Abstract
Animal papillomaviruses are widely used as models to study papillomavirus infection in humans despite differences in genome organization and tissue tropism. Here, we have investigated the extent to which animal models of papillomavirus infection resemble human disease by comparing the life cycles of 10 different papillomavirus types. Three phases in the life cycles of all viruses were apparent using antibodies that distinguish between early events, the onset of viral genome amplification, and the expression of capsid proteins. The initiation of these phases follows a highly ordered pattern that appears important for the production of virus particles. The viruses examined included canine oral papillomavirus, rabbit oral papillomavirus (ROPV), cottontail rabbit papillomavirus (CRPV), bovine papillomavirus type 1, and human papillomavirus types 1, 2, 11, and 16. Each papillomavirus type showed a distinctive gene expression pattern that could be explained in part by differences in tissue tropism, transmission route, and persistence. As the timing of life cycle events affects the accessibility of viral antigens to the immune system, the ideal model system should resemble human mucosal infection if vaccine design is to be effective. Of the model systems examined here, only ROPV had a tissue tropism and a life cycle organization that resembled those of the human mucosal types. ROPV appears most appropriate for studies of the life cycles of mucosal papillomavirus types and for the development of prophylactic vaccines. The persistence of abortive infections caused by CRPV offers advantages for the development of therapeutic vaccines.
Human papillomaviruses (HPVs) are associated with epithelial lesions that can progress to high-grade neoplasia and cancer (22, 103). Cancer of the cervix is caused by high-risk papillomavirus types, such as HPV type 16 (HPV16), and is the second-most-common female cancer worldwide. Low-risk human papillomavirus types, such as HPV11, cause genital warts (88). Genital warts afflict ∼1 in 200 young adults and in many countries are the most common sexually transmitted disease (51). They are difficult to treat and often recur.
The prevalence of HPV infections, and the severity to which some lesions can progress, has led to the development of model systems in which to study virus infection. Papillomaviruses infect epithelial tissue, and their life cycle is regulated as the infected epithelial cell differentiates. This leads to vegetative viral genome amplification in the intermediate epithelial layers and the assembly of infectious virions closer to the epithelial surface. Model systems enable the roles of viral proteins to be examined during productive infection and allow the development of strategies for the prevention and treatment of HPV-associated disease.
Two types of model system are currently in use. The first uses organotypic raft culture or xenotransplantation to support the growth of differentiating human keratinocytes and productive infection (50, 77). Such approaches approximate in vivo infection but are technically demanding and depend on the availability of human tissue. The second type of model makes use of animal papillomaviruses that cause lesions similar to those found in humans (11, 90). This approach is suited to both therapeutic- and prophylactic-vaccine development, as the course of infection can be monitored in an immunocompetent host. Cottontail rabbit papillomavirus (CRPV) has been used extensively for this purpose and has also provided insights into the roles of viral gene products during productive and latent infection and during the progression to cancer (12-14, 33, 69, 104, 107). More recently, canine oral papillomavirus (COPV) has been used for prophylactic-vaccination studies despite the cost of housing large animals and the inability of COPV to infect genital tissue (91, 105).
Although animal models have been used to study infection in humans, there has been no molecular assessment of the extent to which these model systems resemble the HPV infections they are supposed to mimic. This is particularly relevant, as recent work has suggested that rabbit oral papillomavirus (ROPV) may be a more appropriate model of productive human genital infections than COPV or CRPV (28, 29). ROPV infects oral and genital mucosa and does not contain the 1.5-kb noncoding region 2 that is unique to COPV (29, 34). COPV and CRPV do not cause lesions at genital sites (34, 97).
In this study, we have used three markers of the virus life cycle to compare lesions caused by the main animal papillomaviruses used to study infection in humans. In the lower layers of infected tissue, expression of E6 and E7 (and maybe also E5) leads to the up-regulation of E2F-activated genes, such as those for PCNA and cyclin A. Such proteins are surrogate markers of viral gene expression, and their presence identifies cells supporting early events during the virus life cycle (60). E4 expression follows activation of the differentiation-dependent promoter (26, 71). Its appearance has been reported to coincide with the onset of vegetative viral genome amplification and marks the initiation of late events (42). Only upon further differentiation is the major capsid protein expressed, allowing the assembly of infectious virions and their release from the epithelial surface. By mapping life cycle events in lesions caused by animal and human papillomaviruses, we have revealed an unexpected degree of heterogeneity among the viruses. Late events begin in the basal layer in lesions caused by COPV but can be retarded until the superficial layers in lesions caused by HPV16. Of the animal models examined, the productive life cycle of ROPV appears to resemble those of HPV16 and HPV11 most closely.
MATERIALS AND METHODS
Production of expression constructs.
E1∧E4 sequences were predicted from alignment data (ROPV, COPV, HPV63, and HPV65) (44, 108) or from the results of RNA-mapping experiments (HPV11, CRPV, and bovine papillomavirus type 1 [BPV1]) (3, 26, 45, 102). DNA comprising the full-length spliced E1∧E4 gene was cloned between the SalI and EcoRI sites of pGex4T-1 (Amersham, Little Chalfont, United Kingdom) or pMalc (New England Biolabs) using standard methods. The primers used to prepare the E1∧E4 genes of each virus type are listed below. For BPV1 E1∧E4, the forward primer was BPVE14P1 (AGG ATC CGA ATT CGC AAA CGA TAA AGA GAT CGC CCA GAC GGA) and the reverse primer was BPVE14P2 (GCA ACC CGG GTC GAC CTA TCA CTG GTT CTT CCT CTG TG). For CRPV, the forward primer was CRE4P1 (AGG ATC CGA ATT CGC TGA AGC TCC CCC CAG CCG CTG GTC A) and the reverse primer was CRE4P2 (ACC CGG GTC GAC CTA TTA TAA GCT CGC GAA GCC GTC TAT). For COPV, the forward primer was COE14P1 (AGG ATC CGA ATT CGC GGC TAG AAA AGT GCC GCC GGA ACC TCC G) and the reverse primer was COE14P2 (GCA ACC CGG GTC GAC CTA CTA AAA CAA CAA CTG GGG GAT CGT). For HPV11, the forward primer was H11E14P1 (AGG ATC CGA ATT CGC GGA CGA TTC AGC ACT GTA CGA GAA GTA TCC) and the reverse primer was H11E14P2 (GCA ACC CGG GTC GAC CTA CTA TAG GCG TAG CTG CAC TGT GAC). For HPV63, the forward primer was H63E14P1 (AGG ATC CGA ATT CAC CGA CAG AGCT CCC CAC TAC GGA CTT CTG GG) and the reverse primer was H63E14P2 (GCA ACC CGG GTC GAC CTA TTA TGG GTG GAT CCC GAG CCT). For HPV65, the forward primer was H65E14P1 (AGG ATC CGA ATT CGC AGA TAA AGC TCA ACA AAC ACC CCC TCC TTC) and the reverse primer was H65E14P2 (GCA ACC CGG GTC GAC CTA TCA TGG GTG GAT CCC TAG CTT). For ROPV, the forward primer was ROE14P1 (AGG ATC CGA ATT CGC TGA AGC TCA ACC CCC CTA CGG C) and the reverse primer was ROE14P2 (GCA ACC CGG GTC GAC CTA TCA CTG GCG GAT CCC GAG). The integrity of the clones was established by automatic DNA sequencing using an ABI machine.
Generation of E4-specific antibodies.
Glutathione S-transferase (GST)-E1∧E4 fusion proteins were used as antigens to generate specific E1∧E4 antibodies in rabbits (BPV-1, COPV, HPV-11, HPV63, and HPV65) or rats (CRPV and ROPV). Rabbits were immunized at multiple subcutaneous sites with 250 μg of protein in Freund's complete adjuvant, followed by booster immunizations at 14-day intervals with the 250-μg antigen in Freund's incomplete adjuvant. Antibody titers were evaluated by ELISA 14 days after the third injection, and immunization was continued if low antibody titers were found. Rat immunization was carried out in the same way except that 50 μg of protein was used per immunization. Preimmune serum was collected before the immunization protocol was started and was used as a control in the immunostaining experiments. Antibodies were also generated against keyhole limpet hemocyanin-conjugated peptides in rabbits and rats using the immunization protocol described above. Peptide conjugation was carried out using m-maleimidobenzoyl-N-hydroxysuccinimide ester (Sulfo-MBS; Perbio) using an established protocol (55). Antibodies were made against the first 12 N-terminal amino acids of the CRPV (ac-AEAPPSRWSVPLC∗amide) and BPV-1 E1Ê4 (ac-ANDKEIAQTESGC∗amide) proteins and the last 12 C-terminal amino acids of BPV-1 E4 (RSSRPTPQRKNQC∗amide). The methionine residue encoded by the initiation codon was omitted from the N-terminal peptides and replaced by an acetyl group (ac) in order to mimic the predicted in vivo modification of the protein.
The specificities of antibodies were established by ELISA and Western blotting using maltose binding protein-E1Ê4 fusion proteins or bovine serum albumin-conjugated peptides (conjugation was carried out using m-maleimidobenzoyl-N-hydroxysuccinimide ester (Sulfo-MBS; Perbio) as outlined above [55]). For Western blots, 0.5 μg of purified fusion protein (per well) was run on a sodium dodecyl sulfate-10% polyacrylamide gel prior to being transferred to a nitrocellulose membrane by semidry blotting (Bio-Rad). Test antisera were diluted 1:250 in phosphate-buffered saline (PBS) containing 2% nonfat dry milk powder (Marvel), and blotting was carried out as described previously (55). ELISAs were carried out using 0.5 μg of protein (Falcon) per well essentially as described by Harlow and Lane (55). Species-specific (anti-rabbit or anti-rat) horseradish peroxidase-conjugated secondary antibodies (Amersham) were used for detection, and horseradish peroxidase activity was visualized using FAST DAB tablets (Sigma, St. Louis, Mo.). Maltose binding protein or bovine serum albumin was used as a negative control to establish that the reactivity was specific for E4.
Lesions caused by human and animal papillomaviruses.
Tissue biopsy specimens fixed in 10% neutral buffered formalin and embedded in paraffin wax were cut into 5-μm-thick sections and mounted on electrostatically charged slides (BDH). Experimental lesions induced in cottontail rabbits (five were examined) and in New Zealand White rabbits (nine were examined) were provided by two of the authors (Neil Christensen and Janet Brandsma, respectively). Experimental lesions were induced at the cutaneous epithelium on the rabbits' backs using established procedures (12, 13). Naturally occurring COPV-induced oral papillomas (four were examined) were obtained from diagnostic material submitted to Philip Nicholls. Naturally occurring BPV-induced fibropapillomas from cutaneous sites on the head and neck (four total) were provided by Wen Jun Liu. Human verrucas caused by HPV1 (22 were examined for E1∧E4 expression), HPV63 (3 were examined), and HPV65 (6 were examined) were provided by Kiyofumi Egawa or were obtained from previously described sources (38, 42, 43). Cutaneous warts caused by HPV2 (25 were examined for E1∧E4 expression) were also obtained from these sources (38, 42, 43) or were provided by Alan Percival. Low-grade squamous intraepithelial lesions (LSIL) caused by HPV16 (seven were examined) and HPV11 (five were examined) were provided by Karl Sotlar or were obtained from previously described sources. Foreskin tissue infected with HPV11 virions and propagated as skin grafts on SCID mice (seven were examined) or under the kidney capsule of nude mice (six were examined) was provided by John Lewis. Experimental COPV-induced lesions (8 oral lesions were examined) and ROPV-induced lesions (22 oral and 2 penile lesions were examined) were produced in dogs (beagles) or rabbits by tissue scarification and inoculation of virions (29, 75). The propagation of HPV11 (six lesions were examined), ROPV (two lesions were examined), and CRPV (two lesions were examined) in infected tissue implanted under the kidney capsule of nude mice was carried out according to established protocols (9, 28).
