Abstract
Bacteriophage φ6 has a segmented double-stranded RNA genome. The genomic single-stranded RNA (ssRNA) precursors are packaged into a preformed protein capsid, the polymerase complex, composed of viral proteins P1, P2, P4, and P7. Packaging of the genomic precursors is an energy-dependent process requiring nucleoside triphosphates. Protein P4, a nonspecific nucleoside triphosphatase, has previously been suggested to be the prime candidate for the viral packaging engine, based on its location at the vertices of the viral capsid and its biochemical characteristics. In this study we were able to obtain stable polymerase complex particles that are completely devoid of P4. Such particles were not able to package ssRNA segments and did not display RNA polymerase (either minus- or plus-strand synthesis) activity. Surprisingly, a mutation in P4, S250Q, which reduced the level of P4 in the particles to about 10% of the wild-type level, did not affect RNA packaging activity or change the kinetics of packaging. Moreover, such particles displayed minus-strand synthesis activity. However, no plus-strand synthesis was observed, suggesting that P4 has a role in the plus-strand synthesis reaction also.
The assembly of functional virus particles requires specific recognition of the viral genome and its subsequent packaging into the capsid. Two different packaging principles have been described. The viral genome can be encapsidated by concomitant assembly of the capsid and condensation of the genome. A classic example of this mechanism is the packaging of the single-stranded RNA (ssRNA) genome of tobacco mosaic virus into a helical capsid (2). The other alternative is packaging of the viral genome into a preformed capsid. Double-stranded DNA (dsDNA) bacteriophages provide the best-studied examples of this packaging mechanism. In this system the dsDNA genome is translocated into the capsid through a special portal vertex (for a review, see reference 34). Since the packaging of the dsDNA genome into a capsid is energetically unfavorable, this process is driven by a virus-specific ATPase (reviewed in reference 1).
Packaging of the three genomic segments of the dsRNA bacteriophage φ6 follows the mechanism of the dsDNA bacteriophages: the viral genome is packaged into a preformed capsid in an energy-dependent manner. The plus-sense ssRNA genomic precursors corresponding to the three genomic dsRNA segments are encapsidated in a reaction requiring any of the ribo-, deoxyribo-, or dideoxyribonucleoside triphosphates (rNTPs, dNTPs, or ddNTPs, respectively) (6). The genome of φ6 is enclosed in an icosahedral particle composed of four protein species, P1, P2, P4, and P7, which constitute the inner viral particle (core) but also provide all the enzymatic activities required for the viral RNA metabolism. Protein P1 forms the structural framework (17, 23), P2 is the RNA-dependent RNA polymerase subunit (19), P4 is a nonspecific NTPase (11, 27), and P7 is a packaging factor (14, 15). In mature virus particles two additional layers surround the polymerase complex particles: a shell composed of protein P8 surrounds the core, and a lipid-protein envelope forms the outermost layer of the virion.
The genomic RNA segments of φ6 are designated S, M, and L according to their sizes (2,948, 4,063, and 6,374 bp, respectively) (9, 20, 21). The proteins of the polymerase complex are encoded by the l segment (21). Expression of these proteins from a cDNA copy of the l segment in Escherichia coli leads to assembly of empty polymerase complex particles, designated procapsids (10). Such particles are capable of packaging the plus strands, synthesizing the minus strands inside the particle, and actively producing plus strands under in vitro conditions (12).
Protein P4 is a nonspecific NTPase hydrolyzing rNTPs, dNTPs, and ddNTPs. Thus, the substrate specificity of P4 resembles the NTP requirements of the RNA packaging reaction (27). Purified protein P4 forms homohexamers in the presence of divalent cations and ATP or ADP (16). The enzymatic activity is associated only with the multimeric form of the protein. It is enhanced by calcium and zinc ions as well as ssRNA but is down-regulated by magnesium ions (16, 27). Protein P4 can be assembled onto preformed incomplete polymerase complex particles, which lack this protein. The assembly is dependent on the 13 C-terminal amino acids of P4. However, these amino acids are not involved in formation of the multimers and are not required for the enzymatic activity of the protein (26). P4 hexamers are located at the fivefold symmetry positions on the polymerase complex. There is a symmetry mismatch between the location (fivefold) and the multimericity (hexamer) of protein P4 (3). Besides the polymerase, the P4 NTPase is the only NTPase activity detected in the polymerase particle. In this study we used P4 null mutant particles to show that RNA packaging is dependent on the presence of P4 on the procapsid. We also analyzed particles that contain significantly reduced amounts of P4. These analyses indicate that P4 also has a role in plus-strand synthesis and affects the packaging order of the genomic segments.
MATERIALS AND METHODS
Plasmids and bacterial strains.
The plasmids and host strains used in this study are listed in Table 1. Plasmid pLM687 was used to produce complete polymerase complex particles (procapsids). Particles lacking the P4 NTPase were produced by using plasmid pAP6, which encodes proteins P1, P2, and P7 (26). The mutant particles (S250Q particles) produced by using plasmid pLM1224 contain about 10% of the normal amount of P4 (see Fig. 1). The P4 in these particles has serine 250 changed to glutamine (26).
TABLE 1.
