Abstract
Bovine enteric caliciviruses (BEC) are associated with diarrhea in young calves. The BEC strains detected in Europe form a third genogroup within the genus “Norwalk-like viruses” (NLV) of the family Caliciviridae. In this report, we present sequence, clinical, and histological data characterizing a novel enteropathogenic BEC strain, NB, detected in fecal specimens from calves in the United States. The complete RNA genome of the NB virus is 7,453 bases long and is organized into two open reading frames (ORFs). ORF-1 is 2,210 amino acids long and encodes a large nonstructural polyprotein contiguous with the major capsid protein (VP1), similar to the lagoviruses and “Sapporo-like viruses” (SLV). The conserved calicivirus motifs were identified in the nonstructural proteins. ORF-2 is located at the 3′ end of the genome and encodes a small basic protein (VP2) of 225 amino acids. The 5′ and 3′ untranslated regions are 74 and 67 bases long, respectively. Among caliciviruses, NB virus shows amino acid identities of 14.1 to 22.6% over the entire ORF-1 nonstructural-protein sequence with NLV, SLV, vesivirus, and lagovirus strains, while the overall sequence identity of the complete NB VP-1 with other caliciviruses is low, varying between 14.6 and 26.7%. Phylogenetic analysis of the complete VP1 protein, including strains from all four calicivirus genera, showed the closest grouping of NB virus to be with viruses in the genus Lagovirus, which cause liver infections and systemic hemorrhage in rabbits. In gnotobiotic calves, however, NB virus elicited only diarrhea and intestinal lesions that were most severe in the upper small intestine (duodenum and jejunum), similar to the NLV BEC strains. The tissues of major organs, including the lung, liver, kidney, and spleen, had no visible microscopic lesions.
Caliciviruses cause a wide spectrum of diseases in animals and are important etiologic agents of viral gastroenteritis in humans. Members of the family Caliciviridae are small nonenveloped viruses 27 to 35 nm in diameter. They possess a single-stranded (ss) polyadenylated RNA genome of 7.4 to 8.3 kb (14, 42). The genome codes for one major structural capsid protein (VP1) of 55 to 80 kDa, a minor capsid protein (VP2), and a large polyprotein that is posttranslationally processed into nonstructural (NS) proteins, which contain conserved motifs similar to those of the picornavirus 2C helicase/ATPase, 3C protease, and 3D RNA-dependent RNA polymerase (RdRp) proteins (8, 12, 22, 64). In animals, caliciviruses are suspected or confirmed causes of a wide range of diseases that include gastroenteritis, vesicular lesions, respiratory infections, reproductive failure, and a fatal hemorrhagic disease in rabbits (10, 16, 18, 28, 30, 32, 59, 66). In humans, “Norwalk-like viruses” (NLVs) are the leading cause of epidemic, nonbacterial gastroenteritis in all ages, worldwide (15, 34, 56).
Caliciviruses can be divided phylogenetically into four genera: (i) Vesivirus, (ii) Lagovirus, (iii) NLVs, and (iv) “Sapporo-like viruses” (SLVs) (25). Within each genus, the member viruses share a common genomic organization and a high degree of sequence similarity. The lagoviruses and SLVs differ from the vesiviruses and NLVs in that their genomes contain two major open reading frames (ORFs), whereas the genomes of NLVs and vesiviruses contain three ORFs (8). ORF-1 of the lagoviruses and SLVs codes for both the NS and capsid proteins, which differs from NLVs and vesiviruses, in which the major capsid protein is coded for by a separate ORF (ORF-2) (8). ORF-2 of lagoviruses and SLVs, and ORF-3 of NLVs and vesiviruses, codes for a minor viral structural protein (VP2) that is basic (pH) in nature and is reported to be a minor capsid protein in feline calicivirus (FCV) and Norwalk virus (NV) (21, 37, 74).
The complete genomes of 18 calicivirus strains, representing all four genera, have been sequenced and serve as prototypes defining the genomic organization and sequence diversity of caliciviruses (7, 29, 35, 40, 44, 46, 47, 49, 51, 52, 58-60, 65, 67). Recent phylogenetic analyses of NLVs and SLVs place members of each genus into three distinct genogroups (23, 70). Prototype viruses of the NLV genogroups include NV (genogroup I) (40), Lordsdale virus (genogroup II) (11), and the bovine enteric calicivirus (BEC) Jena (genogroup III) (52). Similarly, SLV prototypes include viruses with the typical (cup-shaped) calicivirus morphology: Manchester virus (genogroup I) (49, 51), London/92 virus (genogroup II) (38), and a third genogroup defined by the porcine enteric calicivirus Cowden strain (PEC/Cowden; genogroup III) (29). Both the NLV and SLV genogroups further segregate into distinct clusters or genotypes (2, 70).
The detection of NLVs and SLVs in fecal specimens from cattle and swine has raised concerns about the potential existence of animal reservoirs (9, 29, 52, 77, 78). Previously, using electron microscopy (EM), enteric caliciviruses with typical NLV or small round structured virus (SRSV) morphology (24) were observed in fecal samples from dairy and veal calves in the United States (J. R. Smiley and L. J. Saif, Proc. 81st Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 78, 2000). One such BEC strain collected from a dairy calf in Nebraska in 1980, and designated Nebraska (NB) strain, was shown to elicit diarrhea in orally inoculated gnotobiotic (Gn) calves given filtered, bacterium-free fecal suspensions (K. O. Chang and L. J. Saif, unpublished data). Because little was known about the genetic diversity of BECs circulating in the United States, and because of concerns about the zoonotic potential of such animal-origin enteric caliciviruses, the NB virus genome was sequenced. In this paper, we report the complete genomic sequence of the BEC-NB strain and the partial genomic sequence of a second closely related BEC strain, CV23-OH, that was collected from an Ohio veal calf in 2000. We also further document the enteropathogenicity of the BEC-NB strain in Gn calves.
MATERIALS AND METHODS
Origin of the BEC-NB strain.
The BEC-NB strain was originally identified in a diarrheic stool sample collected from a dairy calf in Nebraska in 1980. A bovine coronavirus (BCV) was also observed in the original sample. Following oral inoculation of Gn calves with filtered (0.45-μm pore size) preparations of either the original field sample or samples collected from Gn calves inoculated with the original sample through Gn calf passage 2, both the BEC-NB and BCV were observed by EM in postinoculation fecal samples. The NB strain was separated from BCV by administration of a 0.22-μm-pore-size-filtered, Gn calf passage 2 fecal suspension to a 32-day-old Gn calf previously exposed to BCV at 18 days of age. The inoculated calf developed diarrhea and was euthanized on the second postinoculation day (PID 2). Immune electron microscopic (IEM) examinations of fecal- and intestinal-content samples (68), and the results of indirect fluorescent antibody staining of intestinal mucosal smears (5), were negative for BCV. Only virus particles with NLV-like SRSV morphology were observed in stool samples and intestinal contents from the euthanized calf. The NB strain from Gn calf passage 3 was subsequently freeze-thawed, diluted, and filtered (0.05-μm pore size) and then passaged an additional two times (passages 4 and 5) in Gn calves and shown to be free of BCV, using the methods described above and by a BCV antigen enzyme-linked immunosorbent assay (72). Lack of seroconversion to BCV (73) was also confirmed in several Gn calves exposed to the BEC inocula. The BEC inocula continued to elicit diarrhea and BEC shedding in exposed calves at each passage level.
Molecular analysis of BEC-NB strain.
For molecular studies, NB viral RNA was isolated from a prediarrheic fecal sample collected from a 9-day-old Gn calf inoculated 24 h previously by intravenous (i.v.) injection of a filtered (0.22-μm pore size) inoculum of a 10% fecal suspension (in Eagle's minimal essential medium; starting volume, 10 ml of feces) containing NB at the fifth Gn-calf passage level. Viral RNA was purified using Trizol reagent (Gibco BRL) from 200 μl of a 20% postinoculation fecal suspension prepared with phosphate-buffered saline (pH 7.2) and was dissolved in 40 μl of RNase-free water. Initial attempts to amplify the virus genome using a number of different published primer pairs in reverse transcription (RT)-PCRs prepared using the One-Step RT-PCR kit (Qiagen Inc., Valencia, Calif.) were unsuccessful. The evaluated primers included NV35-36 (71), JV12-13 (81), GLPSG1-YGDD1 (26), P289-P290 (39), and BEC-Pol 5′ (5′-TAT GAG CCA GCC TAC CTT GG-3′)-BEC-Pol 3′ (5′-ACC TGG GAC GTG CAT GGG A-3′) (Chang and Saif, unpublished). Because no products of the expected size were obtained, a new series of reactions was run using mixed combinations of the listed primers. RT was completed at 50°C for 30 min, and PCR consisted of 35 cycles at 94°C for 30 s, 49°C for 60 s, and 72°C for 1 min. A target reaction product of 188 bp and a strong 344-bp nonspecific product produced in the P290F-BEC-Pol 3′R reaction were gel purified (Qiaex Gel Extraction kit; Qiagen Inc.), cloned into pCR-2.1 T/A cloning vectors (Invitrogen Corp., Carlsbad, Calif.), and sequenced using an ABI-377 automated sequencer (Applied Biosystems, Foster City, Calif.). BlastN and BlastP searches of the GenBank database for the determined nucleotide and predicted polypeptide sequences of the 188- and 344-bp products indicated significant identity with calicivirus RdRps. Each product was forward primed with the P290F primer at positions 4227 to 4249 in the NB virus genome and reverse primed with the BEC-Pol 3′ primer.
