Abstract
Analysis of the nucleotide sequence of the genome of the lactococcal bacteriophage r1t showed that it may encode at least two proteins involved in DNA replication. On the basis of its similarity with the G38P protein encoded by the Bacillus subtilis phage SPP1, the product of orf11 (Pro11) is thought to be involved in the initiation of phage DNA replication. This protein was overexpressed in Lactococcus lactis and partially purified. Gel retardation analysis using various r1t DNA fragments indicates that Pro11 specifically binds to a sequence located within its cognate gene. DNase I footprinting showed that Pro11 protects a stretch of DNA of 47 bp. This region spans four 6-bp short direct repeats, which suggests that the region contains four binding sites for Pro11. 1,10-Phenanthroline-copper footprinting confirmed the protection of the hexamers. An asymmetric protection pattern of each strand was observed, suggesting that Pro11 contacts each DNA strand separately at contiguous hexamers. We propose a model for the binding of Pro11 to its target sites that may account for the torsion strain required for strand opening at the origin of replication.
Lactococcus lactis is a gram-positive obligately fermentative bacterium belonging to the group of lactic acid bacteria. Due to its simple metabolism and the availability of advanced genetic tools, it has been used in numerous basic studies. L. lactis is extremely important in the dairy, and therefore, its bacteriophages have received considerable attention. These studies have shown that many strains of L. lactis are lysogenic (18) and are a likely source of bacteriophages in fermentation processes. Research on lactococcal phages has been generally focused on the development of phage resistance mechanisms enabling the host strains to overcome an infection (for a review, see reference 12). In recent years the study of the molecular genetics of lactococcal phages has also received considerable attention. The complete nucleotide sequences of several lactococcal phage genomes are currently available. Moreover, the regulatory switches as well as the integrase genes and the lytic cassettes of a number of phages have been thoroughly studied.
Notwithstanding these advances, little is known about the molecular events leading to the replication of lactococcal phage DNA, a critical step in the phage life cycle and an obvious target for the development of phage resistance strategies. Several origins of replication from lactococcal phages have been isolated on the basis of their ability, when cloned in a high-copy-number vector, to confer enhanced resistance on the host cell against the phage from which the origin was isolated (15, 24, 25, 27, 30). This Per (phage-encoded resistance) phenotype is characterized by a low rate of phage DNA replication. An origin of replication from the prolate phage c2 was able to drive DNA replication in L. lactis via a theta, and another as-yet-uncharacterized mechanism (6, 44). Noticeably, no phage-encoded protein was required to initiate DNA replication.
Phage r1t is a small isometric lysogenic bacteriophage from L. lactis subsp. cremoris R1 (14, 23) that belongs to the lactococcal phage species P335 (19). Phage r1t possesses a double-stranded linear DNA genome with cohesive termini, and its complete sequence has been previously determined (43). The r1t genome is composed of 33,350 bp, and 50 open reading frames (ORFs) have been identified. On the basis of sequence comparisons, at least two of these ORFs are probably involved in the replication of the phage DNA. orf11 encodes a protein (Pro11) with significant similarity to G38P of the Bacillus subtilis bacteriophage SPP1 (31) and Rep2009 of the lactococcal phage Tuc2009 (27). G38P has been shown to be essential for initiation of replication of phage DNA. It acts as a functional analog of DnaA (29). Rep2009 has been shown to bind specifically to a DNA region within its cognate gene containing a set of direct repeats (27). Several direct repeats within the coding sequence of orf11 share significant structural similarity to theta-type iteron-containing origins of replication (8). The product of orf12 shares significant similarity with DnaC of Escherichia coli. DnaC recruits the essential helicase DnaB and loads it in the preprimosome complex formed by DnaA and the origin of replication (4). DnaC is subsequently released from the complex, whereupon DnaB is activated. The arrangement of the Pro11 and Pro12 genes is similar to that found in phage λ, which encodes a DnaA analog (protein O) and a DnaC analog (P). Among phage-encoded proteins, only O and P are required for λ DNA replication (for a review, see reference 40).
In order to determine the possible role of the orf11 gene product in phage DNA replication, the protein was overproduced and partially purified, and its activity was studied. Results obtained show that Pro11 is a DNA-binding protein that specifically binds to a set of direct repeats located within its own coding sequence. Occupancy of all of the binding sites leads to a major conformational change of the complex. We propose a model for this complex that may account for the driving force required for the melting of the r1t origin of replication.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. L. lactis subsp. lactis strains were grown at 30°C in twofold-diluted M17 broth (41) supplemented with 0.5% glucose (G[1/2]M17). L. lactis subsp. cremoris was grown in M17 broth supplemented with 0.5% lactose (LM17). Erythromycin and chloramphenicol were used at final concentrations of 5 and 10 μg/ml for L. lactis subsp. lactis and at 2.5 and 5 μg/ml for L. lactis subsp. cremoris strains. E. coli was grown in TY broth (33) at 37°C with aeration. Chloramphenicol, ampicillin, and erythromycin were used at final concentrations of 10, 100, and 100 μg/ml for E. coli, respectively.
TABLE 1.
Strains and plasmids used in this study
| Species or plasmid name | Strain | Relevant property(ies) | Reference or source |
|---|---|---|---|
| L. lactis subsp. cremoris | R1 | rlt host strain | 23 |
| R1K10 | Phage-cured derivative of R1 | 43 | |
| MG1363 | Plasmid-free derivative of NCDO712 | 13 | |
| NZ9000 | Plasmid-free MG1363 derivative harboring nisK and nisR in the chromosome | 9 | |
| NZ9400 | Nisin producer | 9 | |
| E. coli | C41 | Mutant of E. coli BL21 with enhanced tolerance for toxic proteins | 28 |
| XL1Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB laclqZΔM15 Tn10 (Tetr)] | Stratagene | |
| pET32a | AmprE. coli expression vector containing the T7lac promoter | Novagen Inc. | |
| pETorf11 | pET32a containing orf11 cloned in frame with a thioredoredoxin moiety and a His tag | This work | |
| pNG8048 | Cmr EmrL. lactis expression vector containing the nisA promoter | Laboratory collection | |
| pNG11 | Cmr; pNG8048 containing orf11 | This work |
Molecular cloning, transformation, and nucleotide sequencing and analysis.
DNA techniques were performed essentially as described by Sambrook et al. (34). Enzymes were purchased from Roche Molecular Biochemicals (Indianapolis, Ind.) and were used according to the instructions of the supplier. DNA was isolated as described before (2) or with the plasmid miniprep isolation kit of Roche Molecular Biochemicals. Total DNA of L. lactis was obtained as described before (22).
Electrotransformation of L. lactis was performed as described earlier (17). E. coli was transformed as described by Mandel and Higa (26). Nucleotide sequences were determined using a Vistra DNA Labstation 625 in combination with a Vistra DNA sequencer 725 (Amersham International, Little Chalfont, United Kingdom). Similarity searches were performed using the Blastp algorithm provided by the National Center for Biotechnology Information. Amino acid sequences were aligned using CLUSTAL W (42).