Plasmids used in this study
| Plasmid | Host | Particle produced | Relevant propertiesa | Reference |
|---|---|---|---|---|
| pLM656 | JM109 | Contains an exact copy of the m segment; plus-strand production with T7 RNA polymerase | 25 | |
| pLM659 | JM109 | Contains an exact copy of the s segment; plus-strand production with T7 RNA polymerase | 7 | |
| pLM682 | JM109 | Contains a copy of the l segment, whose second nucleotide has been changed from U to G; plus-strand production with T7 RNA polymerase | 7 | |
| pLM687 | JM109 | Procapsid | Contains an exact copy of the l segment; plus-strand production with T7 RNA polymerase; produces complete polymerase complex particles (proteins 1, 2, 4, and 7) | 22 |
| pLM1224 | JM109 | S250Q | Produces P1, P2, P4, and P7 particles with reduced P4 amounts (proteins 1, 2, 4*, and 7) | 26 |
| pLM1773 | JM109 | Contains a copy of the s segment with a deletion in the packaging signal; plus-strand production with T7 RNA polymerase | 32 | |
| pAP6 | HMS174(DE3) | P4 null | Produces P1, P2, and P7 particles (proteins 1, 2, and 7) | 26 |
∗, S250Q mutation in P4.
FIG. 1.
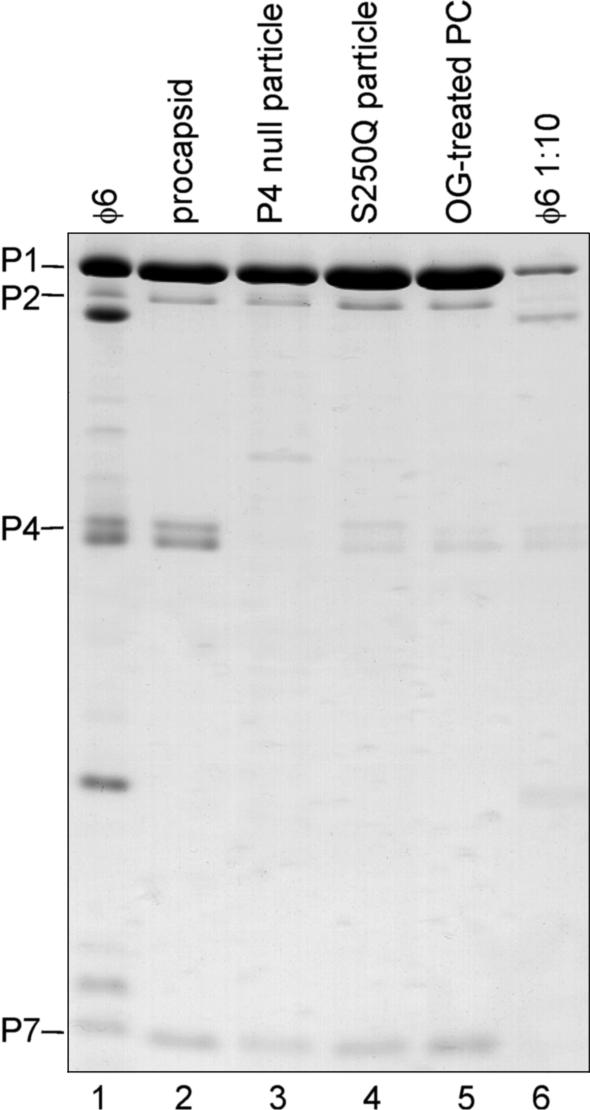
SDS-PAGE gel stained with Coomassie blue showing the protein composition of purified procapsid, P4-null, S250Q, and OG-treated procapsid particles. The φ6 polymerase complex proteins are indicated on the left. Rightmost lane shows 1:10-diluted φ6 virus to demonstrate the amount of P4 in the S250Q and detergent-treated particle preparations.
Preparation of particles.
Complete and incomplete procapsid particles were produced and purified basically as previously described (6, 14). E. coli JM109 or HMS174 cells containing the appropriate expression plasmid(s) were grown overnight in Luria-Bertani medium at 28 or 37°C, diluted in 200 to 300 ml of the same medium to obtain a cell density of 1 × 108/ml, and further incubated under aeration until the cell density reached 4 × 108/ml. Subsequently, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 or 2 mM, and incubation was continued overnight at 18°C. Cells were collected by centrifugation, washed once with 20 mM Tris (pH 8.0)-150 mM NaCl, resuspended in approximately 2 to 3 ml of the same buffer, and either stored at −80°C or used immediately for particle purification. For particle purification, cells were lysed by being passed twice through a cold (4°C) French pressure cell at 20,000 lb/in2. The lysate was extracted with 5% Triton X-114 (6), and the particles in the resulting aqueous phase were sedimented through a sucrose gradient (Sorvall TH641 rotor) (1 h, 50 min, at 10°C and 27,000 rpm or 1 h at 15°C and 35,000 rpm; 5 to 20% sucrose, 20 mM Tris [pH 8.0], 150 mM NaCl). The light-scattering particle zone was collected. The protein composition of the particle preparation was analyzed by sodium dodecyl sulfate-16% polyacrylamide gel electrophoresis (SDS-16% PAGE) (24). Detergent-treated particles were obtained by adding n-octyl-β-d-glucopyranoside (OG) (Sigma) to a final concentration of 1.5% to a fresh procapsid preparation, followed by sedimentation through a sucrose gradient (Sorvall TH641 rotor) (1 h at 15°C and 35,000 rpm; 10 to 25% sucrose, 20 mM Tris [pH 8.0], 150 mM NaCl). The light-scattering particle zone was collected.
