Abstract
A systematic analysis of immune responses on a population level is critical for a human immunodeficiency virus type 1 (HIV-1) vaccine design. Our studies in Botswana on (i) molecular analysis of the HIV-1 subtype C (HIV-1C) epidemic, (ii) frequencies of major histocompatibility complex class I HLA types, and (iii) cytotoxic T-lymphocyte (CTL) responses in the course of natural infection allowed us to address HIV-1C-specific immune responses on a population level. We analyzed the magnitude and frequency of the gamma interferon ELISPOT-based CTL responses and translated them into normalized cumulative CTL responses. The introduction of population-based cumulative CTL responses reflected both (i) essentials of the predominant virus circulating locally in Botswana and (ii) specificities of the genetic background of the Botswana population, and it allowed the identification of immunodominant regions across the entire HIV-1C. The most robust and vigorous immune responses were found within the HIV-1C proteins Gag p24, Vpr, Tat, and Nef. In addition, moderately strong responses were scattered across Gag p24, Pol reverse transcriptase and integrase, Vif, Tat, Env gp120 and gp41, and Nef. Assuming that at least some of the immune responses are protective, these identified immunodominant regions could be utilized in designing an HIV vaccine candidate for the population of southern Africa. Targeting multiple immunodominant regions should improve the overall vaccine immunogenicity in the local population and minimize viral escape from immune recognition. Furthermore, the analysis of HIV-1C-specific immune responses on a population level represents a comprehensive systematic approach in HIV vaccine design and should be considered for other HIV-1 subtypes and/or different geographic areas.
Human immunodeficiency virus type 1 subtype C (HIV-1C) has become the most prevalent subtype in the current AIDS epidemic (32, 76) and is predicted to dominate in the coming years (32, 76). Responsible for the largest proportion of HIV-1 infections worldwide, HIV-1C apparently accounted for 47.2% of new HIV-1 infections in the year 2000 (76). Distinguishing features of the HIV-1C epidemics include but are not limited to (i) a high prevalence rate (up to 20 to 40%) in the adult population (93, 94), (ii) higher odds of vertical transmission (81), (iii) high viral loads (68), (iv) a high level of viral diversity (24, 72, 95), (v) preferential CCR5 coreceptor usage (1, 19, 78, 92), and (vi) a number of unique subtype signatures across the viral genome (39, 72, 82, 84).
Vaccine protection against pathogenic simian immunodeficiency virus or simian-human immunodeficiency virus was demonstrated in a series of experiments with nonhuman primates (reviewed in references 6, 49, and 65). The induction of strong cytotoxic T-lymphocyte (CTL) responses that are able to control viral replication and prevent clinical disease progression was shown to be a necessary component of successful vaccination in a rhesus macaque model (8, 10, 12, 13, 77, 91; T. M. Fu, W. Trigona, M.-E. Davies, Z.-Q. Zhang, D. Casimiro, S. Dubey, D. C. Freed, J. Joyce, K. Grimm, W. A. Schleif, N. L. Letvin, E. A. Emini, and J. W. Shiver, AIDS Vaccine 2001, p. 35, 2001; J. Shiver, AIDS Vaccine 2001, p. 139, 2001). A variety of viral (5, 10, 14, 25, 34, 40, 44, 45, 64, 86, 87) and bacterial (3, 29, 37, 46, 47, 55-57, 62, 73, 80, 89, 90) vectors expressing HIV-1 antigens were reported to elicit potent HIV-1-specific CTL responses, including efficient mucosal immunity. The role of CTLs in inducing and maintaining efficient immune responses was addressed in a number of HIV-1 vaccine trials (43). However, viral escape from CTL recognition during the course of experimental infection (4, 33) or from vaccine-generated CTL protection (11) in the rhesus macaque model suggested a functional impairment of CTLs and highlighted the critical role of stimulation of both neutralizing antibodies and cellular immune responses in achieving vaccine protection.
Ideally, vaccine formulation should target the induction of protective immune responses against HIV-1 and, in particular, broadly protective CTL responses. However, correlates of immune protection are still unknown, and a reliable mechanism to distinguish and select protective immune responses is not yet available. The ability of the immune system to focus T-cell responses to a distinct profile of epitopes, defined here as immunodominance (17, 98), raises interest in the protection potency of the epitopes. Identification of immunodominant epitopes or epitope-rich immunodominant regions that can stimulate a broad range of HIV-specific CTLs may offer the best mode of protection (38) and is a legitimate approach to vaccine design. Assuming that the overall CTL responses in the natural course of HIV-1 infection include protective immune responses, we attempted to identify HIV-1C-specific CTL responses across the entire viral genome on a population level.
The identification of immunodominant regions and fine mapping of epitopes are intimate parts of preparedness for vaccine trials, including vaccine design and development. However, within the extensive list of identified HIV-1-specific epitopes (53), there is an obvious lack of non-B subtype epitopes representative of the areas or populations most affected by the AIDS epidemic. Despite a demonstration of cross-clade immunity in HIV-1 infection (18, 22, 31, 63, 97), a number of subtype-specific immune responses have been reported (23, 30, 83) and warrant further studies to address the comparative significance of cross-reactive versus clade-specific HIV-1 immunity.
Recently we characterized virus-specific CTL responses within four HIV-1C proteins, namely, Gag, Tat, Rev, and Nef (70). In this study we extended screening of CTL responses to all HIV-1C proteins in the cohort of HIV-infected but asymptomatic blood donors in Botswana, a southern African country with a high prevalence of HIV-1C infection. We focused on the comparative analysis of the magnitude and frequency of CTL responses in the course of natural HIV-1C infection. The main question addressed in this study was comparative cell-mediated responses across the HIV-1C proteins on the population level. CTL responses were analyzed by using the gamma interferon (IFN-γ) ELISPOT and overlapping synthetic peptides that spanned all HIV-1C proteins. CTL responses were ranked by their magnitude and frequency and were expressed as normalized cumulative HIV-1C-specific CTL responses, which reflected both (i) the essentials of the predominant circulating virus and (ii) specificities of the genetic background of the Botswana population through the major histocompatibility complex (MHC) class I restriction of CTL responses. These identified immunodominant regions throughout HIV-1C should be considered in the design of a vaccine for the population of southern Africa.
MATERIALS AND METHODS
Study subjects.
Sample collection was performed according to the guidelines of the Institutional Review Boards of the Ministry of Health of Botswana and the Harvard School of Public Health as described previously (70). Briefly, peripheral blood mononuclear cells (PBMC) were isolated from the discarded blood units obtained anonymously from asymptomatic donors who tested HIV seropositive at the National Blood Transfusion Centre in Gaborone, Botswana, during the period from February 2000 to July 2001. Details of the HIV testing and sample selection were described previously (70). The total number of study subjects was 105. Plasma viral load data were available for 103 cases (median, 37,769 copies/ml; range, <400 to >750,000 copies/ml). CD4 and CD8 data were available for 98 study subjects. The medians for CD4 and CD8 counts were 434 and 984, and mean values were 451 and 1,006, respectively. As described previously (70), samples were assigned for the ELISPOT screening of HIV-1C-specific CTL responses based on the actual PBMC viability and availability prior to when any relevant information, including viral load, CD4, CD8, viral sequence, or HLA typing data, had become available. Plasma viral load, CD4 and CD8 counts, and HLA type were determined as described previously (70).
Synthetic peptides.
PBMC were screened for CTL responses in the IFN-γ ELISPOT assay within HIV-1C Gag, Pol, Vif, Vpr, Tat, Rev, Vpu, Env, and Nef by using overlapping peptides of 15 to 20 amino acids (aa) in length that overlapped by 10 aa. Forty-nine HIV-1C Gag synthetic peptides that corresponded to the sequence of isolate 96ZM651.8 (accession number AF286224) were received from the National Institutes of Health AIDS Research and Reference Reagent Program. HIV-1C consensus amino acid sequences were generated based on the full-length genome sequences (67, 71, 72). A program, PeptGen (21), was employed for the design of most of the synthetic peptides. Peptides spanning variable regions were represented by two or three variants. Peptides were commercially synthesized using 9-fluorenylmethoxy carbonyl chemistry. The purity of peptides was established by high-pressure liquid chromatography and in most cases was >85%.
ELISPOT assay.
HIV-1C-specific CTL responses were measured by quantification of the IFN-γ release in a screening ELISPOT assay as described previously (70) with overlapping synthetic peptides representing the HIV-1C consensus sequence (21), a standard method widely used in CTL epitope screening studies. Briefly, MultiScreen 96-well membrane plates (MAIP S45; Millipore) were coated with 100 μl (0.5 μg/ml in phosphate-buffered saline) of an anti-IFN-γ monoclonal antibody, 1-D1K (Mabtech AB, Nacka, Sweden). Synthetic peptides were added directly to wells at a final concentration of 10 μM following an extensive phosphate-buffered saline wash. Frozen PBMC were thawed, washed in R10 twice, and plated into the wells at a concentration of 25,000 to 100,000 cells/well. The plates were incubated at 37°C with 5% CO2 for 20 to 40 h. Incubation with biotinylated anti-IFN-γ monoclonal antibody 7-B6-1 (Mabtech AB) and streptavidin-alkaline phosphatase conjugate (Mabtech AB) was followed by color development with an alkaline phosphatase conjugate substrate kit (Bio-Rad, Hercules, Calif.). IFN-γ-producing cells were counted by direct visualization or by using a stereomicroscope and were expressed as spot-forming cells (SFC) per million PBMC. The number of specific IFN-γ-secreting T cells was calculated by subtracting the negative control value. The negative controls were <30 SFC per million PBMC. Only responses with a magnitude of >100 SFC per million PBMC were considered positive responses in all screening tests, which corresponded to >10 spots at the PBMC concentration of 100,000 per well. The CD8+ T-cell specificity of the IFN-γ ELISPOT-based responses to synthetic peptides was demonstrated previously by others (41, 42) and us (70) in a series of CD8 and CD4 depletion and enrichment experiments. Cumulative CTL responses were measured as described previously (70) by summarizing the responses of per-patient analyses. Thus, the responses were cumulative because they represent summed immune responses, which reflect both magnitude and frequency. Comparison of cumulative CTL responses among HIV-1C proteins was performed by normalizing the data for the number of study subjects screened (the cumulative CTL responses observed per particular peptide were divided by the number of study subjects screened). Thus, the responses were normalized to avoid influence of the number of samples tested, which allowed us to compare and rank them between and across the viral proteins.
Amino acid diversity.
To compute pairwise distances within the regions, an alignment of the HIV-1C sequences was used (71). Amino acid distances were calculated by the PROTDIST program with the PAM model (35, 36).
Statistical analysis.