An NB virus-specific primer pair, NB(F) (5′ 4312GTT TGC GAG TTG CGG TGT GC-3′) and NB(R) (5′ 4547GTA AAG CCC ATC GTC CCC ATA GG-3′), was designed from the 344-bp sequence data and used to identify NB virus in postexposure fecal samples of BEC-NB-inoculated Gn calves. Sequence information for the 3′ end of the virus genome was obtained by preparing cDNA using the SuperScript First-Strand cDNA kit (Gibco BRL-Life Technologies, Rockville, Md.) with the primer 5′-T25VN (T25-A/G/C-A/G/C/T) (1, 52). The prepared cDNA was used in a hot-start PCR with Taq polymerase (Promega) and the NB(F) and T25VN primers to generate 3,169-bp amplicons of the 3′-terminal NB virus genome. The thermocycling conditions for the reaction consisted of denaturation at 94°C for 3 min; 40 cycles of 94°C for 12 s, 50°C for 30 s, and 69°C for 4 min; and a final extension of 69°C for 10 min. The 3′-terminal NB reaction product was then cloned in the T/A vector and sequenced using the M13F/R priming sites to obtain the 5′- and 3′-terminal sequences of the amplified fragment. New 5′ and 3′ primers were designed and used in successive rounds of RT-PCR amplification, cloning, and sequencing to determine the entire 3′-terminal NB virus genome.
The sequence of the 5′ virus genome was concurrently determined using the random-PCR method (20) and adaptations of the procedure as described by others (11, 27, 52). Briefly, the SuperScript cDNA kit (Gibco) was used to make sscDNA using virus-specific antisense primers, with the NB(R) primer used for the first round of cDNA synthesis. sscDNA was converted to double-stranded (ds) cDNA using Klenow enzyme and the random Linker primer LINK-N7 (5′-TAG TAC ATA GTG GAT CCA GCT N7-3′). Random-primed viral dscDNA was amplified in one or two rounds of nested PCR using 3′ virus-specific primers and the 5′ forward LINK primer, which is complementary to the LINK component of primer LINK-N7. Using this technique, 5′ forward extension on the virus genome varied between 400 and 1,400 bp per round of dscDNA synthesis and produced sequence data to within 110 bases of the 5′ terminus of the NB virus genome.
The authentic 5′ genome terminus was determined using the 5′ rapid amplification of cDNA ends (RACE) procedure (19, 54). The NB-specific primer GSP-1 (5′-GGG GGT CCA CCG ATA GTA-3′) was used to prime first-strand cDNA synthesis, after which the cDNA was homopolymeric dC or dA tailed, in separate reactions, using terminal deoxynucleotidyl transferase. The 5′ NB virus terminus was amplified in successive nested PCRs using the 5′ RACE abridged or universal anchor primers in conjunction with the NB gene-specific primer GSP-2 (5′-CTG GTC CGG GGC ATA GTC ACG-3′) or GSP-3 (5′-GGG CCG TCC GTA GCA GGG TCT T-3′) for homopolymeric dC-tailed cDNA, while a single nested PCR was completed for the dA-tailed cDNA using the T25VN and GSP-2 primers. The products were cloned into PCR 2.1 T/A cloning vectors, or sequenced directly, to determine the 5′-terminal sequence. The final NB virus sequence is a consensus sequence based on a minimum of three cloned or directly sequenced RT-PCR amplicons from individual reactions of each genome segment analyzed.
Origin and genomic analysis of BEC CV23-OH virus strain.
A second BEC with SRSV morphology, designated the CV23-OH strain, was identified by EM and RT-PCR testing using the P289-P290 primer set (annealing temperature, 49°C) in May 2000 (J. R. Smiley and L. J. Saif, Proc. 81st Annu. Meet. Conf. Res. Workers Anim. Dis., abstr. 78, 2000). The virus was initially detected by RT-PCR in a pooled fecal sample (48 samples; 10 pools) of Ohio veal calves and was subsequently traced to a single calf sample (CV23-OH). Interestingly, the CV23-OH sample was the sole RT-PCR-positive BEC sample collected on the farm, which was populated with 270 calves 3 to 3.5 weeks of age. Sequence analysis of the cloned P289-P290 reaction products revealed that the CV23-OH virus was closely related to the NB virus, showing 86.6% nucleotide and 97.0% amino acid similarity in the 277-bp virus-specific RdRp target sequence. Attempts to amplify the complete 3′-terminal virus genome using the primers CV23-OH(F) (5′-TGA GCT TGC CCC CAT TTC CTA AC-3′) and T25VN were unsuccessful in producing an expected full-length product. However, sequence analysis of a strong ∼1,000-bp reaction product amplified in a PCR using sscDNA prepared with the T25VN primer and amplified with the CV23-OH(F)-T25VN primer pair revealed it to be a truncated CV23-OH virus-specific product primed as expected on the 5′ and 3′ ends of the virus genome but comprised of nucleotides (5′) 1 to 245 of the expected product conjoined to a 3′-terminal sequence of 701 bp, as predicted by alignment with the previously determined NB virus sequence. The 3′-terminal genome sequence of the CV23-OH virus (3,205 bp) was subsequently determined by primer walking as described for the NB virus. The complete CV23-OH sequence is based on results obtained for two or three cloned or directly sequenced RT-PCR amplicons. Editing, alignment, and analysis of the NB and CV23-OH virus sequences were performed using the Lasergene software package (DNASTAR Inc., Madison, Wis.).
Phylogenetic analyses.
Human and animal calicivirus strains for which the complete genomes have been characterized were used to perform a phylogenetic comparison of the NB virus. Collectively, the virus strains represent all four known calicivirus genera (25). The GenBank accession numbers, genus designations, and virus strains are summarized in Table 1. For phylogenetic comparison of the capsid (VP1) gene, additional virus sequences, representative of the three NLV and SLV genogroups (2, 23, 70), were also included.
TABLE 1.