In vitro synthesis of synthetic plus-sense ssRNAs.
Unlabeled and 32P-labeled ssRNA segments were produced in vitro with T7 RNA polymerase by using plasmids pLM659, pLM656, pLM682, pLM687, and pLM1773 (see Table 1) as templates and were purified as described previously (6).
RNA packaging reactions.
Unless otherwise indicated, previously published φ6 RNA packaging reaction conditions were used in all packaging experiments, namely, 50 mM Tris (pH 8.9), 80 mM ammonium acetate (NH4Ac), 5 mM MgCl2, 2 mM dithiothreitol (DTT), 0.1 mM EDTA, 6% polyethylene glycol 4000, 1 mM ATP, 1 U of rRNasin (Promega)/μl, 0.5 to 1.1 μg of each 32P-labeled ssRNA segment in equimolar amounts (6), and freshly prepared procapsid preparations. Packaging reaction mixtures (25 μl) were incubated at 30°C for 30 to 90 min. Unpackaged RNA molecules were subsequently hydrolyzed by addition of 10 U of RNase ONE (Promega) (30 min at 30°C). RNase treatment was stopped by addition of 3 volumes of stop buffer, giving final concentrations of 1% SDS, 10 mM EDTA, and 20 μg of yeast tRNA. Samples were extracted with phenol, precipitated with ethanol, and analyzed in ethidium bromide (EtBr)-containing 1% agarose gels (28). The gels were dried, analyzed by autoradiography, and quantitated with a phosphorimager (Fuji BAS1500).
Minus- and plus-strand synthesis.
Minus-strand synthesis reaction conditions were as previously described (35), namely, 50 mM Tris (pH 8.9), 3 mM MgCl2, 120 mM NH4Ac, 6% polyethylene glycol 4000, 2 mM DTT, 0.1 mM EDTA, 1 U of rRNasin/μl, 0.2 mM (each) ATP, GTP, CTP, and UTP, and 5 μCi of [α-32P]UTP (Amersham catalog number PB10203). Similar conditions were used for plus-strand synthesis reactions, except that the concentrations of MgCl2 and NH4Ac were 5 and 80 mM, respectively, and 1 mM (each) GTP and ATP were used to activate transcription (35). All polymerase reaction mixtures contained approximately 0.5 to 1 μg of complete or incomplete polymerase complex particles and 0.5 to 1.1 μg of each of the ssRNA segments in equimolar amounts in a 25-μl reaction volume. After 90 min of incubation at 30°C, RNA was extracted from the reaction products as previously described (8) and analyzed as for packaging reactions. Strand separation was achieved by electrophoresis as described elsewhere (28).
Sedimentation assay.
For the sedimentation assay, freshly prepared S250Q particles were concentrated by a Beckman airfuge (A-95 rotor; 15 min at 178,000 × g) to 2 mg/ml. Two 500-μl reactions were set up under modified plus-strand synthesis conditions: the NTP concentration was increased (280 μM [each] CTP and UPT, 1.4 mM [each] ATP and GTP), no radioactive label was added, and the particle concentration was increased to 3 μg per 25-μl reaction volume. For one reaction 4.5 μg of ssRNA segments per 25-μl reaction volume was added, and no RNA was added to the control reaction mixture. After 90 min of incubation at 30°C, unpackaged and unprotected ss- and dsRNAs were hydrolyzed by addition of 80 U of RNase ONE and 4 U of dsRNA-specific cobra venom nuclease V1 (Pharmacia), and incubation was continued for 30 min. Both samples were sedimented through a sucrose gradient (Sorvall TH641 rotor; 45 min at 15°C and 35,000 rpm; 5 to 20% sucrose, 20 mM Tris [pH 8.0], 150 mM NaCl, 2 mM ADP, 1 mM DTT). Samples were fractionated by use of a BioComp Gradient fractionator and were precipitated with acetone. The RNA content of each fraction was analyzed on a 1% agarose gel, and protein composition was analyzed by SDS-16% PAGE.
RESULTS
In earlier studies the purification of stable viral polymerase complex particles that either have a reduced amount of P4 NTPase or are completely devoid of this protein was described (26). Such particles provide a way to investigate the role of P4 in the RNA packaging process. Figure 1 shows the protein composition of the purified particles. P4-null particles contain no P4 (Fig. 1, lane 3), and S250Q particles contain a reduced amount of P4, about 10% of the wild-type (wt) level (compare lanes 4 and 6). Additionally, we observed that detergent treatment of the wt procapsids (with OG) yields polymerase complexes that resemble S250Q particles in that they also have only ∼10% of the P4 present (Fig. 1, lane 5).
Reduced levels of P4 NTPase support packaging.