Statistical analyses and basic graphical delineations were performed using SigmaPlot 2001 (SPSS Inc.), Splus version 6.0 (Insightful Corp.), and Microsoft Excel 2000 (Microsoft Corp.) enhanced by the package Analyze-it (Analyze-it Software, Ltd.). Adobe Illustrator version 8.0 software was used for final graphical presentation. The frequency of CTL responses was compared between HIV-1C proteins by using a chi-square test. For the samples with a positive response, the magnitude of CTL responses was compared between HIV-1C proteins by using a Wilcoxon rank sum test. A rank-based test was used rather than a two-sample t test because the CTL responses tended to be skewed and nonnormal and because it appropriately accounted for the censoring of within-well ELISPOT responses of >50 spots/well. Because CTL responses to different proteins were measured with samples from the same individuals, the independent-samples assumption of the chi-square and Wilcoxon tests could potentially be violated. However, the CTL responses to a variety of synthetic peptides that represented distinct viral proteins and expressed different amino acid sequences were measured. Also, CTL responses were grouped into sets on the peptide basis but not on the sample/individual basis or CTL response basis. Therefore, CTL responses to each peptide can be considered independent sets, and thus the tests are valid, at least approximately.
For both CTL frequency and magnitude, a total of 91 pairs of proteins were compared. To adjust for the large number of tests, the false discovery rate procedure was used (15), with the rate of false-positive tests controlled to less than 5%. Therefore, of the findings identified as statistically significant, fewer than 5% are expected to be false.
To assess whether mean pairwise diversity in an immunodominant region was different than that in the entire corresponding protein, a modification of a one-sample t test that accounted for nonindependence of the differences in pairwise distances was used. The test is a one-sample analogue of the two-sample test introduced previously (71). Briefly, the numerator of the Z-statistic is the average of the differences in pairwise distances between subregions and complete regions. The denominator is the standard error of the numerator. To compute it, with n as the number of sequences, there are m = n(n − 1)/2 differences in pairwise distances, and a calculation described in reference 71 shows that c = m(m − 1) − m(n − 2)(n − 3)/2 of the m(m − 1) pairs of differences of pairwise distances have positive correlation due to sharing of a common sequence. Following reference 71, by assuming a common correlation and estimating it by Pearson's correlation, r, the standard error equals the square root of [(1/m) + (c × r/m2)] multiplied by the standard deviation calculated by using all differences in pairwise distances. The resulting Z-statistic was compared to a standard normal distribution to test the null hypothesis. Since many tests were conducted, the false discovery rate procedure was used to determine the cutoff P value that implies statistical significance. All tests were two tailed.
RESULTS
Magnitude of HIV-1C-specific CTL responses.
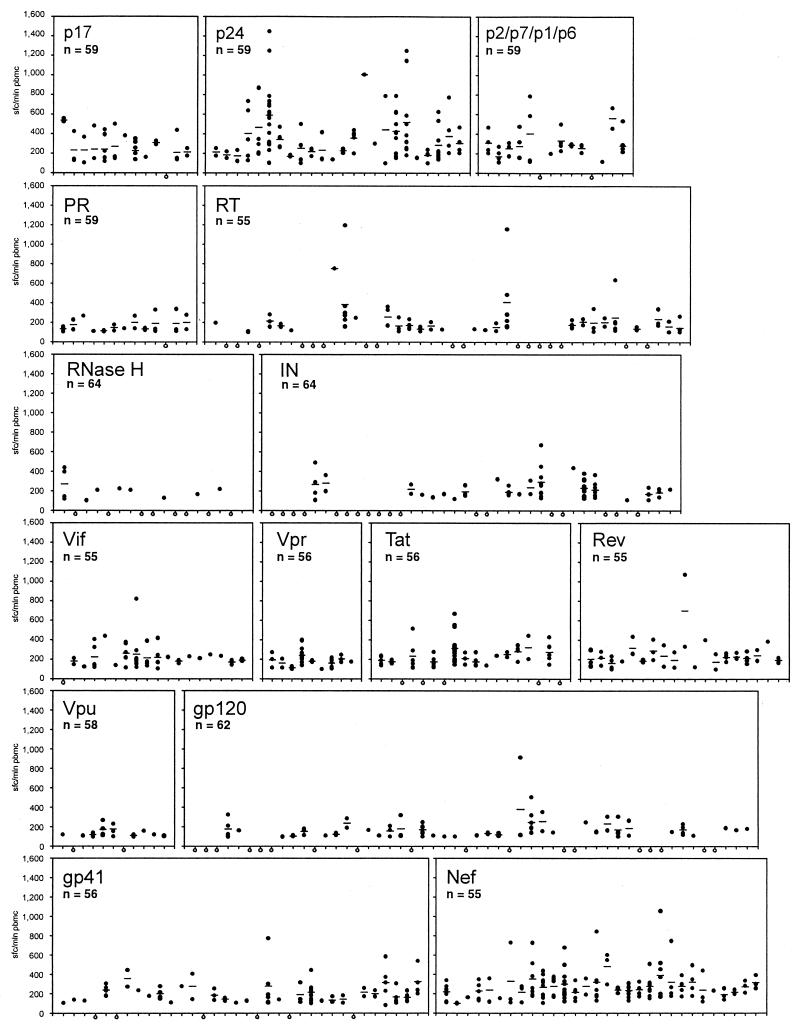
The magnitude of the HIV-1C-specific CTL responses was addressed on a population level. The CTL profiles throughout the HIV-1C proteins are presented in Fig. 1 as a digest of individual IFN-γ ELISPOT-based CTL responses to the overlapping synthetic peptides that span the HIV-1C proteins. The corresponding statistics are introduced in Tables 1 and 2. Figure 1 highlights profiles, magnitudes, and densities of CTL responses across the HIV-1C proteins. Overall, a wide range of CTL response magnitudes was detected throughout HIV-1C, both within and between HIV-1C proteins (Table 1). The highest magnitude was detected within the subset of synthetic peptides straddling the Gag p24 (mean value of 388 SFC/106 PBMC; median, 300 SFC/106 PBMC), followed by Gag p2/p7/p1/p6 (mean value of 292 SFC/106 PBMC; median, 271 SFC/106 PBMC) and Nef (mean value of 278 SFC/106 PBMC; median, 241 SFC/106 PBMC). The lowest magnitude was in the Vpu (mean value of 142 SFC/106 PBMC; median, 122 SFC/106 PBMC). Comparison of the magnitudes of the CTL responses between the HIV-1C proteins revealed a number of significant differences (Table 2). The magnitude within Gag p24 was significantly higher than that in any other HIV-1C protein, except for the Gag region that corresponded to the nucleocapsid and p6 domain, in which case the difference was not statistically significant. Nef demonstrated a higher magnitude than the Pol (except RNase H) or Env proteins, as well as Vif and Vpr. Differences between Nef and p17, Nef and p2/p7/p1/p6, Nef and RNase H, Nef and Tat, or Nef and Rev were not statistically significant, indicating that CTL epitopes that originated in these regions can induce comparable magnitudes. The average magnitude of the HIV-1C-specific CTL responses in responders was 344 SFC/106 PBMC (95% confidence interval [CI], 310 to 378) within the entire Gag protein, 215 SFC/106 PBMC (95% CI, 193 to 237) in Pol, and 197 SFC/106 PBMC (95% CI, 178 to 216) in Env.
FIG. 1.
Magnitude of HIV-1C-specific CTL responses. The x axis of each graph was scaled according to the number of HIV-1C synthetic peptides used for a particular viral protein, and the length of the graph does not necessarily correspond to the actual size of the viral protein because of the differences in the lengths of the synthetic peptides. The y axis was scaled equally for each viral protein. Filled dots represent individual HIV-1C-specific CTL responses to particular synthetic peptides. CTL responses were expressed as SFC per million PBMC (sfc/mln pbmc). Only responses equal to or higher than 100 SFC/106 PBMC were taken into account. Open dots represent nonresponsive synthetic peptides. n represents the number of samples tested with a particular set of synthetic peptides. PR, protease; IN, integrase.
TABLE 1.
Magnitude and density of HIV-1C-specific CTL responsesa
| Protein | No. of synthetic peptides | Magnitude of response (SFC/106 PBMC)
|
Density of responseb | Frequency of high CTL responses (%)c | No. (%d) of nonresponsive peptides | ||
|---|---|---|---|---|---|---|---|
| Mean ± SD | Median | Range | |||||
| Gag p17 | 13 | 258 ± 143 | 200 | 100-556 | 0.046 | 5.7 | 1 (7.7) |
| Gag p24 | 24 | 388 ± 270 | 300 | 100-1,447 | 0.078 | 23.4 | 0 |
| Gag p2/p7/p1/p6 | 14 | 292 ± 162 | 271 | 100-786 | 0.046 | 10.5 | 2 (14.3) |
| Pol protease | 13 | 163 ± 67 | 135 | 107-338 | 0.040 | 0 | 1 (7.7) |
| Pol RT | 44 | 226 ± 192 | 168 | 100-1,195 | 0.031 | 5.3 | 17 (38.6) |
| Pol RNase H | 17 | 214 ± 110 | 210 | 102-438 | 0.010 | 0 | 9 (52.9) |
| Pol integrase | 38 | 227 ± 106 | 190 | 108-672 | 0.025 | 1.6 | 18 (47.4) |
| Vif | 18 | 223 ± 122 | 187 | 104-817 | 0.048 | 2.1 | 1 (5.6) |
| Vpr | 9 | 196 ± 73 | 196 | 101-403 | 0.067 | 0 | 0 |
| Tat | 18 | 249 ± 115 | 218 | 117-667 | 0.061 | 6.6 | 5 (27.8) |
| Rev | 19 | 244 ± 143 | 220 | 100-1,074 | 0.049 | 2.0 | 0 |
| Vpu | 11 | 142 ± 51 | 122 | 100-269 | 0.030 | 0 | 2 (18.2) |
| Env gp120 | 52 | 180 ± 115 | 146 | 100-916 | 0.023 | 2.7 | 18 (34.6) |
| Env gp41 | 34 | 213 ± 117 | 179 | 101-769 | 0.041 | 3.8 | 6 (17.6) |
| Nef | 30 | 278 ± 152 | 241 | 100-1,057 | 0.085 | 7.2 | 0 |
The magnitudes and densities were computed using samples with a positive response only.
Density was computed as the number of CTL responses higher than 100 SFC/106 PBMC within the protein normalized by the number of peptides used and the number of samples tested [density = number of CTL responses/(number of peptides × number of samples)].
Percentage of responses that were equal to or higher than 500 SFC/106 PBMC out of the total number of responses within a particular viral protein.
Percentage of peptides with no responses out of the total number of synthetic peptides within a particular viral protein.
TABLE 2.