Virus strains and GenBank accession numbers, tree notations, and ORF-1 NS protein and capsid sequences used for phylogenetic comparison
| Calicivirus genus | Virus strain | Genbank accession no. | Tree notationa | CGb (bp) | Length (location in ORF-1) of aligned amino acid sequencesc
|
|||
|---|---|---|---|---|---|---|---|---|
| 2C helicase | 3C protease | 3D RdRp | Capsid | |||||
| SLV | HuCV/Manchester/93/UK | X86560 | Manchester | 7,431 | 153 (470-622) | 127 (1069-1195) | 208 (1452-1659) | 561 |
| HuCV/Stockholm/317/97/SE | AF194182 | Stockholm | 565 | |||||
| HuCV/Parkville/94 | U73124 | Parkville | 571 | |||||
| HuCV/Sapporo/82/JP | U65427 | Sapporo82 | 561 | |||||
| HuCV/Houston/7-1181/US | AF435814 | Hou7-1811 | 553 | |||||
| HuCV/Mexico/340/90/MX | AF435812 | Mex340 | 558 | |||||
| HuCV/Lond/29845/92/UK | U95645 | London292 | 557 | |||||
| PEC/Cowden/80/US | AF182760 | PEC Cowden | 7,320 | 153 (454-606) | 127 (1063-1189) | 208 (1444-1651) | 544 | |
| NLV | HuCV/Norwalk/8FIIa/76/US | M87661 | Norwalk | 7,654 | 163 (472-712) | 153 (1115-1267) | 205 (1526-1730) | 530 |
| HuCV/Southampton/91/UK | L07418 | SOV | 7,708 | 163 (551-713) | 153 (1114-1266) | 205 (1525-1729) | 546 | |
| HuCV/DSV/90/SA | U04469 | DSV | 544 | |||||
| BEC/CALFNL/98/NLd | Unpublished | CALFNL98 | 522 | |||||
| BEC/Jena/80/DE | AJ011099 | Jena | 7,338 | 163 (472-634) | 153 (1008-1160) | 205 (1418-1622) | 519 | |
| HuCV/Toronto/91/CA | U02030 | Toronto | 550 | |||||
| HuCV/Hawaii/71/US | U07611 | Hawaii | 7,513 | 163 (485-647) | 153 (1023-1175) | 205 (1435-1639) | 535 | |
| HuCV/Lordsdale/93/UK | X86557 | Lordsdale | 7,555 | 163 (485-647) | 153 (1023-1175) | 205 (1435-1639) | 539 | |
| Hu/NLV/Alphatron/98-2/98/NL | AF195847 | Alphatron | 556 | |||||
| Vesivirus | FCV-F4 | D31836 | FCV-4 | 7,681 | 155 (474-628) | 127 (1095-1221) | 212 (1488-1699) | 668 |
| FCV-F9 | M86739 | FCV-9 | 7,690 | 155 (474-628) | 127 (1095-1221) | 212 (1488-1699) | 671 | |
| Walrus calicivirus | AF321298 | WCV | 8,289 | 155 (580-734) | 127 (1207-1333) | 213 (1602-1814) | 708 | |
| San Miguel sea lion virus-1 | AF181081 | SMSV-1 | 8,284 | 155 (580-734) | 127 (1207-1333) | 213 (1601-1813) | 702 | |
| San Miguel sea lion virus-4 | M87842 | SMSV-4 | 703 | |||||
| Vesicular exanthema of swine A48 | AF181082 | VESV-A48 | 8,284 | 155 (580-734) | 127 (1207-1333) | 213 (1603-1815) | ||
| Primate calicivirus-1 | AF019736 | PAN-1 | 8,304 | 155 (580-734) | 127 (1208-1334) | 213 (1603-1815) | 709 | |
| Lagovirus | European brown hare syndrome virus | Z69620 | EBHSV-GD | 7,442 | 155 (507-661) | 112 (1113-1224) | 210 (1498-1707) | 576 |
| RHDV-Iowa | AF258618 | RHDV-Iowa | 7,467 | 156 (512-667) | 112 (1120-1231) | 210 (1505-1714) | 579 | |
| RHDV-FRG | M67473 | RHDV-GH | 7,437 | 579 | ||||
| Unknowne | BEC/NB/80/US | AY082891 | NB | 7,453 | 154 (446-599) | 120 (1010-1128) | 215 (1389-1603) | 549 |
| BEC/CV23-OH/00/US | AY082890 | CV23-OH | 549 | |||||
Virus notation used in phylogenetic trees (Fig. 3).
CG, complete genome length of reference calicivirus strains.
Lengths and locations of amino acid residues in their respective predicted ORF-1 polypeptide sequences.
NLV BEC isolate from The Netherlands (van der Poel et al. [78]).
Genus designation for NB and CV23-OH is unclear.
Phylogenetic analyses of multiple sequence alignments of the deduced amino acid sequences of the complete capsid and partial 2C helicase/ATPase, 3C protease, and 3D RdRp NS-protein genes were performed. Alignment of the capsid genes (ORF-2 [NLV and vesiviruses] or predicted ORF-1 [SLV, lagoviruses, and NB] coding regions) was completed with the CLUSTAL-W (version 1.4) alignment program in the Bioedit (version 5.0.9) sequence alignment editor and analysis package (33), using its default settings. To create the individual NS-protein alignments, an initial global alignment of the complete NS-protein ORF-1 sequences was completed for the prototype viruses. The sequences were analyzed for the presence of conserved ORF-1 NS-protein sequence motifs, and anchor residues were selected to delimit the partial NS-protein alignments. Anchors were chosen so as to be bounded by proteolytic cleavage sites reported for Southampton virus (SOV), FCV, and rabbit hemorrhagic disease virus (RHDV) (36, 41, 50, 53, 74-76, 82). After being edited, the partial NS-protein sequence alignments were realigned. The lengths and ORF-1 locations of the aligned NS-protein sequences are specified in Table 1.
Trees were constructed for the predicted capsid and NS-protein genes using the maximum-likelihood (ProML), parsimony (Protpar), and distance (Prodist) methods of analysis of PHYLIP (version 3.6a2.1) (17). For each analysis, 100 bootstrapped data sets were evaluated using the default settings for each program, with the following specific selections or exceptions: (i) selection of the JTT (Jones-Taylor-Thornton) amino acid substitution matrix for Prodist and ProML analyses, (ii) randomization of sequence input order, (iii) switching off the S-option (speedier but rougher) of ProML, and (iv) use of the FITCH program (Fitch-Margolish method) to build final distance trees for distance analysis. The CONSENSE program of PHYLIP was used to construct a single majority-rule tree of all trees calculated for bootstrapped data sets, and the tree-drawing program Treeview (version 1.6.6) (62) was used to prepare the final tree diagrams.
Experimental inoculation of Gn calves with BEC-NB strain. (i) Calf challenge.
Fecal suspensions (10 or 20% in phosphate-buffered saline, pH 7.2 to 7.4) were prepared from frozen (−70°C) aliquots of BEC-NB-infected feces (10.0 ml) at Gn-calf passage 5. The prepared suspensions were sonicated (80 mHz for 30 s [two cycles]), centrifuged (8,470 × g; 30 min; 4°C), and filtered (0.22-μm pore size) to prepare inocula for calf exposures. Four Gn calves (3 to 7 days old) were administered the prepared inocula orally (n = 2) or by slow i.v. injection via the jugular vein (n = 2). A single Gn calf was mock inoculated at 4 days of age and euthanized at 6 days of age for use as an age-matched control for histopathologic evaluations. Feces were collected daily, scored for diarrhea (scale, 0 to 4; diarrhea, ≥2) and calf appetite and alertness were assessed. The BEC-NB-inoculated calves were euthanized on day 4 (n = 3) or day 5 (n = 1) postexposure coincident with or following the onset of diarrhea.
(ii) Histopathology.
Tissues of major organs (kidney, liver, spleen, and lung) and intestinal sections of the duodenum (midpoint-pylorous and sphincter of Oddi), jejunum (mid), ileum (∼15 cm anterior to the ileocecal junction), cecum, spiral colon, and rectum were collected and fixed in Prefer (Anatech Ltd., Battle Creek, Mich.) or 10% neutral-buffered formalin. The preserved tissues were dehydrated, embedded in paraffin, transversely sectioned, and stained with hematoxylin and eosin. Histopathologic analysis of tissues was completed using light microscopy.
(iii) RT-PCR and EM testing of fecal samples for BEC-NB.
Available Gn-calf fecal samples, including prechallenge, PID 0 samples from all calves but calf 2, were tested in a 50.0-μl one-step RT-PCR assay utilizing the NB(F)-NB(R) primer set. Selected samples were also tested by IEM using procedures and techniques described previously (69).
Nucleotide sequence accession numbers.
The sequences of the BEC-NB and CV23-OH viruses have been submitted to the EMBL and GenBank databases under accession numbers AY082891 and AY082890, respectively.
RESULTS
Genomic organization of BEC-NB virus.
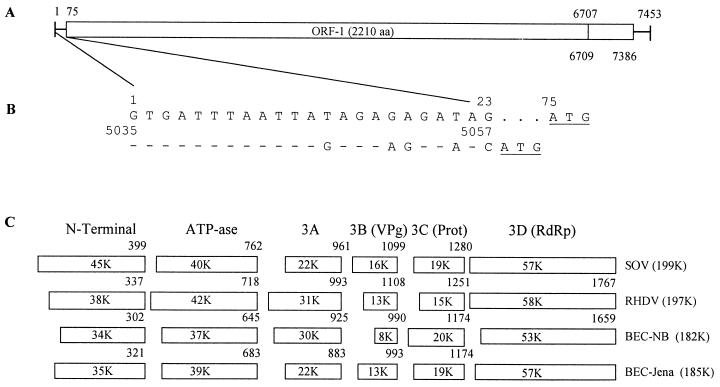
The complete RNA genome of the BEC-NB strain consists of 7,453 bases, excluding its 3′ poly(A) tail, and it has a ribonucleoside composition of 27.1% G, 21.6% A, 21.8% U, and 29.5% C. The 5′ untranslated region (UTR) of the NB virus is 74 bases long and begins with the 5′-terminal triribonucleoside sequence GUG. Sequence analysis of the determined genome predicts the presence of two ORFs (Fig. 1A), similar in organization to the lagoviruses and SLVs. The first initiation codon, also the start of ORF-1, begins at nucleotide 75. ORF-1 contains 6,633 bases (nucleotides 75 to 6707), whose predicted translated sequence of 2,210 amino acids (aa) includes the virus NS and major capsid (VP1) proteins. ORF-2 consists of 678 bases (nucleotides 6709 to 7386), is frameshifted +2 relative to ORF-1, and is predicted to encode a 225-aa protein. The 3′ UTR, which excludes the poly(A) tail, is 67 bases long and contains a preponderance (38 of 67) of uridine residues.