Construction of plasmids.
To overexpress Pro11 in E. coli under control of the T7lac promoter, NcoI and SacI restriction enzyme sites were introduced upstream and downstream of orf11, respectively, by PCR using the primers ORF11-F2 and ORF11-R1 (Table 2) and Pwo polymerase (Roche Molecular Biochemicals). The PCR product was digested with NcoI and SalI and cloned in pET32a digested with the same enzymes, resulting in plasmid pET-ORF11. The introduction of the NcoI site changed the GTG start codon present in orf11 into ATG. The protein was overexpressed as a fusion with a thioredoxin moiety and an internal histidine tag. To overexpress Pro11 in L. lactis under control of the nisA promoter, plasmid pET-ORF11 was digested with NcoI and SalI. The fragment spanning orf11 was isolated from an agarose gel using the QIAquick gel vextraction kit (Qiagen GmbH, Hilden, Germany) and ligated to pNG8048 digested with the same enzymes. This resulted in the plasmid pNG11.
TABLE 2.
Primers used in this study
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| ORF11F2 | AGGAGTATCCATGGCACAAAGAAG |
| ORF11F3 | GATCGATATAAAGCTAG |
| ORF11F4 | CATATTCTGAAATTCTTG |
| ORF11F5 | TGGTCTACTTCATAATTG |
| ORF11F6 | TGAAGAATTTGGAACCGAAATGG |
| ORF11F7 | AATTTGAAATGGAACCGAAACGG |
| ORF11F8 | AATTTGAAACGGAACCAACTTGG |
| ORF11F9 | AATTTAACTTGGAACCTCAGATAAG |
| ORF11F10 | AATTTGAAACGGAACCAACTTAACTTGGAACCTCAGATAAG |
| ORF11F11 | AATTTGAAACGGAACCAACTTAACTTCAACTTGGAACCTCAGATAAG |
| ORF11R1 | AGGAGTATCCATGGCACAAAGAAG |
| ORF11R2 | CAAGAATTTCAGAATATG |
| ORF11R3 | CTAGCTTTATATCGATC |
The hexamers are indicated in boldface type.
Purification of Pro11 fusion protein and production of antibodies against Pro11.
E. coli C41(pET-ORF11) was grown at 37°C with vigorous shaking in TY broth with ampicillin (100 μg/ml) until an optical density at 600 nm of 0.6 was reached. Then, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to 0.1 mM and incubation was continued for 2 h. All subsequent operations were performed at 4°C. The culture was centrifuged, and the cells were resuspended in TNM buffer (50 mM Tris HCl [pH 8.0], 200 mM NaCl, 5 mM MgCl2). Cells were lysed by several passages through a French pressure cell, and the lysate was centrifuged at 12,000 × g for 20 min. The supernatant was removed, and the pellet was resuspended in TNM buffer containing 5% Triton X-100. The suspension was centrifuged at 30,000 rpm in an SW50 rotor (Beckman Coulter Inc., Fullerton, Calif.) for 30 min. The pellet was resuspended in TNM buffer containing 8 M urea (TNMU) and centrifuged under the same conditions. The supernatant was applied to a Talon spin column (Clontech, Palo Alto, Calif.). After passage of the sample, the column was washed with 1 volume of TNMU and the protein was eluted with 1.5 volumes of TNMU containing 100 mM imidazole. The eluant was dialyzed against PBS buffer (34). At this stage the protein aggregated. The suspension was recovered from the dialysis bag and centrifuged, and the pellet was used to raise polyclonal antibodies in New Zealand White rabbits (Eurogentec Bel SA, Seraing, Belgium). The specificity of the antiserum was demonstrated by Western hybridization using crude extracts of E. coli and L. lactis and purified Pro11.
Partial purification of Pro11.
L. lactis NZ9000 (pNG11) was grown in G[1/2]M17 at 30°C until an optical density at 600 nm of 0.6 was reached. Then, 1/1,000 volume of supernatant from an overnight culture of L. lactis NZ9400 (a nisin producer) was added and incubation was continued for 2 h. Cells were centrifuged, resuspended in TNM buffer, and lysed by several passages through a French pressure cell. The lysate was centrifuged at 12,000 × g for 20 min. The supernatant was removed, and the pellet was resuspended in TNM buffer and centrifuged under the same conditions. This washing step was performed three times, with increasing concentrations of NaCl (250 mM, 500 mM, and 1 M, respectively). The pellet was resuspended in TNM containing 5% Triton X-100 and centrifuged at 30,000 rpm in an SW50 rotor (Beckman Coulter Inc.) for 30 min. The pellet was resuspended in TNMU and centrifuged under the same conditions. The supernatant was dialysed against TNM containing 4 M urea and then against several changes of 25 mM sodium phosphate buffer (pH 7.0) with 5 mM MgCl2, each wash containing increasing concentrations of NaCl and decreasing concentrations of urea (500 mM NaCl, 2 M urea; 750 mM NaCl, 1 M urea; 750 mM NaCl, 0.5 M urea; 1 M NaCl, 0 M urea). The solution was transferred to a 2-ml Eppendorf tube and centrifuged in a bench-top centrifuge at maximum speed for 15 min at 4°C. One volume of 80% sterile glycerol was added to the supernatant, which was subsequently divided into 100-μl aliquots and stored at −20°C. This preparation was used for the DNA-binding assays described below.
Preparation of cell extracts.
Cells from a 100-ml culture were harvested at the late exponential growth phase, unless otherwise stated, and washed twice with 1 volume of 0.9% NaCl. The pellet was resuspended in 700 μl of sample buffer (50 mM Tris HCl [pH 8.0], 0.3% sodium dodecyl sulfate [SDS], 20 mM dithiothreitol [DTT]), and 7 μl of Prefabloc (Roche Molecular Biochemicals) was added. The cell suspension was mixed with 1 g of glass beads (100-μm diameter) and lysed in a BeadBeater (Biospec Products, Bartlesville, Okla.) for 8 min at 4°C. The mixture was centrifuged at 12,000 × g for 5 min, and the supernatant was recovered.
Protein electrophoresis and Western hybridization.
Samples were prepared for SDS-polyacrylamide gel electrophoresis (SDS-PAGE) by diluting aliquots of the suspensions twice in 2× sample buffer (20) containing 2.5% SDS and subsequent boiling for 5 min. SDS-PAGE was carried out as described earlier (20). SDS-PAGE gels were stained with Coomassie brilliant blue. The concentration of protein was determined by the method of Bradford (3). Proteins were electroblotted onto polyvinylidene difluoride membranes (Schleicher & Schuell, Dassel, Germany) according to standard protocols (34). Monoclonal antibodies against six-His tag were purchased from Clontech. Monoclonal horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were obtained from Amersham International and detected using the ECL detection kit (Amersham International).
Gel retardation analysis.