Magnitude of CTL responses: relationship between HIV-1C proteinsa
| Protein |
P value (Wilcoxon rank sum test) or median difference in magnitudeb:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag p17 | Gag p24 | Gag p2/p7/p1/p6 | Pol protease | Pol RT | Pol RNase H | Pol integrase | Vif | Vpr | Tat | Rev | Vpu | Env gp120 | Env gp41 | Nef | |
| Gag p17 | 0.005 | 0.340 | 0.004 | 0.168 | 0.520 | 0.819 | 0.638 | 0.294 | 0.706 | 0.976 | 0.001 | 0.004 | 0.227 | 0.248 | |
| Gag p24 | −76.0 | 0.093 | <0.001 | <0.001 | 0.014 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | 0.004 | |
| Gag p2/p7/p1/p6 | −30.0 | 46.0 | <0.001 | 0.002 | 0.103 | 0.041 | 0.021 | 0.004 | 0.269 | 0.128 | <0.001 | <0.001 | 0.003 | 0.774 | |
| Pol protease | 46.1 | 140.0 | 101.4 | 0.020 | 0.170 | <0.001 | 0.001 | 0.045 | <0.001 | <0.001 | 0.087 | 0.752 | 0.017 | <0.001 | |
| Pol RT | 23.6 | 106.4 | 70.2 | −24.2 | 0.769 | 0.051 | 0.179 | 0.630 | 0.003 | 0.010 | 0.001 | 0.022 | 0.687 | <0.001 | |
| Pol RNase H | 22.1 | 101.6 | 64.5 | −36.7 | −8.2 | 0.511 | 0.785 | 0.741 | 0.240 | 0.281 | 0.037 | 0.157 | 0.931 | 0.089 | |
| Pol integrase | 4.5 | 87.7 | 47.0 | −46.5 | −22.7 | −16.2 | 0.652 | 0.374 | 0.207 | 0.341 | <0.001 | <0.001 | 0.145 | 0.008 | |
| Vif | 9.7 | 93.8 | 57.4 | −43.8 | −16.1 | −5.2 | 6.6 | 0.563 | 0.089 | 0.149 | <0.001 | 0.001 | 0.433 | 0.004 | |
| Vpr | 30.5 | 109.5 | 69.1 | −44.7 | −7.4 | 5.3 | 16.4 | 7.5 | 0.026 | 0.048 | 0.004 | 0.025 | 0.965 | <0.001 | |
| Tat | −10.0 | 70.0 | 24.0 | −68.0 | −40.0 | −33.6 | −18.5 | −24.7 | −36.0 | 0.698 | <0.001 | <0.001 | 0.009 | 0.200 | |
| Rev | 0.0 | 77.0 | 32.4 | −68.5 | −38.4 | −27.6 | −16.2 | −21.2 | −29.2 | 6.4 | <0.001 | <0.001 | 0.040 | 0.125 | |
| Vpu | 61.2 | 157.8 | 119.0 | 13.0 | 39.4 | 51.3 | 61.8 | 56.6 | 58.9 | 84.6 | 82.9 | 0.071 | <0.001 | <0.001 | |
| Env gp120 | 43.3 | 132.1 | 96.0 | −3.1 | 20.2 | 27.4 | 44.5 | 37.5 | 30.0 | 60.0 | 62.8 | −16.8 | 0.010 | <0.001 | |
| Env gp41 | 23.6 | 104.1 | 59.3 | −30.7 | −4.0 | −1.6 | 17.1 | 10.5 | −0.3 | 35.0 | 31.2 | −46.3 | −24.8 | <0.001 | |
| Nef | −25.0 | 53.1 | 7.9 | −88.6 | −58.7 | −50.6 | −37.4 | −44.4 | −56.5 | −19.4 | −25.0 | −105.4 | −80.7 | −54.8 | |
The magnitude was computed using samples with a positive response only.
The P values from the Wilcoxon rank sum test are shown in the upper triangle. According to the false discovery rate adjustment method (see Materials and Methods), only P values of <0.024 (in boldface) are significant. The median differences in magnitude between the proteins are shown in the Tower triangle.
The density of HIV-1C-specific CTL responses, measured as a number of significant CTL responses normalized by both a number of peptides and a number of samples, was addressed (Table 1). Nef and Gag p24 demonstrated the highest density of CTL responses (0.085 and 0.078, respectively). The lowest density was observed within the Pol RNase H (0.010), Env gp120 (0.023), Pol integrase (0.025), Vpu (0.030), and Pol reverse transcriptase (RT) (0.031). A high frequency of the top-level CTL responses (higher than 500 SFC/106 PBMC) was observed predominantly in Gag p24 (frequency of 23.4% among all p24 CTL responses), although Gag p2/p7/p1/p6, Nef, and Tat also demonstrated a substantial amount of high CTL responses (frequencies of 10.5, 7.2, and 6.6%, respectively). A relatively high number of nonresponsive synthetic peptides were noticed in Pol RNase H (52.9% of the peptides used), integrase (47.4%), RT (38.6%), Env gp120 (34.6%), and Tat (27.8%).
Frequency of HIV-1C-specific CTL responses.
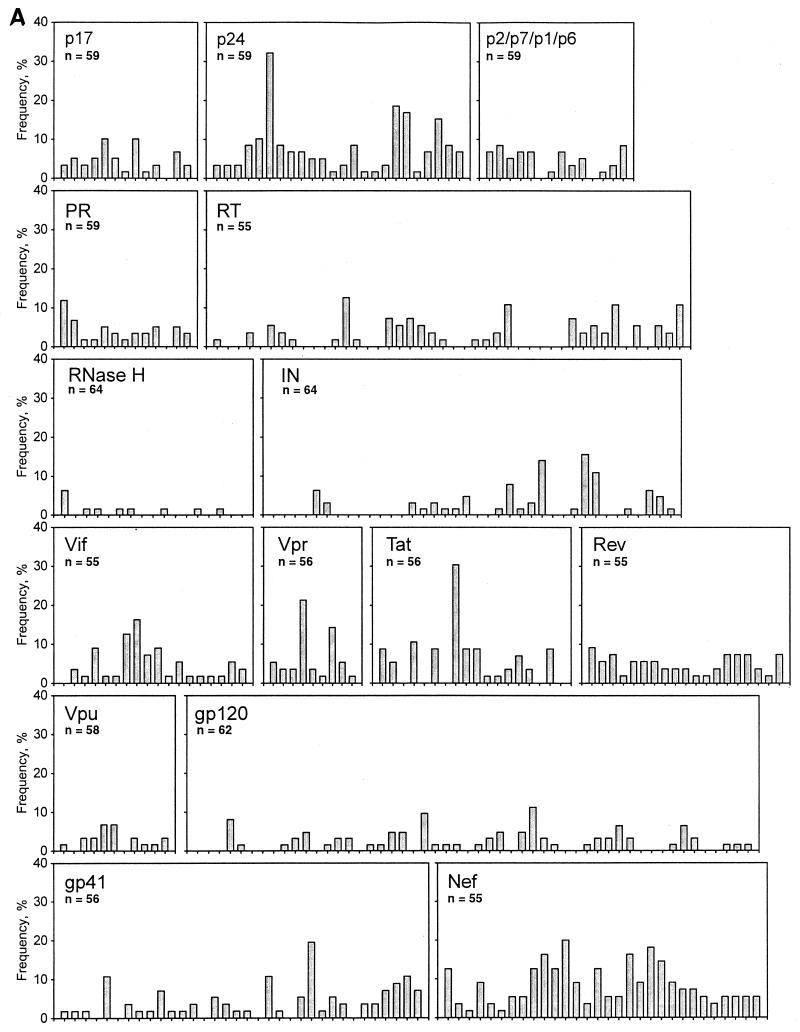
To address the frequency of the HIV-1C-specific CTL responses and their profiles, the ELISPOT-based CTL responses to each synthetic peptide were presented in the form of the fraction of tested samples responding to a particular peptide for all HIV-1C proteins (Fig. 2A). The average frequency of CTL responses across all viral proteins was relatively low (mean value, 4.3%; 95% CI, 3.8 to 4.8%; median, 3.4%). The frequency varied in a wide range from 0 to 32%, which illuminated a dramatically uneven distribution of CTL responses across and within the HIV-1C proteins. The frequencies of CTL responses to a number of synthetic peptides within Gag p24, Vpr, Tat, Env gp41, and Nef were at the level of 20%, while in two cases the frequencies reached 32.2 and 30.4% (Gag p24 and Tat, respectively).
FIG. 2.
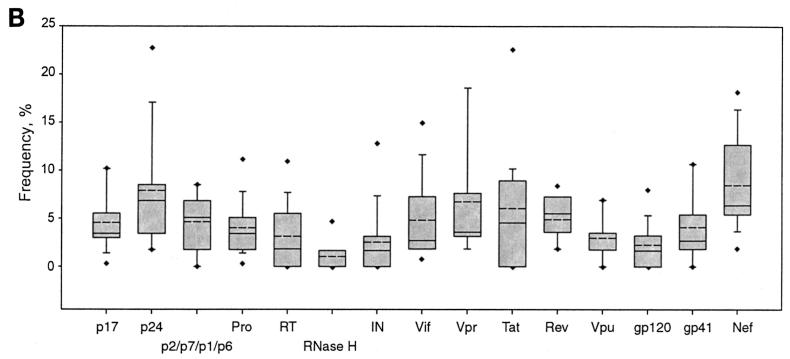
(A) Frequency of HIV-1C-specific CTL responses. The x axis of each graph was scaled according to the number of HIV-1C synthetic peptides used for a particular viral protein, and the length of the graph does not necessarily correspond to the actual size of the viral protein because of the differences in the lengths of the synthetic peptides. The y axis was scaled equally for each viral protein. Bars represent percent frequency for a particular synthetic peptide. n represents the number of samples tested with a particular set of synthetic peptides. PR, protease; IN, integrase. (B) Frequency distribution of HIV-1C-specific CTL responses to the synthetic peptides representing viral proteins. The boundary of the box closest to zero indicates the 25th percentile, a solid line within the box marks the median, a dashed line within the box shows the mean value, and the boundary of the box farthest from zero indicates the 75th percentile. Bars below and above the boxes indicate the 10th and 90th percentiles, respectively. Points below and above the bars indicate the 5th and 95th percentiles, respectively, when the sample size permitted these calculations. Pro, protease; IN, integrase.
The spectrum of frequencies of CTL responses within and between HIV-1C proteins is presented in Fig. 2B. Table 3 highlights significant differences in the CTL frequency between different proteins. Overall, Gag p24 and Nef demonstrated higher frequencies, while Pol proteins, Vpu, and Env gp120 showed the lowest frequencies. The frequencies of CTL responses in Gag p24 and Nef were significantly higher than those in Gag p17, Pol protease, Pol RT, Pol RNase H, Pol integrase, Vif, Rev, Vpu, Env gp120, and Env gp41. The Nef frequency was also higher than those in Gag p2/p7/p1/p6 and Tat. Peptides spanning Pol RNase H, Pol integrase, and Env gp120 demonstrated a significantly lower frequency of CTL responses than any other HIV-1C protein, except for the Pol RT and Vpu.
TABLE 3.