FIG. 1.
Genome organization, 5′ UTR and subgenomic conserved sequences, and predicted ORF-1 NS proteins of the BEC-NB strain. (A) ORFs and UTR sequences in the complete 7,453-bp genome. (B) Locations and sequence conservation of the 5′ UTR (nucleotides 1 to 23) and subgenomic (nucleotides 5035 to 5057) sequence segments in BEC-NB. Only differences are indicated. (C) Predicted ORF-1 NS proteins of BEC-NB and the NLV BEC-Jena based on sequence comparison with maps of characterized cleavage products of a lagovirus, RHDV, and a human NLV, SOV. The calculated protein molecular weights are shown, and the COOH-terminal amino acid residue in the predicted virus ORF-1 sequence is shown above each box.
Alignment of the 5′ UTR (1 to 74 nucleotides) with the downstream NB virus genome revealed a significant alignment (81%) between nucleotides 1 to 23 and nucleotides 5035 to 5057 (Fig. 1B). Interestingly, the initiation codons for ORF-1 (nucleotides 75 to 77) and the predicted subgenomic transcript at positions 5058 to 5060 of the NB virus are each located downstream of the conserved sequence motif, which differs from all other characterized caliciviruses, in which the initiation codons are integral to the conserved sequence motif (45). Each of the predicted initiation codons of the NB virus is found in a favorable context for translation initiation of eukaryotic mRNA, as predicted by the ribosome-scanning model (43).
ORF-1 NS proteins.
The calculated molecular mass of the ORF-1 polyprotein is 239 kDa and is predicted to include both the viral NS (182,000-molecular-weight [182K]) and VP1 (57K) proteins. Conserved amino acid sequence motifs characteristic of picornaviruses and caliciviruses, including the 2C-like helicase-nucleoside triphosphatase DNA-binding (GXXGXGKS/T) and ATP hydrolysis (KXXXFXSXXXXXS/TTN) motifs, the 3-D RdRp (GLPSG and YGDD) motifs, and the VP1 capsid (PPG) motifs, are present in the ORF-1 polyprotein (8, 55, 63, 64). In the 3C-like protease of NB virus, tyrosine (Y) replaces aspartic acid (D; underlined) in the conserved GDCG motif, which is found in all other caliciviruses. However, histidine (H) residues at positions 1025 and 1118 and aspartic acid at position 1039 in the NB ORF-1 protease sequence are found spaced similarly to the same conserved residues in the 3C-like proteases of other caliciviruses (64). Because these residues are postulated to form the catalytic core of the protease enzyme, it is likely that this function is similarly conserved in the NB virus. The calicivirus VPg protein (D/E)EY(D/E)E motif was also found (positions 938 to 943) in the predicted NB ORF-1 NS-protein sequence (13). However, the KGK(N/T)K VPg motif and its 5′-juxtaposed 3C protease cleavage site, which defines the NH2 protein terminus, was replaced by 914KTANK in the NB virus, while the predicted 3C dipeptide (925QG) cleavage site was found 3′, rather than 5′, of the conserved motif (13).
Potential ORF-1 NS-protein cleavage sites of the NB virus were predicted from a multiple alignment completed for NB and other representative viruses of the four calicivirus genera, using NS-protein sequence motifs and defined 3C protease cleavage sites for FCV, RHDV, and SOV (Fig. 1C). From comparison with these viruses, the predicted ORF-1 protein map of the NB virus is NH2-P34-P37(2C)-P30-P7.6(VPg)-P20(3C)-P53(3D)-P57(VP1)-COOH. While this map requires further experimental verification, both the linear arrangement of the conserved protein motifs and their predicted sizes closely parallel those of other caliciviruses, with the exception of the VPg protein, which at 7.6 kDA is smaller than any previously described calicivirus VPg.
The sequence identities of the NS proteins of NB virus with caliciviruses of other genera overall were low and varied from protein to protein (Table 2). The sequence identity of the NB virus with other caliciviruses in the complete ORF-1 NS-protein sequence varied between 13.8 and 22.6% and was distinctly lower with viruses of the genus NLV (Table 2). As recognized for other caliciviruses, there is a low sequence identity in the 5′ N-terminal protein of ORF-1 of the NB virus with other caliciviruses. For individual alignments completed for the partial coding sequences of the 2C-like helicase/ATPase, 3C-like protease, and 3D-like RdRp NS protein, sequence identities varied between 29.4 and 41.9, 14.2 and 35.4, and 27.5 and 41.0%, respectively, for NB and other caliciviruses (Table 2 and Fig. 1). As in the ORF-1 comparison, NB showed the lowest overall identity with viruses of the genus NLV.
TABLE 2.
Percent amino acid identity of BEC-NB virus with other caliciviruses in regions aligned for phylogenetic comparisona
| Genus | Virusb | % Identity
|
|||||
|---|---|---|---|---|---|---|---|
| ORF-1 NS | 2C helicase | 3C protease | RdRp | VP1 | VP2c | ||
| SLV | Manchester | 21.1 | 37.4 | 35.4 | 33.0 | 21.0 | 16.9 |
| PEC | 21.7 | 40.0 | 30.7 | 34.8 | 19.9 | 13.9 | |
| NLV | Jena | 15.0 | 31.2 | 14.2 | 29.8 | 14.7 | 17.0 |
| Norwalk | 14.7 | 31.2 | 17.5 | 30.7 | 15.7 | 15.5 | |
| SOV | 13.8 | 31.9 | 16.2 | 29.8 | 15.4 | 16.1 | |
| Hawaii | 14.3 | 29.4 | 14.9 | 27.5 | 14.6 | 12.9 | |
| Lordsdale | 14.1 | 29.4 | 14.2 | 28.4 | 14.8 | 17.3 | |
| Vesivirus | VESV-A48 | 21.5 | 40.6 | 30.4 | 32.5 | 17.8 | 16.2 |
| SMSV-1 | 22.3 | 41.9 | 30.4 | 33.0 | 17.8 | 16.2 | |
| PAN-1 | 22.0 | 40.6 | 32.0 | 32.1 | 17.8 | 15.3 | |
| WCV | 22.3 | 41.2 | 30.4 | 33.0 | 18.2 | 15.3 | |
| FCV-F4 | 21.8 | 40.6 | 25.0 | 34.8 | 17.8 | 16.8 | |
| FCV-F9 | 22.6 | 40.6 | 24.2 | 34.8 | 18.0 | 15.0 | |
| Lagovirus | EBHSV | 21.9 | 37.8 | 24.0 | 41.0 | 26.6 | 20.0 |
| RHDV | 21.6 | 36.5 | 22.4 | 40.5 | 26.7 | 21.2 | |
See Table 1 and footnotes for descriptions of aligned regions.
EBHSV, European brown have syndrome virus; VESV, vesicular exanthema of swine virus; SMSV, San Miguel sea lion virus; WCV, walrus calicivirus.
Sequence identities calculated for complete VP2 ORF alignment.
Major (VP-1) and minor (VP-2) capsid proteins.
The NB virus VP1 is predicted to be 549 aa long (ORF-1 residues 1662 to 2210) and to have a molecular mass of 57.5 kDa, which is similar to the major capsid protein of other caliciviruses. The predicted initiation codon of the NB virus VP1 (1662M) in ORF-1, as described previously, is close to, but distinct from, the presumptive subgenomic transcription initiation site. A second unusual feature of the NB virus in this sequence region is the occurrence of a potential 3C-like protease cleavage site (1659EA) 5′ of the VP1 initiation codon, which contrasts with the SLVs and lagoviruses, in which a similar cleavage dipeptide is found directly 3′ of the initiation codon (29, 58). As noted for other caliciviruses, the N-terminal region (aa 1 to 280) of VP1 of the NB virus is the most highly conserved (21.8 to 32.7%) of the entire capsid-coding region, as determined in completed regional (I, II, and III) sequence alignments of representative caliciviruses similar to that described in a previous study (data not shown and reference 48). The overall sequence identity of the complete NB VP1 with other caliciviruses was low, varying between 14.6 and 26.7% (Table 2), with the highest identity being with the lagoviruses.