PCR-generated DNA probes were end labeled with [γ-32P]ATP using T4 polynucleotide kinase. The specific activities of the probes used in this study were similar. Unless otherwise stated, assays were performed in binding buffer [20 mM bis-Tris HCl at pH 6.5, 250 mM NaCl, 5 mM MgCl2, 0.5 mM DTT, 10% glycerol, 0.05 mg/ml poly(I-C), bovine serum albumin (0.05 mg/ml)]. Binding reactions were performed using 5,000 cpm of radiolabeled probe. Since the actual amount of active protein after renaturation is unknown, protein amounts are given in relative units (RU) throughout this study. 1 RU corresponds to 0.8 μg of protein. After 20 min of incubation at 30°C the samples were loaded on a nondenaturing 4% polyacrylamide gel. Gels were run in either TAE buffer (40 mM Tris-acetate, pH 8; 2 mM EDTA) or Tris-glycine buffer (50 mM Tris, 0.38 M glycine, 2 mM EDTA) at 100 V, dried, and subjected to autoradiography. For detection of protein-DNA complexes by Western hybridization, PAA gels were incubated in 1% SDS for 15 min and washed several times with water. Transfer of proteins to polyvinylidene difluoride membranes and subsequent steps were performed as described above. To study the inhibition by specific DNA-groove binding drugs, probes were incubated in binding buffer with the indicated concentrations of actinomycin D, chromomycin A3, distamycin A, or methyl-green for 20 min at room temperature prior to incubation with Pro11. For assays with methyl-green, DTT was omitted from the reaction buffer since it may interfere with the binding of the drug to DNA (Leendert Hamoen, personal communication). After the addition of the protein, the complexes were allowed to form for 30 min at room temperature.
DNase I footprinting.
DNase I footprinting was performed following the description supplied with the Sure Track Footprinting kit (Pharmacia, Uppsala, Sweden). The DNA probes were obtained by PCR using primers which were end labeled with T4 polynucleotide kinase using [γ-32P]ATP. Binding reactions were performed as described for the gel retardation experiments except that the NaCl concentration was 125 mM, in a total volume of 40 μl, and that 50,000 cpm of radiolabeled probe was used. In order to determine whether Pro11 would aggregate under our experimental conditions, a mock assay was performed in which the concentration of protein remaining in the supernatant after centrifugation of the mixture was determined for the highest concentration used in these assays (8 RU, corresponding to 160 μg/ml). No significant differences were observed between the expected concentration and the actual measurements. Therefore, we concluded that no significant aggregation occurred. Complexes were allowed to form for 20 min at room temperature, after which 10 μl of 10 mM CaCl2 and 0.1 U of DNase I were added. Digestion was allowed to proceed for 1 min, after which the reactions were terminated by adding 140 μl of stop solution (192 mM Na-acetate, 32 mM EDTA, 0.14% SDS, yeast tRNA [64 μg/ml]). Samples were extracted twice with phenol-chloroform, and the DNase I digestion products were precipitated with ethanol. The precipitates were resuspended in 6 μl of loading buffer and loaded on a 6% polyacrylamide-urea gel. A+G Maxam and Gilbert reactions were run on the same gel to locate sequence positions and protected regions (34).
Phenanthroline-copper footprinting.
Binding reactions were performed as described above for the DNase I footprinting. Phenanthroline-copper footprinting was performed in solution as described by Sigman et al. (37) with some modifications. Four microliters of 2 mM 1,10-phenanthroline-0.46 mM CuSO4 and 4 μl of 58 mM 3-mercaptopropinoic acid were added to 40 μl of Pro11/orf11F5R2 mixture and incubated for 10 min at 37°C. The reaction was quenched by adding 4 μl of 28 mM 2,9-dimethyl-phenanthroline. To precipitate DNA, 120 μl of water, 10 μl of 5 M NaCl, and 1 μl of yeast tRNA (10 mg/ml) were added, and the total was mixed with 3 volumes of cold ethanol. Samples were incubated at −20°C for 2 h, centrifuged, washed with 200 μl of ethanol, air dried, and dissolved in 6 μl of loading buffer. Analysis of the digestion products was performed as described above for the DNase I footprinting assays.
RESULTS
Partial purification of Pro11.
From the analysis of the DNA sequence of the bacteriophage r1t it was inferred that orf11 could encode a replisome organizer. This supposition comes from both comparisons of the structural organization of the genomes of other temperate bacteriophages (Fig. 1) and sequence similarities between the product of orf11 and the previously characterized replisome organizer protein G38P of the B. subtilis bacteriophage SPP1.
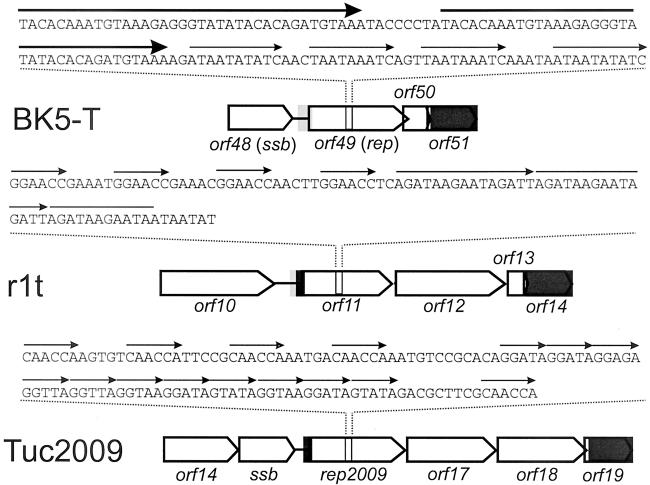
FIG. 1.
Structure of the replication modules of three lactococcal bacteriophages. ORFs are indicated as block arrows. Grey rectangles indicate DNA regions with >90% sequence identity. Thin arrows above the nucleotide sequence indicate direct repeats.
In order to obtain a relatively pure fraction of Pro11 protein for protein-DNA interaction studies, the gene orf11 was cloned in pNG8048 to overexpress the protein in L. lactis NZ9000. Upon induction with nisin, Pro11 could be readily detected by SDS-PAGE and Western hybridization. Fractionation studies showed that Pro11 was mostly present in the insoluble fraction. The protein could only be dissolved in 8 M urea, resulting in a fraction in which Pro11 accounted for approximately 95% of the total protein (results not shown). In order to renature the protein, the sample was dialyzed stepwise against buffers containing decreasing amounts of urea and increasing amounts of salt. The protein was finally obtained in a buffer containing 1 M NaCl. Although further lowering of the salt concentration resulted in aggregation of Pro11, addition of 1 volume of 80% glycerol was possible (final NaCl concentration, 0.5 M). At a low NaCl concentration (100 mM) with or without glycerol, protein concentrations higher than 40 to 50 μg/ml resulted in protein aggregation.
Pro11 binds specifically to its own coding sequence.