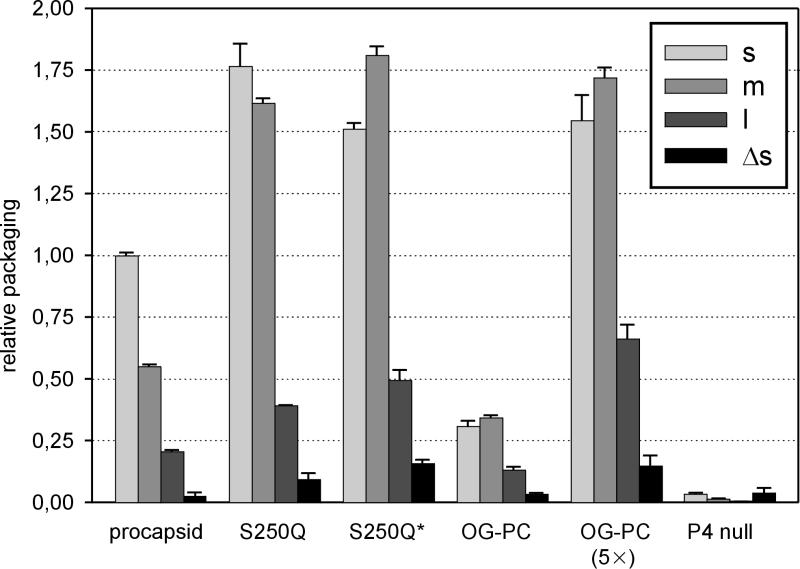
The packaging activity of each freshly prepared polymerase complex particle shown in Fig. 1 was determined by packaging each of the three ssRNA segments independently (Fig. 2). Additionally, we investigated the packaging activity of S250Q particles that had been frozen and thawed (S250Q*) (Fig. 2). Normal procapsids packaged efficiently, while the activity of P4-null particles was at the background level. S250Q particles were very efficient at packaging, displaying activity almost twice as high as that of normal procapsids. The packaging efficiency of OG-treated particles was about 50% of that of normal particles. Both procapsid and S250Q particles packaged the s segment most efficiently, followed by the m and l segments. Freezing and thawing of the S250Q particles enhanced packaging of the m segment, with a packaging efficiency order of m > s > l, as was also the case with OG-treated particles. The specificity of the packaging reaction was determined by using an s segment (Δs) in which deletion of nucleotides 11 to 43 had destroyed the packaging signal (32). Use of the Δs segment with procapsid, OG-treated, and S250Q particles resulted in only background levels of packaging activity, indicating an identical packaging specificity among these particles.
FIG. 2.
Relative molar RNA packaging efficiencies of procapsid, S250Q, OG-treated, and P4-null particles. Individual packaging of the s, m, and l segments is analyzed as indicated in the figure. Packaging of the l segment was enhanced by addition of a nonradioactive m segment to the reaction mixtures (6). The negative control, Δs, is a derivative of the s segment containing a deletion that destroys the packaging signal and abolishes packaging activity. Results were normalized first to the amount of protein P1 in each reaction and then against the level of s-segment packaging with procapsids, which was set to 1. S250Q*, frozen and thawed S250Q particles. For better comparison of the packaging efficiencies of the OG-treated particles, a 5× multiplied version is also included.
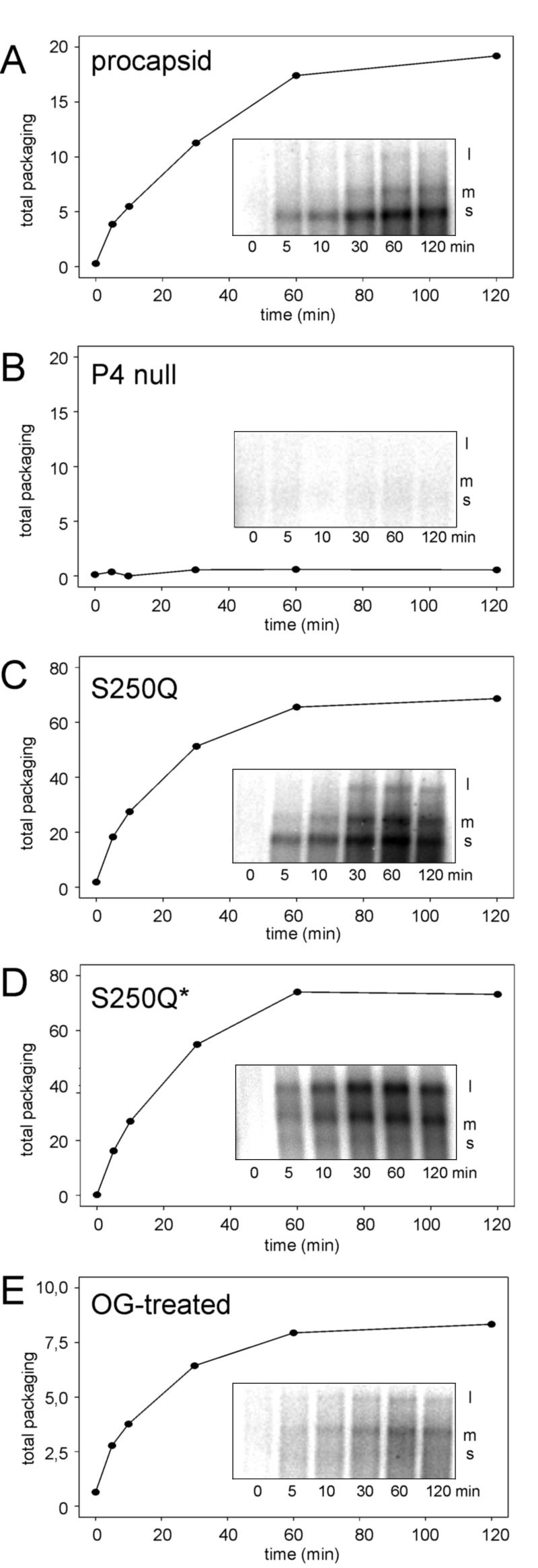
The kinetics of the packaging reaction was determined by following the time course of packaging of genomic ssRNA segments (Fig. 3). Again, P4-null particles showed no packaging activity. Qualitatively, wt and S250Q particles behaved similarly, showing a packaging order of m>s>l. However, the total amounts of RNA packaged by S250Q particles were about three times higher than those packaged by wt particles. Freezing and thawing of the S250Q particles led to reduced packaging of the s segment. This is not due to an inability of the particles to package s, as seen in Fig. 2; rather, the m segment competed with the s segment. If the reaction was started by adding only the s segment, and the other segments were added later, the s segment was packaged normally (data not shown). Again, the behavior of the OG-treated particles resembled that of frozen S250Q particles in that the m segment was favored. These analyses show that while the presence of P4 is necessary for packaging activity, its stoichiometric amount is not crucial (see below).