VP2 of the NB virus, coded by the small ORF-2, is comprised of 225 aa and has a calculated molecular mass of 23.6 kDA. Biophysically, the protein is predicted to be hydrophilic and basic in nature, with a calculated isoelectric point of 10.5, which is comparable to the predicted properties of this protein in other caliciviruses. The start codon (6709AUG) of NB ORF-2 begins at the second nucleotide (+2 position) downstream of the stop codon (UAG6707) for ORF-1, after a skip of 1 nucleotide. Sequence identities for the NB virus VP2 with caliciviruses of other genera are low and vary from 12.9 to 21.2% (Table 2), which is typical of cross-genus comparisons of this reading frame in caliciviruses. The highest overall similarity in the complete ORF is with the lagoviruses (20.0 to 21.2%).
Sequence comparison of NB and CV-23 virus strains.
A 3,205-bp sequence corresponding to the 3′ terminus of the BEC CV23-OH virus genome was determined. The region sequenced included the 3′-terminal 805 bp of the predicted 3D RdRp and complete VP1, VP2, and 3′ UTR regions, as described for the NB virus. Combined alignment and ORF analyses demonstrated their shared genomic organization, including the complete conservation of the internal sequence motif preceding the start codon of VP1 and a single-nucleotide gap between the terminus of ORF-1 and the start codon of ORF-2. A nucleotide identity of 89.1% was observed over the entire aligned sequences, while separate identities for the 3′-terminal RdRP, VP1, and VP2 were 86.8, 91.3, and 87.0%, respectively. The corresponding identities of the predicted polypeptide sequences for the 3′-terminal RdRp (269 aa), VP1 (549 aa), and VP2 (225 aa) were 97.0, 98.5, and 93.4%, respectively.
Phylogenetic analyses.
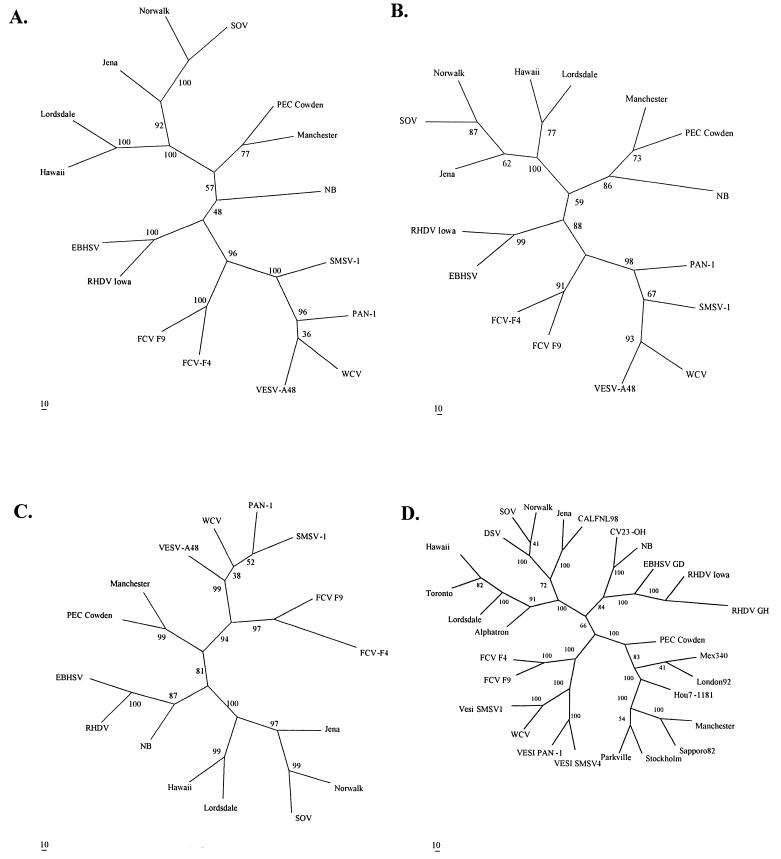
The results of the phylogenetic analyses of the NB virus 2C-like helicase/ATPase, 3C-like protease, 3D RdRp, and complete VP1 multiple alignments using parsimony, distance, and maximum-likelihood methods of analysis predicted similar consensus trees in each genomic region examined. Unrooted consensus trees prepared using the more rigorous maximum-likelihood method are presented in Fig. 2. Trees produced for each of the genomic regions evaluated grouped the characterized prototype calicivirus strains similarly to reports of previous phylogenetic analyses (2, 4, 70). However, phylogenetic classification of the NB virus varied with the genome region evaluated and included placements as a new genus (2C helicase/ATPase), a new SLV genogroup (3C protease), and a new lagovirus genogroup (3D RdRP and VP1 capsid). Such disparate classification of the NB virus was not unexpected due to its generally low level of sequence identity in the aligned regions evaluated (Table 2). The bootstrap values of the branch points for the genera Lagovirus and SLV compared to NB in the 2C and 3C consensus trees are also low, indicating caution in interpreting the significance of these branch placements in the analyses. Similarly, consensus distance and parsimony trees determined for 100 bootstrapped data sets of a complete ORF-NS-protein alignment classified NB in a genogroup of the lagoviruses by distance analysis (bootstrap = 88%) or as a new calicivirus genus using parsimony (bootstrap = 44%) (data not shown).
FIG. 2.
Unrooted consensus trees of 2C-like helicase/ATPase (A), 3C-like protease (B), 3D-like RdRp (C), and major capsid (VP-1) proteins (D) of BEC-NB. The lengths and ORF-1 locations of the amino acid sequences used to prepare the multiple alignments of the viruses used in the comparisons are shown in Table 1. The final trees depicted represent a consensus of trees for 100 bootstrapped data sets analyzed using the maximum-likelihood algorithm. The bootstrap values indicate the percentage of replicates supporting the branch points predicted in the consensus trees.
Gn-calf challenge with BEC-NB.
Each of the BEC-NB-inoculated Gn calves developed diarrhea on PID 3 or 4 (Table 3). Fecal changes postexposure followed a progression from a normal thick brown-green paste to a thin tan paste to a yellow liquid with flocculent debris. In all challenged calves, changes in appetite and alertness were mild and were most commonly observed on PID 2 and 3. A summary of diarrhea and RT-PCR and IEM results for calf fecal samples and intestinal histopathologic lesions is shown in Table 3. Of note, no major differences occurred between orally and i.v.-exposed calves in virus shedding or histopathologic lesions. For samples tested by RT-PCR, all calves were negative prechallenge but positive by PID 1 or 2 (Table 3). For three of the four NB virus-exposed calves for which samples were available, the results of RT-PCR tests remained positive through the time of euthanasia on PID 4 or 5 (data not shown).
TABLE 3.
Onset of diarrhea and fecal virus shedding and histopathologic findings in the small intestines of Gn calves after inoculation with BEC-NB strain
| Inoculation route | Calf no. | Age exposed (days) | Age euthanized (days) | Onset of diarrhea (PID) | Onset of fecal virus shedding (PID)
|
Intestinal lesion scorea
|
|||
|---|---|---|---|---|---|---|---|---|---|
| IEM | RT-PCRb | Duodenum | Jejunum | Ileum | |||||
| Oral | 1 | 4 | 8 | 4 | 2 | 1 | 2.5 | 1.5 | 1.0 |
| Oral | 2 | 5 | 10 | 4 | 2 | 2 | 2.5 | 2.0 | 0 |
| i.v. | 3 | 3 | 7 | 3 | 2 | 2 | 3.0 | 1.5 | 0 |
| i.v. | 4 | 7 | 11 | 3 | 2 | 2 | 1.0 | 2.5 | 0 |
| Mock | 5 | 4 | 6 | NDc | ND | 0 | 0 | 0 | |
0, normal; 1, mild villous atrophy; 2, moderate villous atrophy; 3, severe villous atrophy.
All calves (except no. 2 [specimens discarded]) were tested for BEC by RT-PCR preexposure (PID 0) and were negative.
ND, not determined; specimens discarded (freezer failure); other mock-inoculated Gn calves (age matched) tested negative for BEC in feces by IEM and RT-PCR.
Histopathologic lesions were limited to the small intestine and followed a general pattern in which pathological changes were most severe in the anterior small intestine (duodenum) and milder distally (ileum), where lesions, if present, were mild and diffuse in nature (Table 3). In calf 4, intestinal lesions were more severe in the jejunum, while pathological changes were observed in the ileum of one calf, number 1.
