Abstract
Bacteriophage with linear, double-stranded DNA genomes package DNA into preassembled protein shells called procapsids. Located at one vertex in the procapsid is a portal complex composed of a ring of 12 subunits of portal protein. The portal complex serves as a docking site for the DNA packaging enzymes, a conduit for the passage of DNA, and a binding site for the phage tail. An excess of the P22 portal protein alters the assembly pathway of the procapsid, giving rise to defective procapsid-like particles and aberrant heads. In the present study, we report the isolation of escape mutant phage that are able to replicate more efficiently than wild-type phage in the presence of excess portal protein. The escape mutations all mapped to the same phage genome segment spanning the portal, scaffold, coat, and open reading frame 69 genes. The mutations present in five of the escape mutants were determined by DNA sequencing. Interestingly, each mutant contained the same mutation in the scaffold gene, which changes the glycine at position 287 to glutamate. This mutation alone conferred an escape phenotype, and the heads assembled by phage harboring only this mutation had reduced levels of portal protein and exhibited increased head assembly fidelity in the presence of excess portal protein. Because this mutation resides in a region of scaffold protein necessary for coat protein binding, these findings suggest that the P22 scaffold protein may define the portal vertices in an indirect manner, possibly by regulating the fidelity of coat protein polymerization.
Spherical viruses possess icosahedral symmetry, the simplest form of which contains 60 subunits arranged as 12 pentamers (9, 26). Larger capsids contain subtriangulated faces in which subunits are positioned in nonequivalent bonding environments (9, 26). Remarkably, many viral capsid proteins are capable of conformational switching during assembly to allow for subtriangulation with a single protein. This variability in conformational states renders capsid assembly pathway dependent, and so information needs to be imparted to the incoming subunits during assembly to ensure that the proper global icosahedral symmetry is obtained (26). To accomplish this, most double-stranded DNA (dsDNA) bacteriophage as well as herpesviruses first assemble a procapsid devoid of nucleic acid by using catalytic scaffold protein to guide the assembly process (10). Scaffold proteins bind to capsid proteins and are required for assembly; however, they are not present in the mature virion. Interestingly, scaffold proteins do not appear to form icosahedral cores, and so the mechanism by which icosahedral symmetry is imparted remains uncertain (29).
Procapsid assembly is further complicated in that 1 of the 12 fivefold rotationally symmetric vertices is differentiated, comprising the location of the portal vertex (1, 31). The portal vertex contains a dodecamer of portal protein that serves as a recognition site for the DNA packaging enzymes, a conduit for the passage of DNA, and a site of tail attachment (1, 31). Because of these functions, portal proteins are essential phage head components. The assembly of a portal ring into a capsid requires symmetry to be broken in two ways. First, the global icosahedral symmetry of the procapsid is disrupted because the portal ring replaces a pentamer of coat protein (22, 27). Second, the 12-fold rotationally symmetric portal ring is located in a fivefold rotationally symmetric capsid position (15, 22, 27). This “symmetry mismatch” has been postulated to allow for easier portal rotation during DNA packaging (15). The incorporation of portals into procapsids represents developmental bifurcation at the molecular level.
How might phage proteins self-assemble such that each procapsid gets one, and only one, portal vertex? Murialdo and Becker (24) proposed that the portals form an initiation complex for head assembly. By starting the assembly of the procapsid at the portal vertex, each structure would acquire only one portal. This “nucleation” model was supported by experiments with the prolate bacteriophage T4, in which it was determined that portal protein was required for the formation of prolate heads (17, 31). Additional support for a portal initiation model came from observations that the phage lambda portal protein promoted head assembly in cell lysates (20, 24, 31).
For the nucleation mechanism to be efficient, the initiation of procapsid assembly would need to be rate limiting, thereby reducing the possibility of procapsids assembling without portals. Bazinet and King (2) tested the nucleation mechanism for portal incorporation in the T=7 bacteriophage P22 by monitoring the rate of procapsid appearance in infected cells in the presence and absence of portal protein. Surprisingly, the rate of head assembly appeared to be unaffected by portal protein, suggesting that portal protein was not incorporated by a nucleation mechanism. Similar observations have also been made for the assembly of phage SPP1 procapsids (11). Ideally, portal vertex formation could be monitored in vitro with purified components to identify essential players and to establish a formal mechanism; unfortunately, this assembly process has not been successfully reproduced for any phage system.