Use of polyclonal and monoclonal antibodies.
Paraffin-embedded sections were dewaxed in xylene (one time for 10 min followed by one time for 5 min) and rehydrated by passage through graded alcohols (twice for 3 min each time in 100% ethanol and then for 2 min each in 90, 80, 50, and 30% ethanol) before being placed in PBS (twice for 5 min each time). For the detection of E1Ê4 in lesions caused by BPV, CRPV, ROPV, HPV11, HPV16, HPV63, and HPV65, formalin-fixed tissue sections were either microwaved three times for 5 min each time (at 650 W) in citrate buffer (pH 6.0 for all E4 detection except CRPV, which was carried out at pH 8.0) or heated in a domestic pressure cooker in the same buffer for 3 to 6 min. The citrate buffer was prepared by adding 9 ml of 0.1 M citric acid and 45 ml of 0.1 M sodium citrate to 500 ml of water. The sections were subsequently blocked for 1 h at room temperature using 10% fetal bovine serum or 10% goat serum in PBS before being incubated with the monoclonal or polyclonal antibodies. Epitope exposure was not necessary in order to visualize E1Ê4 in lesions caused by COPV, HPV1, and HPV2. Polyclonal antibodies to the E4 protein of BPV were used at dilutions of 1:100 to 1:150 (anti-GST-E1∧E4) or 1:200 to 1:400 (anti-peptide), while those to the E4 protein of COPV were used at a dilution of 1:600. Polyclonal antibodies to the E4 proteins of CRPV and HPV11 were used at 1:100 dilution (anti-peptide and anti-GST-E1Ê4), while those to the E4 protein of ROPV were used at a dilution of 1:400 (anti-GST-E1∧E4). Preimmune serum was used as a control.
Double staining was carried out following the incubation of tissue sections with two different antibodies. In lesions caused by BPV, COPV, and HPV-11, E4 was detected using E4-specific rabbit polyclonal serum and L1 was detected using the mouse monoclonal antibody CAMVIR-1 (1:100 dilution; Pharmingen). Rabbit polyclonal antibodies raised against BPV particles (1:100 dilution; DAKO) were used to detect L1 in lesions caused by ROPV, while the L1 protein of CRPV was detected using NZ264 serum (1:100 dilution; kindly provided by Francoise Breitburd, Institut Pasteur, Paris, France). PCNA was detected using the mouse monoclonal antibody PC10 (Neomarkers). Antibodies to cyclin A and MCM7 were obtained from commercial sources (Novocastra and Neomarkers). Incubation with primary antibodies was carried out for 1 h at room temperature or overnight at 4°C before the sections were washed (three times for 5 min each time) in PBS containing 0.05% Tween 20.
Immunodetection of antibodies.
Primary antibodies were detected using species-specific secondary antibodies (1:50 dilution) conjugated to fluorescein or Texas red (Amersham), except where signal amplification was necessary using streptavidin ABC-alkaline phosphatase or horse radish peroxidase (DAKO). In such instances, the section was incubated for 1 h in the presence of a biotinylated species-specific secondary antibody before being washed (three times for 5 min each time) in Tris-buffered saline containing 0.05% Tween 20. A biotin-streptavidin-alkaline phosphatase or horseradish peroxidase complex (DAKO) was then added along with the nucleic acid stain Sytox green (125 nM in PBS) or DAPI (4′,6′-diamidino-2-phenylindole; 500 nM) (Molecular Probes). The section was then incubated for 30 min at room temperature before being washed (three times for 5 min each time) in Tris-buffered saline containing 0.05% Tween 20 and developed using Fast Red TR-Naphthol AS-MX (Sigma) or tyramide substrate (NEN Life Science Products) both prepared according to the manufacturer's instructions. The colorimetric reaction was stopped when development was complete by washing the section for 5 min in PBS.
DNA labeling and in situ hybridization.
DNA probes for in situ hybridization were labeled with either biotin (BioProbe Random Primed DNA Labeling system; Enzo) or digoxigenin (DIG DNA Labeling and Detection kit; Roche Molecular Biochemicals) according to the manufacturer's instructions. Full-length linearized papillomavirus DNA was used as a template for random primer labeling. In situ hybridization reactions were performed on paraffin-embedded formalin-fixed tissue sections after dewaxing and rehydration (as described above). The sections were then digested for 15 min at 37°C using proteinase K (50 μg/ml) before being washed in PBS (twice for 2 min each time) and air dried at room temperature. The labeled DNA probes were diluted 1:25 in hybridization buffer (50% deionized formamide, 1× Denhardt's, 5% dextran sulfate, 200 μg of salmon sperm DNA/ml, 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) before being added to the sections. The sections were then covered with glass coverslips, and sealed with cow gum (Cow Proofings Ltd.), and heated at 93°C for 5 min. The slides were immediately quenched in ice for 5 min and were then incubated overnight at 37°C in a humid chamber. After removal of the cow gum, the coverslips were shaken off in 0.5× SSC and the slides were washed in stringent wash buffers (50% formamide, 2× SSC, 0.05% Tween 20 [twice for 5 min each time] and 2× SSC [twice for 5 min each time]) at 42°C. DIG-labeled DNA was detected using a Tyramide Signal Amplification-Direct system (BlueFISH or RedFISH) (NEN Life Science Products) according to the manufacturer's instructions. Endogenous peroxidase activity was removed by incubation in 3% hydrogen peroxide at room temperature for 10 min. Detection of the DIG-conjugated probe was carried out using a horseradish peroxidase-conjugated antibody to DIG followed by development (8 min) in the presence of tyramide substrate and propidium iodide. Biotin-labeled DNA was visualized using streptavidin ABC-alkaline phosphatase and Fast Red substrate deposition as described above.
RESULTS
Variation in the extent of early gene activity during the life cycles of human and animal papillomaviruses.
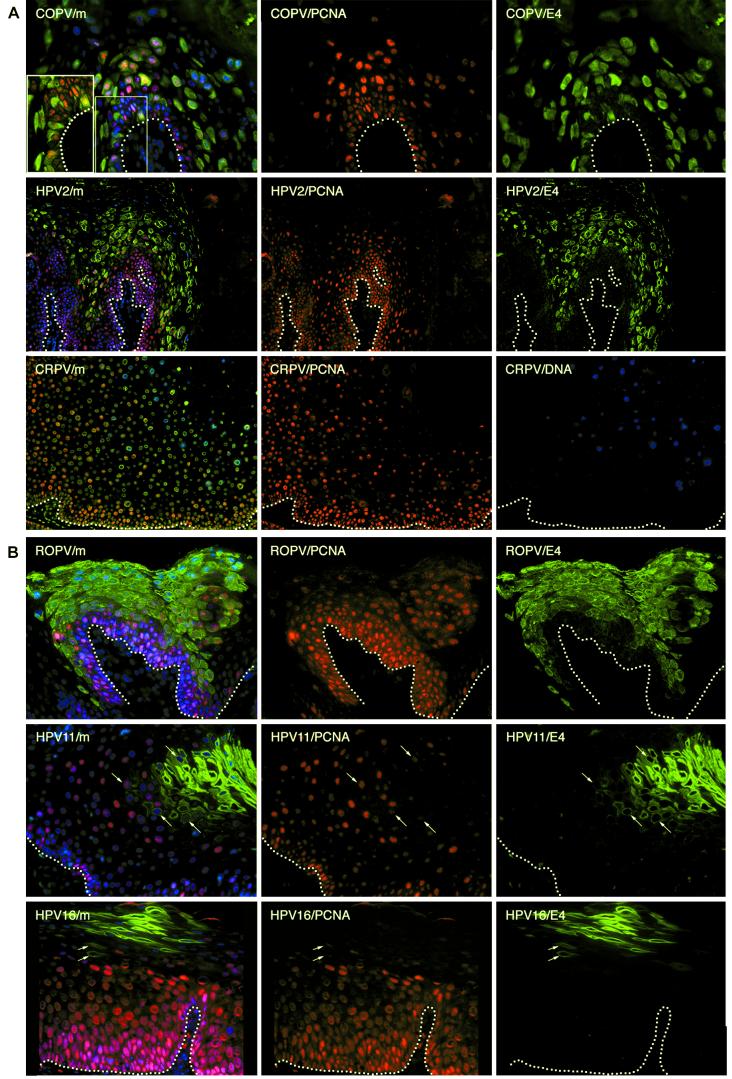
Following the infection of basal keratinocytes, the viral early genes (E6, E7, and maybe E5) are expressed from a promoter in the upstream regulatory region (reviewed in references 58 and 65a). Although these viral proteins are difficult to detect because of their low levels of expression, the cellular gene products they induce can be readily identified (60). Such proteins are normally absent from differentiating epithelium, and their presence in warts is an indication of early viral gene activity. Three E2F-activated gene products (cyclin A, MCM7, and PCNA) were examined for their usefulness as markers of cells expressing E6, E7, and E5 in human and animal tissues. Antibodies to all three proteins gave similar patterns of staining, but that to PCNA (PC10; Neomarkers) showed good cross-reactivity between species and could detect the protein in bovine, canine, and rabbit tissue. PCNA was present in the basal layers of infected and uninfected epithelium but extended into the upper epithelial layers in papilloma tissue (Fig. 1). Early gene activity, as revealed by the presence of surrogate markers, never persisted to the epithelial surface in areas of productive viral infection but showed a pattern that was characteristic of each infecting papillomavirus type (Fig. 1). Oral lesions caused by COPV typically expressed PCNA in basal cells and in sporadic cells immediately above them—a pattern of expression similar to that seen in human cutaneous lesions (verrucas) caused by HPV1 (data not shown and reference 42) and HPV2 (Fig. 1A). Oral and penile lesions caused by ROPV (17) usually showed more extensive early gene activity, with PCNA being detected throughout the epithelium (an oral papilloma is shown in Fig. 1B). This pattern is similar to that seen in human mucosal lesions caused by HPV11 and HPV16 (an LSIL is shown in Fig. 1B). Lesions caused by HPV16 do not always support the productive stages of the virus life cycle, however, and under these circumstances, markers of early gene expression can be found in cells at the epithelial surface (data not shown and K. Middleton, L. Morris, W. Peh, A. El-Sherif, K. Sotlar, D. Jenkins, R. Seth, H. Griffin, H. Hibma, R. Laskey, N. Coleman, and J. Doorbar, 19th Int. Papillomavirus Conf., abstr. P-21, 2001). This is a characteristic of high-grade squamous intraepithelial lesions (HSIL) caused by HPV16 and was not seen during productive infection.
FIG.1.
Heterogeneity in the timing of events during the life cycles of human and animal papillomaviruses. (A) Tissue sections from a mucosal lesion caused by COPV (top row) and a cutaneous lesion caused by HPV2 (middle row) were double stained using antibodies to E4 (green) andPCNA (red) before being counterstained with DAPI (blue) to visualize cell nuclei. Surrogate markers of E7 expression (PCNA) were confined to sporadic cells in the lowest parabasal layers. E4 (green) was first detected in these cells as they migrated through the lower layers of the epithelium. E4 was expressed in the basal layer in lesions caused by COPV and in a few cells above the basal layer in lesions caused by HPV2. Surrogate markers of E7 were lost soon after the appearance of E4. The merged images (m) are shown at the left. A section through a cutaneous lesion caused by CRPV is shown in the bottom row after immunostaining to detect PCNA (red) and in situ hybridization to detect viral genome amplification (blue). The nuclei were counterstained using Sytox green. Surrogate markers of E7 expression (PCNA) did not persist to the epithelial surface and were lost soon after the onset of viral genome amplification (blue). Genome amplification began in the mid-spinous layers in lesions caused by CRPV. The broken lines indicate the positions of the basal layers. The images were taken using a 10× (COPV and HPV2) or 20× (COPV/m inset) objective. (B) Tissue sections from mucosal lesions caused by ROPV (top row), HPV11 (middle row), and HPV16 (bottom row) were double stained with antibodies to E4 (green) and PCNA (red) before being counterstained with DAPI (blue) to visualize cell nuclei. The merged images are shown at the left. In such lesions, surrogate markers of E7 expression (PCNA; red) usually persisted into the upper epithelial layers but were lost following the expression of E4. E4 was rarely detected in the lower epithelial layers in lesions caused by HPV11 and -16. Cells expressing both E4 and PCNA are indicated by arrows in the HPV11 and HPV16 images. The broken lines indicate the positions of the basal layers. The images were taken using a 20× objective.