Frequency of CTL responses: relationship between HIV-1C proteins
| Protein |
P value (chi-square test) or difference in frequency (%)a:
|
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag p17 | Gag p24 | Gag p2/p7/p1/p6 | Pol protease | Pol RT | Pol RNase H | Pol integrase | Vif | Vpr | Tat | Rev | Vpu | Env gp120 | Env gp41 | Nef | |
| Gag p17 | 0.005 | 0.933 | 0.706 | 0.079 | <0.001 | 0.005 | 0.862 | 0.121 | 0.204 | 0.841 | 0.118 | <0.001 | 0.663 | <0.001 | |
| Gag p24 | −3.3 | 0.004 | <0.001 | <0.001 | <0.001 | <0.001 | 0.005 | 0.484 | 0.108 | 0.005 | <0.001 | <0.001 | <0.001 | 0.560 | |
| Gag p2/p7/p1/p6 | 0.0 | 3.2 | 0.671 | 0.063 | <0.001 | 0.004 | 0.888 | 0.121 | 0.206 | 0.863 | 0.105 | <0.001 | 0.617 | <0.001 | |
| Pol protease | 0.5 | 3.8 | 0.6 | 0.275 | <0.001 | 0.036 | 0.488 | 0.044 | 0.075 | 0.462 | 0.273 | 0.010 | 1.000 | <0.001 | |
| Pol RT | 1.4 | 4.7 | 1.5 | 0.9 | <0.001 | 0.215 | 0.020 | <0.001 | <0.001 | 0.016 | 0.777 | 0.061 | 0.110 | <0.001 | |
| Pol RNase H | 3.6 | 6.8 | 3.6 | 3.0 | 2.1 | 0.006 | <0.001 | <0.001 | <0.001 | <0.001 | 0.009 | 0.012 | <0.001 | <0.001 | |
| Pol integrase | 2.1 | 5.3 | 2.1 | 1.5 | 0.6 | −1.5 | <0.001 | <0.001 | <0.001 | <0.001 | 0.764 | 0.663 | 0.004 | <0.001 | |
| Vif | −0.3 | 3.0 | −0.2 | −0.8 | −1.7 | −3.8 | −2.3 | 0.160 | 0.277 | 1.000 | 0.058 | <0.001 | 0.399 | <0.001 | |
| Vpr | −2.2 | 1.1 | −2.1 | −2.7 | −3.6 | −5.7 | −4.2 | −1.9 | 0.680 | 0.164 | 0.003 | <0.001 | 0.017 | 0.245 | |
| Tat | −1.5 | 1.8 | −1.5 | −2.0 | −2.9 | −5.0 | −3.5 | −1.2 | 0.7 | 0.284 | 0.004 | <0.001 | 0.024 | 0.026 | |
| Rev | −0.3 | 3.0 | −0.3 | −0.8 | −1.7 | −3.9 | −2.4 | 0.0 | 1.9 | 1.2 | 0.052 | <0.001 | 0.368 | <0.001 | |
| Vpu | 1.7 | 5.0 | 1.8 | 1.2 | 0.3 | −1.8 | −0.3 | 2.0 | 3.9 | 3.2 | 2.1 | 0.512 | 0.180 | <0.001 | |
| Env gp120 | 2.3 | 5.5 | 2.3 | 1.7 | 0.8 | −1.3 | 0.2 | 2.6 | 4.5 | 3.8 | 2.6 | 0.5 | <0.001 | <0.001 | |
| Env gp41 | 0.5 | 3.7 | 0.5 | −0.1 | −1.0 | −3.1 | −1.6 | 0.8 | 2.6 | 2.0 | 0.8 | −1.3 | −1.8 | <0.001 | |
| Nef | −3.9 | −0.6 | −3.9 | −4.4 | −5.3 | −7.5 | −6.0 | −3.6 | −1.7 | −2.4 | −3.6 | −5.7 | −6.2 | −4.4 | |
The P values from the chi-square test are shown in the upper triangle. According to the false discovery rate adjustment method (see Materials and Methods), only P values of <0.027 (in boldface) are significant. The differences in frequencies between the proteins are shown in the lower triangle.
Cumulative HIV-1C-specific CTL responses.
By combining data on CTL magnitude and CTL frequency, we introduced normalized cumulative CTL responses as a focal feature of the HIV-1C-specific immune response on a population level. The nature of normalized cumulative HIV-specific CTL responses reflects both the viral and the host components in a local AIDS epidemic. The predominant circulating virus in a given geographic area, Botswana in particular, was mirrored through the synthetic peptides that were synthesized based on tangible viral sequences analyzed recently in Botswana (66, 67, 71, 72). The MHC class I HLA types or alleles that are common locally in Botswana were echoed in the normalized cumulative CTL responses through the restrictive nature of the viral antigens presented on the cell surface for recognition by CD8+ T cells. Thus, the reported normalized cumulative HIV-1C-specific CTL responses might represent an instantaneous trait of the population-based immune response in the current HIV-1C epidemic in Botswana.
The profiles of normalized cumulative HIV-1C-specific CTL responses across the viral proteins are presented in Fig. 3. Based on the magnitude and frequency of CTL responses, the profiles highlighted responses with both high magnitude and high frequency. The normalized cumulative HIV-1C-specific CTL responses were spread out extremely unevenly both between and within viral proteins. Placed on the same scale (Fig. 3), normalized cumulative CTL responses (i) demonstrated the relative contribution of each HIV-1C protein to the CTL responses and (ii) highlighted particular regions with high and low levels of CTL responses across HIV-1C proteins. Gag p24 contained the highest peaks of cumulative HIV-1C-specific CTL responses, followed by a number of relatively high peaks in Nef and fewer peaks in RT, integrase, Vif, Vpr, Tat, gp120, and gp41.
FIG. 3.
Normalized cumulative HIV-1C-specific CTL responses. The x axis of each graph was scaled according to the number of HIV-1C synthetic peptides used for a particular viral protein, and the length of the graph does not necessarily correspond to the actual size of the viral protein because of the differences in the lengths of the synthetic peptides. The y axis was scaled equally for each viral protein. Bars represent normalized cumulative HIV-1C-specific CTL responses to a particular synthetic peptide across the viral genome. n represents the number of samples tested with a particular set of synthetic peptides. PR, protease; IN, integrase.
The normalized cumulative HIV-1C-specific CTL responses were ranked according to their value and frequency. The locations and sequences of the top 28 synthetic peptides associated with the strongest CTL responses are shown in Table 4. These peptides were located in Gag (five peptides, including two pairs of overlapping peptides), Pol (five peptides), Vif (two peptides), Vpr (one peptide), Tat (two peptides), Env (four peptides), and Nef (nine peptides, including eight overlapping). The peptides demonstrated a frequency of CTL responses higher than 10% and a value of the cumulative CTL responses higher than 25. Eight synthetic peptides were found to represent the uppermost CTL responses in the HIV-1C (three in Gag, one in Vpr, one in Tat, and three in Nef; the value of cumulative the CTL responses is above 50) (Table 4).
TABLE 4.
Immunodominant regions within HIV-1C
| HIV-1C protein | aa
|
Amino acid sequencea | Frequency of CTL responses (% of samples) | Normalized cumulative CTL responsesb | Amino acid diversityc
|
|||
|---|---|---|---|---|---|---|---|---|
| Start | End | % | P value | Mean difference, % (95% CI) | ||||
| Gag | 161 | 180 | EKAFSPEVIPMFTALSEGAT | 10.2 | 47.3 | 3.2 | <0.0001 | 6.2 (4.8, 7.6) |
| 171 | 190 | MFTALSEGATPQDLNTMLNT | 32.2 | 190.5 | 2.5 | <0.0001 | 6.9 (5.5, 8.4) | |
| 291 | 310 | EPFRDYVDRFFKTLRAEQAT | 18.6 | 80.2 | 3.2 | 0.0062 | 1.9 (0.5, 3.2) | |
| 301 | 320 | FKTLRAEQATQDVKNWMTDT | 17.0 | 87.7 | 5.5 | <0.0001 | 4.0 (2.1, 5.9) | |
| 331 | 350 | KTILRALGPGATLEEMMTAC | 15.3 | 43.7 | 7.2 | 0.0043 | 2.2 (0.7, 3.7) | |
| Pol | 271 | 290 | FSVPLDEDFRKYTAFTIPSI | 12.7 | 48.8 | 9.3 | 0.0064 | −2.8 (−4.9, −0.8) |
| 421 | 440 | WASQIYPGIKVRQLCKLLRG | 10.9 | 44.8 | 9.9 | 0.12 | −3.5 (−8.0, 1.0) | |
| 521 | 540 | KQLTEAVOKIAMESIVIWGK | 10.9 | 27.3 | 11.2 | <0.0001 | −4.8 (−6.8, −2.9) | |
| 894 | 911 | AVFIHNFKRKGGIGGYSA | 14.1 | 41.4 | 3.0 | <0.0001 | 3.4 (2.5, 4.4) | |
| 925 | 942 | TKELQKQIIKIQNFRVYY | 15.6 | 36.7 | 9.8 | 0.00025 | −3.4 (−5.3, −1.6) | |
| Vif | 61 | 80 | EARLVIKTYWGLOTGERDWH | 12.7 | 32.8 | 18.4 | <0.0001 | −6.8 (−9.5, −4.0) |
| 71 | 90 | GLQTGERDWHLGHGVSIEWR | 16.4 | 41.1 | 9.4 | 0.016 | 2.2 (0.4, 3.9) | |
| Vpr | 31 | 50 | VRHFPRPWLHSLGQYIYETY | 21.4 | 52.6 | 19.4 | <0.0001 | −7.5 (−9.9, −5.0) |
| Tat | 16 | 30 | SQPKTACNKCYCKRC | 10.7 | 25.0 | 31.8 | <0.0001 | −13.5 (−16.5, −10.4) |
| 36 | 50 | VCFQTKGLGISYGRK | 30.4 | 96.0 | 11.9 | <0.0001 | 5.9 (3.9, 7.9) | |
| Env | 299 | 319 | PNNNTRKSIRIGPGQTFYA | 11.3 | 28.1 | 10.7 | <0.0001 | 9.4 (7.6, 11.1) |
| 552 | 571 | QSNLLRAIEAQQHMLQLTVW | 10.7 | 25.2 | 4.9 | <0.0001 | 15.1 (14.0, 16.2) | |
| 702 | 721 | LSIVNRVRQGYSPLSFQTLT | 10.7 | 29.3 | 8.7 | <0.0001 | 11.3 (9.3, 13.4) | |
| 742 | 761 | RDRSIRLVSGFLALAWDDLR | 19.6 | 42.2 | 17.4 | 0.028 | 2.6 (0.3, 5.0) | |
| Nef | 1 | 16 | MGGKWSKSSIVGWPAV | 12.7 | 27.9 | 25.8 | 0.0059 | −7.2 (−12.3, −2.1) |
| 67 | 81 | GFPVRPQVPLRPMTY | 12.7 | 45.1 | 9.2 | <0.0001 | 9.4 (5.5, 13.3) | |
| 72 | 86 | PQVPLRPMTYKGAFD | 16.4 | 43.8 | 16.1 | 0.20 | 2.5 (−1.3, 6.2) | |
| 77 | 91 | RPMTYKGAFDLSFFL | 12.7 | 35.2 | 20.1 | 0.35 | −0.5 (−4.7, 1.6) | |
| 82 | 96 | KGAFDLSFFLKEKGG | 20.0 | 60.4 | 16.3 | 0.064 | 2.3 (−0.1, 4.7) | |
| 97 | 111 | LEGLIYSKKRQEILD | 12.7 | 40.7 | 15.2 | 0.0095 | 3.4 (0.8, 6.0) | |
| 112 | 126 | LWVYHTQGYFPDWQN | 16.4 | 38.4 | 11.1 | <0.0001 | 7.5 (5.5, 9.6) | |
| 122 | 136 | PDWQNYTPGPGVRYP | 18.2 | 50.6 | 9.0 | <0.0001 | 9.7 (7.4, 11.9) | |
| 127 | 141 | YTPGPGVRYPLTFGW | 14.5 | 56.9 | 8.5 | <0.0001 | 10.1 (8.0, 12.2) | |
The top eight immunodominant regions are in boldface.