Several genes have been implicated in the process of portal vertex formation during P22 procapsid assembly. Extragenic suppressors of a conditionally defective portal gene (1−csH137) map in the coat and tailspike genes (3, 16). Because tailspike protein is not required for procapsid assembly or DNA packaging, Bazinet et al. proposed that the mRNA of the tailspike gene was capable of forming an initiation complex for procapsid assembly (3). This mechanism has not been experimentally verified. What has been clearly demonstrated is that the P22 scaffold protein is required for portal protein incorporation into procapsids (12, 30). In the absence of scaffold protein, coat protein assembles into aberrant structures as well as T=7 shells (12, 30). However, these assemblages do not contain portal protein or any of the minor head proteins normally associated with procapsids (5). More evidence for the necessity of scaffold protein in recruiting portal protein into procapsids came from studies by Greene and King (13), in which some pseudorevertants of amber mutant P22 scaffold genes were found to assemble procapsid-like particles (PLPs) that lacked portal and/or minor head proteins. Although these findings suggest that the P22 scaffold protein directly interacts with portal protein, no such interactions have been demonstrated biochemically. To our knowledge, the only physical scaffold protein-portal protein interaction that has been demonstrated is a weak interaction between phage φ29 scaffold and portal proteins (14).
P22 is rather unique among the dsDNA bacteriophages in that the fidelity of head assembly is not affected by the absence of portal protein (23). In most other instances, when portal proteins are absent during assembly, coat and scaffold proteins assemble into aberrant spiral structures or long open-ended tubes (17, 31). Therefore, portal proteins appear to play some role in head form determination. Interestingly, when wild-type P22 portal protein is overexpressed in trans during a P22 infection, the fidelity of procapsid assembly is greatly reduced, resulting in the formation of heads with the wrong triangulation (T) number and spiral shells (23). This finding suggests that the P22 portal protein can also play a role in form determination under certain circumstances.
The aberrant procapsids that form in the presence of excess P22 portal protein apparently contain extra portal vertices (23). This finding provided a unique opportunity to investigate the process of portal vertex formation in vivo. Because both excess portal protein and phage were wild type, the interactions that promoted the formation of the extra portal vertices were probably also wild type in nature. If P22 escape mutants could be isolated that were capable of replicating more efficiently in the presence of excess portal protein, then the genes responsible for the decision-making process of portal vertex formation could be identified. In this article, we report the isolation, identification, and characterization of P22 mutants that are resistant to excess wild-type portal protein.
MATERIALS AND METHODS
Selection of excess portal escape mutants.
A single-plaque isolate of bacteriophage P22 containing a clear-plaque mutation (cI-7) (4) was picked from a lawn of Salmonella enterica serovar Typhimurium strain DB7000 (4) with a capillary tube and vortexed in 100 μl of Luria-Bertani (LB) broth with 5 μl of chloroform. After standing for several hours with intermittent vortexing, the titer of the phage solution was determined on a lawn of DB7000. This plaque stock served as the parental, wild-type phage stock for the evolution experiments.
The β-galactosidase (β-Gal) and portal protein hosts were previously described in detail (23). Briefly, each host carries an expression plasmid that contains either the Escherichia coli β-Gal or P22 portal gene under the control of the lac-UV5 promoter in plasmid pNFPW. The β-Gal host was induced with 0.07 mM isopropyl-β-d-galactopyranoside (IPTG) and the portal protein host was induced with 1 mM IPTG to achieve induction levels that did not reduce the doubling time of the cultures (23). The plasmids were maintained under selection with 100 μg of ampicillin/ml. Mid-log-phase stocks of the hosts were prepared by growing the bacteria from washed overnight cultures to mid-log phase (∼3 × 108 to 5 × 108/ml) in LB broth with ampicillin, chilling the cultures on ice, pelleting then, and resuspending them at 1/10 the original culture volume in ice-cold LB broth. The concentrated cultures were then stored at 4°C for up to 1 week.
For each round of selection, the portal protein host was diluted from a stock to 7 × 107 bacteria per ml, induced to express portal protein, and grown for 1 h at 37°C. The culture (now generally containing ∼1 × 108 bacteria per ml) was infected at a multiplicity of infection (MOI) of 0.1. After 8 to 10 h of growth at 20°C, chloroform was added to the culture to kill uninfected bacteria and to aid in the lysis of the remaining infected host. The amount of phage in the sample was assessed by titer determination. Because some phage were made during each infection (∼5% of normal) and because multiple rounds of infection occurred in the liquid culture, the resulting titer of virus in the medium was ∼1 × 1010 per ml in the initial rounds of selection. The selection was repeated by infecting another preinduced portal protein host culture with 107 phage from the previous round. As a control, parallel infections were performed with hosts expressing β-Gal from the same plasmid. The titer of phage after each round of growth in the control culture was generally ∼2 × 1011 per ml. The escape mutant phenotype was scored as the ability to form plaques on lawns of the induced portal protein host grown overnight at room temperature (∼20°C).