If Pro11 is the replisome organizer of the bacteriophage r1t replication apparatus the protein should bind to the putative origin of replication identified in the orf11 gene. In order to test this possibility, gel retardation experiments were performed using a PCR fragment spanning the entire orf11 gene and several nonspecific DNA fragments. Pro11 was able to bind to its own gene but did not bind to an unspecific fragment. Several partial fragments of orf11 were produced by PCR and tested in gel retardation assays (Fig. 2A). Only those fragments spanning the putative origin of r1t replication were retarded, further demonstrating the specificity of the binding reaction. Similar results were obtained when the 184-bp fragment orf11F3R2 was used (data not shown, but see Fig. 3). Therefore, the binding site for Pro11 should lie within this nucleotide sequence of 184 bp.
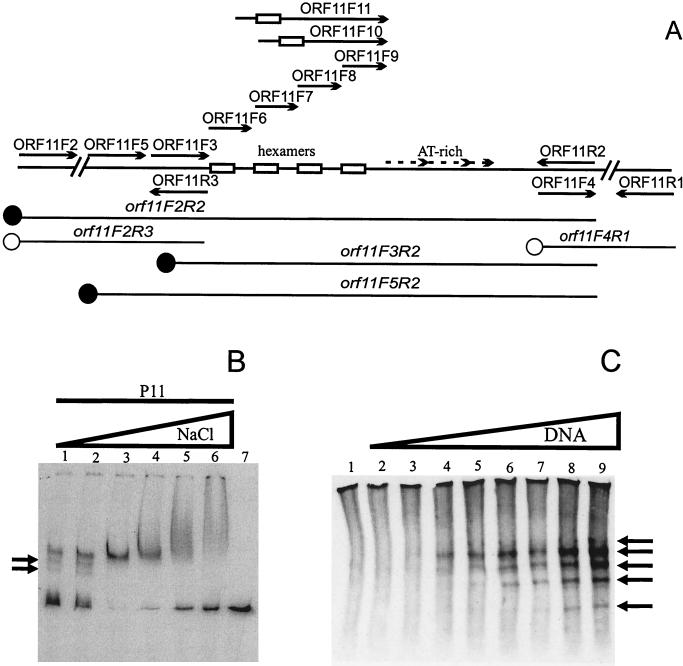
FIG. 2.
Pro11 binding to orf11 DNA. (A) Schematic representation of the different probes used in band mobility shift assays and the different primers used to obtain the DNA probes for the experiments presented in this study. Hexamers and A+T-rich repeats are indicated. The position of the first hexamer in primers ORF11F10 and ORF11F11 is likewise indicated. Solid circles represent DNA fragments retarded by Pro11; empty circles indicate DNA probes not bound by Pro11. (B) Effect of NaCl concentration on Pro11 binding of the probe orf11F2R2. Reactions were performed as described in Materials and Methods except that the NaCl concentration was varied as indicated. The gel was run in Tris-glycine buffer. Lanes: 1, 25 mM NaCl; 2, 125 mM; 3, 250 mM; 4, 500 mM; 5, 750 mM; 6, 1 M; 7, free probe (at 500 mM NaCl). Arrows indicate additional retarded bands detected at low NaCl concentrations. (C) Gel mobility shift assay using increasing amounts of DNA. Complexes (indicated by arrows) were detected by Western blotting using rabbit polyclonal antibodies raised against His-tagged Pro11. The standard binding buffer was used (NaCl: 25 mM; 1 RU of Pro11; probe: orf11F3R2). The gel was run in Tris-glycine buffer. Lanes, 1, no probe DNA; 2, 0.01 μg of probe; 3, 0.05 μg; 4, 0.1 μg; 5, 0.5 μg; 6, 1 μg; 7, 2.5 μg; 8, 5 μg; 9, 10 μg.
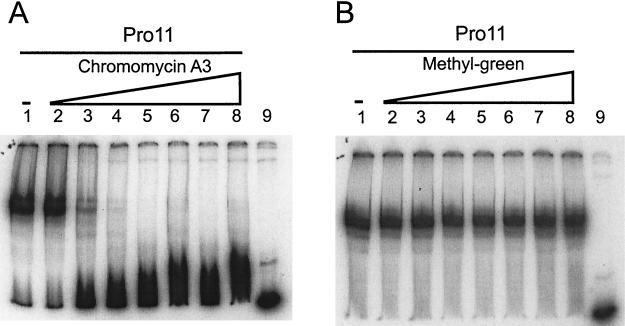
FIG. 3.
Effects in gel mobility shift assays of different DNA-binding drugs on Pro11 binding to the DNA fragment orf11F3R2 (Fig. 2A). (A) Chromomycin A3. Lanes: 2 to 8, 0.5, 1, 2.5, 5, 12.5, 50, and 75 μM, respectively. (B) Methyl-green. Lanes 2 to 8, 1, 2, 5, 10, 25, 50, 100, and 125 μM, respectively. (A and B) Lanes 1, no drug; lanes 9, free probe (no protein or drug added).
When TAE buffer was used during electrophoresis, the retarded complexes hardly entered the gel. A high-ionic-strength Tris-glycine buffer allowed complexes to enter the gel and was used for further assays.
In order to optimize the binding reaction, the effects of several variables were studied. No significant differences were observed over a range of pH values and different buffering compounds employed, using four different buffers: 20 mM bis-Tris HCl (pH 6.5), 20 mM sodium phosphate (pH 7.0), 20 mM HEPES (pH 7.5), and 20 mM Tris HCl (pH 8.0). As less label was retained in the well and a sharper band was observed in the gel with bis-Tris (pH 6.5), this buffer was used throughout the rest of the study.
The effect on the binding reaction of divalent cations such as Ca2+ and Mg2+ was subsequently studied. Increasing the CaCl2 concentration had a moderate inhibitory effect on the binding of Pro11 as judged from the increasing signal of free probe. Moreover, at concentrations ranging from 10 to 50 mM (the highest concentration assayed) the band corresponding to the retarded complex became less sharp and more signal was observed in the well. The effect of MgCl2 was different: concentrations of up to 5 mM favored the binding of Pro11, while higher concentrations were slightly inhibitory. Only some smearing of the retarded band was observed at 100 mM MgCl2 (data not shown).
NaCl concentration had a major effect on Pro11 binding under our experimental conditions (Fig. 2B). Optimal binding was observed at 250 mM of NaCl, a concentration that was used in all further assays unless otherwise noted. Higher concentrations resulted in a less effective binding, while the retarded band became less sharp. At 1 M NaCl, a smear from the well to the expected position of the band was observed, suggesting that additional subunits of Pro11 bind to the complex under these conditions. Pro11 binding was also less efficient at NaCl concentrations lower than 250 mM. Additional retarded bands were detected under these conditions, suggesting that more than one Pro11 binding site is present in the orf11F2R2 DNA fragment. The band corresponding to the free probe was less sharp, indicating that some dissociation of Pro11 occurred during electrophoresis.