CRPV and BPV infect cutaneous epithelium, making comparison with human mucosal infections more difficult. BPV-induced papillomas from cattle and CRPV-induced papillomas from cottontail (natural host) and New Zealand White (artificial host) rabbits express markers of viral oncogene expression above the basal layer (a cottontail rabbit papilloma is shown in Fig. 1A). The distributions of PCNA were broadly similar in papillomas induced in both rabbit hosts, even though New Zealand White rabbits do not support the full CRPV life cycle. In Fig. 1, DNA in situ hybridization was used to identify cells supporting late events during the life cycle of CRPV, as the CRPV E4 antibodies gave relatively high levels of background staining (Fig. 2B). The distribution of surrogate markers of early gene expression in CRPV-induced papillomas in their natural host resembled that seen in LSIL caused by HPV11 and -16 and in oral papillomas caused by ROPV (Fig. 1). The expression of such markers without the subsequent onset of late events was seen only in lesions produced in New Zealand White rabbits (artificial host) and HSIL caused by HPV16 (data not shown).
FIG.2.
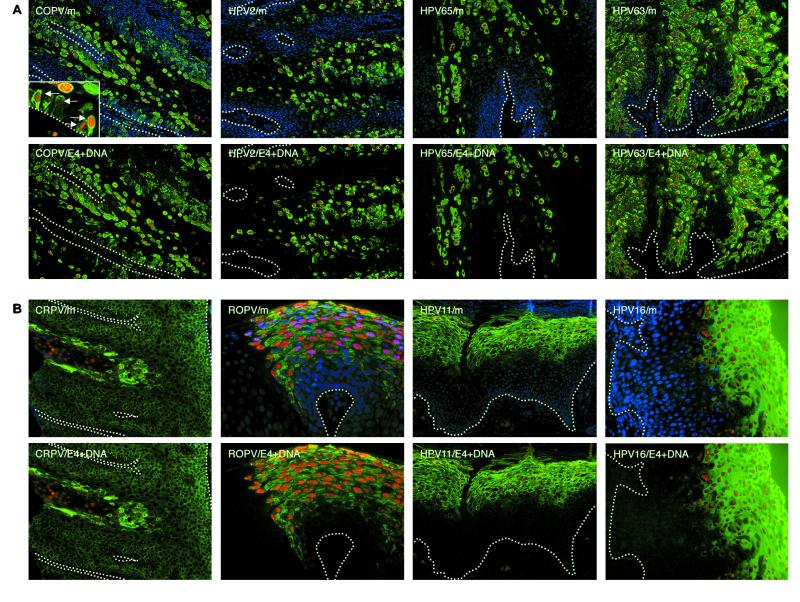
The onset of vegetative viral genome amplification coincides closely with the expression of E4 in all papillomavirus infections. (A) Tissue sections from a mucosal lesion caused by COPV and cutaneous lesions caused by HPV2, HPV65, and HPV63 were probed to detect the presence of amplified viral DNA (red) and E4 (green). The nuclei were counterstained using DAPI (blue). Genome amplification was generally confined to cells expressing E4 (green). In warts caused by COPV, HPV2, and HPV65, such cells were distributed sporadically throughout the lesion. The broken lines indicate the positions of the basal layers. The images were taken using a 10× objective. The inset shows COPV E4 and viral genome amplification in the basal layer (green, E4; red, amplified viral DNA; blue, DAPI counterstain). The images were taken using a 40× objective. (B) Tissue section from a cutaneous lesion caused by CRPV and from mucosal lesions caused by ROPV, HPV11, and HPV16 were probed by in situ hybridization to detect cells that had amplified viral DNA (red). E4 was detected by using specific antibodies to the E4 proteins of each virus type (green), and the nuclei were counterstained using DAPI (blue). E4 expression and viral genome amplification were restricted to a band of cells in the middle and upper epithelial layers. The broken lines indicate the positions of the basal layers. The images were taken using a 10× objective.
Heterogeneity in the timing of late-stage onset in lesions caused by animal and human papillomaviruses.
The late stage of the virus life cycle is triggered only as the infected cell migrates towards the epithelial surface. Although E6 and E7 are difficult to detect by immunostaining, E4 is abundantly expressed and can be detected during epithelial cell differentiation by using type-specific antibodies. The expression of E4 has previously been shown to coincide precisely with the onset of vegetative viral genome amplification in lesions caused by HPV1 and HPV16 (42), and in the first instance we sought to establish that this was a common event in the life cycles of all papillomavirus types. To establish this, antibodies were generated to the E1Ê4 major late proteins of ROPV, CRPV, COPV, and BPV after expression as fusion proteins in bacteria. Antibodies to the E1Ê4 proteins of HPV2, HPV11, HPV63, and HPV65 were also generated for comparison. In all cases, high-affinity antibodies were obtained which allowed detection of the appropriate E4 protein in biopsy material. With the exception of antibodies to the BPV1 and HPV11 E4 proteins, which cross-reacted with the E4 proteins of BPV2 and HPV6, respectively, the antibodies did not cross-react between virus types. Antibodies to the E1Ê4 proteins of HPV1 and HPV16 have been described previously (39, 41, 42). No staining was apparent using preimmune serum.
In all the human and animal papillomaviruses examined, genome amplification coincided with the presence of detectable levels of E4, although as with early events, the life cycles of the different viruses showed variation. The most unexpected observation was that viral genome amplification and the expression of E4 begins in cells of the basal layer in oral lesions caused by COPV (Fig. 2A). Most basal cells express PCNA (Fig. 1A), but in lesions caused by COPV, a subset of these express E4 (Fig. 1A) and support viral genome amplification (Fig. 2A) (73). Viral genome amplification in the basal layer was not observed in lesions caused by other papillomavirus types, although HPV1 and -63 (to which COPV is closely related [24, 34]) and HPV65 do initiate genome amplification in cells of the lowest parabasal layers (Fig. 2A) (42). These viruses cause verrucas rather than oral papillomas. By contrast, ROPV, HPV11, and HPV16 (LSIL) showed similarity in the timing of their late events (Fig. 2B). In both cases, genome amplification and E4 expression coincided in the mid-spinous layers, with E4 being detectable to the epithelial surface (Fig. 2B). A correlation between the detection of E4 and the onset of genome amplification was also observed in lesions caused by BPV1 (data not shown) and CRPV (Fig. 2B). The timing of late events in CRPV-induced lesions generated in cottontail rabbits resembled that seen in productive human lesions caused by HPV11 and HPV16 (LSIL) and was distinct from the “sporadic” pattern seen in human cutaneous papillomas caused by related viruses, such as HPV63 (24) (Fig. 2).
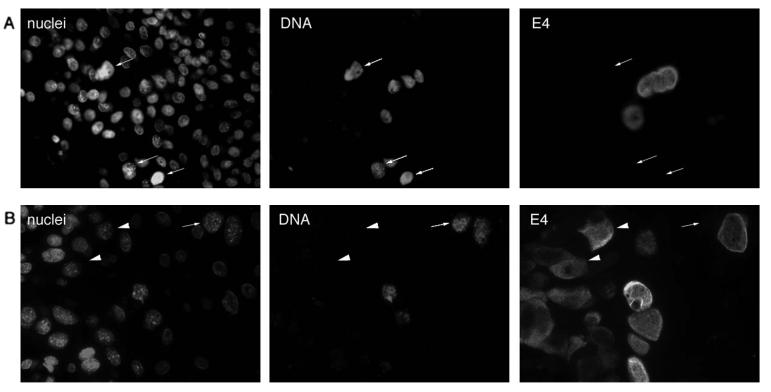
Although the association between the appearance of detectable levels of E4 and the onset of viral genome amplification was a common feature of all the lesions examined here, high levels of viral DNA were occasionally seen in the absence of E4 in lesions caused by COPV, as well as in lesions caused by CRPV (Fig. 2B and 3). In CRPV-induced lesions (Fig. 2B), such cells appear to arise from E4-expressing cells (present in the spinous layer) that are supporting viral genome amplification, and we assume that E4 does not persist to the epithelial surface or that the antibody is not able to detect it in the upper epithelial layers. Although this may also be true of the cells that contain amplified viral genomes (but not E4) in lesions caused by COPV, the sporadic distribution of E4-containing cells in these lesions makes this less evident. We do not know for certain whether the in situ signals seen in the E4-negative cells in COPV-induced lesions represent true genome amplification or a partial relaxation of the copy number control that operates in the basal layer. Cells containing amplified viral genomes in the absence of E4 were not seen in lesions caused by other papillomavirus types.
FIG. 3.
E4 expression and the onset of genome amplification are not necessarily coincident. (A and B) Tissue sections taken from two different COPV-infected oral lesions. Tissue sections were double stained for the presence of viral genome amplification (DNA) and for E4 expression (E4). Viral DNA was occasionally detected in the absence of E4 (cells indicated by arrows). The nuclei were counterstained using Sytox green (nuclei). (B) E4 expression in cells (indicated by arrowheads) that are not supporting viral genome amplification. A cell (indicated by an arrow) supporting viral genome amplification but lacking E4 is shown in the same field of view. The images were taken using a 40× objective.
The onset of late events is closely linked to the completion of early events during the papillomavirus life cycle.