FIG. 3.
Time course of s, m, and l RNA packaging of procapsid (A), P4-null (B), S250Q (C), frozen and thawed S250Q (S250Q*) (D), and detergent-treated (E) particles. Insets in every panel show the appearance of individual ssRNA segments during the course of packaging. Plots show, at different time points, the total amounts of radioactivity in the RNA bands, expressed in arbitrary units. Note the different scaling in panels C, D, and E.
Particles with reduced amounts of P4 synthesize minus strands but not plus strands.
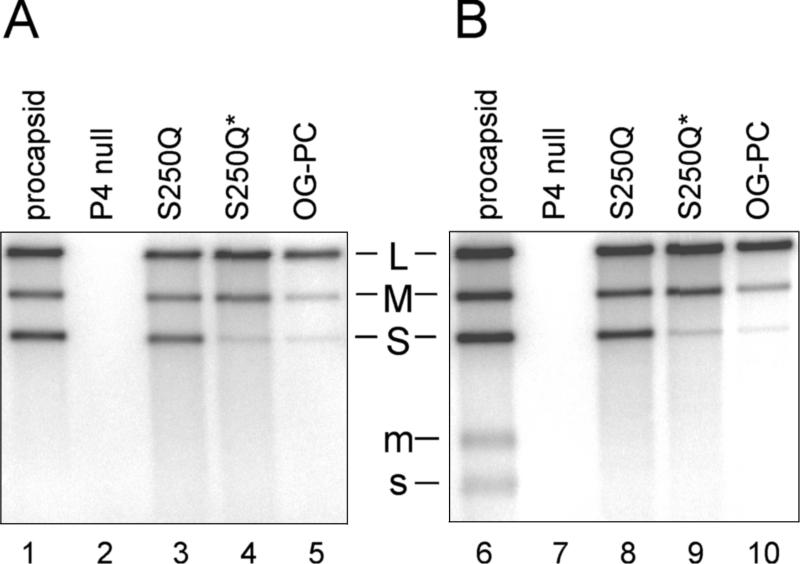
Minus- and plus-strand synthesis activities of the particles were analyzed by incubating all three segments simultaneously in a combined RNA packaging and synthesis reaction (Fig. 4). In such reactions the polymerase activity is dependent on a prior packaging of the l segment (8). P4-null particles did not synthesize minus strands at all, although they contained the RNA polymerase protein P2. Minus-strand synthesis by freshly prepared S250Q particles was similar to that by the procapsid, since the three complementary strands were synthesized effectively. Similar results were also obtained with frozen and thawed procapsids (data not shown). Frozen and thawed S250Q particles and OG-treated particles showed reduced minus-strand synthesis of the s segment. These results are in agreement with the packaging results (see Fig. 3).
FIG. 4.
Qualitative RNA polymerase activities of procapsid, P4-null, S250Q, and detergent-treated particles under minus-strand (A) and plus-strand (B) synthesis conditions (particle amount is not adjusted). An asterisk indicates that frozen and thawed particles were used. Positions of double-stranded (capital letters) and single-stranded (lowercase letters) segments are indicated. The wild-type segment L is not transcribed under the conditions used (see reference 8).
Under reaction conditions containing elevated levels of purine nucleotides, the particles switched to the plus-strand synthesis mode after finishing minus-strand synthesis (35). Labeled ssRNA segments, indicative of plus-strand synthesis, were seen with the wt procapsid (Fig. 4, lane 6). Interestingly, no labeled ssRNA molecules were detected with particles containing reduced amounts of P4 (Fig. 4, lanes 8 to 10), suggesting a deficiency in the switch to the plus-strand synthesis mode. However, because of the semiconservative nature of φ6 replication, the first round of plus-strand synthesis (where transcripts are not labeled) is not detected in conventional EtBr-containing agarose gels, which only separate dsRNA and ssRNA segments from each other. The complete lack of transcription was confirmed by using EtBr-free strand separation gels and a modified l segment (8) with enhanced transcription efficiency (data not shown).
Mutant P4 is distributed evenly among S250Q particles.