In order to investigate the latter point further, gel retardation was done using decreasing amounts of protein. At low protein concentrations, additional retarded bands are observed using the DNA fragment orf11F2R2. Moreover, these bands appear both above and below the single band observed at high protein concentrations. Similar results were obtained with DNA fragments orf11F3R1 or orf11F3R2 (data not shown). As the number of different complexes could not be estimated under the conditions used in these assays, gel retardation assays were performed using a great excess of DNA. Unlabeled DNA fragment orf11F3R2 was used in Western hybridization to detect Pro11-DNA complexes (Fig. 2C). Without the DNA fragment, Pro11 does not enter the gel, although a smear throughout the lane was observed. After incubation of Pro11 with the DNA fragment, five distinct bands were detected upon electrophoresis. One band migrated slower than the major band and disappeared as the amount of DNA became limiting. These observations are in agreement with the results previously obtained using labeled probe. The results show that Pro11 is present in the complexes, and that the DNA binding activity is not caused by a contaminant protein in the sample.
Pro11 binds through the minor groove.
The effects of several major- and minor-groove-specific binding ligands on DNA binding of Pro11 were examined in gel mobility shift assays (Fig. 3 and results not shown). Methyl-green, which binds to the major groove, did not significantly affect Pro11 binding at concentrations up to 100 μM. On the other hand, all three minor-groove-binding molecules affected the binding of Pro11 to the orf11F3R2 fragment, albeit to different extents. Chromomycin A3 at 2.5 μM inhibited Pro11 binding, while a 5 μM concentration almost prevented Pro11 binding. Actinomycin D was less effective: at 10 μM some binding was still detected, while a concentration of 25 μM completely prevented Pro11 binding. Distamycin A was the least effective inhibitor. Pro11 was still able to bind to the orf11F3R2 fragment even at a distamycin A concentration of 100 μM. These results indicate that Pro11 binds through the minor groove and probably does not make contact in the major groove.
Mapping of the Pro11 binding sites indicates that Pro11 recognizes the region containing the four hexamers GGAACC.
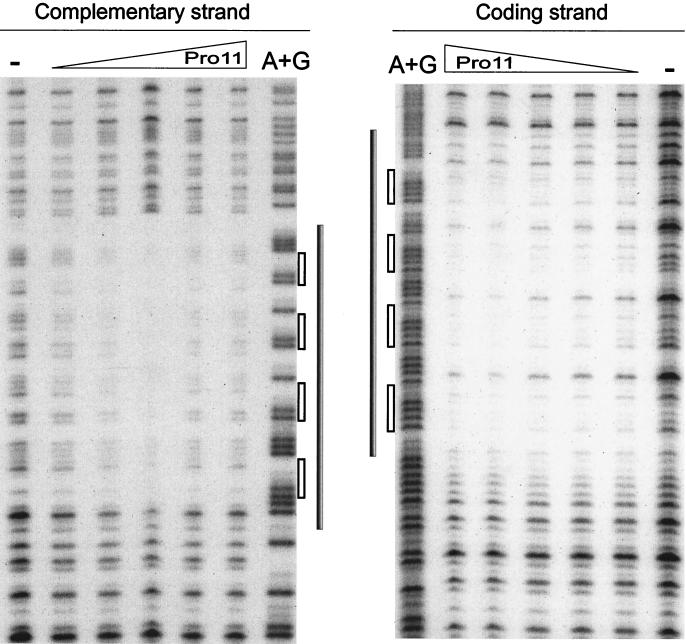
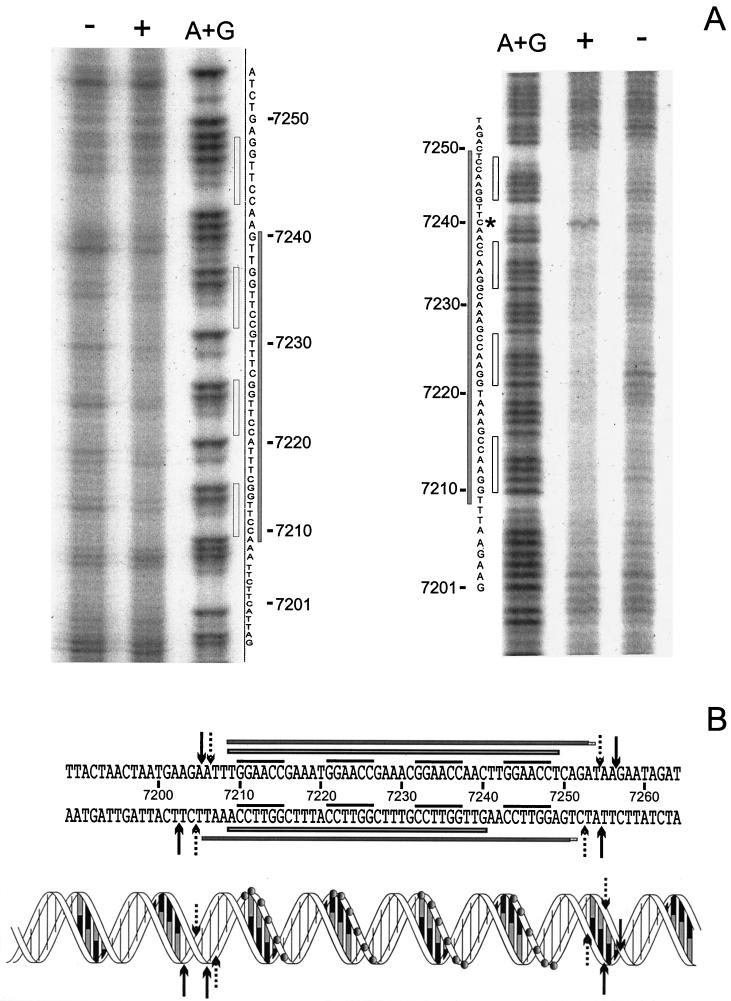
DNase I footprinting on DNA fragment orf11F5R2 was carried out to determine the sequences recognized by Pro11. Figure 4 shows that Pro11 protects a single region of 47 bp within orf11F5R2. This region contains four repeats of the hexamer GGAACC, each separated from the next one by 5 bp (see also Fig. 6B). The protected area does not extend to the A+T-rich region located immediately downstream of the four hexamers. Different amounts of Pro11 did not result in the preferential loss of protection of any site (Fig. 4). Binding of DNase I to its target requires contacts in the DNA backbone extending two steps upstream and downstream of the cleavage site in the same strand and two phosphate groups three and four steps upstream of the cleaved bond in the opposite strand (39). Taking into account the contacts required by DNase I to cut the DNA at a certain position, it can be inferred that Pro11 protects one step upstream of the first hexamer, since the step AT (positions 7206 to 7207 [see Fig. 6B]) is protected in the coding strand, and the protection extends to the step CT (positions 7205 to 7204) in the noncoding strand. On the 3′ side the protection extends at least five steps downstream of the last hexamer in the forward strand, since the step TA at positions 7254 to 7255 is protected, while three steps are protected in the opposite strand (until step TG, positions 7253 to 7252). These data indicate that both strands are unequally protected on this side of the binding region.
FIG. 4.
DNase I footprinting analysis of the Pro11-orf11F5R2 complex. The left panel shows complementary strand, and the right panel shows coding strand. A dash above a lane indicates no Pro11 added. The triangles indicate the increase in the amounts of Pro11 present in the binding reaction (0.5, 1, 2, 4, and 8 RU). A+G: lane with the A+G Maxam and Gilbert reaction products of the same DNA fragment. Grey bars indicate the protected regions. Empty bars indicate the position of hexamers.
