corey, and A. S. Fauci. 1998. Early establishment of a pool of latently infected, resting CD4+ T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 95:8869-8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chun, T. W., and A. S. Fauci. 1999. Latent reservoirs of HIV: obstacles to the eradication of virus. Proc. Natl. Acad. Sci. USA 96:10958-10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cone, R. W., P. Gowland, M. Opravil, P. Grob, B. Ledergerber, and the Swiss HIV Cohort Study. 1998. Levels of HIV-infected peripheral blood cells remain stable throughout the natural history of HIV-1 infection. AIDS 12:2253-2260. [DOI] [PubMed] [Google Scholar]
- 11.Desire, N., A. Dehee, V. Schneider, C. Jacomet, C. Goujon, P. M. Girard, W. Rozenbaum, and J. C. Nicolas. 2001. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J. Clin. Microbiol. 39:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dewar, R. L., H. C. Highbarger, M. D. Sarmiento, J. A. Todd, M. B. Vasudevachari, R. T. Davey, Jr., J. A. Kovacs, N. P. Salzman, H. C. Lane, and M. S. Urdea. 1994. Application of branched DNA signal amplification to monitor human immunodeficiency virus type 1 burden in human plasma. J. Infect. Dis. 170:1172-1179. [DOI] [PubMed] [Google Scholar]
- 13.Eyster, M. E., M. H. Gail, J. O. Ballard, H. Al-Mondhiry, and J. J. Goedert. 1987. Natural history of human immunodeficiency virus infections in hemophiliacs: effects of T-cell subsets, platelet counts, and age. Ann. Intern. Med. 107:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 15.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 16.Furtado, M. R., R. Murphy, and S. M. Wolinsky. 1993. Quantification of human immunodeficiency virus type 1 tat mRNA as a marker for assessing the efficacy of antiretroviral therapy. J. Infect. Dis. 167:213-216. [DOI] [PubMed] [Google Scholar]
- 17.Goedert, J. J., T. R. O'Brien, A. Hatzakis, and L. G. Kostrikis for the Multicenter Hemophilia Cohort Study. 2001. T cell receptor excision circles and HIV-1 2-LTR episomal DNA to predict AIDS in patients not receiving effective therapy. AIDS 15:2245-2250. [DOI] [PubMed] [Google Scholar]
- 18.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. Valentine, L. Jonas, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734-739. [DOI] [PubMed] [Google Scholar]
- 19.Gupta, P., M. Ding, M. Cottrill, C. Rinaldo, L. Kingsley, S. Wolinsky, and J. Mellors. 1995. Quantitation of human immunodeficiency virus type 1 DNA and RNA by a novel internally controlled PCR assay. J. Clin. Microbiol. 33:1670-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, P., L. Kingsley, J. Armstrong, M. Ding, M. Cottrill, and C. Rinaldo. 1993. Enhanced expression of human immunodeficiency virus type 1 correlates with development of AIDS. Virology 196:586-595. [DOI] [PubMed] [Google Scholar]
- 21.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, J. J. Eron, Jr., J. E. Feinberg, H. H. Balfour, Jr., L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl for the AIDS Clinical Trials Group 320 Study Team. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. N. Engl. J. Med. 337:725-733. [DOI] [PubMed] [Google Scholar]
- 22.Hatzakis, A., G. Touloumi, R. Karanicolas, A. Karafoulidou, T. Mandalaki, C. Anastassopoulou, L. Zhang, J. J. Goedert, D. D. Ho, and L. G. Kostrikis. 2000. Effect of recent thymic emigrants on progression of HIV-1 disease. Lancet 355:599-604. [DOI] [PubMed] [Google Scholar]