Cumulative CTL responses were normalized by the number of samples screened with a particular set of synthetic peptides.
The amino acid diversity for a particular immunodominant region was compared with the amino acid diversity of the entire corresponding viral protein (P value and mean difference).
Ranking the cumulative CTL responses allowed us to map 6 primary and 15 secondary regions across HIV-1C proteins that corresponded to both high magnitudes and high frequencies of CTL responses. Two out of six primary (immunodominant) regions (Fig. 4) were identified in Gag and corresponded to aa 171 to 190 (according to the HXB2 numbering system [54]) and 291 to 320. Both Vpr and Tat contained one primary region that extended over aa 31 to 50 and 36 to 50, respectively. Two primary regions were also identified in Nef, at aa 82 to 96 and 122 to 141. Altogether, the identified six primary regions spanned a region of 120 aa. Combined together (Fig. 4), 6 primary (immunodominant) and 15 secondary (subdominant) regions extended over 426 aa: 80 aa in Gag, 96 aa in Pol, 30 aa in Vif, 20 aa in Vpr, 30 aa in Tat, 79 aa in Env, and 91 aa in Nef.
FIG. 4.
Immunodominant regions in HIV-1C. Immunodominant and subdominant regions in the HIV-1C genome in the context of CTL responses are shown. The locations of immunodominant and subdominant regions are scaled to their actual position in the HIV-1C genome. For details, see Table 4. LTR, long terminal repeat; PR, protease; IN, integrase.
The uneven distribution of HIV-1C-specific cumulative CTL responses across the viral proteins might provide valuable data for vaccine design. Regions that demonstrated relatively high cumulative CTL responses on a population level might be of particular interest to be included into vaccine constructs. In contrast, localities with relatively low CTL responses or without CTL responses could be irrelevant in regard to potency for CTL induction and elicitation.
Diversity within immunodominant regions.
To address diversity within the identified immunodominant regions, a set of 73 nonrecombinant HIV-1C genome sequences (71) was utilized. Translated amino acid sequences spanning each of the immunodominant regions were aligned, and amino acid distances within the immunodominant regions were computed. Diversity within most of the immunodominant regions was lower than that within the entire corresponding protein (Table 4), and significant differences (with adjustment for multiple tests, a P value of <0.03 implies statistical significance) were observed in 17 out of 28 regions studied. All immunodominant regions within Env and Gag were less diverse than the corresponding entire region, most strongly for Env aa 552 to 571 (Env 552-571) (mean difference, 15.1%; 95% CI, 14.0 to 16.2%) and Env 702-721 (mean difference, 11.3%; 95% CI, 9.3 to 13.4%). In addition, three of five immunodominant regions in Gag p24 showed significantly less diversity than Gag p24, and five of nine immunodominant regions in Nef showed significantly restricted diversity. Furthermore Pol 894-911, Tat 36-50, and Vif 71-90 showed significantly less diversity. In contrast, seven immunodominant regions showed significantly greater diversity, most notably three of five immunodominant regions in Pol, Nef 1-16 (mean difference, −7.2%; 95% CI, −12.3 to −2.1%), Tat 16-30 (mean difference, −13.5%; 95% CI, −16.5 to −10.4%), Vif 61-80 (mean difference, −6.8%; 95% CI, −9.5 to −4.0%), and Vpr 31-50 (mean difference, −7.5%; 95% CI, −9.9 to −5.0%).
DISCUSSION
Although the correlates of immune protection remain to be elucidated, eliciting HIV-1-specific CD8+ T-cell responses remains a critical goal of vaccine design.
We have focused on the antigenic component of an HIV vaccine and have approached strengthening of the vaccine immunogenicity through the identification of the immune responses specific to the prevailing HIV-1 subtype on a population level. Our goal was to identify and rank potential CTL epitope-rich regions throughout HIV-1C that could be considered in a vaccine design. We analyzed the magnitude and frequency of CTL responses across the viral genome as essential elements and vital characteristics of the overall cell-mediated immune responses. Amplified on a population level, this measurement of virus-specific CTL responses has become an important attribute of the regional HIV-1C epidemic and provided an overview of the distribution and density of HIV-1C-specific CTL responses in the Botswana population (Fig. 1, 2A, and 3).
In this study we addressed the magnitude, frequency and normalized cumulative HIV-1C-specific CTL responses in Botswana, a country with a very high prevalence of HIV-1C. Our previous study provided data for the profiles of CTL responses in four HIV-1C proteins (70), while in this study we presented a comparative analysis and ranking of CTL responses throughout the viral genome. The CTL responses were analyzed for natural HIV-1 infections in asymptomatic HIV-1-infected blood donors, a cohort that fairly represented the general population in Botswana.
The extent to which the results of this study could be extrapolated to the general population in Botswana or to the nearby area would depend on how representative a chosen cohort of blood donors was in relation to the general population in the same geographic area. A comparative analysis of the MHC class I HLA allele frequencies within the general population in Botswana (69) and within the blood donors (70) revealed a similar distribution of the HLA alleles and antigen specificities within the HLA-A, -B, and -C loci. Taken in the context of the nature of MHC class I restriction of CD8+ T-cell responses, similarities in the HLA allele frequencies between blood donors and the general population provided evidence that the profiles of CTL responses and identified immunodominant regions across HIV-1C might fairly represent the overall cell-mediated immune responses in the HIV epidemic in Botswana. Moreover, the predominant virus in the local HIV-1 epidemic was represented by the HIV-1C-specific synthetic peptides that were synthesized based on the actual sequences from Botswana and used in the study to quantify virus-specific CTL responses. This match provides an additional link between circulating virus and identified CTL responses in Botswana. The overall HIV vaccine design and development would benefit if a similar methodology for the analysis of virus-specific immune responses on a population level would be considered in other studies that target discrete HIV-1 subtypes and/or different populations in distinct geographic areas.
We observed an uneven distribution and disproportionate density of CTL responses in HIV-1C. An introduction of the normalized cumulative HIV-1C-specific CTL responses allowed us to identify a number of regions across HIV-1C that were associated with an increased level of virus-specific CTL responses in the Botswana population.
Differences in the magnitude and frequency of CTL responses between and within HIV-1C proteins highlighted the uneven character of CTL responses across the viral genome (Tables 1 to 3; Fig. 2B). A phenomenon of CTL epitope clustering can be found within the HIV-1 proteins, namely, Gag, Pol, Env, and Nef (20, 27, 42, 53, 99; A. Bansal, S. Sabbaj, B. Edwards, D. Ritter, J. Tang, B. Korber, R. Kaslow, C. Wilson, P. Goepfert, and M. Mulligan, AIDS Vaccine 2001, p. 39, 2001). Although CTL epitopes were shown to be associated with more conserved regions in viral proteins (28, 85), no preference for epitope clusters to be in either conserved or variable regions was reported (100).
A profile of CTL epitope clustering might reflect specifics of immune responses to the predominant HIV variant in a particular population. The results of our study were consistent with the data of Goulder et al. (42), who showed that the HIV-1C-specific CTL responses in Africans clustered within Gag p24, while HIV-1B-specific CTL responses in Caucasians grouped in Gag p17. In addition, profiles of HIV-1C-specific CTL responses in this study differed from the dominant HIV-1B-specific CTL responses in the regulatory HIV proteins Tat and Rev (2) and in the accessory proteins Vif, Vpr, and Vpu (7), highlighting subtype- and host-specific aspects of immune responses in HIV-1 infection. In contrast, CTL responses in Nef clustered in the central region of the protein in both HIV-1C- and HIV-1B-infected cohorts (27, 61, 70; A. Bansal, D. Ritter, S. Sabbaj, C. Perkins, B. Edwards, B. Korber, D. Kaslow, C. Wilson, P. Goepfert, and M. Mulligan, AIDS Vaccine 2001, p. 39, 2001). We analyzed patterns of CTL epitope clustering between HIV-1B and -1C by comparing the HIV-1B data from the HIV Molecular Immunology 2000 database (53) with the locations of HIV-1C immunodominant regions in this study. The profiles of CTL epitopes and epitope-rich regions within most viral proteins, except gp41 and Nef, were noticeably dissimilar (data not shown). Taken together, differences in the distribution of CTL responses and a relatively unique nature of CTL profiles in Botswana might suggest that CTL responses depend on the predominant circulating virus and genetic background of the population. This might be particularly accentuated between distinct geographic areas that have a predominance of a particular viral subtype or variant and relatively homogeneous ethnicity of the population. In contrast, an absence of clustering and a deceptively even spread of CTL epitopes might be observed in geographic areas with multiple HIV-1 subtypes and/or circulating recombinant forms and heterogeneous hosts. Whether the profiles of virus-specific CTL responses differ between HIV-1 subtypes and/or between geographic areas and the extent of the difference are subjects for future studies.
Although a detailed analysis of the factors that can affect virus-specific CTL responses was beyond the scope of this study, it is noteworthy that molecular modification and enhancement might improve immunogenicity and efficiency of vaccine constructs. For example, a deletion of 19 N-terminal amino acids in Nef that included the myristoylation site was shown to completely eliminate both MHC class I and CD4 down-regulation (79), which might suggest that the first subdominant region in Nef (aa 1 to 16) should be omitted from the vaccine construct. Moreover, such factors as intracellular processing, binding affinity, and presentation efficiency of viral antigens can influence and alter CTL responses and should be considered in a vaccine design. The expression level of the MHC class I-peptide complex on the cell surface and the frequency of CTL precursors might impinge on the magnitude of virus-specific CTL responses (38). Epitopes in a natural viral protein might not necessarily be optimal for binding to MHC molecules (74). The flanking residues of the CTL epitopes may (16, 88) or may not (58) affect proper processing and recognition of the presented epitopes. Yield and availability (26, 50, 96) together with efficiency of CTL epitope processing (59) are believed to be major determinants of immunogenicity. Moreover, the use of cytokines and costimulatory molecules can enhance and steer CTL responses (9, 13, 48). Therefore, the immunodominant regions identified across HIV-1C in this study warrant further research to address coherent engineering of vaccine constructs in order to achieve optimal processing and immunogenicity. Fine mapping of CTL epitopes within the identified immunodominant and subdominant regions could further improve the design of an HIV-1C vaccine. In addition, an extreme HIV-1 variability might hamper efficiency of immune responses and CTL competence in particular. Allen et al. reported that viral escape variants associated with mutations in Tat CTL epitopes emerged during acute simian immunodeficiency virus infection (4). In addition, Kelleher et al. described viral escape variants of HIV-1 Gag following the loss of the specific CTL response to Gag and the subsequent detection of partial reversion to the original wild-type sequence (51). These findings do support our conclusion that given the high variability of HIV, the vaccine approach of incorporating consensus sequences rich in CTL epitopes of HIV should have a better chance to provide a broader coverage of HIV variants and to minimize the escape variants than those approaches that rely on limited viral genes derived from a single variant.