FIG. 6.
(A) Phenanthroline-copper footprinting analysis of the Pro11-orf11F5R2 complex. The left panel shows noncoding strand (inverted in order to facilitate the comparison); the right panel shows coding strand. + and −, presence or absence of Pro11, respectively; A+G, lane containing the products of an A+G Maxam and Gilbert sequence reaction; grey bars, protected regions; empty bars, position of hexamers; asterisk, hypersensitive residue C7240. (B) Summary of DNase I and phenanthroline-copper footprinting results. The sequence of orf11 from position 7189 to 7264 (base numbering is according to the numbering of the published r1t sequence [GenBank accession number U38906]) is shown above, while a schematic representation of the double helix is given below. In the latter, grey circles indicate the phosphates of the hexamers. An empty bar on each strand indicates nucleotides protected from phenanthroline-copper cleavage. Filled bars on each strand indicate the region protected from DNase I digestion. The four hexamers are indicated by the overlining. DNase I digestion sites important to define the boundaries of the protected regions are indicated by either dotted (protected sites) or black (unprotected sites) arrows.
Two properly positioned Pro11 binding sites are required for stable binding of Pro11.
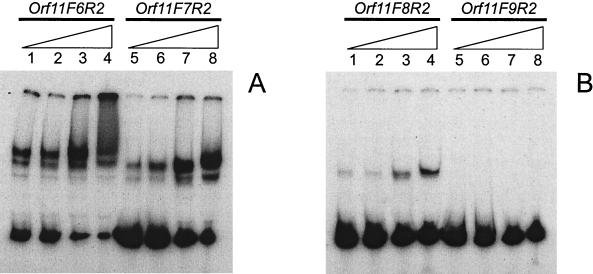
In order to determine the minimal number of Pro11 sites required to form a stable complex, a series of fragments containing four, three, two or one hexamer of the binding region were prepared using the primers ORF11-F6 to ORF11-F9, respectively, in combination with primer ORF11-R2 (Fig. 2A). Results of gel retardation assays (Fig. 5) show that Pro11 fails to bind to a DNA fragment harboring only one binding site. When two binding sites were present, a band corresponding to a retarded complex was detected, while binding was increasingly more effective as more binding sites were present in the DNA fragment. As this effect could also result from complex dissociation during electrophoresis, a series of binding reactions were performed using increasing amounts of protein (from 0 to 8 RU) and the fragment orf11F9R2 in TAE buffer. Again, no retarded complexes could be detected (data not shown). NaCl concentrations of up to 500 mM enhanced the binding of Pro11 for all fragments except orf11F9R2. Interestingly, only with fragment orf11F6R2 (four hexamers) did we observe a smear at 500 mM NaCl. This suggests that binding of additional Pro11 subunits to the complex requires at least four binding sites, and shows that this smearing is not due to unspecific binding of Pro11 to DNA. To determine the effect of the relative position of the binding sites on Pro11 binding activity, binding of Pro11 to fragments orf11F10R2 and orf11F11R2 was examined. Both fragments harbor two binding sites for Pro11. In orf11F10R2 10 bp have been inserted between the two hexamers, thus positioning them on opposite sides of the double helix, whereas they are one helix turn apart and on the same side of the helix in orf11F11R2. Since additional contacts within the intervening sequence between hexamers cannot be ruled out, the insertions consisted of repeats of the intervening sequence (Table 2). Binding of Pro11 is abolished in both cases, indicating that proper spacing of the hexamers (5 bp in the wild-type situation) is essential for the stability of the complex (results not shown).
FIG. 5.
Formation of complexes between Pro11 and DNA fragments containing different numbers of Pro11 binding sites. (A) Lanes 1 to 4, DNA fragment orf11F6R2 (four binding sites); lanes 5 to 8, DNA fragment orf11F7R2 (three binding sites). (B) Lanes 1 to 4, fragment orf11F8R2 (two binding sites); lanes 5 to 8, fragment orf11F9R2 (one binding site). The binding reactions were carried out with 1 RU of Pro11 in standard buffer containing increasing amounts of NaCl (50, 125, 250, and 500 mM), as indicated by the triangles.
Phenanthroline-copper footprinting reveals an asymmetric protection pattern of each DNA strand.
In order to examine the Pro11-DNA complex in more detail, phenanthroline-Cu footprinting was carried out. The 1,10-phenanthroline-copper complex is a chemical nuclease that nicks DNA and RNA under physiological conditions (32, 37). The reaction occurs in the minor groove, so that only if access to the C-1 of the ribose ring is blocked (either directly or indirectly), protection will be observed. Moreover, the small size of the reactant allows mapping the binding site for a protein with greater detail. The results (Fig. 6A) reveal a stretch of protected sequence spanning the four binding sites plus one base at each end in the coding strand. This suggests that the binding site for each monomer spans eight base pairs centered around the GGAACC hexamer. This result is supported by the fact that the C in position 7240 (Fig. 6A) is unprotected; in fact this position is more sensitive to the reactant than in the control. The footprint of the opposite strand shows a shorter protected area, from position 7240 to position 7209, leaving the fourth hexamer fully unprotected. These results suggest that every Pro11 subunit makes asymmetrical contacts with the DNA backbone.
DISCUSSION
Comparative genomics has evidenced the modular genetic organization of bacteriophages infecting gram-positive and gram-negative bacteria. Chopin et al. (7) reported that L. lactis bacteriophages belonging to the species P335 share the same modular genetic organization. In this way, a set of gene modules encoding proteins putatively involved in replication of bacteriophage DNA could be identified within the genomes of these bacteriophages. Interestingly, a similar organization can be found in several bacteriophages infecting other lactic acid bacteria as well as in bacteriophages from streptococci (5, 10). These modules typically specify a single strand binding protein and a putative replisome organizer (bIL285, BK5-T, TP901-1, and Tuc2009), a DnaC analog and a putative replisome organizer (r1t and bIL309), or all three genes (bIL286). They are located in the same region within the genomes of these temperate bacteriophages (7, 10). Most putative replisome organizers described in lactococcal bacteriophages contain a set of direct repeats structurally similar to iteron-containing origins of replication. Moreover, indirect evidence of the presence of functional iteron-containing origins of replication, based on the so-called Per effect, has been reported for some of these bacteriophages (24, 25, 27, 30). In bacteriophage TP901-1, mutational analysis of orf13 has shown that both the product of orf13 and the repeats located within orf13 are essential for replication of DNA in vivo (30). Bacteriophage r1t protein Pro11 shares significant similarity with previously characterized (putative) replisome organizers. The N-terminal of Pro11 is significantly similar to the N-terminal part of Rep2009, the presumed replisome organizer of the lactococcal bacteriophage Tuc2009 (27), and to that of G38P (Fig. 1). In its C-terminal part it is also similar to G38P. G38P binds to two regions within the genome of the bacteriophage SPP1, one located within the coding sequence of G38P and the other one in a noncoding region between late structural genes (29, 31). These regions share significant structural similarity to that found in r1t within orf11: they all consist of two sets of directly repeated sequences termed Box AB and a 70-bp A+T-rich region. The repeat units are ten bp in length and separated by one bp, so that a binding site is placed every 11 bp. This arrangement is also found in r1t.