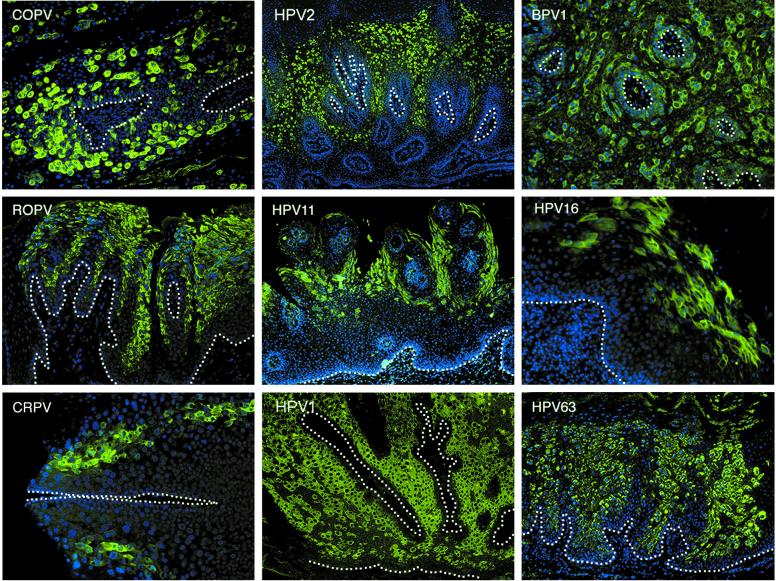
Although our data show that the timing of the late-event activation is divergent among papillomavirus types, a high degree of consistency was apparent when lesions caused by the same virus type were compared. This was most evident in papillomas induced by the evolutionarily related viruses (24) COPV, HPV1, and HPV63, which trigger late gene expression in the basal and parabasal cell layers (Fig. 1 and 4). Over 60% of the cells in the upper epithelial layers of such lesions express E4, with such cells usually appearing in clusters—a distribution that is also seen in lesions caused by BPV1 (Fig. 4). No variation in this pattern was apparent in >8 COPV-induced oral papillomas and 25 HPV1-HPV63-induced verruca lesions examined. Lesions caused by CRPV in its natural host, ROPV, HPV11, and HPV16 (LSIL) also showed consistency in their patterns of late gene expression (which began in the intermediate layers), but these were distinct from the patterns seen in oral lesions caused by COPV and in cutaneous lesions caused by HPV1, HPV2, or HPV63 (Fig. 4). The expression pattern characteristic of each papillomavirus type is shown in Fig. 4. Despite the heterogeneity among viruses, the disappearance of early markers always occurred after the onset of late events, suggesting that the two stages in the virus life cycle are closely coordinated (Fig. 1). Cells expressing both E4 and viral oncogenes, such as E7 (as determined by the presence of surrogate markers), were found in a narrow region of overlap, which could be as little as one cell layer thick in LSIL caused by HPV16 (Fig. 1B) to over five cell layers thick in COPV-induced oral papillomas or cutaneous warts caused by HPV2 (Fig. 1A). The overlap between surrogate markers of viral early gene activity (e.g., PCNA and cyclin A) and late markers (e.g., E4 and genome amplification) shown in Fig. 1 was a common feature of all the productive papillomavirus infections examined here. Only during abortive infection, such as in HPV16-associated HSIL or in lesions produced by CRPV in New Zealand White rabbits (artificial host), is this pattern of expression disrupted.
FIG. 4.
Timing of late events is highly conserved among lesions caused by the same papillomavirus type. Similarities in the patterns of E4 expression and genome amplification confirmed that E4 was an effective marker of the late stages of the papillomavirus life cycle. Tissue biopsies infected by different papillomaviruses were sectioned and stained using E4-specific antibodies (green) before being counterstained with DAPI (blue). The staining patterns illustrated were typical of those seen following the examination of five or more independent biopsy specimens infected by each virus type (except for HPV63, for which three lesions were examined [see Materials and Methods]). The timing of late-stage onset was a characteristic feature of each infecting virus type. All images were taken using a 4× objective except for the HPV16 and CRPV images, which were taken using a 10× objective lens. The basal layers are indicated by the broken lines.
Intracellular distribution of E4 in lesions caused by animal papillomaviruses.
The E4 proteins of many papillomaviruses form intracellular structures that are characteristic of the infecting virus type. While this is particularly evident in lesions caused by HPV1 and HPV4 (38), it has been suggested that most cutaneous viruses form some sort of cytoplasmic inclusion (32, 46, 47, 70). By contrast, the E4 proteins of human mucosal viruses do not produce inclusions (19, 21, 40, 42, 81). HPV16 E4 associates with the cellular intermediate filament network, whereas HPV11 E4 localizes to the cornified envelope of the cell. E4 is one of the few viral proteins that can easily be detected during productive infection, and the generation of antibodies allows its intracellular distribution to be examined. Although the role of E4 is uncertain, similarities in intracellular localization may suggest functional conservation.
Of the viruses examined here, only ROPV E4 had an in vivo intracellular distribution that broadly resembled that of the human mucosal viruses. The E4 proteins of HPV16 and HPV11 are predominantly cytoplasmic, and this was also true of ROPV E4 (Fig. 5A). Unlike the E4 proteins of HPV16 and HPV11, however, ROPV E4 assembled into cytoplasmic inclusion granules similar to those seen in warts caused by HPV1 (82, 83), and these increased in size as the cells approached the epithelial surface (Fig. 5A). Granule formation is an inherent property of the E4 proteins of HPV1 and ROPV and also occurred following their expression in vitro (references 46, 48, 82, and 83 and our unpublished results).
FIG.5.
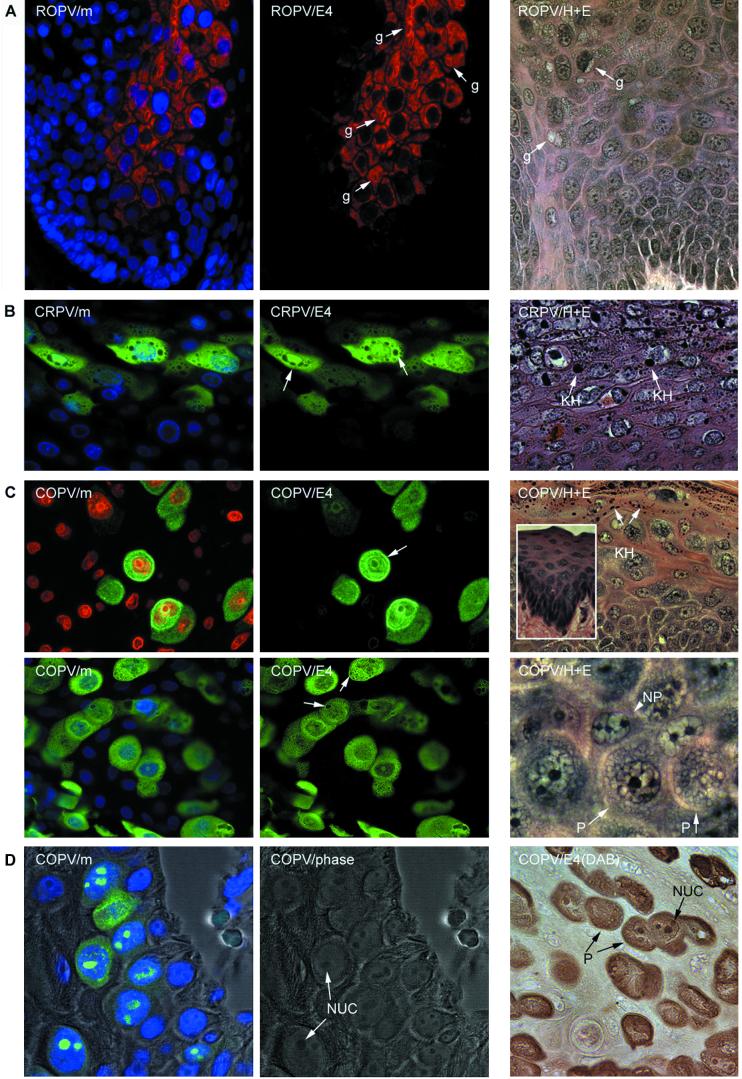
Intracellular distribution of the E4 proteins of animal papillomaviruses. (A) The E4 protein of ROPV (red) is predominantly cytoplasmic and is associated with inclusion granules (center image, g) similar to those seen in cutaneous lesions caused by HPV1. The nuclei werecounterstained with DAPI (blue) and are visible in the merged image at the left (ROPV/m). The E4 granules (g) are also shown in the hematoxylin- and eosin-stained image at the right (labeled ROPV/H+E). The images were taken using a 40× objective. (B) The E4 protein of CRPV (green) is distributed throughout the nucleus and the cytoplasm. The nuclei were counterstained with DAPI (blue) and are visible in the merged image shown on the left (CRPV/m). The cytoplasmic structures that did not stain with antibodies to E4 are keratohyalin granules (center image, arrows). They are also shown (KH) in the hematoxylin- and eosin-stained image (CRPV/H+E) on the right. (C) The E4 protein of COPV (green) was cytoplasmic and nuclear but was also associated with the nuclear and cellular periphery (upper center image, arrow). The nuclei were counterstained with either propidium iodide (upper left image, red) or DAPI (lower left image, blue). A granular pattern was apparent in the cytoplasm of some cells (granule-like structures [arrows] in lower center image). Keratohyalin granules (KH) are abundant in COPV-induced warts and are shown in the hematoxylin- and eosin-stained image (COPV/H+E; upper right). The surrounding mucosal epithelium is devoid of keratohyalin (inset). Cells expressing COPV E4 had a characteristic morphology that may result from the presence of E4 inclusion granules. The permissive cells (P) that express E4 and the nonpermissive cells (NP) that do not express E4 are shown in the hematoxylin- and eosin-stained image on the lower right. (D) In the lower epithelial layers of experimental warts caused by COPV, nuclear E4 protein (green) was found associated with the nucleoli. At the left (COPV/m), the E4 and DAPI (blue) stain is shown as an overlay of the phase-contrast image. The phase-contrast image is shown in the center panel to indicate the presence of the nucleoli (NUC). The presence of the permissive “granular” cells (P) and the nucleoli is clearly visualized in the immunoperoxidase stain (brown) shown on the right [COPV/E4 (DAB)].
Although E4 is predominantly cytoplasmic in all the human papillomavirus infections examined to date (17, 38, 42), in lesions caused by COPV and CRPV the protein was distributed throughout the cell (Fig. 5). Cells expressing COPV E4 contained cytoplasmic structures that were similar in appearance to the inclusions produced by ROPV (Fig. 5A and C). COPV E4 protein was also associated with the nucleoli (Fig. 5D) and with cell membranes (Fig. 5C). Although CRPV E4 was not associated with any obvious structures, lesions caused by CRPV and COPV contained abundant keratohyalin granules. In CRPV-induced warts, these were found in the cells expressing E4 (Fig. 5B). Mucosal epithelium, such as that infected by COPV and ROPV, does not normally contain abundant keratohyalin granules (Fig. 5C, inset), making their presence in COPV-infected tissue surprising. Keratohyalin granules were not, however, present in cells expressing COPV E4. This is similar to the situation seen in lesions caused by HPV1, where E4-expressing cells lack filaggrin and do not produce visible keratohyalin (42). Although ROPV and COPV infect similar epithelial sites, keratohyalin granules were never detected in lesions caused by ROPV and were only occasionally present in human mucosal infections caused by HPV11 and -16 (data not shown).
Expression of E4 always precedes expression of the L1 capsid protein.
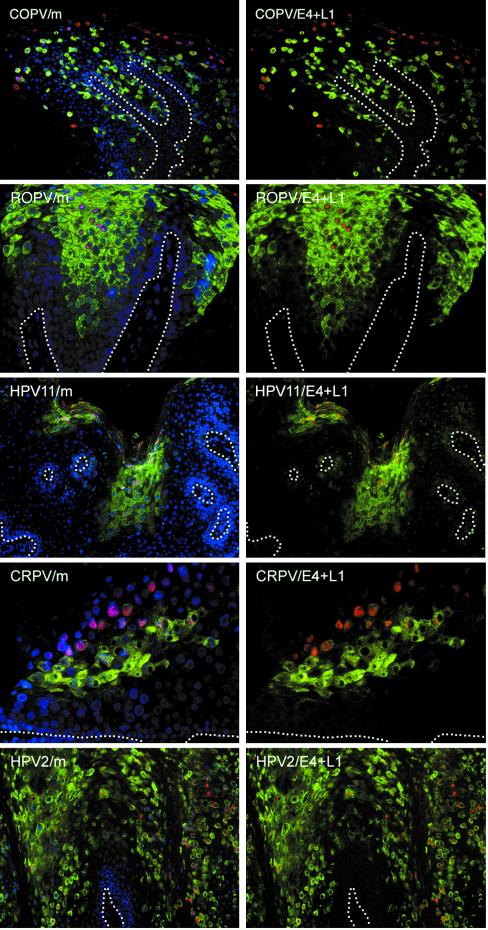
The major virus coat protein was expressed in the nuclei of terminally differentiating cells near the surface of the epidermis. In all the lesions examined here, L1 expression followed that of E4, and a gap was apparent between the first appearance of E4 and the first appearance of L1 (Fig. 6). Although L1 expression was first detected in E4-containing cells, L1 expression was not always supported, and cells expressing only E4 could occasionally be found at the epithelial surface. This was sometimes apparent in productive infections (e.g., HPV11 [Fig. 6]) but was seen more regularly in HSIL caused by HPV16. Such lesions resemble those produced in New Zealand White rabbits by CRPV and do not support the full life cycle of the virus (data not shown and Middleton et al., 19th Int. Papillomavirus Conf.). While the coordinated pattern of E4 and L1 expression was conserved in all productive lesions examined, the gap between the first appearance of E4 and the onset of L1 expression varied considerably. Viruses that initiate their late events in the lower epithelial layers, such as COPV, HPV1, and HPV63 (which are evolutionarily related [24]), typically showed a greater number of L1-expressing cells than those that initiate late gene expression in the upper epithelial layers (compare COPV and HPV11 in Fig. 6). LSIL caused by HPV16 and cutaneous papillomas caused by CRPV (in the cottontail rabbit host [Fig. 6]) had intervals as small as one or two cell layers between the site of E4 expression and that of L1. Cutaneous lesions caused by HPV1 and HPV2 (Fig. 6) usually initiated L1 expression four or five cell layers after the first appearance of E4. By contrast, L1 expression in HPV63- or COPV-induced lesions may be separated from the onset of E4 expression by as many as 20 cell layers (42) (Fig. 6). Of the lesions examined here, those caused by HPV11 and ROPV showed the greatest similarity in the expression patterns of their E4 and L1 proteins, although E4 expression in ROPV lesions usually began earlier than in lesions caused by HPV11 (Fig. 6).