- 23.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 24.Hu, W. S., and H. M. Temin. 1990. Retroviral recombination and reverse transcription. Science 250:1227-1233. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, M. D., V. A. Johnson, M. S. Hirsch, J. W. Bremer, T. Elbeik, A. Erice, D. R. Kuritzkes, W. A. Scott, S. A. Spector, N. Basgoz, M. A. Fischl, and R. T. D'Aquila for the AIDS Clinical Trials Group 24 Protocol Virology Substudy Team. 1997. Monitoring plasma HIV-1 RNA levels in addition to CD4+ lymphocyte count improves assessment of antiretroviral therapeutic response. Ann. Intern. Med. 126:929-938. [DOI] [PubMed] [Google Scholar]
- 26.Ibanez, A., T. Puig, J. Elias, B. Clotet, L. Ruiz, and M. A. Martinez. 1999. Quantification of integrated and total HIV-1 DNA after long-term highly active antiretroviral therapy in HIV-1-infected patients. AIDS 13:1045-1049. [DOI] [PubMed] [Google Scholar]
- 27.Ioannidis, J. P., P. S. Rosenberg, J. J. Goedert, L. J. Ashton, T. L. Benfield, S. P. Buchbinder, R. A. Coutinho, J. Eugen-Olsen, T. Gallart, T. L. Katzenstein, L. G. Kostrikis, H. Kuipers, L. G. Louie, S. A. Mallal, J. B. Margolick, O. P. Martinez, L. Meyer, N. L. Michael, E. Operskalski, G. Pantaleo, G. P. Rizzardi, H. Schuitemaker, H. W. Sheppard, G. J. Stewart, I. D. Theodorou, H. Ullum, E. Vicenzi, D. Vlahov, D. Wilkinson, C. Workman, J. F. Zagury, and T. R. O'Brien for the International Meta-Analysis. 2001. Effects of CCR5-Δ32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann. Intern. Med. 135:782-795. [DOI] [PubMed] [Google Scholar]
- 28.Izopet, J., C. Tamalet, C. Pasquier, K. Sandres, B. Marchou, P. Massip, and J. Puel. 1998. Quantification of HIV-1 proviral DNA by a standardized colorimetric PCR-based assay. J. Med. Virol. 54:54-59. [DOI] [PubMed] [Google Scholar]
- 29.Katzenstein, D. A., S. M. Hammer, M. D. Hughes, H. Gundacker, J. B. Jackson, S. Fiscus, S. Rasheed, T. Elbeik, R. Reichman, A. Japour, T. C. Merigan, and M. S. Hirsch for the AIDS Clinical Trials Group Study 175 Virology Study Team. 1996. The relation of virologic and immunologic markers to clinical outcomes after nucleoside therapy in HIV-infected adults with 200 to 500 CD4 cells per cubic millimeter. N. Engl. J. Med. 335:1091-1098. [DOI] [PubMed] [Google Scholar]
- 30.Kempf, D. J., R. A. Rode, Y. Xu, E. Sun, M. E. Heath-Chiozzi, J. Valdes, A. J. Japour, S. Danner, C. Boucher, A. Molla, and J. M. Leonard. 1998. The duration of viral suppression during protease inhibitor therapy for HIV-1 infection is predicted by plasma HIV-1 RNA at the nadir. AIDS 12:F9-F14. [DOI] [PubMed] [Google Scholar]
- 31.Kostrikis, L. G., Y. Huang, J. P. Moore, S. M. Wolinsky, L. Zhang, Y. Guo, L. Deutsch, J. Phair, A. U. Neumann, and D. D. Ho. 1998. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 4:350-353. [DOI] [PubMed] [Google Scholar]
- 32.Kostrikis, L. G., A. U. Neumann, B. Thomson, B. T. Korber, P. McHardy, R. Karanicolas, L. Deutsch, Y. Huang, J. F. Lew, K. McIntosh, H. Pollack, W. Borkowsky, H. M. Spiegel, P. Palumbo, J. Oleske, A. Bardeguez, K. Luzuriaga, J. Sullivan, S. M. Wolinsky, R. A. Koup, D. D. Ho, and J. P. Moore. 1999. A polymorphism in the regulatory region of the CC-chemokine receptor 5 gene influences perinatal transmission of human immunodeficiency virus type 1 to African-American infants. J. Virol. 73:10264-10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kostrikis, L. G., S. Tyagi, M. M. Mhlanga, D. D. Ho, and F. R. Kramer. 1998. Spectral genotyping of human alleles. Science 279:1228-1229. [DOI] [PubMed] [Google Scholar]