The results presented in this work show that Pro11 binds specifically to target sites in its own gene, orf11. On the basis of band mobility shift assays using probes containing different numbers of binding sites, as well as footprinting analysis of the Pro11/DNA complex, it was shown that Pro11 recognizes a region in orf11 containing a quadruplet of the hexamer GGAACC that is present four times in orf11. The combined results of the DNase I and phenanthroline-copper footprinting are presented in Fig. 6B.
Binding of Pro11 to its target was less stable at low NaCl concentrations, so that several complexes were observed in gel mobility shift assays due to dissociation during electrophoresis, while only one complex could be detected at 250 mM NaCl (saturated complex). This effect was exploited to estimate the number of Pro11 binding sites by using limiting amounts of DNA or protein. The results suggested that there are four binding sites for Pro11. Moreover, the saturated complex migrates faster than some of the intermediates. This observation can be explained by assuming that a conformational change takes place in the complex that leads to a more condensed form that would migrate faster. Similar results have been previously reported for HMf, a DNA binding protein from Methanothermus fervidus, which were explained by protein-induced DNA condensation (35).
The effects that different groove-specific DNA binding drugs have on the binding of Pro11 to its binding site indicate that Pro11-DNA contacts are limited to the minor groove. This is further substantiated by the phenanthroline-copper footprinting data, since the nick reaction occurs in the minor groove (37).
DNase I footprinting assays showed that Pro11 protects a single region of 47 bp spanning the four hexamers GGAACC but did not protect the A+T-rich region located immediately downstream thereof, indicating that Pro11 does not interact with this region, at least when a linear substrate is used. The DNase I digestion pattern showed an appreciable asymmetry between the two strands. The coding strand was relatively resistant to digestion when compared with the other regions of the molecule, but four positions were highly sensitive. These positions were located two steps downstream of each hexamer. This pattern was not observed in the noncoding strand where, instead, a more even digestion pattern was obtained. DNase I is particularly sensitive to structural variations of DNA and there is evidence that the intrinsic bendability of DNA is a major factor determining DNase I rate of cleavage (16, 38). Resolution of the structure of DNase I-DNA complexes has shown that the enzyme distorts the DNA structure upon binding, widening the minor groove and bending the DNA towards the major groove. A major contribution to the alteration on the DNA structure comes from the stacking interaction between a tyrosine and the base located two positions upstream of the cleaved bond (39). Our results suggest that the step C-C in the hexamer is intrinsically but not freely flexible and should have a preferred direction of bending in order to explain the differences observed in the digestion pattern of each strand.
Phenanthroline-copper footprints displayed a remarkably asymmetric pattern. While the four hexamers GGAACC were protected in the coding strand, the complementary sequence of the fourth hexamer was completely unprotected in the noncoding strand. These results indicate that Pro11 contacts the DNA backbone at a hexamer in the coding strand but can also contact the DNA backbone on the minor groove side in the noncoding strand at the complementary sequence of the next upstream hexamer. In this way, the Pro11 molecule bound to the first hexamer would contact only this hexamer, while the other three would contact two hexamers each. This could explain why at least two properly spaced hexamers are needed to form a stable complex and how Pro11 can specifically recognize its target sequence from the minor groove. Such an arrangement of binding sites is found only once in the entire r1t genome, namely, in orf11. Although detailed structural data are not available, it seems reasonable to assume that in order to establish such an array of contacts, the DNA must be heavily distorted to avoid steric hindrance of the protein molecules bound to adjacent hexamers. Several minor-groove-binding proteins, such as HMG box proteins, IHF, or HU induce severe distortions on the DNA structure within their cognate sequences (for a review, see reference 1). The enhanced sensitivity of C at position 52 provides evidence for an alteration of the DNA structure. The asymmetry of the DNase I footprint, albeit to a lesser extent due to the bulkiness of the protein, provides some additional evidence to support the model. Moreover, the DNase I digestion pattern of the binding region indicates that the step CC of the hexamers possesses an intrinsic directed high bendability, allowing for the suggested bending of the DNA.
The Pro11/DNA complex put forward here has an asymmetric structure. We propose that this asymmetry induced by Pro11 binding is propagated directionally, i.e., towards the AT-rich region immediately downstream of the binding site, which could result in destabilization of the double helix structure there. Such origin melting by the putative replisome organizer Pro11 would subsequently allow the loading of additional proteins required for the initiation of r1t DNA replication. Considerable work has to be done in order to confirm the model. The effect of supercoiling has not been addressed at all in this study. Our footprinting data indicate that Pro11 does not interact directly with the AT-rich region, and no significant alterations of the secondary structure in this region are evidenced either by DNase I or phenanthroline-copper footprinting.
This picture may change considerably when a supercoiled DNA is considered. Schnoss et al. (36) have shown that binding of lambda protein O to its target site leads to the destabilization of the adjacent AT-rich region, but this structural transition was absolutely dependent on supercoiling of the DNA substrate. Electron microscopy data indicated that the G38P protein of phage SPP1 bends DNA, although no data are available regarding the effect of supercoiling (29). Interestingly, G38P forms a complex in which the DNA apparently is not wrapped around the G38P particle (29). Protein O forms a nucleoprotein complex, the O-some. Dodson et al. (11) proposed a structure of four dimers of protein O with the DNA wrapped around this protein core. Evidence for this model relied on electron microscopic observations and has not been confirmed by other experimental approaches. A comparative study between these three analogous proteins, lambda O, G38P, and Pro11, may provide a more detailed picture of the steps leading to the initiation of the replication in this kind of replicons. Although the initiation of replication of phages lambda and SPP1 has received considerable attention, a detailed characterization of the DNA-protein complexes formed by their initiation proteins is lacking. This is partly due to the fact that replisome organizer proteins are not easily studied by band mobility shift assays; the complexes they form with DNA do not enter the gel under the usual conditions employed for these assays (21, 29). Most knowledge about these proteins has come from footprinting assays and electron microscopy. The gel retardation conditions employed in this work have made Pro11 amenable for gel retardation analysis. Results obtained by this technique, combined with the footprinting evidence, have allowed proposing a preliminary model to explain how binding of Pro11 protein may provide the driving force for phage r1t origin melting
Acknowledgments
M. Zúñiga was supported by a Marie Curie training grant within the Biotech Programme of the European Union (contract ERBFMBICT975027) and a Postdoctoral Fellowship of the Spanish Ministry of Culture, Education, and Science.
We thank Leendert Hamoen for technical assistance, critical reading of the manuscript, and helpful suggestions; Juan Evaristo Suárez for helpful suggestions regarding the renaturation of Pro11; and Juan Carlos Alonso and Miquel Coll for critical reading of the manuscript.