Our study suggested that inclusion of the identified immunodominant and subdominant regions into vaccine constructs may improve the immunogenicity of a vaccine designed specifically for the Botswana population. Multiple immunodominant regions should minimize viral escape from immune recognition due to simultaneous targeting of several viral proteins, even in the case when some of the immunodominant regions could not elicit sufficient immune responses. A CTL-based vaccine construct could include immunodominant and subdominant regions, for example, as a DNA plasmid encoding a tandem of CTL-rich regions across HIV-1C. The notion that tandemly repeated sequences can significantly potentiate B- and T-cell-mediated immunogenicity (52, 60, 75, 101) suggests that the use of tandem arrays of immunodominant CTL regions might be advantageous in HIV vaccine design. In contrast, immunologically silent regions with low magnitudes and frequencies of CTL responses might be omitted from the vaccine constructs.
Recently we identified consensus sequences that might be utilized for an AIDS vaccine design and demonstrated relative distance for the consensus sequences that represent different subsets of HIV-1C sequences (71). Thus, a vaccine construct based on the consensus sequences of the identified immunodominant regions in Botswana could presumably be immunogenic in other southern African countries, assuming a relatively great overlap of common MHC class I HLA alleles between ethnic groups in southern Africa. Further molecular monitoring of the HIV epidemic might be required to guide additional modifications of the vaccine constructs to correspond to the current virus in the epidemic.
In summary, this study focused on T-cell responses in natural HIV-1C infection in Botswana, a country severely affected by the AIDS epidemic. Comparative analysis revealed that the magnitudes and frequencies of the HIV-1C-specific CTL responses were spread unevenly across the viral genome, within and between most of the viral proteins. Regions with increased magnitude and/or frequency, as well as regions with low CTL responses or without CTL responses, were identified throughout HIV-1C. Profiles of cumulative HIV-1C-specific immune responses allowed us to identify and rank immunodominant and subdominant regions within viral proteins. The strongest CTL responses were identified within Gag p24, Vpr, Tat, and Nef. Subdominant CTL responses were also distributed in Gag p24, Pol RT and integrase, Vif, Tat, Env gp120 and gp41, and Nef. The study represented a comprehensive, systematic approach for an HIV vaccine design on a population level and suggested the inclusion of identified immunodominant regions into vaccine constructs to restrain the HIV-1C epidemic in Botswana.
Acknowledgments
We thank the Botswana Ministry of Health for encouragement; S. Y. Chang, S. Gaseitsiwe, E. Sepako, G. Sebetso, N. Monametsi, A. Reich, and Y. Wu for sample processing and HIV-1 diagnostics; personnel of the National Blood Transfusion Center in Botswana for collaboration; and Chanc E. VanWinkle for editorial assistance.
This research was supported in part by grants AI47067, AI43255, and HD37793 from the National Institutes of Health and grant TW00004 from the Fogarty International Center, National Institutes of Health.
REFERENCES
- 1.Abebe, A., D. Demissie, J. Goudsmit, M. Brouwer, C. L. Kuiken, G. Pollakis, H. Schuitemaker, A. L. Fontanet, and T. F. Rinke de Wit. 1999. HIV-1 subtype C syncytium- and non-syncytium-inducing phenotypes and coreceptor usage among Ethiopian patients with AIDS. AIDS 13:1305-1311. [DOI] [PubMed] [Google Scholar]
- 2.Addo, M. M., M. Altfeld, E. S. Rosenberg, R. L. Eldridge, M. N. Philips, K. Habeeb, A. Khatri, C. Brander, G. K. Robbins, G. P. Mazzara, P. J. Goulder, B. D. Walker, and the HIV Controller Study Collaboration. 2001. The HIV-1 regulatory proteins Tat and Rev are frequently targeted by cytotoxic T lymphocytes derived from HIV-1-infected individuals. Proc. Natl. Acad. Sci. USA 98:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldovini, A., and R. A. Young. 1991. Humoral and cell-mediated immune responses to live recombinant BCG-HIV vaccines. Nature 351:479-482. [DOI] [PubMed] [Google Scholar]
- 4.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 5.Allen, T. M., T. U. Vogel, D. H. Fuller, B. R. Mothe, S. Steffen, J. E. Boyson, T. Shipley, J. Fuller, T. Hanke, A. Sette, J. D. Altman, B. Moss, A. J. McMichael, and D. I. Watkins. 2000. Induction of AIDS virus-specific CTL activity in fresh, unstimulated peripheral blood lymphocytes from rhesus macaques vaccinated with a DNA prime/modified vaccinia virus Ankara boost regimen. J. Immunol. 164:4968-4978. [DOI] [PubMed] [Google Scholar]
- 6.Almond, N. M., and J. L. Heeney. 1998. AIDS vaccine development in primate models. AIDS 12(Suppl. A):S133-S140. [PubMed] [Google Scholar]
- 7.Altfeld, M., M. M. Addo, R. L. Eldridge, X. G. Yu, S. Thomas, A. Khatri, D. Strick, M. N. Phillips, G. B. Cohen, S. A. Islam, S. A. Kalams, C. Brander, P. J. Goulder, E. S. Rosenberg, and B. D. Walker. 2001. Vpr is preferentially targeted by CTL during HIV-1 infection. J. Immunol. 167:2743-2752. [DOI] [PubMed] [Google Scholar]
- 8.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 9.Barouch, D. H., A. Craiu, M. J. Kuroda, J. E. Schmitz, X. X. Zheng, S. Santra, J. D. Frost, G. R. Krivulka, M. A. Lifton, C. L. Crabbs, G. Heidecker, H. C. Perry, M. E. Davies, H. Xie, C. E. Nickerson, T. D. Steenbeke, C. I. Lord, D. C. Montefiori, T. B. Strom, J. W. Shiver, M. G. Lewis, and N. L. Letvin. 2000. Augmentation of immune responses to HIV-1 and simian immunodeficiency virus DNA vaccines by IL-2/Ig plasmid administration in rhesus monkeys. Proc. Natl. Acad. Sci. USA 97:4192-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barouch, D. H., A. Craiu, S. Santra, M. A. Egan, J. E. Schmitz, M. J. Kuroda, T. M. Fu, J. H. Nam, L. S. Wyatt, M. A. Lifton, G. R. Krivulka, C. E. Nickerson, C. I. Lord, B. Moss, M. G. Lewis, V. M. Hirsch, J. W. Shiver, and N. L. Letvin. 2001. Elicitation of high-frequency cytotoxic T-lymphocyte responses against both dominant and subdominant simian-human immunodeficiency virus epitopes by DNA vaccination of rhesus monkeys. J. Virol. 75:2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 12.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 14.Belyakov, I. M., L. S. Wyatt, J. D. Ahlers, P. Earl, C. D. Pendleton, B. L. Kelsall, W. Strober, B. Moss, and J. A. Berzofsky. 1998. Induction of a mucosal cytotoxic T-lymphocyte response by intrarectal immunization with a replication-deficient recombinant vaccinia virus expressing human immunodeficiency virus 89.6 envelope protein. J. Virol. 72:8264-8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. Ser. B 57:289-300. [Google Scholar]
- 16.Bergmann, C. C., Q. Yao, C. K. Ho, and S. L. Buckwold. 1996. Flanking residues alter antigenicity and immunogenicity of multi-unit CTL epitopes. J. Immunol. 157:3242-3249. [PubMed] [Google Scholar]
- 17.Berzofsky, J. A. 1988. Immunodominance in T lymphocyte recognition. Immunol. Lett. 18:83-92. [DOI] [PubMed] [Google Scholar]
- 18.Betts, M. R., J. Krowka, C. Santamaria, K. Balsamo, F. Gao, G. Mulundu, C. Luo, N. N′Gandu, H. Sheppard, B. H. Hahn, S. Allen, and J. A. Frelinger. 1997. Cross-clade human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte responses in HIV-infected Zambians. J. Virol. 71:8908-8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Björnal, Å., A. Sönnerborg, C. Tschering, J. Albert, and E. M. Fenyö. 1999. Phenotypic characteristics of human immunodeficiency virus type 1 subtype C isolates of Ethiopian AIDS patients. AIDS Res. Hum. Retroviruses 15:647-653. [DOI] [PubMed] [Google Scholar]
- 20.Buseyne, F., M. McChesney, F. Porrot, S. Kovarik, B. Guy, and Y. Riviere. 1993. Gag-specific cytotoxic T lymphocytes from human immunodeficiency virus type 1-infected individuals: Gag epitopes are clustered in three regions of the p24gag protein. J. Virol. 67:694-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calef, C., R. Thakallapally, D. Lang, C. Brander, P. Goulder, O. Yang, and B. Korber. 2000. PeptGen: designing peptides for immunological studies and application to HIV consensus sequences, p. I-63-I-100. In B. Korber, C. Brander, B. Haynes, R. Koup, C. Kuiken, J. Moore, B. Walker, and D. Watkins (ed.), HIV molecular immunology 2000. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 22.Cao, H., P. Kanki, J. L. Sankale, A. Dieng-Sarr, G. P. Mazzara, S. A. Kalams, B. Korber, S. Mboup, and B. D. Walker. 1997. Cytotoxic T-lymphocyte cross-reactivity among different human immunodeficiency virus type 1 clades: implications for vaccine development. J. Virol. 71:8615-8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao, H., I. Mani, R. Vincent, R. Mugerwa, P. Mugyenyi, P. Kanki, J. Ellner, and B. D. Walker. 2000. Cellular immunity to human immunodeficiency virus type 1 (HIV-1) clades: relevance to HIV-1 vaccine trials in Uganda. J. Infect. Dis. 182:1350-1356. [DOI] [PubMed] [Google Scholar]
- 24.Choudhury, S., M. A. Montano, C. Womack, J. T. Blackard, J. K. Maniar, D. G. Saple, S. Tripathy, S. Sahni, S. Shah, G. P. Babu, and M. Essex. 2000. Increased promoter diversity reveals a complex phylogeny of human immunodeficiency virus type 1 subtype C in India. J. Hum. Virol. 3:35-43. [PubMed] [Google Scholar]
- 25.Colmenero, P., P. Berglund, T. Kambayashi, P. Biberfeld, P. Liljestrom, and M. Jondal. 2001. Recombinant Semliki Forest virus vaccine vectors: the route of injection determines the localization of vector RNA and subsequent T cell response. Gene Ther. 8:1307-1314. [DOI] [PubMed] [Google Scholar]
- 26.Crotzer, V. L., R. E. Christian, J. M. Brooks, J. Shabanowitz, R. E. Settlage, J. A. Marto, F. M. White, A. B. Rickinson, D. F. Hunt, and V. H. Engelhard. 2000. Immunodominance among EBV-derived epitopes restricted by HLA-B27 does not correlate with epitope abundance in EBV-transformed B-lymphoblastoid cell lines. J. Immunol. 164:6120-6129. [DOI] [PubMed] [Google Scholar]
- 27.Culmann-Penciolelli, B., S. Lamhamedi-Cherradi, I. Couillin, N. Guegan, J. P. Levy, J. G. Guillet, and E. Gomard. 1994. Identification of multirestricted immunodominant regions recognized by cytolytic T lymphocytes in the human immunodeficiency virus type 1 Nef protein. J. Virol. 68:7336-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Da Silva, J., and A. L. Hughes. 1998. Conservation of cytotoxic T lymphocyte (CTL) epitopes as a host strategy to constrain parasite adaptation: evidence from the nef gene of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 15:1259-1268. [DOI] [PubMed] [Google Scholar]
- 29.Di Fabio, S., D. Medaglini, C. M. Rush, F. Corrias, G. L. Panzini, M. Pace, P. Verani, G. Pozzi, and F. Titti. 1998. Vaginal immunization of Cynomolgus monkeys with Streptococcus gordonii expressing HIV-1 and HPV 16 antigens. Vaccine 16:485-492. [DOI] [PubMed] [Google Scholar]
- 30.Dorrell, L., T. Dong, G. S. Ogg, S. Lister, S. McAdam, T. Rostron, C. Conlon, A. J. McMichael, and S. L. Rowland-Jones. 1999. Distinct recognition of non-clade B human immunodeficiency virus type 1 epitopes by cytotoxic T lymphocytes generated from donors infected in Africa. J. Virol. 73:1708-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Durali, D., J. Morvan, F. Letourneur, D. Schmitt, N. Guegan, M. Dalod, S. Saragosti, D. Sicard, J. P. Levy, and E. Gomard. 1998. Cross-reactions between the cytotoxic T-lymphocyte responses of human immunodeficiency virus-infected African and European patients. J. Virol. 72:3547-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Esparza, J., and N. Bhamarapravati. 2000. Accelerating the development and future availability of HIV-1 vaccines: why, when, where, and how? Lancet 355:2061-2066. [DOI] [PubMed] [Google Scholar]
- 33.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 34.Evans, T. G., M. C. Keefer, K. J. Weinhold, M. Wolff, D. Montefiori, G. J. Gorse, B. S. Graham, M. J. McElrath, M. L. Clements-Mann, M. J. Mulligan, P. Fast, M. C. Walker, J. L. Excler, A. M. Duliege, and J. Tartaglia. 1999. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes given alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J. Infect. Dis. 180:290-298. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein, J. 1993. PHYLIP: phylogeny inference package, 3.52c ed. University of Washington, Seattle.