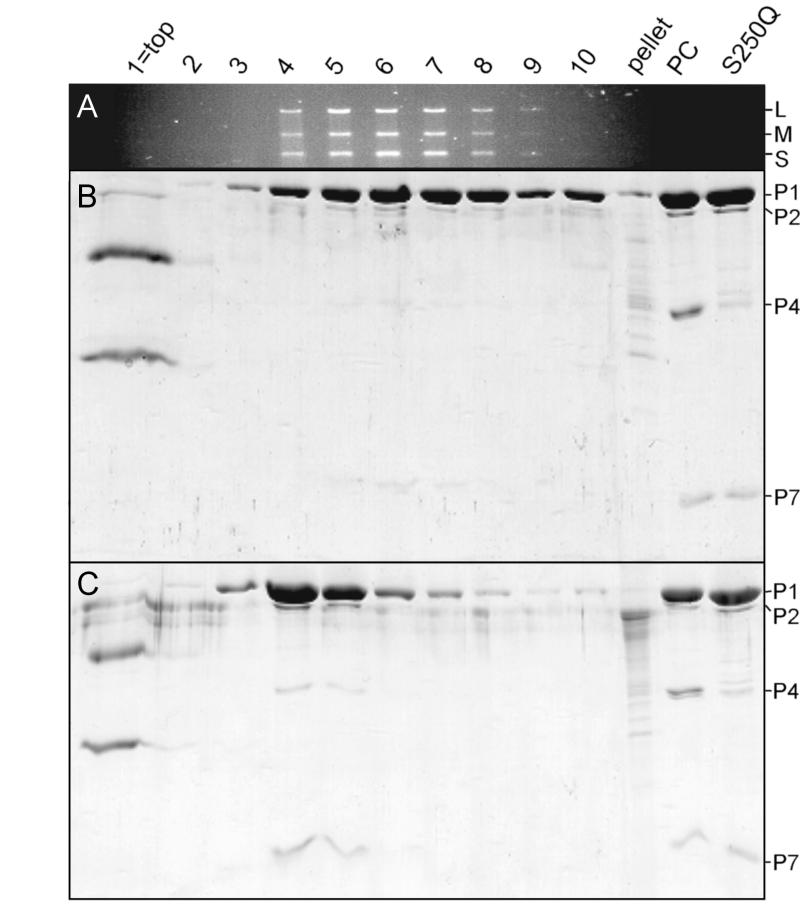
The data described above provided a surprising result, indicating that S250Q particles containing only ∼10% of P4 are virtually identical in packaging and minus-strand synthesis activities to the wt procapsid. A possible explanation could be the heterogeneity of the particle preparation, i.e., 10% of the particles contain a full complement of P4 while 90% of the particles totally lack this protein. To address this question, we incubated S250Q particles under plus-strand synthesis reaction conditions with and without RNA addition and analyzed the reactions in sucrose gradients (Fig. 5). In the control reaction without RNA addition, most of the S250Q particles were in fractions 4 and 5. In the reaction containing RNA, S250Q particles were shifted down in the gradient, to fractions that also contained dsRNA (fractions 4 to 9), showing that the majority of the particles in the reaction packaged RNA efficiently and continued with minus-strand synthesis. Careful analysis of the P4 content in the fractions revealed that there was no enrichment in the amount of P4 in any of the fractions regardless of whether dsRNA was present or not.
FIG. 5.
Sedimentation assay of S250Q particles. A combined packaging and minus-strand synthesis reaction with particles containing reduced amounts of P4 (S250Q) was analyzed by sedimentation through a linear 5-to-20% sucrose gradient. (A) One percent agarose gel showing the RNase-protected dsRNA content in the fractions (from top to bottom) and pellet. RNA segments are indicated at the right. (B) SDS-16% PAGE gel showing protein distribution in the same fractions. Procapsid proteins are indicated at the right. (C) SDS-16% PAGE gel from a similarly treated reaction but without RNA addition. Top fractions also contain RNase inhibitor and RNase added to the reactions. Lanes PC (procapsid) and S250Q, particle markers.
DISCUSSION
Protein P4 is the only NTPase detected in the φ6 procapsid. This observation and the similar requirements for packaging and NTPase activities has led to the proposal that P4 is responsible for generating the energy for translocation of the genomic ssRNA precursors into the procapsid. Success in production of stable P4-deficient particles (26) provided an opportunity to directly examine the role of P4 in the φ6 life cycle. Here we demonstrate that P4 is essential for the packaging of the genomic precursors of φ6. Particles devoid of P4 showed no detectable packaging activity. In contrast, particles containing reduced amounts (1/10) of P4 (either mutant or wt) packaged the ssRNA segments and replicated them to a double-stranded form but were not able to carry out the plus-strand synthesis reaction.
Hexameric P4 has been observed at all or most of the 12 fivefold positions on the particle surface (3). Since the mutant particle preparation seems to be homogeneous (Fig. 5), the amount of P4 observed suggests that only about 1 fivefold position per particle is occupied by a P4 hexamer (1/12 ≈ 8.3%). Quite surprisingly, such particles have a higher packaging efficiency than the procapsids (Fig. 2 and 3). Furthermore, they display normal minus-strand synthesis activity (Fig. 4). These observations have several consequences. Obviously, a single vertex is capable of selecting all three segments and translocating them in to the particle in an ordered fashion with approximately the same kinetics as the normal procapsid (Fig. 3). Earlier studies, however, have suggested that more than one vertex could function in packaging (31). Currently we do not have a model explaining both of these results.
We also observe that P4 occupies two chemically distinct positions. The majority of P4 is sensitive to removal with OG, and a minority is resistant. S250Q particles seem to contain P4 in amounts equivalent to the amount of OG-resistant P4. Our preliminary results suggest that these P4 hexamers are also resistant to OG. Procapsid assembly initiation is dependent on the presence of hexameric P4 (29). It is conceivable that the P4 hexamer in the initiation complex is associated with the capsid differently from the other 11 hexamers, explaining the two chemically different positions of P4.