In the duodenum and jejunum of calves 1 and 2, pathological changes consisted of a diffuse pattern of villous atrophy, moderate to severe in nature, accompanied by exfoliation and loss of villous enterocytes. Exposed cores of the lamina propria of denuded villi in both calves appeared hypercellular due to stromal condensation. There was coagulative necrosis of the distal one-third of multiple villi demonstrated by a loss of differential staining and pyknosis of leukocytic nuclei. Enterocytes on the lateral aspects of villi often showed moderate attenuation with a low columnar or cuboidal morphology. Villous fusion, when present, was generally mild and focal in appearance. Villous-to-crypt ratios were reduced and fell in a range, measured or estimated, between 1:1 and 3:1 (normal, 4:1 to 6:1). Crypt hyperplasia was demonstrated by elongation, increased numbers of mitotic figures, cytoplasmic basophilia, and increased numbers of irregularly arranged nuclei. In calf 1, changes in the ileum included a patchy detachment of strips of villous enterocytes, with subsequent exposure of cores of the lamina propria and moderate attenuation of lateral villous enterocytes. Most villi were normal in length, but multiple crypts appeared mildly elongated and had increased basophilia indicative of epithelial hyperplasia. Tissues of major organs, including the lung, liver, kidney, and spleen, and lower intestinal sections, including the cecum, spiral colon, and rectum, had no visible microscopic lesions in either the control or BEC-NB-exposed calves.
DISCUSSION
Molecular and phylogenetic studies of human caliciviruses (HuCV) demonstrate that they are genetically diverse (2, 70). Similar studies of caliciviruses from feces or intestinal contents of pigs (29, 77, 78) and calves (9, 52; Smiley and Saif, Proc. 81st Annu. Meet. Conf. Res. Workers Anim. Dis.) indicate that they are genetically more closely related to HuCVs than to other animal caliciviruses. Bovine caliciviruses enteropathogenic for Gn and colostrum-deprived calves, with small round structured morphology like that of human NLVs, have also been reported in Europe and the United States (6, 28, 32, 83). Although these viruses appear to be species specific, it has not yet been determined if such animal enteric caliciviruses can infect humans. Because the characterized BEC sequence database to evaluate such zoonotic potential is small (9, 52, 78), our goal was to determine the NB virus genome sequence, both to enhance the data set and to establish a BEC reference strain of North American origin.
The genetic distinctness of the BEC-NB and CV23-OH strains, compared to the Jena and Newbury agent (NA)-2 NLV BEC strains, was apparent from the initial acquisition of their RdRp sequence data. The BEC-NB strain, a historical isolate originally identified by EM, was initially tested in Gn calves to confirm its enteropathogenicity but could not be detected using either standard BEC or human-NLV primer sets. At 7,453 bp, the genomic sequence of the NB virus strain is closest in size to the NLV genogroup I BEC-Jena strain and to caliciviruses in the genera Lagovirus and SLV (Table 1). The genome of the NB virus also has some unusual features (Fig. 1). At 74 bp, the 5′ UTR of the NB virus is more than three times longer than the 5′UTR of any other calicivirus. There is also a unique separation between the predicted initiation codon of ORF-1 and the putative 5′ UTR and subgenomic conserved sequence segments, which are characteristic of the caliciviruses and have been hypothesized to be potential signal sequences or sites of transcription initiation (8, 37, 57). Evidence for the correct resolution of the 5′ UTR and conserved subgenomic sequences is the localization of a conserved region (nucleotides 5035 to 5057) in ORF-1 at a site 5′ of and contiguous with the initiation codon (nucleotides 5058 to 5060) for the predicted 57.5-kDa protein corresponding to VP1. While it is possible that these results could be due to sequencing artifacts or premature termination of transcription, these possibilities are unlikely due to the determination of similar results for two independent RACE procedures.
Alignment of the translated ORF-1 polyprotein of NB virus with those of caliciviruses of the other genera predicted the NB virus protein map shown in Fig. 1C. Identifying defined and conserved calicivirus structural-functional motifs and proteolytic cleavage sites predicted seven potential cleavage products of NB virus ORF-1. Their predicted order, NH2-P34-P37(2C)-P30-P7.6(VPg)-P20(3C)-P53(3D)-P57(VP1)-COOH, and projected functions (ATPase, VPg, 3C protease, RdRp, and capsid) confirm the NB virus as a member of the family Caliciviridae (8, 13, 55, 64, 74, 79, 80). An unusual feature of the predicted polypeptide of the 3C protease of the NB virus was the substitution of tyrosine (Y) for an aspartic acid (D) residue (underlined) in the characteristic GDCG motif that is found in all other caliciviruses. The cysteine (C) residue has previously been shown to be essential for catalytic activity of the protease (50). Comparison of this same conserved motif in different picornaviruses shows that the composition of the motif can vary as G(D/M/Q/W/Y)CG (3), which suggests that this area of the NB protein has most likely undergone functional or divergent evolution.
Based on our finding that the NB virus is genetically most similar to SLVs and lagoviruses, we also investigated its pathogenicity in Gn calves inoculated orally or i.v. with NB virus fecal filtrates. In particular, others have shown that RHDV, a lagovirus, induces viremia with subsequent spread to the liver, resulting in fatal systemic hemorrhage in rabbits exposed via various routes (61). Moreover, a recent report from our laboratory also confirmed that the SLV PEC/Cowden induces viremia and intestinal, but not systemic, lesions after oral or i.v. exposure of Gn pigs (31).
We found that the type and distribution pattern of histopathologic lesions in the small intestines of NB virus-exposed calves were similar to those described previously for NA-1 (32). Moreover, unlike lagoviruses that induce systemic lesions (61), no lesions were observed in any extraintestinal organs examined (liver, kidney, spleen, and lung) of either i.v.- or orally inoculated Gn calves. These findings are consistent with those reported by Guo et al. (31) after oral or i.v. exposure of Gn pigs to PEC/Cowden. There was no clear indication that the route of inoculation (oral versus i.v.) influenced the onset or distribution of infection, but this issue should be addressed further using a larger number of calves. In addition, since the calves were all euthanized near the onset of diarrhea to examine histopathologic lesions, we did not examine whether viremia occurred, as detected in Gn pigs exposed to PEC/Cowden (31). Our findings, similar to those reported for the genetically uncharacterized BEC NA-1, showed an anterior distribution of lesions in the small intestine which mostly spared the ileum (33). The ileal changes identified in calf 1 were mild in nature and had some features, crypt hyperplasia and elongation, that were also found in the NA-1 study and were postulated to be due to mitotic stimulation by enteroglucagon (32). While lesions produced by the NB virus were judged to be most severe in the duodenum, these results will need to be further confirmed in a temporal study to control for bias introduced in these trials by using the onset of diarrhea as the termination point for euthanizing exposed calves.
The NB strain shares a common genomic organization, based on two ORFs, with SLVs and lagoviruses. The low overall amino acid sequence identities calculated for the NB virus with members of other calicivirus genera (Table 2) and its divergent placement in different genera in phylogenetic analyses suggests that it has undergone a significant period of divergent evolution from a putative ancestral calicivirus progenitor based on a genome organized into two ORFs. Interestingly, the characterization of the genome of the NB virus presents a second example of the identification of an animal species that is host to infection with enteric caliciviruses classified in two of the presently defined calicivirus genera and which are also distinguished on the basis of their genomic organization into either two or three ORFs. These caliciviruses include the human NLVs and SLVs and the BEC Jena and NB strains. A presumptive third pair is represented by the porcine viruses PEC/Cowden and partially characterized NLV genogroup II-like viruses detected in pigs (78). If similar type I (two-ORF) and type II (three-ORF) infectious caliciviruses are found in other species, it may be useful to integrate these patterns of genome organization into schemes for future taxonomic classification and nomenclature systems for the family Caliciviridae.
Acknowledgments
This work was supported in part by the National Institute of Allergy and Infectious Diseases, NIH (grant R01 AI 49716), and USDA, NRI, CGP Grant 1999 02009. Salaries and partial research support were provided by state and federal grants provided to the Ohio Agricultural Research and Development Center (OARDC) of The Ohio State University.
We thank Juliette Hanson, Rich McCormick, Paul Nielsen, and Don Westfall for technical assistance and the staff of the OARDC Molecular and Cellular Imaging Center for DNA sequencing.