- 36.Felsenstein, J. 1996. PHYLIP: phylogeny inference package, 3.572c ed. University of Washington, Seattle.
- 37.Friedman, R. S., F. R. Frankel, Z. Xu, and J. Lieberman. 2000. Induction of human immunodeficiency virus (HIV)-specific CD8 T-cell responses by Listeria monocytogenes and a hyperattenuated Listeria strain engineered to express HIV antigens. J. Virol. 74:9987-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallimore, A., H. Hengartner, and R. Zinkernagel. 1998. Hierarchies of antigen-specific cytotoxic T-cell responses. Immunol Rev. 164:29-36. [DOI] [PubMed] [Google Scholar]
- 39.Gao, F., D. L. Robertson, C. D. Carruthers, S. G. Morrison, B. Jian, Y. Chen, F. Barre-Sinoussi, M. Girard, A. Srinivasan, A. G. Abimiku, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1998. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J. Virol. 72:5680-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalo, R. M., D. Rodriguez, A. Garcia-Sastre, J. R. Rodriguez, P. Palese, and M. Esteban. 1999. Enhanced CD8+ T cell response to HIV-1 env by combined immunization with influenza and vaccinia virus recombinants. Vaccine 17:887-892. [DOI] [PubMed] [Google Scholar]
- 41.Goulder, P. J., M. M. Addo, M. A. Altfeld, E. S. Rosenberg, Y. Tang, U. Govender, N. Mngqundaniso, K. Annamalai, T. U. Vogel, M. Hammond, M. Bunce, H. M. Coovadia, and B. D. Walker. 2001. Rapid definition of five novel HLA-A∗3002-restricted human immunodeficiency virus-specific cytotoxic T-lymphocyte epitopes by Elispot and intracellular cytokine staining assays. J. Virol. 75:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goulder, P. J., C. Brander, K. Annamalai, N. Mngqundaniso, U. Govender, Y. Tang, S. He, K. E. Hartman, C. A. O'Callaghan, G. S. Ogg, M. A. Altfeld, E. S. Rosenberg, H. Cao, S. A. Kalams, M. Hammond, M. Bunce, S. I. Pelton, S. A. Burchett, K. McIntosh, H. M. Coovadia, and B. D. Walker. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children. J. Virol. 74:5679-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Graham, B. S. 2000. Clinical trials of HIV vaccines, p. 82-105. In C. Kuiken, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, J. Mullins, S. Wolinsky, and B. Korber (ed.), HIV sequence compendium 2000. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 44.Hanke, T., and A. J. McMichael. 2000. Design and construction of an experimental HIV-1 vaccine for a year-2000 clinical trial in Kenya. Nat. Med. 6:951-955. [DOI] [PubMed] [Google Scholar]
- 45.Hanke, T., V. C. Neumann, T. J. Blanchard, P. Sweeney, A. V. Hill, G. L. Smith, and A. McMichael. 1999. Effective induction of HIV-specific CTL by multi-epitope using gene gun in a combined vaccination regime. Vaccine 17:589-596. [DOI] [PubMed] [Google Scholar]
- 46.Honda, M., K. Matsuo, T. Nakasone, Y. Okamoto, H. Yoshizaki, K. Kitamura, W. Sugiura, K. Watanabe, Y. Fukushima, S. Haga, Y. Katsura, H. Tasaka, K. Komuro, T. Yamada, T. Asano, A. Yamazak, and S. Yamazaki. 1995. Protective immune responses induced by secretion of a chimeric soluble protein from a recombinant Mycobacterium bovis bacillus Calmette-Guerin vector candidate vaccine for human immunodeficiency virus type 1 in small animals. Proc. Natl. Acad. Sci. USA 92:10693-10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hone, D. M., S. Wu, R. J. Powell, D. W. Pascual, J. Van Cott, J. McGhee, T. R. Fouts, R. G. Tuskan, and G. K. Lewis. 1996. Optimization of live oral Salmonella-HIV-1 vaccine vectors for the induction of HIV-specific mucosal and systemic immune responses. J. Biotechnol. 44:203-207. [DOI] [PubMed] [Google Scholar]
- 48.Iwasaki, A., B. J. Stiernholm, A. K. Chan, N. L. Berinstein, and B. H. Barber. 1997. Enhanced CTL responses mediated by plasmid DNA immunogens encoding costimulatory molecules and cytokines. J. Immunol. 158:4591-4601. [PubMed] [Google Scholar]
- 49.Johnston, M. I. 2000. The role of nonhuman primate models in AIDS vaccine development. Mol. Med. Today 6:267-270. [DOI] [PubMed] [Google Scholar]
- 50.Kageyama, S., T. J. Tsomides, Y. Sykulev, and H. N. Eisen. 1995. Variations in the number of peptide-MHC class I complexes required to activate cytotoxic T cell responses. J. Immunol. 154:567-576. [PubMed] [Google Scholar]
- 51.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan, C. M., B. Villarreal-Ramos, R. J. Pierce, R. Demarco de Hormaeche, H. McNeill, T. Ali, S. Chatfield, A. Capron, G. Dougan, and C. E. Hormaeche. 1994. Construction, expression, and immunogenicity of multiple tandem copies of the Schistosoma mansoni peptide 115-131 of the P28 glutathione S-transferase expressed as C-terminal fusions to tetanus toxin fragment C in a live aro-attenuated vaccine strain of Salmonella. J. Immunol. 153:5634-5642. [PubMed] [Google Scholar]
- 53.Korber, B., C. Brander, B. Haynes, R. Koup, C. Kuiken, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.). 2000. HIV molecular immunology 2000. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 54.Korber, B. T., B. T. Foley, C. L. Kuiken, S. K. Pillai, and J. G. Sodroski. 1998. Numbering positions in HIV relative to HXB2CG, p. III-102-III-111. In B. Korber, C. Kuiken, B. Foley, B. Hahn, F. McCutchan, J. Mellors, and J. Sodroski (ed.), Human retroviruses and AIDS 1998. Theoretical Biology and Biophysics, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 55.Lagranderie, M., A. M. Balazuc, B. Gicquel, and M. Gheorghiu. 1997. Oral immunization with recombinant Mycobacterium bovis BCG simian immunodeficiency virus Nef induces local and systemic cytotoxic T-lymphocyte responses in mice. J. Virol. 71:2303-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lagranderie, M., N. Winter, A. M. Balazuc, B. Gicquel, and M. Gheorghiu. 1998. A cocktail of Mycobacterium bovis BCG recombinants expressing the SIV Nef, Env, and Gag antigens induces antibody and cytotoxic responses in mice vaccinated by different mucosal routes. AIDS Res. Hum. Retroviruses 14:1625-1633. [DOI] [PubMed] [Google Scholar]
- 57.Lim, E. M., M. Lagranderie, R. Le Grand, J. Rauzier, M. Gheorghiu, B. Gicquel, and N. Winter. 1997. Recombinant Mycobacterium bovis BCG producing the N-terminal half of SIVmac251 Env antigen induces neutralizing antibodies and cytotoxic T lymphocyte responses in mice and guinea pigs. AIDS Res. Hum. Retroviruses 13:1573-1581. [DOI] [PubMed] [Google Scholar]
- 58.Lippolis, J. D., L. M. Mylin, D. T. Simmons, and S. S. Tevethia. 1995. Functional analysis of amino acid residues encompassing and surrounding two neighboring H-2Db-restricted cytotoxic T-lymphocyte epitopes in simian virus 40 tumor antigen. J. Virol. 69:3134-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Livingston, B. D., M. Newman, C. Crimi, D. McKinney, R. Chesnut, and A. Sette. 2001. Optimization of epitope processing enhances immunogenicity of multiepitope DNA vaccines. Vaccine 19:4652-4660. [DOI] [PubMed] [Google Scholar]
- 60.Lo-Man, R., P. Martineau, M. Hofnung, and C. Leclerc. 1993. Induction of T cell responses by chimeric bacterial proteins expressing several copies of a viral T cell epitope. Eur. J. Immunol. 23:2998-3002. [DOI] [PubMed] [Google Scholar]
- 61.Mashishi, T., S. Loubser, W. Hide, G. Hunt, L. Morris, G. Ramjee, S. Abdool-Karim, C. Williamson, and C. M. Gray. 2001. Conserved domains of subtype C Nef from South African HIV type 1-infected individuals include cytotoxic T lymphocyte epitope-rich regions. AIDS Res. Hum. Retroviruses 17:1681-1687. [DOI] [PubMed] [Google Scholar]
- 62.Mata, M., and Y. Paterson. 1999. Th1 T cell responses to HIV-1 Gag protein delivered by a Listeria monocytogenes vaccine are similar to those induced by endogenous listerial antigens. J. Immunol. 163:1449-1456. [PubMed] [Google Scholar]
- 63.McAdam, S., P. Kaleebu, P. Krausa, P. Goulder, N. French, B. Collin, T. Blanchard, J. Whitworth, A. McMichael, and F. Gotch. 1998. Cross-clade recognition of p55 by cytotoxic T lymphocytes in HIV-1 infection. AIDS 12:571-579. [DOI] [PubMed] [Google Scholar]
- 64.McGettigan, J. P., H. D. Foley, I. M. Belyakov, J. A. Berzofsky, R. J. Pomerantz, and M. J. Schnell. 2001. Rabies virus-based vectors expressing human immunodeficiency virus type 1 (HIV-1) envelope protein induce a strong cross-reactive cytotoxic T-lymphocyte response against envelope proteins from different HIV-1 isolates. J. Virol. 75:4430-4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nathanson, N., V. M. Hirsch, and B. J. Mathieson. 1999. The role of nonhuman primates in the development of an AIDS vaccine. AIDS 13(Suppl. A):S113-S120. [PubMed] [Google Scholar]
- 66.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ndung'u, T., B. Renjifo, V. A. Novitsky, M. F. McLane, S. Gaolekwe, and M. Essex. 2000. Molecular cloning and biological characterization of full-length HIV-1 subtype C from Botswana. Virology 278:390-399. [DOI] [PubMed] [Google Scholar]