FIG. 6.
Expression of capsid proteins follows expression of E4 in lesions caused by different papillomavirus types. Tissue sections of lesions caused by COPV, ROPV, HPV11, CRPV, and HPV2 were double stained using antibodies to the L1 capsid protein (red) and E4 (green) before being counterstained with DAPI (blue). The merged images (m) are shown on the left. Although E4 expression always precedes the expression of L1, the distance between the first appearance of E4 and the first appearance of L1 varied considerably. The positions of the basal layers are indicated by broken lines. The images were taken using a 10× (HPV2, HPV11, and COPV) or 20× (CRPV) objective.
Viral gene expression in xenografts resembles that seen during infection of the natural host.
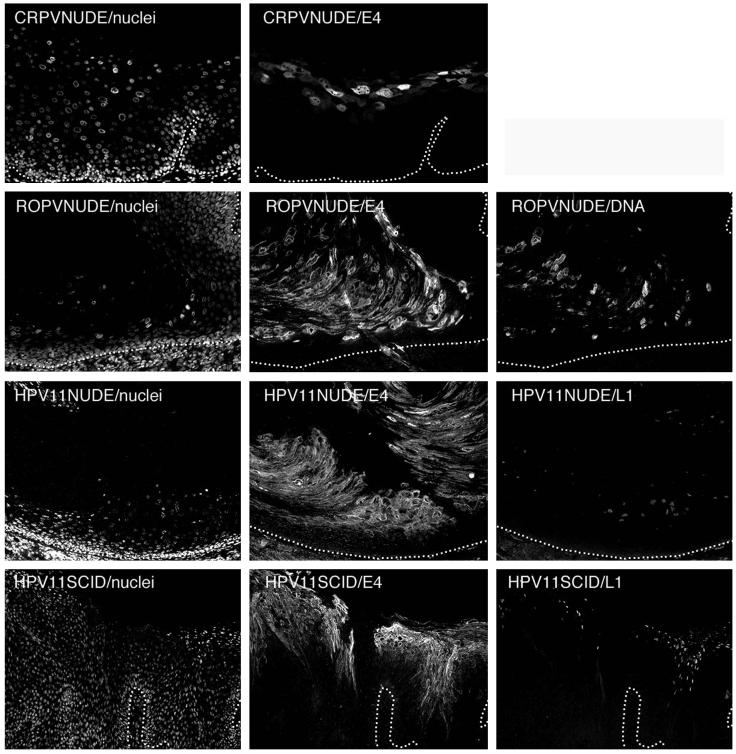
Several papillomaviruses, including ROPV (28), CRPV (30), and HPV11 (36, 63), can be propagated in epithelial tissue implanted under the renal capsule. This approach has been used to generate stocks of animal (28) and human papillomaviruses and to study the HPV life cycle in vivo (18, 20). Renal-capsule xenografts infected with CRPV or ROPV were compared to experimental infections in their natural hosts with regard to the timing of initiation of late events described above. The lesions produced in both systems were broadly similar, and the timing of E4 expression, genome amplification, and virus synthesis (described here) was preserved (Fig. 7). Detectable E4 expression began in the intermediate and spinous cell layers and coincided with the onset of viral genome amplification (ROPV is shown in Fig. 7). Although genome amplification was not examined in CRPV-infected xenografts, the expression of CRPV E4 followed a pattern that resembled that seen during natural infection (Fig. 4 and 7). In all xenografts examined, irrespective of the infecting papillomavirus type, the L1 protein was first expressed in only a subset of E4-positive cells in the upper layers of the epidermis (HPV11 is shown in Fig. 7). Xenografts propagated under the kidney capsule, however, did show a lower degree of papillomatosis than lesions produced at the natural site of infection, as reported previously (8). It appears that L1 expression always follows that of E4, in contrast to previous reports, which have indicated that the expression of E4 and L1 is coincident (18, 20). HPV11 xenografts propagated on the skin (in SCID mice) showed similarity to naturally occurring human genital lesions both in their morphology and in their pattern of late gene expression (compare Fig. 1, 2, 4, and 6 and HPV11SCID/E4 and -L1 in Fig. 7).
FIG. 7.
The timing of late gene expression in xenografts resembles that seen during natural infection. Epithelial tissue infected by CRPV or ROPV was propagated under the kidney capsule of nude mice (images labeled CRPVNUDE and ROPVNUDE) before being stained for E4 and (for ROPV) genome amplification. The nuclei were counterstained with DAPI and are shown on the left. In all cases, the timing of late-stage activation (as determined by E4 expression) was similar to that seen following natural and experimental infection of the natural host (Fig. 4). Genome amplification and E4 expression coincided closely (images labeled ROPVNUDE/DNA and ROPVNUDE/E4). Epithelial tissue infected by HPV11 was propagated under the kidney capsule of nude mice or as a skin graft in SCID mice. L1 expression followed that of E4 in renal capsule xenografts infected with HPV11 (images labeled HPV11NUDE/E4 and HPV11NUDE/L1). This pattern of expression was also seen in skin grafts propagated on SCID mice (HPV11SCID/E4 and HPV11SCID/L1). The morphology of the skin xenografts closely resembled that seen in HPV11-induced genital lesions (Fig. 2 and 4). The images were taken using a 10× (ROPV and HPV11) or 20× objective. The broken lines indicate the positions of the epithelial basal layers.
DISCUSSION
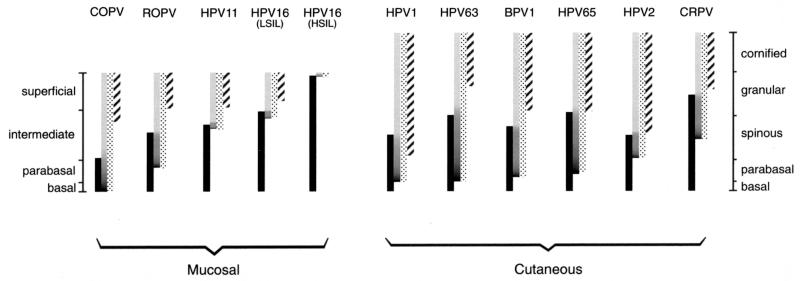
Papillomaviruses are highly host specific, making the use of animal models a valuable strategy for the in vivo analysis of infection. CRPV has been extensively used for the development of vaccines (16, 53, 54, 65, 87, 94) and has provided insight into the roles of viral proteins during papilloma formation (13, 14, 33, 72, 104). Vaccination studies have also been carried out using COPV, which, unlike CRPV, infects oral epithelium (5, 91, 95, 105). Although the mucosal tropism of COPV has led to its use as a model of genital HPV infections, the COPV life cycle differs in several ways from those of HPV11 and HPV16. At the DNA and protein sequence levels, COPV shows homology with HPV types that cause plantar and palmar warts, and this is reflected in similarities among lesions caused by these viruses (24). Of the papillomaviruses examined here, the COPV E4 protein resembles the E4 proteins of HPV4 and HPV65 most closely, while its E6 protein shows the greatest sequence homology with the E6 proteins of HPV1 and -63. The initiation of late events in basal and parabasal cell layers is a characteristic of these related virus types and is an obvious difference between lesions caused by COPV and those caused by CRPV, ROPV, HPV11, and HPV16 (summarized in Fig. 8). Interestingly, COPV-infected cells contained inclusions that were similar in appearance to those seen in lesions caused by HPV1 and ROPV. The E4 proteins of COPV and ROPV contain variants of the C-terminal DLXDYW motif that is found in cutaneous human papillomaviruses and which is involved in E4 multimerization (2). This motif has not previously been identified in the E4 proteins of mucosal papillomavirus types (44). If vaccination studies using model systems are to be relevant in the prevention and treatment of human disease, the chosen model should closely mimic infection in humans. COPV and HPV11 have similar tissue tropisms but differ in the timing of their life cycle events (Fig. 8). By contrast, the life cycles of HPV11, HPV16, and ROPV (which all infect genital tissue) are organized similarly. The ability of New Zealand White rabbits to support the full ROPV life cycle makes this system useful for the study of late gene function, but the short duration of the experimental ROPV-induced papillomas may limit its use to the development of prophylactic vaccines. Although experimental papillomas caused by CRPV do not support late events, they are persistent and offer some advantages in the development of therapeutic vaccines and antivirals.
FIG. 8.
The regulation of early and late events in lesions caused by different papillomavirus types. The timing of life cycle events in lesions caused by animal and human papillomaviruses is indicated by the bars. The shaded bars show the presence of amplified viral DNA, while the stippled bars show the extent of E4 expression. The expression pattern of surrogate markers of E7 is shown by the solid bars, whereas the L1 expression pattern is indicated by the hatched bars. The darker region at the bottom of the shaded bars indicates the region where vegetative viral genome amplification is thought to occur. Mucosal epithelial tissue infected by COPV, ROPV, HPV11, and HPV16 is divided into four layers, shown on the left. Cutaneous tissue infected by HPV1, HPV63, BPV1, HPV65, HPV2, and CRPV is divided into five layers, shown on the right. For each group of papillomaviruses, those that initiate their late events in the lower epithelial layers are shown on the left. Viruses that initiate their late events close to the epithelial surface are shown on the right. COPV triggers late events in the lowest epithelial layers. Lesions caused by HPV11 and HPV16 (LSIL) usually support late gene expression only in the upper half of the epidermis, whereas late events are not always supported in HSIL. In all instances, the loss of surrogate markers of E7 does not occur until after E4 has accumulated to detectable levels.