The reduced amount of P4 had no influence on minus-strand synthesis but completely abolished plus-strand synthesis activity. Based on its stoichiometry (∼12 per particle) and data from other, similar virus systems, it is thought that the RNA polymerase subunit (P2) is also located at every fivefold position (3). Therefore, it is conceivable that both P4 and P2 could bind the entering ssRNA molecules, thus leading to efficient minus-strand synthesis after packaging. The subsequent plus-strand synthesis reaction requires the opening of the dsRNA molecule and the exit of the newly synthesized ssRNA molecule through an exit pore, which is most probably located at the particle vertices. We envisage that the P4 hexamers are active in both ssRNA entry and exit and that the transcripts may exit through the same portal where the synthesizing RNA polymerase is located. S250Q mutant and detergent-treated particles each have only a single P4-containing vertex, suggesting that this single vertex is not active in plus-strand synthesis.
The amount of P4 in the particles also had an effect on the packaging order of the segments. Frozen and thawed S250Q particles, as well as fresh detergent-treated procapsids, favored packaging of the m segment over that of the s segment. These particles seem to be in the “m-segment packaging mode” proposed by Qiao et al. (32). Loss of most of the P4 from the particle, together with freezing or detergent treatment, could change the conformation of the particle to favor m-segment or even l-segment packaging, as seen with OG-treated particles (see Fig. 2 to 4). It has been observed previously that treatment of in vivo-derived procapsids with Sarkosyl removed about 90% of protein P4 (5), making the particles equivalent to S250Q and OG-treated particles. P4 removal had no significant effect on minus-strand synthesis, but it also completely abolished plus-strand synthesis (5). These results are in agreement with the in vitro results obtained here by using recombinant particles.
The picture emerging from our present work and previous studies describing symmetry mismatch between the hexameric P4 and its fivefold location is reminiscent of the portal complexes of dsDNA phages that assist both the packaging and the exit of nucleic acid. The most distinct difference in the φ6 system is the occupancy of this portal at every fivefold position. However, it was shown here that only one vertex is sufficient for packaging. In the cores of Reoviridae, the enzymatically active non-shell-forming proteins have been localized at the fivefold position (13, 30, 33). The same fivefold position is also used for exit of the ssRNA transcripts (4, 13, 18). Therefore, in all these complex dsRNA viruses, the molecular engines responsible for RNA synthesis and translocation seem to occupy a similar position.
Acknowledgments
M. J. Pirttimaa and A. O. Paatero contributed equally to this work.
The technical assistance of Marja-Leena Perälä and Riitta Tarkiainen is greatly appreciated.
This work was supported by the Academy of Finland (Finnish Centre of Excellence Programme 2000-2005, grants 168694, 164298, and 172621 to D.H.B.). M.J.F. was supported by grants 50116 and 50527 from the Academy of Finland. M.J.P. is a fellow of the Viikki Graduate School in Biosciences.
REFERENCES
- 1.Black, L. W. 1989. DNA packaging in dsDNA bacteriophages. Annu. Rev. Microbiol. 43:267-292. [DOI] [PubMed] [Google Scholar]
- 2.Caspar, D. L., and K. Namba. 1990. Switching in the self-assembly of tobacco mosaic virus. Adv. Biophys. 26:157-185. [DOI] [PubMed] [Google Scholar]
- 3.de Haas, F., A. O. Paatero, L. Mindich, D. H. Bamford, and S. D. Fuller. 1999. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage φ6. J. Mol. Biol. 294:357-372. [DOI] [PubMed] [Google Scholar]
- 4.Diprose, J. M., J. N. Burroughs, G. C. Sutton, A. Goldsmith, P. Gouet, R. Malby, I. Overton, S. Zientara, P. P. Mertens, D. I. Stuart, and J. M. Grimes. 2001. Translocation portals for the substrates and products of a viral transcription complex: the bluetongue virus core. EMBO J. 20:7229-7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ewen, M. E., and H. R. Revel. 1990. RNA-protein complexes responsible for replication and transcription of the double-stranded RNA bacteriophage φ6. Virology 178:509-519. [DOI] [PubMed] [Google Scholar]
- 6.Frilander, M., and D. H. Bamford. 1995. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage φ6: the three segments modulate each other's packaging efficiency. J. Mol. Biol. 246:418-428. [DOI] [PubMed] [Google Scholar]
- 7.Frilander, M., P. Gottlieb, J. Strassman, D. H. Bamford, and L. Mindich. 1992. Dependence of minus-strand synthesis on complete genomic packaging in the double-stranded RNA bacteriophage φ6. J. Virol. 66:5013-5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frilander, M., M. Poranen, and D. H. Bamford. 1995. The large genome segment of dsRNA bacteriophage φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA 1:510-518. [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlieb, P., S. Metzger, M. Romantschuk, J. Carton, J. Strassman, D. H. Bamford, N. Kalkkinen, and L. Mindich. 1988. Nucleotide sequence of the middle dsRNA segment of bacteriophage φ6: placement of the genes of membrane-associated proteins. Virology 163:183-190. [DOI] [PubMed] [Google Scholar]