- 68.Neilson, J. R., G. C. John, J. K. Carr, P. Lewis, J. K. Kreiss, S. Jackson, R. W. Nduati, D. Mbori-Ngacha, D. D. Panteleeff, S. Bodrug, C. Giachetti, M. A. Bott, B. A. Richardson, J. Bwayo, J. Ndinya-Achola, and J. Overbaugh. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Novitsky, V., P. O. Flores-Villanueva, P. Chigwedere, S. Gaolekwe, H. Bussman, G. Sebetso, R. Marlink, E. J. Yunis, and M. Essex. 2001. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum. Immunol. 62:146-156. [DOI] [PubMed] [Google Scholar]
- 70.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific Elispot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Novitsky, V., U. R. Smith, P. Gilbert, M. F. McLane, P. Chigwedere, C. Williamson, T. Ndung'u, I. Klein, S. Y. Chang, T. Peter, I. Thior, B. T. Foley, S. Gaolekwe, N. Rybak, S. Gaseitsiwe, F. Vannberg, R. Marlink, T. H. Lee, and M. Essex. 2002. HIV-1 subtype C molecular phylogeny: consensus sequence for an AIDS vaccine design? J. Virol. 76:5435-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novitsky, V. A., M. A. Montano, M. F. McLane, B. Renjifo, F. Vannberg, B. T. Foley, T. P. Ndung'u, M. Rahman, M. Makhema, J., R. Marlink, and M. Essex. 1999. Molecular cloning and phylogenetic analysis of human immunodeficiency virus type 1 subtype C: a set of 23 full-length clones from Botswana. J. Virol. 73:4427-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oggioni, M. R., D. Medaglini, L. Romano, F. Peruzzi, T. Maggi, L. Lozzi, L. Bracci, M. Zazzi, F. Manca, P. E. Valensin, and G. Pozzi. 1999. Antigenicity and immunogenicity of the V3 domain of HIV type 1 glycoprotein 120 expressed on the surface of Streptococcus gordonii. AIDS Res. Hum. Retroviruses 15:451-459. [DOI] [PubMed] [Google Scholar]
- 74.Oscherwitz, J., F. M. Gotch, K. B. Cease, and J. A. Berzofsky. 1999. New insights and approaches regarding B- and T-cell epitopes in HIV vaccine design. AIDS 13(Suppl. A):S163-S174. [PubMed] [Google Scholar]
- 75.Oscherwitz, J., M. E. Zeigler, T. E. Gribbin, and K. B. Cease. 1999. A V3 loop haptenic peptide sequence, when tandemly repeated, enhances immunogenicity by facilitating helper T-cell responses to a covalently linked carrier protein. Vaccine 17:2392-2399. [DOI] [PubMed] [Google Scholar]
- 76.Osmanov, S., C. Pattou, N. Walker, B. Schwardlander, and J. Esparza. 2002. Estimated global distribution and regional spread of HIV-1 genetic subtypes in the year 2000. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 29:184-190. [DOI] [PubMed] [Google Scholar]
- 77.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-Gag-Pol-Env-based vaccination and macaque major histocompatibility complex class I (A∗01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peeters, M., R. Vincent, J. L. Perret, M. Lasky, D. Patrel, F. Liegeois, V. Courgnaud, R. Seng, T. Matton, S. Molinier, and E. Delaporte. 1999. Evidence for differences in MT2 cell tropism according to genetic subtypes of HIV-1: syncytium-inducing variants seem rare among subtype C HIV-1 viruses. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:115-121. [DOI] [PubMed] [Google Scholar]
- 79.Peng, B., and M. Robert-Guroff. 2001. Deletion of N-terminal myristoylation site of HIV Nef abrogates both MHC-1 and CD4 down-regulation. Immunol. Lett. 78:195-200. [DOI] [PubMed] [Google Scholar]
- 80.Rayevskaya, M. V., and F. R. Frankel. 2001. Systemic immunity and mucosal immunity are induced against human immunodeficiency virus Gag protein in mice by a new hyperattenuated strain of Listeria monocytogenes. J. Virol. 75:2786-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Renjifo, B., W. Fawzi, D. Mwakagile, D. Hunter, G. Msamanga, D. Spiegelman, M. Garland, C. Kagoma, A. Kim, B. Chaplin, E. Hertzmark, and M. Essex. 2001. Differences in perinatal transmission among human immunodeficiency virus type 1 genotypes. J. Hum. Virol. 4:16-25. [PubMed] [Google Scholar]
- 82.Rodenburg, C. M., Y. Li, S. A. Trask, Y. Chen, J. Decker, D. L. Robertson, M. L. Kalish, G. M. Shaw, S. Allen, B. H. Hahn, and F. Gao. 2001. Near full-length clones and reference sequences for subtype C isolates of HIV type 1 from three different continents. AIDS Res. Hum. Retroviruses 17:161-168. [DOI] [PubMed] [Google Scholar]
- 83.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. J. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Invest. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Salminen, M. O., B. Johansson, A. Sonnerborg, S. Ayehunie, D. Gotte, P. Leinikki, D. S. Burke, and F. E. McCutchan. 1996. Full-length sequence of an Ethiopian human immunodeficiency virus type 1 (HIV-1) isolate of genetic subtype C. AIDS Res. Hum. Retroviruses 12:1329-1339. [DOI] [PubMed] [Google Scholar]
- 85.Seibert, S. A., C. Y. Howell, M. K. Hughes, and A. L. Hughes. 1995. Natural selection on the gag, pol, and env genes of human immunodeficiency virus 1 (HIV-1). Mol. Biol. Evol. 12:803-813. [DOI] [PubMed] [Google Scholar]
- 86.Seth, A., I. Ourmanov, M. J. Kuroda, J. E. Schmitz, M. W. Carroll, L. S. Wyatt, B. Moss, M. A. Forman, V. M. Hirsch, and N. L. Letvin. 1998. Recombinant modified vaccinia virus Ankara-simian immunodeficiency virus gag pol elicits cytotoxic T lymphocytes in rhesus monkeys detected by a major histocompatibility complex class I/peptide tetramer. Proc. Natl. Acad. Sci. USA 95:10112-10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Seth, A., I. Ourmanov, J. E. Schmitz, M. J. Kuroda, M. A. Lifton, C. E. Nickerson, L. Wyatt, M. Carroll, B. Moss, D. Venzon, N. L. Letvin, and V. M. Hirsch. 2000. Immunization with a modified vaccinia virus expressing simian immunodeficiency virus (SIV) Gag-Pol primes for an anamnestic Gag-specific cytotoxic T-lymphocyte response and is associated with reduction of viremia after SIV challenge. J. Virol. 74:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shastri, N., T. Serwold, and F. Gonzalez. 1995. Presentation of endogenous peptide/MHC class I complexes is profoundly influenced by specific C-terminal flanking residues. J. Immunol. 155:4339-4346. [PubMed] [Google Scholar]
- 89.Shata, M. T., and D. M. Hone. 2001. Vaccination with a Shigella DNA vaccine vector induces antigen-specific CD8+ T cells and antiviral protective immunity. J. Virol. 75:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shata, M. T., M. S. J. Reitz, A. L. DeVico, G. K. Lewis, and D. M. Hone. 2001. Mucosal and systemic HIV-1 Env-specific CD8(+) T-cells develop after intragastric vaccination with a Salmonella Env DNA vaccine vector. Vaccine 20:623-629. [DOI] [PubMed] [Google Scholar]
- 91.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 92.Tscherning, C., A. Alaeus, R. Fredriksson, A. Bjorndal, H. Deng, D. R. Littman, E. M. Fenyo, and J. Albert. 1998. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 241:181-188. [DOI] [PubMed] [Google Scholar]
- 93.UNAIDS. 2001, posting date. AIDS epidemic update. December 2001. [Online.] http://www.unaids.org/worldaidsday/2001/Epiupdate2001/Epiupdate2001_en.pdf. UNAIDS.
- 94.UNAIDS and W. H. O. 2000, posting date. Global HIV/AIDS & STD surveillance. Epidemiological fact sheets by country. [Online.] http://www.who.int/emc-hiv/fact_sheets/.
- 95.van Harmelen, J., C. Williamson, B. Kim, L. Morris, J. Carr, S. S. Abdool Karim, and F. McCutchan. 2001. Characterization of full length HIV-1 subtype C sequences from South Africa. AIDS Res. Hum. Retroviruses 17:1527-1531. [DOI] [PubMed] [Google Scholar]
- 96.Wherry, E. J., K. A. Puorro, A. Porgador, and L. C. Eisenlohr. 1999. The induction of virus-specific CTL as a function of increasing epitope expression: responses rise steadily until excessively high levels of epitope are attained. J. Immunol. 163:3735-3745. [PubMed] [Google Scholar]
- 97.Wilson, S. E., S. L. Pedersen, J. C. Kunich, V. L. Wilkins, D. L. Mann, G. P. Mazzara, J. Tartaglia, C. L. Celum, and H. W. Sheppard. 1998. Cross-clade envelope glycoprotein 160-specific CD8+ cytotoxic T lymphocyte responses in early HIV type 1 clade B infection. AIDS Res. Hum. Retroviruses 14:925-937. [DOI] [PubMed] [Google Scholar]
- 98.Yewdell, J. W., and J. R. Bennink. 1999. Immunodominance in major histocompatibility complex class I-restricted T lymphocyte responses. Annu. Rev. Immunol. 17:51-88. [DOI] [PubMed] [Google Scholar]
- 99.Yu, X. G., H. Shang, M. M. Addo, R. L. Eldridge, M. N. Phillips, M. E. Feeney, D. Strick, C. Brander, P. J. Goulder, E. S. Rosenberg, B. D. Walker, and M. Altfeld. 2002. Important contribution of p15 Gag-specific responses to the total Gag-specific CTL responses. AIDS 16:321-328. [DOI] [PubMed] [Google Scholar]
- 100.Zhang, C., J. L. Cornette, J. A. Berzofsky, and C. DeLisi. 1997. The organization of human leucocyte antigen class I epitopes in HIV genome products: implications for HIV evolution and vaccine design. Vaccine 15:1291-1302. [DOI] [PubMed] [Google Scholar]
- 101.Zheng, B., F. L. Graham, D. C. Johnson, T. Hanke, M. R. McDermott, and L. Prevec. 1993. Immunogenicity in mice of tandem repeats of an epitope from herpes simplex gD protein when expressed by recombinant adenovirus vectors. Vaccine 11:1191-1198. [DOI] [PubMed] [Google Scholar]