REFERENCES
- 1.Bewley, C. A., A. M. Gronenborn, and G. M. Clore. 1998. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 27:105-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 7:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of proteins utilizing the principle of protein dye-binding. Anal. Biochem. 72:248-252. [DOI] [PubMed] [Google Scholar]
- 4.Bramhill, D., and A. Kornberg. 1988. A model for initiation at origins of DNA replication. Cell 54:915-918. [DOI] [PubMed] [Google Scholar]
- 5.Brondsted, L., S. Ostergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 6.Callanan, M. J., P. W. O'Toole, M. W. Lubbers, and K. M., Polzin. 2001. Examination of lactococcal bacteriophage c2 DNA replication using two-dimensional agarose gel electrophoresis. Gene 278:101-106. [DOI] [PubMed] [Google Scholar]
- 7.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Del Solar, G., R. Giraldo, M. J. Ruiz-Echavarría, M. Espinosa, and R. Díaz-Orejas. 1998. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 62:434-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desière, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brussow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 11.Dodson, M., R. McMacken, and H. Echols. 1989. Specialized nucleoprotein structures at the origin of replication of bacteriophage λ. Protein association and disassociation reactions responsible for localized initiation of replication. J. Biol. Chem. 264:10719-10725. [PubMed] [Google Scholar]
- 12.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defense systems in lactic acid bacteria. Antonie Leeuwenhoek 76:89-113. [PubMed] [Google Scholar]
- 13.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Georghiou, D., S. H. Phua, and E. Terzaghi. 1981. Curing of a lysogenic strain of Streptococcus cremoris and characterization of the temperate bacteriophage. J. Gen. Microbiol. 122:295-303. [Google Scholar]
- 15.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1990. Cloning, expression, and sequence determination of a bacteriophage fragment encoding bacteriophage resistance in Lactococcus lactis. J. Bacteriol. 172:6419-6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan, M. E., M. W. Robertson, and R. H. Austin. 1989. DNA flexibility variation may dominate DNaseI cleavage. Proc. Natl. Acad. Sci. USA 86:9273-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holo, H., and I. F. Nes. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huggins, A. R., and W. E. Sandine. 1977. Incidence and properties of temperate bacteriophages induced from lactic streptococci. Appl. Environ. Microbiol. 33:184-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Learn, B. A., S. J. Um, L. Huang, and R. McMacken. 1997. Cryptic single-stranded-DNA binding activities of the phage lambda P and Escherichia coli DnaC replication initiation proteins facilitate the transfer of E. coli DnaB helicase onto DNA. Proc. Natl. Acad. Sci. USA 94:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leenhouts, K. J., J. Kok, and G. Venema. 1990. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl. Environ. Microbiol. 56:2726-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowrie, R. J. 1974. Lysogenic strains of group N lactic streptococci. Appl. Microbiol. 27:210-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen, S. M., D. Mills, G. Djordjevic, H. Israelsen, and T. R. Klaenhammer. 2001. Analysis of the genetic switch and replication region of a P335-type bacteriophage with an obligate lytic lifestyle on Lactococcus lactis. Appl. Environ. Microbiol. 67:1128-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahanivong, C., J. D. Boyce, B. E. Davidson, and A. J. Hillier. 2001. Sequence analysis and molecular characterization of the Lactococcus lactis temperate bacteriophage BK5-T. Appl. Environ. Microbiol. 67:3564-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandel, M., and A. Higa. 1970. Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53:159-162. [DOI] [PubMed] [Google Scholar]
- 27.McGrath, S., J. F. M. L. Seegers, G. F. Fitzgerald, and D. Van Sinderen. 1999. Molecular characterization of a phage-encoded resistance system in Lactococcus lactis. Appl. Environ. Microbiol. 65:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 29.Missich, R., F. Weise, S. Chai, R. Lurz, X. Pedré, and J. C. Alonso. 1997. The replisome organizer (G38P) of Bacillus subtilis bacteriophage SPP1 forms specialized nucleoprotein complexes with two discrete distant regions of the SPP1 genome. J. Mol. Biol. 270:50-64. [DOI] [PubMed] [Google Scholar]
- 30.Ostergaard, S., L. Brondsted, and F. K. Vogensen. 2001. Identification of a replication protein and repeats essential for DNA replication of the temperate lactococcal bacteriophage TP901-1. Appl. Environ. Microbiol. 67:774-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedré, X., F. Weise, S. Chai, G. Lüder, and J. C. Alonso. 1994. Analysis of cis and trans elements required for the initiation of DNA replication in the Bacillus subtilis bacteriophage SPP1. J. Mol. Biol. 236:1324-1340. [DOI] [PubMed] [Google Scholar]
- 32.Pope, L. M., K. A. Reich, D. R. Graham, and D. S. Sigman. 1982. Products of DNA cleavage by the 1,10-phenanthroline-copper complex. Inhibitors of Escherichia coli DNA polymerase I. J. Biol. Chem. 257:12121-12128. [PubMed] [Google Scholar]
- 33.Rottlander, E., and T. A. Trautner. 1970. Genetic and transfection studies with B. subtilis phage SP 50. I. Phage mutants with restricted growth on B. subtilis strain 168. Mol. Gen. Genet. 108:47-60. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Sandman, K., J. A. Krzycki, B. Dobrinkski, R. Lurz, and J. N. Reeve. 1990. HMf, a DNA-binding protein isolated from the hyperthermophylic archaeon Methanothermus fervidus, is most closely related to histones. Proc. Natl. Acad. Sci. USA 87:5788-5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnoss, M., K. Zahn, R. B. Inman, and F. R. Blattner. 1988. Initiation protein induced helix destabilization at the λ origin: a prepriming step in DNA replication. Cell 52:385-395. [DOI] [PubMed] [Google Scholar]
- 37.Sigman, D. S., M. D. Kuwabara, C.-H. B. Chen, and T. W. Bruice. 1991. Nuclease activity of 1,10-phenanthroline-copper in study of protein-DNA interactions. Methods Enzymol. 208:414-433. [DOI] [PubMed] [Google Scholar]
- 38.Suck, D. 1994. DNA recognition by DNaseI. J. Mol. Recog. 7:65-70. [DOI] [PubMed] [Google Scholar]
- 39.Suck, D., A. Lahm, and C. Oefner. 1988. Structure refined to 2 Å of a nicked DNA octanucleotide complex with DNaseI. Nature 332:464-468. [DOI] [PubMed] [Google Scholar]
- 40.Taylor, K., and G. Wegrzyn. 1995. Replication of coliphage lambda DNA. FEMS Microbiol. Rev. 17:109-119. [DOI] [PubMed] [Google Scholar]
- 41.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. J. Ruiters, G. Venema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 44.Waterfield, N. R., M. W. Lubbers, K. M. Polzin, R. W. F. Le Page, and A. W. Jarvis. 1996. An origin of DNA replication from Lactococcus lactis bacteriophage c2. Appl. Environ. Microbiol. 62:1452-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]