REFERENCES
- 1.Ando, T., S. S. Monroe, J. S. Noel, and R. I. Glass. 1997. A one-tube method of reverse transcription-PCR to efficiently amplify a 3-kilobase region from the RNA polymerase gene to the poly(A) tail of small round-structured viruses (Norwalk-like viruses). J. Clin. Microbiol. 35:570-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ando, T., J. S. Noel, and R. L. Fankhauser. 2000. Genetic classification of “Norwalk-like viruses.” J. Infect. Dis. 181(Suppl. 2):S336-S348. [DOI] [PubMed] [Google Scholar]
- 3.Bazan, J. F., and R. J. Fletterick. 1988. Viral cysteine proteases are homologous to the trypsin-like family of serine proteases: structural and functional implications. Proc. Natl. Acad. Sci. USA 85:7872-7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berke, T., B. Golding, X. Jiang, D. W. Cubitt, M. Wolfaardt, A. W. Smith, and D. O. Matson. 1997. Phylogenetic analysis of the caliciviruses. J. Med. Virol. 52:419-424. [DOI] [PubMed] [Google Scholar]
- 5.Bohl, E. H., L. J. Saif, K. W. Theil, A. G. Agnes, and R. F. Cross. 1982. Porcine pararotavirus: detection, differentiation from rotavirus, and pathogenesis in gnotobiotic pigs. J. Clin. Microbiol. 15:312-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bridger, J. C., G. A. Hall, and J. Brown. 1984. Characterization of a calici-like virus (Newbury agent) found in association with astrovirus in bovine diarrhea. Infect. Immun. 43:133-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, M. J., I. D. Milton, J. Meanger, M. Bennett, R. M. Gaskell, and P. C. Turner. 1992. The complete nucleotide sequence of a feline calicivirus. Virology 190:443-448. [DOI] [PubMed] [Google Scholar]
- 8.Clarke, I. N., and P. R. Lambden. 2000. Organization and expression of calicivirus genes. J. Infect. Dis. 181(Suppl. 2):S309-S316 [DOI] [PubMed] [Google Scholar]
- 9.Dastjerdi, A. M., J. Green, C. I. Gallimore, D. W. Brown, and J. C. Bridger. 1999. The bovine Newbury agent-2 is genetically more closely related to human SRSVs than to animal caliciviruses. Virology 254:1-5. [DOI] [PubMed] [Google Scholar]
- 10.Dawson, S., D. Bennett, S. D. Carter, M. Bennett, J. Meanger, P. C. Turner, M. J. Carter, I. Milton, and R. M. Gaskell. 1994. Acute arthritis of cats associated with feline calicivirus infection. Res. Vet. Sci. 56:133-143. [DOI] [PubMed] [Google Scholar]
- 11.Dingle, K. E., P. R. Lambden, E. O. Caul, and I. N. Clarke. 1995. Human enteric Caliciviridae: the complete genome sequence and expression of virus-like particles from a genetic group II small round structured virus. J. Gen. Virol. 76:2349-2355. [DOI] [PubMed] [Google Scholar]
- 12.Dougherty, W. G., and B. L. Semler. 1993. Expression of virus-encoded proteinases: functional and structural similarities with cellular enzymes. Microbiol. Rev. 57:781-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham, D. M., X. Jiang, T. Berke, A. W. Smith, and D. O. Matson. 1998. Genomic mapping of a calicivirus VPg. Arch. Virol. 143:2421-2430. [DOI] [PubMed] [Google Scholar]
- 14.Estes, M. K., R. L. Atmar, and M. E. Hardy. 1997. Norwalk and related diarrhea viruses, p. 1073-1095. In D. D. Richman, R. J. Whitley, and F. G. Hayden (ed.), Clinical virology. Churchill Livingstone, New York, N.Y.
- 15.Estes, M. K., and M. E. Hardy. 1995. Norwalk and other enteric caliciviruses, p. 1009-1034. In M. J. Blaser et al. (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 16.Evermann, J. F., A. J. McKeirnan, A. W. Smith, D. E. Skilling, and R. L. Ott. 1985. Isolation and identification of caliciviruses from dogs with enteric infections. Am. J. Vet. Res. 46:218-220. [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 18.Flynn, W. T., L. J. Saif, and P. D. Moorhead. 1988. Pathogenesis of porcine enteric calicivirus-like virus in four-day-old gnotobiotic pigs. Am. J. Vet. Res. 49:819-825. [PubMed] [Google Scholar]
- 19.Frohman, M. A., M. K. Dush, and G. R. Martin. 1988. Rapid production of full-length cDNA's from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85:8988-9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Froussard, P. 1992. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 20:2900.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glass, P., L. J. White, J. M. Ball, I. Leparc-Goffart, M. E. Hardy, and M. K. Estes. 2000. Norwalk virus open reading frame 3 encodes a minor structural protein. J. Virol. 74:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya, A. E., E. V. Koonin, and Y. I. Wolf. 1990. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 262:145-148. [DOI] [PubMed] [Google Scholar]
- 23.Green, J., J. Vinje, C. Gallimore, M. Koopmans, A. Hale, and D. W. G. Brown. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 24.Green, K. Y. 1997. The role of human caliciviruses in epidemic gastroenteritis. Arch. Virol. Suppl. 13:153-165. [DOI] [PubMed] [Google Scholar]
- 25.Green, K. Y., T. Ando, M. S. Balayan, T. Berke, I. N. Clarke, M. K. Estes, D. O. Matson, S. Nakata, J. D. Neill, M. J. Studdert, and H. J. Thiel. 2000. Taxonomy of the caliciviruses. J. Infect. Dis. 181(Suppl. 2):S322-S330 [DOI] [PubMed] [Google Scholar]
- 26.Green, S. M., P. R. Lambden, Y. Deng, J. A. Lowes, S. Lineham, J. Bushell, J. Rogers, E. O. Caul, C. R. Ashley, and I. N. Clarke. 1995. Polymerase chain reaction detection of small round-structured viruses from two related hospital outbreaks of gastroenteritis using inosine-containing primers. J. Med. Virol. 45:197-202. [DOI] [PubMed] [Google Scholar]
- 27.Grothues, D., C. Cantor, and C. Smith. 1993. PCR amplification of megabase DNA with tagged random primers (T-PCR). Nucleic Acids Res. 21:1321-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gunther, H., P. Otto, and P. Heilmann. 1984. Diarrhea in young calves. 6. Determination of the pathogenicity of a bovine coronavirus and an unidentified icosahedral virus. Arch. Exp. Vet. Med. 38:781-792. [PubMed] [Google Scholar]
- 29.Guo, M., K. O. Chang, M. E. Hardy, Q. Zhang, A. V. Parwani, and L. J. Saif. 1999. Molecular characterization of a porcine enteric calicivirus genetically related to Sapporo-like human caliciviruses. J. Virol. 73:9625-9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo, M., J. F. Evermann, and L. J. Saif. 2001. Detection and molecular characterization of cultivable caliciviruses from clinically normal mink and enteric caliciviruses associated with diarrhea in mink. Arch. Virol. 146:479-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo, M., J. Hayes, K. O. Cho, A. V. Parwani, L. M. Lucas, and L. J. Saif. 2001. Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J. Virol. 75:9239-9251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall, G. A., J. C. Bridger, B. E. Brooker, K. R. Parsons, and E. Ormerod. 1984. Lesions of gnotobiotic calves experimentally infected with a calicivirus-like (Newbury) agent. Vet. Pathol. 21:208-215. [DOI] [PubMed] [Google Scholar]
- 33.Hall, T. A. 1999. Bioedit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 34.Hardy, M. E. 1999. Norwalk and “Norwalk-like viruses” in epidemic gastroenteritis. Clin. Lab. Med. 19:675-690. [PubMed] [Google Scholar]
- 35.Hardy, M. E., and M. K. Estes. 1996. Completion of the Norwalk virus genome sequence. Virus Genes 12:287-290. [DOI] [PubMed] [Google Scholar]
- 36.Hardy, M. E., L. J. White, J. M. Ball, and M. K. Estes. 1995. Specific proteolytic cleavage of recombinant Norwalk virus capsid protein. J. Virol. 69:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herbert, T. P., I. Brierley, and T. D. Brown. 1996. Detection of the ORF3 polypeptide of feline calicivirus in infected cells and evidence for its expression from a single, functionally bicistronic, subgenomic mRNA. J. Gen. Virol. 77:123-127. [DOI] [PubMed] [Google Scholar]
- 38.Jiang, X., W. D. Cubitt, T. Berke, W. Zhong, X. Dai, S. Nakata, L. K. Pickering, and D. O. Matson. 1997. Sapporo-like human caliciviruses are genetically and antigenically diverse. Arch. Virol. 142:1813-1827. [DOI] [PubMed] [Google Scholar]
- 39.Jiang, X., P. Huang, W. Zhong, T. Farkas, D. Cubitt, and D. O. Matson. 1999. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT/PCR. J. Virol. Methods 83:145-154. [DOI] [PubMed] [Google Scholar]
- 40.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 41.Joubert, P., C. Pautigny, M. F. Madelaine, and D. Rasschaert. 2000. Identification of a new cleavage site of the 3C-like protease of rabbit haemorrhagic disease virus. J. Gen. Virol. 81:481-488. [DOI] [PubMed] [Google Scholar]