- 34.Levy, J. A. 1993. Pathogenesis of human immunodeficiency virus infection. Microbiol. Rev. 57:183-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin, H. J., M. Haywood, and F. B. Hollinger. 1996. Application of a commercial kit for detection of PCR products to quantification of human immunodeficiency virus type 1 RNA and proviral DNA. J. Clin. Microbiol. 34:329-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mallet, F., C. Hebrard, J. M. Livrozet, O. Lees, F. Tron, J. L. Touraine, and B. Mandrand. 1995. Quantitation of human immunodeficiency virus type 1 DNA by two PCR procedures coupled with enzyme-linked oligosorbent assay. J. Clin. Microbiol. 33:3201-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellors, J. W., L. A. Kingsley, C. R. Rinaldo, Jr., J. A. Todd, B. S. Hoo, R. P. Kokka, and P. Gupta. 1995. Quantitation of HIV-1 RNA in plasma predicts outcome after seroconversion. Ann. Intern. Med. 122:573-579. [DOI] [PubMed] [Google Scholar]
- 38.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 39.Menzo, S., P. Bagnarelli, M. Giacca, A. Manzin, P. E. Varaldo, and M. Clementi. 1992. Absolute quantitation of viremia in human immunodeficiency virus infection by competitive reverse transcription and polymerase chain reaction. J. Clin. Microbiol. 30:1752-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Montoya, J. G., R. Wood, D. Katzenstein, M. Holodny, and T. C. Merigan. 1993. Peripheral blood mononuclear cell human immunodeficiency virus type 1 proviral DNA quantification by polymerase chain reaction: relationship to immunodeficiency and drug effect. J. Clin. Microbiol. 31:2692-2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mulder, J., N. McKinney, C. Christopherson, J. Sninsky, L. Greenfield, and S. Kwok. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ngo-Giang-Huong, N., C. Deveau, I. Da Silva, I. Pellegrin, A. Venet, M. Harzic, M. Sinet, J. F. Delfraissy, L. Meyer, C. Goujard, and C. Rouzioux for the French PRIMO Cohort Study Group. 2001. Proviral HIV-1 DNA in subjects followed since primary HIV-1 infection who suppress plasma viral load after one year of highly active antiretroviral therapy. AIDS 15:665-673. [DOI] [PubMed] [Google Scholar]
- 43.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 44.O'Brien, S. J., G. W. Nelson, C. A. Winkler, and M. W. Smith. 2000. Polygenic and multifactorial disease gene association in man: lessons from AIDS. Annu. Rev. Genet. 34:563-591. [DOI] [PubMed] [Google Scholar]
- 45.O'Brien, T. R., W. A. Blattner, D. Waters, E. Eyster, M. W. Hilgartner, A. R. Cohen, N. Luban, A. Hatzakis, L. M. Aledort, P. S. Rosenberg, W. J. Miley, B. L. Kroner, and J. J. Goedert. 1996. Serum HIV-1 RNA levels and time to development of AIDS in the Multicenter Hemophilia Cohort Study. JAMA 276:105-110. [PubMed] [Google Scholar]
- 46.O'Brien, W. A., P. M. Hartigan, E. S. Daar, M. S. Simberkoff, J. D. Hamilton, and the Veterans Affairs Cooperative Study Group on AIDS. 1997. Changes in plasma HIV RNA levels and CD4+ lymphocyte counts predict both response to antiretroviral therapy and therapeutic failure. Ann. Intern. Med. 126:939-945. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien, W. A., P. M. Hartigan, D. Martin, J. Esinhart, A. Hill, S. Benoit, M. Rubin, M. S. Simberkoff, J. D. Hamilton, and the Veterans Affairs Cooperative Study Group on AIDS. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. N. Engl. J. Med. 334:426-431. [DOI] [PubMed] [Google Scholar]
- 48.Pantaleo, G., C. Graziosi, and A. S. Fauci. 1993. New concepts in the immunopathogenesis of human immunodeficiency virus infection. N. Engl. J. Med. 328:327-335. [DOI] [PubMed] [Google Scholar]
- 49.Polis, M. A., I. A. Sidorov, C. Yoder, S. Jankelevich, J. Metcalf, B. U. Mueller, M. A. Dimitrov, P. Pizzo, R. Yarchoan, and D. S. Dimitrov. 2001. Correlation between reduction in plasma HIV-1 RNA concentration 1 week after start of antiretroviral treatment and longer-term efficacy. Lancet 358:1760-1765. [DOI] [PubMed] [Google Scholar]
- 50.Pomerantz, R. J. 1999. Residual HIV-1 disease in the era of highly active antiretroviral therapy. N. Engl. J. Med. 340:1672-1674. [DOI] [PubMed] [Google Scholar]