It has been suggested that papillomaviruses evade immune detection, at least in part, by expressing their abundant late proteins only in the upper epithelial layers (86, 89, 92). Viral early proteins, such as E6 and E7, are expressed at levels below those necessary to stimulate an effective immune response. While this hypothesis is plausible for human mucosal infections, it does not explain the results obtained here from the analysis of lesions caused by HPV1, HPV65, or COPV, where high levels of E4 are apparent in the basal and parabasal layers (Fig. 4 and 8). Unlike those caused by HPV16 or HPV11, naturally occurring lesions induced by COPV are rarely persistent (74) and usually undergo spontaneous regression within 12 weeks of infection (23). Perhaps significantly, plantar warts caused by HPV1 (myrmecia) are also often short-lived (52). Such lesions usually respond to treatment better than plantar warts caused by HPV2 (mosaic warts) and rarely recur, due to the presence of neutralizing antibodies (64, 80). Little is known of the natural history of verrucas caused by HPV63 and HPV65, but it is possible that wart persistence may be linked to an ability to retard the expression of E4 and capsid proteins until the cell has cleared the basal layer. If this is the case, then infections caused by HPV11 and HPV16 may be more persistent than infections caused by COPV or HPV1 (Fig. 8). The rapid regression of genital lesions caused by HPV6 or -11 is relatively uncommon and usually occurs following a period of wart proliferation, as can often occur during pregnancy (7, 76, 96). HPV16 infections vary greatly in duration (57, 99) and also show variation in the timing of their late gene expression. Persistent infections caused by HPV16 (at cervical epithelial sites) and CRPV (in New Zealand White rabbits) are associated with an increased risk of malignant progression (31, 62, 101). Although there is as yet no firm evidence to link the duration of infection with the extent of virus synthesis, many reports have revealed the importance of the host immune system in restricting papilloma growth and in stimulating regression (reviewed in references 56 and 89). The exposure of viral late proteins to the immune system can prevent papillomavirus spread and can stimulate the early regression of existing warts (reviewed in references 15 and 59).
The rapid regression of experimental warts caused by COPV and ROPV may be related to the fact that they were induced by scarification using concentrated virions. Under these conditions, an immune response to infecting virions is generated (29, 75), and it is not unreasonable to think that this may also occur during natural infection if the titer of the infecting virus is high. Papillomaviruses may thus have reached a balance between the need to produce high numbers of virus particles and the need to evade detection by the host's immune system. The initiation of late events in the basal and parabasal layers is compatible with the high-level production of virions, as occurs in warts caused by HPV1 (4). For viruses such as these (e.g., HPV65 [verrucas] and COPV [canine oral papillomas]), where transmission by intimate physical contact is unlikely, the chance of initiating a new infection is enhanced. Papillomaviruses that are transmitted by intimate physical contact, such as those that infect genital sites (e.g., HPV16), may be required at lower levels. The low abundance of HPV16 virions in many genital lesions may be an evolutionary adaptation that ensures that an immune response to infection does not occur.
Differences in the target tissue and the type of contact necessary for transmission may provide some explanation for the life cycle heterogeneity seen in this study. The four animal and six human papillomaviruses examined have diverse natural histories, and it is perhaps not surprising that they show differences in their patterns of gene expression following infection (Fig. 8). All papillomaviruses, however, must replicate and package their DNA, and certain features were found to be conserved among the life cycles of all the viruses examined. A correlation between E4 expression and viral genome amplification has previously been observed from the analysis of lesions caused by HPV1 and HPV16 (42) and from the analysis of cell lines containing HPV DNA (84). The discovery that E4 expression correlates closely with genome amplification in natural and experimental infections caused by a diverse collection of papillomaviruses supports the assumption that this is a conserved event. In most of the papillomaviruses that have been examined to date, E4 is expressed from a differentiation-dependent promoter late in infection (37). In human papillomaviruses, this promoter is positioned in front of the E1 helicase, which is known to have a direct role in the replication of viral genomes (reviewed in references 25 and 35). The E1Ê4 mRNAs share the same initiation codon, and the relative abundances of the two messages are controlled at the level of splice site selection. This apparent link between the synthesis of full-length E1 transcripts and the production of the primary E1Ê4 message suggests that the two proteins may be required together during the virus life cycle. In fact, E4 appears to be expressed as part of a single transcriptional unit that also includes E1, E2, and E5. E5 reactivates DNA synthesis in quiescent cells (1, 10, 93, 98, 100), while E2 is necessary for the efficient recruitment of E1 to viral origins (49, 67). Interestingly, a role for HPV16 E4 in genome amplification has recently been proposed (C. Davy, D. Jackson, K. Raj, P. Masterson, J. Millar, and J. Doorbar, 19th Int. Papillomavirus Conf., abstr. O-174, 2001), and it has been shown that loss of E4 prevents amplification of the CRPV genome in domestic and cottontail rabbits (W. Peh, J. Brandsma, N. Cladel, N. Christensen, and J. Doorbar, 19th Int. Papillomavirus Conf., abstr. O-146, 2001).
E4 expression and the onset of genome amplification were always found to begin in cells expressing surrogate markers of early gene activity, such as PCNA or cyclin A (Fig. 1 and 8). The viral E6, E7, and E5 proteins drive cells into S phase and stimulate the synthesis of the cellular proteins necessary for viral genome amplification. The region where E4-PCNA double-positive cells were found varied greatly in thickness in different lesions. In HPV1- or HPV2-induced warts, double-positive cells were abundant, while in lesions caused by HPV16 they were scarce. The increase in the levels of the viral replication proteins E1 and E2 that result from the activation of the differentiation-dependent promoter are thought to contribute to genome amplification (6, 61, 78, 79). Our observation that markers of E6 and E7 expression are lost soon after the appearance of E4 can be readily explained if the levels of E2 and E4 rise concomitantly. Both E2 and E4 are expressed from the differentiation-dependent promoter late in infection (26, 27, 42), with E2 showing an expression pattern similar to that of E4 (66). High-level expression of E2 leads to down-regulation of the viral early promoter and inhibition of E6 and E7 expression. Although E2 is necessary for the amplification of viral genomes, its accumulation may eventually restrict the duration of vegetative viral DNA replication by down-regulating the expression of E7.
Our comparison of the life cycles of 10 viruses reveals differences that can to some extent be explained by the divergence in virus transmission routes and infection sites. Key life cycle events, such as the onset of genome amplification and virus assembly, are likely to be similarly regulated in all papillomaviruses, and current theories of protein function effectively explain the expression patterns seen here. The major differences between different viruses appear to lie in the timing of events. As far as we are aware, this is the first comparative study of animal and human papillomavirus life cycles despite the extensive use of animal papillomaviruses to study infection in humans. BPV1 has been used most extensively to study the role of the viral transforming proteins because of its ability to replicate in fibroblasts in vitro and to transform them. Although studies of the BPV life cycle have been carried out (1, 68), it is not a convenient animal model system, and it is generally acknowledged that the results obtained from the study of BPV may not be directly applicable to our understanding of HPVs. CRPV, which, unlike BPV, is not a fibropapillomavirus, has been used more extensively as a model system for human infections. The timing of life cycle events in lesions caused by CRPV closely resembles that seen in human lesions caused by HPV11 and HPV16, and studies of the immune response to viral early proteins in CRPV are likely to be relevant to infection by human viruses (87). This should also be true for studies that use CRPV to investigate the mechanism of malignant progression (62, 106) and the roles of viral early proteins during cell proliferation and genome amplification (14, 104; Peh et al., 19th Int. Papillomavirus Conf.). The inability of CRPV to infect mucosal tissue and to go through its productive life cycle in laboratory animals limits its value as a model of productive genital infections. Of the mucosal models of human papillomavirus infection that are currently available, it appears that ROPV offers advantages over COPV. ROPV infects rabbits rather than dogs and, unlike COPV, can cause lesions at genital sites (90). The timing of events during the life cycle of ROPV is more similar to those of HPV11 and HPV16, and genome amplification in the basal layer does not occur. This may be important for vaccination studies. Vaccination of dogs using virus late proteins (95, 105) stimulates antibody production and partially protects the vaccinated animals from high-dose viral infection. A similar approach is being used to develop a prophylactic vaccine for the treatment of human genital lesions (85). Therapeutic-vaccination strategies however, aim to resolve existing infections and rely on the ability to generate a cell-mediated immune response to virus-infected cells. The site of infection (mucosal versus cutaneous) and the timing of events during the virus life cycle are likely to affect the outcome of such experiments. Similarities in the site of infection and the timing of life cycle events suggest that ROPV may be particularly appropriate for life cycle and prophylactic-vaccine studies. The longer duration of nonproductive infections, such as those caused by CRPV in domestic rabbits, offers advantages for the development of therapeutic vaccines and antiviral therapies.
Acknowledgments
Funding for this work was provided largely by the United Kingdom Medical Research Council and the Royal Society.
We thank John Skehel and Jonathan Stoye for advice and encouragement during the course of this work and Francoise Breitburd (Institut Pasteur, Paris, France) for the gift of the NZ264 serum.
REFERENCES
- 1.Alderborn, A., and S. Burnett. 1994. Regulation of DNA synthesis in division-arrested mouse C127 cells permissive for bovine papillomavirus DNA amplification. J. Virol. 68:4349-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashmole, I., P. H. Gallimore, and S. Roberts. 1998. Identification of conserved hydrophobic C-terminal residues of the human papillomavirus type 1 E1E4 protein necessary for E4 oligomerisation in vivo. Virology 240:221-231. [DOI] [PubMed] [Google Scholar]
- 3.Baker, C. C., and P. M. Howley. 1987. Differential promoter utilization by the bovine papillomavirus in transformed cells and in productively infected wart tissues. EMBO J. 6:1027-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrera-Oro, J. G., K. O. Smith, and J. L. Melnick. 1962. Quantitation of papova virus particles in human warts. J. Natl. Cancer Inst. 29:583-595. [PubMed] [Google Scholar]
- 5.Bell, J. A., J. P. Sundberg, S. J. Ghim, J. Newsome, A. B. Jenson, and R. Schlegel. 1994. A formalin-inactivated vaccine protects against mucosal papillomavirus infection: a canine model. Pathobiology 62:194-198. [DOI] [PubMed] [Google Scholar]
- 6.Bellanger, S., C. Demeret, S. Goyat, and F. Thierry. 2001. Stability of the human papillomavirus type 18 E2 protein is regulated by a proteasome degradation pathway through its amino-terminal transactivation domain. J. Virol. 75:7244-7251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bishop, P. E., A. McMillan, and S. Fletcher. 1990. An immunohistological study of spontaneous regression of condylomata acuminata. Genitourin. Med. 66:79-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonnez, W. 1998. Murine models of human papillomavirus-infected human xenografts. Papillomavirus Rep. 9:27-38. [Google Scholar]
- 9.Bonnez, W., R. C. Rose, C. da Rin, C. Borkhuis, K. L. de Mesy Jensen, and R. C. Reichman. 1993. Propagation of human papillomavirus type 11 in human xenografts using the severe combined immunodeficiency (SCID) mouse and comparison to the nude mouse model. Virology 197:455-458. [DOI] [PubMed] [Google Scholar]
- 10.Bouvard, V., G. Matlashewski, Z. M. Gu, A. Storey, and L. Banks. 1994. The human papillomavirus type 16 E5 gene cooperates with the E7 gene to stimulate proliferation of primary cells and increases viral gene expression. Virology 203:73-80. [DOI] [PubMed] [Google Scholar]
- 11.Brandsma, J. L. 1994. Animal models of human-papillomavirus-associated oncogenesis. Intervirology 37:189-200. [DOI] [PubMed] [Google Scholar]
- 12.Brandsma, J. L., and W. Xiao. 1993. Infectious virus replication in papillomas induced by molecularly cloned cottontail rabbit papillomavirus DNA. J. Virol. 67:567-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandsma, J. L., Z. Yang, S. W. Barthold, and E. A. Johnson. 1991. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc. Natl. Acad. Sci. USA 88:4816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brandsma, J. L., Z. H. Yang, D. DiMaio, S. W. Barthold, E. Johnson, and W. Xiao. 1992. The putative E5 open reading frame of cottontail rabbit papillomavirus is dispensable for papilloma formation in domestic rabbits. J. Virol. 66:6204-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitburd, F., and P. Coursaget. 1999. Human papillomavirus vaccines. Semin. Cancer Biol. 9:431-444. [DOI] [PubMed] [Google Scholar]
- 16.Breitburd, F., R. Kirnbauer, N. L. Hubbert, B. Nonnenmacher, C. Trin-Dinh-Desmarquet, G. Orth, J. T. Schiller, and D. R. Lowy. 1995. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 69:3959-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown, D. R., J. Bryan, M. Rodriguez, R. C. Rose, and D. G. Strike. 1991. Detection of human papillomavirus types 6 and 11 E4 gene products in condylomata acuminatum. J. Med. Virol. 34:20-28. [DOI] [PubMed] [Google Scholar]
- 18.Brown, D. R., J. T. Bryan, L. Pratt, V. Handy, K. H. Fife, and M. H. Stoler. 1995. Human papillomavirus type 11 E1Ê4 and L1 proteins colocalize in the mouse xenograft system at multiple time points. Virology 214:259-263. [DOI] [PubMed] [Google Scholar]
- 19.Brown, D. R., J. T. Bryan, M. Rodriguez, and B. P. Katz. 1992. Factors associated with detection of human papillomvirus E4 and L1 proteins in condylomata acuminata. J. Infect. Dis. 166:512-517. [DOI] [PubMed] [Google Scholar]
- 20.Brown, D. R., L. Fan, J. Jones, and J. Bryan. 1994. Colocalization of human papillomavirus type 11 E1Ê4 and L1 proteins in human foreskin implants grown in athymic mice. Virology 201:46-54. [DOI] [PubMed] [Google Scholar]
- 21.Bryan, J. T., K. H. Fife, and D. R. Brown. 1998. The intracellular expression pattern of the human papillomavirus type 11 E1Ê4 protein correlates with its ability to self associate. Virology 241:49-60. [DOI] [PubMed] [Google Scholar]
- 22.Cancer Research Campaign. 1995. Cancer—world perspectives. Factsheet 22.1. CRC, London, United Kingdom.