- 42.Kapikian, A. Z., M. Estes, and R. M. Chanock. 1996. Norwalk group of viruses, p. 783-810. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 43.Kozak, M. 1991. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J. Biol. Chem. 266:19867-19870. [PubMed] [Google Scholar]
- 44.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 45.Lambden, P. R., and I. N. Clarke. 1995. Genome organization of the Caliciviridae. Trends Microbiol. 3:261-265. [DOI] [PubMed] [Google Scholar]
- 46.Lambden, P. R., B. Liu, and I. N. Clarke. 1995. A conserved sequence motif at the 5′ terminus of the Southampton virus genome is characteristic of the Caliciviridae. Virus Genes 10:149-152. [DOI] [PubMed] [Google Scholar]
- 47.Le, G. G., S. Huguet, P. Vende, J. F. Vautherot, and D. Rasschaert. 1996. European brown hare syndrome virus: molecular cloning and sequencing of the genome. J. Gen. Virol. 77:1693-1697. [DOI] [PubMed] [Google Scholar]
- 48.Lew, J. F., A. Z. Kapikian, J. Valdesuso, and K. Y. Green. 1994. Molecular characterization of Hawaii virus and other Norwalk-like viruses: evidence for genetic polymorphism among human caliciviruses. J. Infect. Dis. 170:535-542. [DOI] [PubMed] [Google Scholar]
- 49.Liu, B., I. N. Clarke, E. O. Caul, and P. R. Lambden. 1997. The genomic 5′ terminus of Manchester calicivirus. Virus Genes 15:25-28. [DOI] [PubMed] [Google Scholar]
- 50.Liu, B., I. N. Clarke, and P. R. Lambden. 1996. Polyprotein processing in Southampton virus: identification of 3C-like protease cleavage sites by in vitro mutagenesis. J. Virol. 70:2605-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu, B. L., I. N. Clarke, E. O. Caul, and P. R. Lambden. 1995. Human enteric caliciviruses have a unique genome structure and are distinct from the Norwalk-like viruses. Arch. Virol. 140:1345-1356. [DOI] [PubMed] [Google Scholar]
- 52.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu, B. L., G. J. Viljoen, I. N. Clarke, and P. R. Lambden. 1999. Identification of further proteolytic cleavage sites in the Southampton calicivirus polyprotein by expression of the viral protease in E. coli. J. Gen. Virol. 80:291-296. [DOI] [PubMed] [Google Scholar]
- 54.Loh, E. Y., J. F. Elliot, S. Cwirla, L. L. Lanier, and M. M. Davis. 1989. Polymerase chain reaction with single-sided specificity: analysis of T-cell receptor δ chain. Science 243:217.. [DOI] [PubMed] [Google Scholar]
- 55.Marin Soledad, M., R. Casais, J. Martin Alonso, and F. Parra. 2000. ATP binding and ATPase activities associated with recombinant rabbit hemorrhagic disease virus 2C-like polypeptide. J. Virol. 74:10846-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyers, G., C. Wirblich, and H. J. Thiel. 1991. Genomic and subgenomic RNAs of rabbit hemorrhagic disease virus are both protein-linked and packaged into particles. Virology 184:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyers, G., C. Wirblich, and H. J. Thiel. 1991. Rabbit hemorrhagic disease virus—molecular cloning and nucleotide sequencing of a calicivirus genome. Virology 184:664-676. [DOI] [PubMed] [Google Scholar]
- 59.Neill, J. D., R. F. Meyer, and B. S. Seal. 1995. Genetic relatedness of the caliciviruses: San Miguel sea lion and vesicular exanthema of swine viruses constitute a single genotype within the Caliciviridae. J. Virol. 69:4484-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Neill, J. D., R. F. Meyer, and B. S. Seal. 1998. The capsid protein of vesicular exanthema of swine virus serotype A48: relationship to the capsid protein of other animal caliciviruses. Virus Res. 54:39-50. [DOI] [PubMed] [Google Scholar]
- 61.Ohlinger, V. F., B. Haas, and H. J. Thiel. 1993. Rabbit hemorrhagic disease (RHD): characterization of the causative calicivirus. Vet. Res. 24:103-116. [PubMed] [Google Scholar]
- 62.Page, R. D. 1996. Treeview: an application to display phylogenetic trees on personal computers. Comp. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 63.Palmenberg, A. C. 1989. Sequence alignments of picornaviral capsid proteins, p. 211-241. In B. L. Semler and E. Ehrenfeld (ed.), Molecular aspects of picornavirus infection and detection. American Society for Microbiology, Washington, D.C.
- 64.Pfister, T., and E. Wimmer. 2001. Polypeptide p41 of a Norwalk-like virus is a nucleic acid-independent nucleoside triphosphatase. J. Virol. 75:1611-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pletneva, M. A., S. V. Sosnovtsev, and K. Y. Green. 2001. The genome of Hawaii virus and its relationship with other members of the Caliciviridae. Virus Genes 23:5-16. [DOI] [PubMed] [Google Scholar]
- 66.Reubel, G. H., D. E. Hoffmann, and N. C. Pedersen. 1992. Acute and chronic faucitis of domestic cats. A feline calicivirus-induced disease. Vet. Clin. N. Am. Small Anim. Pract. 22:1347-1360. [DOI] [PubMed] [Google Scholar]
- 67.Rinehart-Kim, J. E., W. M. Zhong, X. Jiang, A. W. Smith, and D. O. Matson. 1999. Complete nucleotide sequence and genomic organization of a primate calicivirus, Pan-1. Arch. Virol. 144:199-208. [DOI] [PubMed] [Google Scholar]
- 68.Saif, L. J., E. H. Bohl, E. M. Kohler, and J. H. Hughes. 1977. Immune electron microscopy of transmissible gastroenteritis virus and rotavirus (reovirus-like agent) of swine. Am. J. Vet. Res. 38:13-20. [PubMed] [Google Scholar]
- 69.Saif, L. J., E. H. Bohl, K. W. Theil, R. F. Cross, and J. A. House. 1980. Rotavirus-like, calicivirus-like and 23 nm virus-like particles associated with diarrhea in young pigs. J. Clin. Microbiol. 12:111.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schuffenecker, I., T. Ando, D. Thouvenot, B. Lina, and M. Aymard. 2001. Genetic classification of “Sapporo-like viruses.” Arch. Virol. 146:2115-2132. [DOI] [PubMed] [Google Scholar]
- 71.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith, D. R., H. Tsunemitsu, R. A. Heckert, and L. J. Saif. 1996. Evaluation of two antigen-capture ELISAs using polyconal or monoclonal antibodies for the detection of bovine coronavirus. J. Vet. Diagn. Investig. 8:99-105. [DOI] [PubMed] [Google Scholar]
- 73.Smith, D. R., P. R. Nielsen, K. L. Gadfield, and L. J. Saif. 1998. Further evaluation of antibody-capture and antigen-capture ELISAs for determining exposure of cattle to bovine coronavirus. Am. J. Vet. Res. 59:956-960. [PubMed] [Google Scholar]
- 74.Sosnovtsev, S. V., and K. Y. Green. 2000. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology 277:193-203. [DOI] [PubMed] [Google Scholar]
- 75.Sosnovtsev, S. V., S. A. Sosnovtseva, and K. Y. Green. 1998. Cleavage of the feline calicivirus capsid precursor is mediated by a virus-encoded proteinase. J. Virol. 72:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sosnovtseva, S. A., S. V. Sosnovtsev, and K. Y. Green. 1999. Mapping of the feline calicivirus proteinase responsible for autocatalytic processing of the nonstructural polyprotein and identification of a stable proteinase-polymerase precursor protein. J. Virol. 73:6626-6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sugieda, M., H. Nagaoka, Y. Kakishima, T. Ohshita, S. Nakamura, and S. Nakajima. 1998. Detection of Norwalk-like virus genes in the caecum contents of pigs. Arch. Virol. 143:1215-1221. [DOI] [PubMed] [Google Scholar]
- 78.van der Poel, W. H. M., J. Vinje, R. van der Heide, M. I. Herrera, A. Vivo, and M. P. Koopmans. 2000. Norwalk-like calicivirus genes in farm animals. Emerg. Infect. Dis. 6:36-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vazquez, A. L., J. M. Alonso, R. Casais, and J. A. Boga. 1998. Expression of enzymatically active rabbit hemorrhagic disease virus RNA-dependent RNA polymerase in Escherichia coli. J. Virol. 72:2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vazquez, A. L., J. M. Alonso, and F. Parra. 2000. Mutation analysis of the GDD sequence motif of a calicivirus RNA-dependent RNA polymerase. J. Virol. 74:3888-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 82.Wirblich, C., H. J. Thiel, and G. Meyers. 1996. Genetic map of the calicivirus rabbit hemorrhagic disease virus as deduced from in vitro translation studies. J. Virol. 70:7974-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Woode, G., and J. C. Bridger. 1978. Isolation of small viruses resembling astroviruses and caliciviruses from acute enteritis of calves. J. Med. Microbiol. 11:441-452. [DOI] [PubMed] [Google Scholar]