- 51.Powderly, W. G., M. S. Saag, S. Chapman, G. Yu, B. Quart, and N. J. Clendeninn. 1999. Predictors of optimal virological response to potent antiretroviral therapy. AIDS 13:1873-1880. [DOI] [PubMed] [Google Scholar]
- 52.Saksela, K., C. E. Stevens, P. Rubinstein, P. E. Taylor, and D. Baltimore. 1995. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS. Ann. Intern. Med. 123:641-648. [DOI] [PubMed] [Google Scholar]
- 53.Sei, S., D. E. Kleiner, J. B. Kopp, R. Chandra, P. E. Klotman, R. Yarchoan, P. A. Pizzo, and H. Mitsuya. 1994. Quantitative analysis of viral burden in tissues from adults and children with symptomatic human immunodeficiency virus type 1 infection assessed by polymerase chain reaction. J. Infect. Dis. 170:325-333. [DOI] [PubMed] [Google Scholar]
- 54.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tamalet, C., A. Lafeuillade, J. Fantini, C. Poggi, and N. Yahi. 1997. Quantification of HIV-1 viral load in lymphoid and blood cells: assessment during four-drug combination therapy. AIDS 11:895-901. [DOI] [PubMed] [Google Scholar]
- 56.Touloumi, G., A. Karafoulidou, A. Gialeraki, O. Katsarou, I. Milona, V. Kapsimali, T. Mandalaki, and A. Hatzakis. 1998. Determinants of progression of HIV infection in a Greek hemophilia cohort followed for up to 16 years after seroconversion. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:89-97. [DOI] [PubMed] [Google Scholar]
- 57.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 58.Valentin, A., H. Trivedi, W. Lu, L. G. Kostrikis, and G. N. Pavlakis. 2000. CXCR4 mediates entry and productive infection of syncytia-inducing (X4) HIV-1 strains in primary macrophages. Virology 269:294-304. [DOI] [PubMed] [Google Scholar]
- 59.Vandegraaff, N., R. Kumar, C. J. Burrell, and P. Li. 2001. Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA. J. Virol. 75:11253-11260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vandegraaff, N., R. Kumar, H. Hocking, T. R. Burke, Jr., J. Mills, D. Rhodes, C. J. Burrell, and P. Li. 2001. Specific inhibition of human immunodeficiency virus type 1 (HIV-1) integration in cell culture: putative inhibitors of HIV-1 integrase. Antimicrob. Agents Chemother. 45:2510-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Gemen, B., R. van Beuningen, A. Nabbe, D. van Strijp, S. Jurriaans, P. Lens, and T. Kievits. 1994. A one-tube quantitative HIV-1 RNA NASBA nucleic acid amplification assay using electrochemiluminescent (ECL) labelled probes. J. Virol. Methods 49:157-167. [DOI] [PubMed] [Google Scholar]
- 62.Vesanen, M., C. E. Stevens, P. E. Taylor, P. Rubinstein, and K. Saksela. 1996. Stability in controlling viral replication identifies long-term nonprogressors as a distinct subgroup among human immunodeficiency virus type 1-infected persons. J. Virol. 70:9035-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wong, J. K., H. F. Gunthard, D. V. Havlir, Z. Q. Zhang, A. T. Haase, C. C. Ignacio, S. Kwok, E. Emini, and D. D. Richman. 1997. Reduction of HIV-1 in blood and lymph nodes following potent antiretroviral therapy and the virologic correlates of treatment failure. Proc. Natl. Acad. Sci. USA 94:12574-12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 65.Yerly, S., L. Kaiser, C. Baumberger, B. Hirschel, and L. H. Perrin. 1995. Early and prolonged decrease of viremia in HIV-1-infected patients treated with didanosine. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:358-364. [PubMed] [Google Scholar]
- 66.Zhang, H., G. Dornadula, M. Beumont, L. Livornese, Jr., B. Van Uitert, K. Henning, and R. J. Pomerantz. 1998. Human immunodeficiency virus type 1 in the semen of men receiving highly active antiretroviral therapy. N. Engl. J. Med. 339:1803-1809. [DOI] [PubMed] [Google Scholar]