- 23.Chambers, V. C., and C. A. Evans. 1959. Canine oral papillomatosis. I. Virus assay and observations on the various stages of the experimental infection. Cancer Res. 19:1188-1195. [PubMed] [Google Scholar]
- 24.Chan, S.-Y., H. Delius, A. L. Halpern, and H.-U. Bernard. 1995. Analysis of genomic sequences of 95 papillomavirus types: Uniting typing, phylogeny and taxonomy. J. Virol. 69:3074-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chow, L. T., and T. R. Broker. 1994. Papillomavirus DNA replication. Intervirology 37:150-158. [DOI] [PubMed] [Google Scholar]
- 26.Chow, L. T., M. Nasseri, S. M. Wolinsky, and T. R. Broker. 1987. Human papillomavirus types 6 and 11 mRNAs from genital condylomata acuminata. J. Virol. 61:2581-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow, L. T., S. S. Reilly, T. R. Broker, and L. B. Taichman. 1987. Identification and mapping of human papillomavirus type 1 RNA transcripts recovered from plantar warts and infected epithelial cell cultures. J. Virol. 61:1913-1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christensen, N. D., N. M. Cladel, C. A. Reed, L. R. Budgeon, P. A. Welsh, S. D. Patrick, and J. W. Kreider. 1996. Laboratory production of infectious stocks of rabbit oral papillomavirus. [DOI] [PubMed]
- 29.Christensen, N. D., N. M. Cladel, C. A. Reed, and R. Han. 2000. Rabbit oral papillomavirus complete genome sequence and immunity following genital infection. Virology 269:451-461. [DOI] [PubMed] [Google Scholar]
- 30.Christensen, N. D., and J. W. Kreider. 1990. Antibody-mediated neutralization in vivo of infectious papillomavirus. J. Virol. 64:3151-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chua, K. L., and A. Hjerpe. 1996. Persistence of human papillomavirus (HPV) infections preceding cervical carcinoma. Cancer 77:121-127. [DOI] [PubMed] [Google Scholar]
- 32.Croissant, O., F. Breitburd, and G. Orth. 1985. Specificity of the cytopathic effect of cutaneous human papillomaviruses. Clin. Dermatol. 3:43-55. [DOI] [PubMed] [Google Scholar]
- 33.Defeo-Jones, D., G. A. Vuocolo, K. M. Haskell, M. G. Hanobik, D. M. Kiefer, E. M. McAvoy, M. Iveyhole, J. L. Brandsma, A. Oliff, and R. E. Jones. 1993. Papillomavirus E7 protein-binding to the retinoblastoma protein is not required for viral induction of warts. J. Virol. 67:716-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delius, H., M. A. van Ranst, A. B. Jenson, H. zur Hausen, and J. P. Sundberg. 1994. Canine oral papillomavirus genomic sequence: a unique 1.5Kb intervening sequence between the E2 and L2 open reading frames. Virology 204:447-452. [DOI] [PubMed] [Google Scholar]
- 35.Desaintes, C., and C. Demeret. 1996. Control of papillomavirus DNA replication and transcription. Semin. Cancer Biol. 7:339-347. [DOI] [PubMed] [Google Scholar]
- 36.Dollard, S. C., L. T. Chow, J. W. Kreider, T. R. Broker, N. L. Lill, and M. K. Howett. 1989. Characterization of an HPV type 11 isolate propagated in human foreskin implants in nude mice. Virology 171:294-297. [DOI] [PubMed]
- 37.Doorbar, J. 1998. Late stages of the papillomavirus life cycle. Papillomavirus Rep. 9:119-123. [Google Scholar]
- 38.Doorbar, J., I. Coneron, and P. H. Gallimore. 1989. Sequence divergence yet conserved physical characteristics among the E4 proteins of cutaneous human papillomaviruses. Virology 172:51-62. [DOI] [PubMed] [Google Scholar]
- 39.Doorbar, J., S. Ely, N. Coleman, M. Hibma, D. H. Davies, and L. Crawford. 1992. Epitope mapped monoclonal antibodies against the HPV16 E1Ê4 protein. Virology 187:353-359. [DOI] [PubMed] [Google Scholar]
- 40.Doorbar, J., S. Ely, J. Sterling, C. McLean, and L. Crawford. 1991. Specific interaction between HPV16 E1Ê4 and cytokeratins results in collapse of the epithelial cell intermediate filament network. Nature 352:824-827. [DOI] [PubMed] [Google Scholar]
- 41.Doorbar, J., H. S. Evans, I. Coneron, L. V. Crawford, and P. H. Gallimore. 1988. Analysis of HPV1 E4 gene expression using epitope-defined antibodies. EMBO J. 7:825-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doorbar, J., C. Foo, N. Coleman, E. Medcalf, O. Hartley, T. Prospero, S. Napthine, J. Sterling, G. Winter, and H. Griffin. 1997. Characterisation of events during the late stages of HPV16 infection in vivo using high affinity synthetic Fabs to E4. Virology 238:40-52. [DOI] [PubMed] [Google Scholar]
- 43.Doorbar, J., M. Medcalf, and S. Napthine. 1996. Analysis of HPV1 E4 complexes and their association with keratins in vivo. Virology 218:114-126. [DOI] [PubMed] [Google Scholar]
- 44.Doorbar, J., and G. Myers. 1996. The E4 protein, p. 58-80. In G. Myers, H. Delius, J. Icenogel, H.-U. Bernard, C. Baker, A. Halpern, and C. Wheeler (ed.), Human papillomaviruses 1996, vol. III. Los Alamos National Laboratory, Los Alamos, N. Mex.
- 45.Dostatni, N., M. Yaniv, O. Danos, and R. C. Mulligan. 1988. Use of retroviral vectors for mapping of splice sites in cottontail rabbit papillomavirus. J. Gen. Virol. 69:3093-3100. [DOI] [PubMed] [Google Scholar]
- 46.Egawa, K. 1994. New types of human papillomaviruses and intracytoplasmic inclusion bodies: a classification of inclusion warts according to clinical features, histology and associated HPV types. Br. J. Dermatol. 130:158-166. [DOI] [PubMed] [Google Scholar]
- 47.Egawa, K., Y. Honda, T. Ono, and H. Kitasato. 2000. A case of viral warts with particular fibrillar intracytoplasmic inclusion bodies. Dermatology 200:275-278. [DOI] [PubMed] [Google Scholar]
- 48.Egawa, K., Y. Inaba, K. Yoshimura, and T. Ono. 1993. Varied clinical morphology of HPV-1-induced warts, depending on anatomical factors. Br. J. Dermatol. 128:271-276. [DOI] [PubMed] [Google Scholar]
- 49.Frattini, M. G., and L. A. Laimins. 1994. Binding of the human papillomavirus E1 origin recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc. Natl. Acad. Sci. USA 91:12398-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frattini, M. G., H. B. Lim, J. Doorbar, and L. A. Laimins. 1997. Induction of human papillomavirus type 18 late gene expression and genomic amplification in organotypic cultures from transfected DNA templates. J. Virol. 71:7068-7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gross, G. E, and R. Barrasso (ed.). 1997. Human papillomavirus infection: a clinical atlas. Ullstein Mosby, Berlin, Germany.
- 52.Gross, G. E., S. Jablonska, and H. Hugel. 1997. Skin diagnosis. In G. E. Gross and R. Barrasso (ed.), Human papillomavirus infection: a clinical atlas. Ullstein Mosby, Berlin, Germany.
- 53.Han, R., N. M. Cladel, C. A. Reed, X. Peng, L. R. Budgeon, M. Pickel, and N. D. Christensen. 2000. DNA vaccination prevents and/or delays carcinoma development of papillomavirus-induced skin papillomas on rabbits. J. Virol. 74:9712-9716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han, R., N. M. Cladel, C. A. Reed, X. Peng, and N. D. Christensen. 1999. Protection of rabbits from viral challenge by gene gun-based intracutaneous vaccination with a combination of cottontail rabbit papillomavirus E1, E2, E6, and E7 genes. J. Virol. 73:7039-7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Harwood, C. A., and C. M. Proby. 2001. Human papillomavirus and immunosuppression, p. 102-117. In J. Sterling and S. K. Tyring (ed.), Human papillomaviruses. Arnold, London, United Kingdom.
- 57.Ho, G. Y., R. Bierman, L. Beardsley, C. J. Chang, and R. D. Burk. 1998. Natural history of cervicovaginal papillomavirus infection in young women. N. Engl. J. Med. 338:423-428. [DOI] [PubMed] [Google Scholar]
- 58.Howley, P. M., and D. R. Lowy. 2001. Papillomaviruses and their replication, p. 2197-2229. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 59.Inglis, S. C., and T. O'Neill. 2001. Vaccines, p. 131-138. In J. Sterling and S. K. Tyring (ed.), Human papillomaviruses. Arnold, London, United Kingdom.