- 10.Gottlieb, P., J. Strassman, D. H. Bamford, and L. Mindich. 1988. Production of a polyhedral particle in Escherichia coli from a cDNA copy of the large genomic segment of bacteriophage φ6. J. Virol. 62:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb, P., J. Strassman, and L. Mindich. 1992. Protein P4 of the bacteriophage φ6 procapsid has a nucleoside triphosphate-binding site with associated nucleoside triphosphate phosphohydrolase activity. J. Virol. 66:6220-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottlieb, P., J. Strassman, X. Y. Qiao, A. Frucht, and L. Mindich. 1990. In vitro replication, packaging, and transcription of the segmented double-stranded RNA genome of bacteriophage φ6: studies with procapsids assembled from plasmid-encoded proteins. J. Bacteriol. 172:5774-5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimes, J. M., J. N. Burroughs, P. Gouet, J. M. Diprose, R. Malby, S. Zientara, P. P. Mertens, and D. I. Stuart. 1998. The atomic structure of the bluetongue virus core. Nature 395:470-478. [DOI] [PubMed] [Google Scholar]
- 14.Juuti, J. T., and D. H. Bamford. 1995. RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage φ6. J. Mol. Biol. 249:545-554. [DOI] [PubMed] [Google Scholar]
- 15.Juuti, J. T., and D. H. Bamford. 1997. Protein P7 of phage φ6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J. Mol. Biol. 266:891-900. [DOI] [PubMed] [Google Scholar]
- 16.Juuti, J. T., D. H. Bamford, R. Tuma, and G. J. Thomas, Jr. 1998. Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ6. J. Mol. Biol. 279:347-359. [DOI] [PubMed] [Google Scholar]
- 17.Ktistakis, N. T., and D. Lang. 1987. The dodecahedral framework of the bacteriophage φ6 nucleocapsid is composed of protein P1. J. Virol. 61:2621-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawton, J. A., C. Q. Zeng, S. K. Mukherjee, J. Cohen, M. K. Estes, and B. V. Prasad. 1997. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer. J. Virol. 71:7353-7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makeyev, E., and D. H. Bamford. 2000. Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage φ6. EMBO J. 19:124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGraw, T., L. Mindich, and B. Frangione. 1986. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage φ6: novel mechanism of natural translational control. J. Virol. 58:142-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mindich, L., I. Nemhauser, P. Gottlieb, M. Romantschuk, J. Carton, S. Frucht, J. Strassman, D. H. Bamford, and N. Kalkkinen. 1988. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: genes specifying the viral replicase and transcriptase. J. Virol. 62:1180-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mindich, L., X. Qiao, S. Onodera, P. Gottlieb, and M. Frilander. 1994. RNA structural requirements for stability and minus-strand synthesis in the dsRNA bacteriophage φ6. Virology 202:258-263. [DOI] [PubMed] [Google Scholar]
- 23.Olkkonen, V. M., and D. H. Bamford. 1987. The nucleocapsid of the lipid-containing double-stranded RNA bacteriophage φ6 contains a protein skeleton consisting of a single polypeptide species. J. Virol. 61:2362-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olkkonen, V. M., and D. H. Bamford. 1989. Quantitation of the adsorption and penetration stages of bacteriophage φ6 infection. Virology 171:229-238. [DOI] [PubMed] [Google Scholar]
- 25.Olkkonen, V. M., P. Gottlieb, J. Strassman, X. Y. Qiao, D. H. Bamford, and L. Mindich. 1990. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc. Natl. Acad. Sci. USA 87:9173-9177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paatero, A. O., L. Mindich, and D. H. Bamford. 1998. Mutational analysis of the role of nucleoside triphosphatase P4 in the assembly of the RNA polymerase complex of bacteriophage φ6. J. Virol. 72:10058-10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paatero, A. O., J. E. Syväoja, and D. H. Bamford. 1995. Double-stranded RNA bacteriophage φ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J. Virol. 69:6729-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pagratis, N., and H. R. Revel. 1990. Detection of bacteriophage φ6 minus-strand RNA and novel mRNA isoconformers synthesized in vivo and in vitro, by strand-separating agarose gels. Virology 177:273-280. [DOI] [PubMed] [Google Scholar]
- 29.Poranen, M. M., A. O. Paatero, R. Tuma, and D. H. Bamford. 2001. Self-assembly of a viral molecular machine from purified protein and RNA constituents. Mol. Cell 7:845-854. [DOI] [PubMed] [Google Scholar]
- 30.Prasad, B. V., R. Rothnagel, C. Q. Zeng, J. Jakana, J. A. Lawton, W. Chiu, and M. K. Estes. 1996. Visualization of ordered genomic RNA and localization of transcriptional complexes in rotavirus. Nature 382:471-473. [DOI] [PubMed] [Google Scholar]
- 31.Qiao, X., J. Qiao, and L. Mindich. 1995. Interference with bacteriophage φ6 genomic RNA packaging by hairpin structures. J. Virol. 69:5502-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao, X., J. Qiao, and L. Mindich. 1997. Stoichiometric packaging of the three genomic segments of double-stranded RNA bacteriophage φ6. Proc. Natl. Acad. Sci. USA 94:4074-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinisch, K. M., M. L. Nibert, and S. C. Harrison. 2000. Structure of the reovirus core at 3.6 Å resolution. Nature 404:960-967. [DOI] [PubMed] [Google Scholar]
- 34.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 35.van Dijk, A. A., M. Frilander, and D. H. Bamford. 1995. Differentiation between minus- and plus-strand synthesis: polymerase activity of dsRNA bacteriophage φ6 in an in vitro packaging and replication system. Virology 211:320-323. [DOI] [PubMed] [Google Scholar]