- 60.Keating, J. T., T. Ince, and C. P. Crum. 2001. Surrogate biomarkers of HPV infection in cervical neoplasia screening and diagnosis. Adv. Anat. Pathol. 8:83-92. [DOI] [PubMed] [Google Scholar]
- 61.Klumpp, D. J., and L. A. Laimins. 1999. Differentiation-induced changes in promoter usage for transcripts encoding the human papillomavirus type 31 replication protein E1. Virology 257:239-246. [DOI] [PubMed] [Google Scholar]
- 62.Kreider, J. W., and G. L. Bartlett. 1981. The Shope papilloma-carcinoma complex of rabbits: a model system of neoplastic progression and spontaneous regression. Adv. Cancer Res. 35:81-110. [DOI] [PubMed] [Google Scholar]
- 63.Kreider, J. W., M. K. Howett, E. E. Leure-Dupree, R. J. Zaino, and J. A. Weber. 1987. Laboratory production in vivo of infectious human papillomavirus type 11 from condylomata accuminata. J. Virol. 61:590-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laurent, R., J. L. Kienzler, O. Croissant, and G. Orth. 1982. two anatomicoclinical types of warts with plantar localisation: specific cytopathogenic effects of papillomavirus type 1 (HPV1) and type 2 (HPV2). Arch. Dermatol. Res. 274:101-111. [DOI] [PubMed] [Google Scholar]
- 65.Leachman, S. A., R. E. Tigelaar, M. Shlyankevich, M. Slade, M. Irwin, E. Chang, T. C. Wu, W. Xiao, S. Pazhani, D. L. Zelterman, and J. L. Brandsma. 2000. GM-CSF priming plus papillomavirus E6 DNA vaccination: effects on papilloma formation and regression in the CRPV-rabbit model. J. Virol. 74:8700-8708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65a.Lowy, D. R., and P. M. Howley. 2001. Papillomaviruses, p. 2231-2264. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 66.Maitland, N. J., S. Conway, N. S. Wilkinson, J. Ramsdale, J. R. Morris, C. M. Sanders, J. E. Burns, P. L. Stern, and M. Wells. 1998. Expression patterns of the human papillomavirus type 16 transcription factor E2 in low- and high-grade cervical intraepithelial neoplasia. J. Pathol. 186:275-280. [DOI] [PubMed] [Google Scholar]
- 67.Masterson, P. J., M. A. Stanley, A. P. Lewis, and M. A. Romanos. 1998. A C-terminal helicase domain of the human papillomavirus E1 protein binds E2 and the DNA polymerase alpha-primase p68 subunit. J. Virol. 72:7407-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McBride, A. A., A. Dlugosz, and C. C. Baker. 2000. Production of infectious bovine papillomavirus from cloned viral DNA by using an organotypic raft/xenograft technique. Proc. Natl. Acad. Sci. USA 97:5534-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyers, C., J. Harry, Y. L. Lin, and F. O. Wettstein. 1992. Identification of three transforming proteins encoded by cottontail rabbit papillomavirus. J. Virol. 66:1655-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mitsuishi, T., M. Kawashima, K. Egawa, and S. T. 2001. Novel intracytoplasmic inclusion bodies in human papillomavirus-associated warts. Br. J. Dermatol. 145:171-173. [DOI] [PubMed] [Google Scholar]
- 71.Nasseri, M., R. Hirochika, T. R. Broker, and L. Chow. 1987. A human papillomavirus type 11 transcript encoding an E1Ê4 protein. Virology 159:433-439. [DOI] [PubMed] [Google Scholar]
- 72.Nasseri, M., C. Meyers, and F. O. Wettstein. 1989. Genetic analysis of CRPV pathogenesis: the L1 open reading frame is dispensable for cellular transformation but is required for papilloma formation. Virology 170:321-325. [DOI] [PubMed] [Google Scholar]
- 73.Nicholls, P., J. Doorbar, R. W. Moore, W. Peh, D. Anderson, and M. Stanley. 2001. Detection of Viral DNA and E4 protein in basal keratinocytes of experimental canine oral papillomavirus lesions. Virology 283:82-98. [DOI] [PubMed] [Google Scholar]
- 74.Nicholls, P., B. Klaunberg, R. A. Moore, E. B. Santos, N. R. Parry, G. W. Gough, and M. A. Stanley. 1999. Naturally occurring, nonregressing canine oral papillomavirus infection: host immunity, virus characterization, and experimental infection. Virology 265:365-374. [DOI] [PubMed] [Google Scholar]
- 75.Nicholls, P. K., P. F. Moore, D. M. Anderson, R. A. Moore, N. R. Parry, G. W. Gough, and M. A. Stanley. 2001. Regression of canine oral papillomas is associated with infiltration of CD4+ and CD8+ lymphocytes. Virology 283:31-39. [DOI] [PubMed] [Google Scholar]
- 76.Oriel, J. D. 1971. Natural History of genital warts. Br. J. Vener. Dis. 47:1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ozbun, M. A., and C. Meyers. 1997. Characterisation of late gene transcripts expressed during vegetative replication of human papillomavirus type 31b. J. Virol. 71:5161-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ozbun, M. A., and C. Meyers. 1998. Human papillomavirus type 31b E1 and E2 transcript expression correlates with vegetative viral genome amplification. Virology 248:218-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Penrose, K. J., and A. A. McBride. 2000. Proteasome-mediated degradation of the papillomavirus E2-TA protein is regulated by phosphorylation and can modulate viral genome copy number. J. Virol. 74:6031-6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pfister, H., and H. zur Hausen. 1978. Seroepidemiological studies of human papillomavirus (HPV1) infections. Int. J. Cancer 21:161-165. [DOI] [PubMed] [Google Scholar]
- 81.Pray, T. R., and L. A. Laimins. 1995. Differentiation-dependent expression of E1Ê4 proteins in cell lines maintaining episomes of human papillomavirus type 31b. Virology 206:679-685. [DOI] [PubMed] [Google Scholar]
- 82.Rogel-Gaillard, C., F. Breitburd, and G. Orth. 1992. Human papillomavirus type 1 E4 proteins differing by their N-terminal ends have distinct cellular locations when transiently expressed in vitro. J. Virol. 66:816-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rogel-Gaillard, C., G. Pehau-Arnaudet, F. Breitburd, and G. Orth. 1993. Cytopathic effect in human papillomavirus type 1-induced inclusion warts: in vitro analysis of the contribution of two forms of the viral E4 protein. J. Investig. Dermatol. 101:843-851. [DOI] [PubMed] [Google Scholar]
- 84.Ruesch, M. N., F. Stubenrauch, and L. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schiller, J. T., and D. R. Lowy. 2001. Papillomavirus-like particle based vaccines: cervical cancer and beyond. Expert Opin. Biol. Ther. 1:571-581. [DOI] [PubMed] [Google Scholar]
- 86.Schwartz, S. 2000. Regulation of human papillomavirus late gene expression. Ups. J. Med. Sci. 105:171-192. [PubMed] [Google Scholar]
- 87.Selvakumar, R., L. A. Borenstein, Y. L. Lin, R. Ahmed, and F. O. Wettstein. 1995. Immunization with nonstructural proteins E1 and E2 of cottontail rabbit papillomavirus stimulates regression of virus-induced papillomas. J. Virol. 69:602-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shah, K. V., and P. M. Howley. 1996. Papillomaviruses, p. 2077-2109. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Virology, 3rd ed., vol. 2. Lippincott-Raven, Philadelphia, Pa..
- 89.Stanley, M. 1998. The immunology of genital human papilloma virus infection. Eur. J. Dermatol. 8:8-12. [PubMed] [Google Scholar]
- 90.Stanley, M. A., P. J. Masterson, and P. K. Nicholls. 1997. In vitro and animal models for antiviral therapy in papillomavirus infections. Antivir. Chem. Chemother. 8:381-400. [Google Scholar]
- 91.Stanley, M. A., R. A. Moore, P. K. Nicholls, E. B. Santos, L. Thomsen, N. Parry, S. Walcott, and G. Gough. 2001. Intra-epithelial vaccination with COPV L1 DNA by particle-mediated DNA delivery protects against mucosal challenge with infectious COPV in beagle dogs. Vaccine 19:2783-2792. [DOI] [PubMed] [Google Scholar]
- 92.Stern, P. L., M. Brown, S. N. Stacey, H. C. Kitchener, I. Hampson, E. S. Abdel-Hady, and J. V. Moore. 2000. Natural HPV immunity and vaccination strategies. J. Clin. Virol. 19:57-66. [DOI] [PubMed] [Google Scholar]
- 93.Straight, S., P. M. Hinckle, R. J. Jewers, and D. J. McCance. 1993. The E5 oncoprotein of HPV16 transforms fibroblasts and effects the downregulation of the EGF receptor in keratinocytes. J. Virol. 69:4521-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sundaram, P., R. E. Tigelaar, and J. L. Brandsma. 1997. Intracutaneous vaccination of rabbits with the cottontail rabbit papillomavirus (CRPV) L1 gene protects against virus challenge. Vaccine 15:664-671. [DOI] [PubMed] [Google Scholar]
- 95.Suzich, J. A., S. J. Ghim, F. J. Palmer-Hill, W. I. White, J. K. Tamura, J. A. Bell, J. A. Newsome, A. B. Jenson, and R. Schlegel. 1995. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc. Natl. Acad. Sci. USA 92:11553-11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Syrjanen, K. 1998. Anogenital human papilloma virus and the problem of persistence. Eur. J. Dermatol. 8:5-7. [PubMed] [Google Scholar]
- 97.Tokita, H., and S. Konishi. 1975. Studies on canine oral papillomatosis. II. Oncogenicity of canine oral papillomavirus to various tissues of dog with special reference to eye tumour. Jpn. J. Vet. Sci. 37:109-120. [DOI] [PubMed] [Google Scholar]
- 98.Valle, G. F., and L. Banks. 1995. The human papillomavirus (HPV)-6 and HPV-16 E5 proteins co-operate with HPV-16 E7 in the transformation of primary rodent cells. J. Gen. Virol. 76:1239-1245. [DOI] [PubMed] [Google Scholar]
- 99.van Belkum, A., L. Juffermans, L. Schrauwen, G. van Doornum, M. Burger, and W. Quint. 1995. Genotyping human papillomavirus type 16 isolates from persistently infected promiscuous individuals and cervical neoplasia patients. J. Clin. Microbiol. 33:2957-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Venuti, A., D. Salani, F. Poggiali, V. Manni, and A. Bagnato. 1998. The E5 oncoprotein of human papillomavirus type 16 enhances endothelin-1-induced keratinocyte growth. Virology 248:1-5. [DOI] [PubMed] [Google Scholar]
- 101.Wallin, K. L., F. Wiklund, T. Angstrom, F. Bergman, U. Stendahl, G. Wadell, G. Hallmans, and J. Dillner. 1999. Type-specific persistence of human papillomavirus DNA before the development of invasive cervical cancer. N. Engl. J. Med. 341:1633-1638. [DOI] [PubMed] [Google Scholar]
- 102.Wettstein, F. O., M. S. Barbosa, and M. Nasseri. 1987. Identification of the major cottontail rabbit papillomavirus late RNA cap site and mapping and quantitation of an E2 and minor E6 coding mRNA in papillomas and carcinomas. Virology 159:321-328. [DOI] [PubMed] [Google Scholar]
- 103.Wright, T. C., R. J. Kurman, and A. Ferenczy. 1994. Precancerous lesions of the cervix, p. 229-277. In R. J. Kurman (ed.), Blaustein's pathology of the female genital tract. Springer-Verlag, New York, N.Y.
- 104.Wu, X., W. Xiao, and J. L. Brandsma. 1994. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J. Virol. 68:6097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yuan, H., P. A. Estes, Y. Chen, J. Newsome, V. A. Olcese, R. L. Garcea, and R. Schlegel. 2001. Immunization with a pentameric L1 fusion protein protects against papillomavirus infection. J. Virol. 75:7848-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zeltner, R., L. A. Borenstein, F. O. Wettstein, and T. Iftner. 1994. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J. Virol. 68:3620-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang, P., M. Nouri, J. Brandsma, T. Iftner, and B. M. Steinberg. 1999. Induction of E6/E7 Expression in cottontail rabbit papillomavirus latency following UV activation. Virology 263:388-394. [DOI] [PubMed] [Google Scholar]
- 108.Zhu, Q.-L., T. F. Smith, E. J. Lefkowitz, L. T. Chow, and T. R. Broker. 1994. Nucleic acid and protein sequence alignments of human and animal papillomaviruses constrained by functional sites. University of Alabama at Birmingham Press, Birmingham.