Marker rescue mapping.
Because excess portal protein interferes with both head assembly and DNA packaging (23), we suspected that the escape mutations would be present in the region of the phage genome associated with these processes. Therefore, PCR was used to amplify a 9.9-kb genome segment from the escape mutant phage by using 1 μl of a plaque isolate resuspended in 100 μl of LB broth with 5 μl of chloroform as a template in a 50-μl reaction mixture. This 9.9-kb segment contained, from left to right, genes 13, 19, and 15 and open reading frame 201 (ORF201), ORF80, ORF1, 3, 2, 1, 8, 5, and most of ORF69 (32). The same region was also cloned from the parental phage stock to serve as a negative control. The PCR products were cloned into vector pCR-XL-TOPO (Invitrogen) according to the manufacturer's instructions.
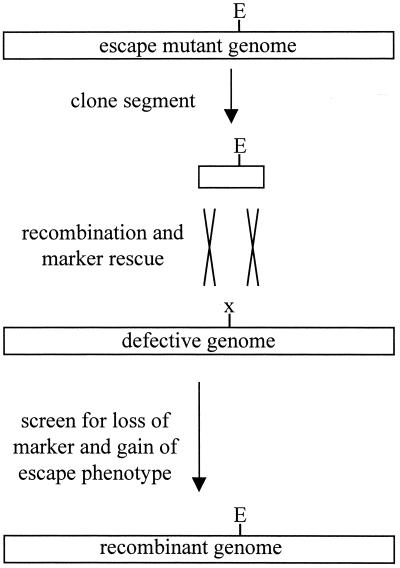
Plasmid DNA was purified from the wild-type and escape mutant clones and transformed by electroporation into Salmonella host strain MS1363 (supE; a gift from M. Susskind, University of Southern California) for mapping. Mid-log-phase liquid cultures growing in LB broth harboring the plasmids were then infected (MOI, 0.1) with P22 containing amber mutations in genes 13, 1, and 5; diluted 10-fold; grown for 1 h at 37°C; and then sterilized with chloroform. The titers of the resulting phage were determined on lawns of both the β-Gal and the portal protein hosts. The bacteria forming the lawn were sup0, and so only recombinant phage that acquired DNA without amber mutations from the cloned inserts could form plaques (Fig. 1). If the resulting recombinant phage were also escape mutants, then the cloned DNA used to rescue the marker must have contained escape mutations.
FIG. 1.
Marker rescue strategy. To map escape mutations (E), a region of the mutant phage genome was cloned into a plasmid and introduced into Salmonella. The cells were then infected with a phage that contains a conditional selectable marker (x) under permissive conditions. During replication, some of the phage genomes undergo homologous recombination with the cloned escape mutant DNA and generate phage that no longer contain the marker. Subsequent plating under restrictive conditions allows the growth of only recombinant phage. If those recombinants exhibit the escape phenotype, then the cloned DNA fragment must have contained escape mutations.
The approximate location of the escape mutations contained in the cloned DNA segments was determined by repeating the marker rescue experiment with defined point mutants. The escape mutations were transferred with the rescue of an amber mutation in gene 13 approximately 20% of the time and with a cs marker in gene 1 (1−csH137) approximately 75% of the time, suggesting that the escape mutations were closer to the 3′ end of gene 1 (7).
To determine the location of the escape mutations more precisely, the cloned genome segments (mutant and wild type) were reduced in size by restriction digestion and ligation, and the resulting plasmids were again moved into supE Salmonella for further rounds of marker rescue with 1−csH137 phage. The use of 1−csH137 for marker rescue allowed the recombinant phage to be screened for the portal escape phenotype in one plating step because 1−csH137 fails to form plaques at room temperature (16). The recombination templates were shortened by cutting with XhoI to remove a fragment containing genes 13 through 3 and ligated closed. After determination that the remaining segments derived from the escape mutant phage still contained the portal escape mutations by marker rescue, they were further shortened by cutting with XhoI and NdeI, end-filled with the Klenow fragment, and religated. The shortened constructs conferred the escape phenotype and contained the latter half of gene 1, all of genes 8 and 5, and a portion of ORF69 (Fig. 2). In subsequent mapping experiments, PCR was used to directly clone 4.8-kb phage DNA segments from each mutant that spanned the 3′ end of gene 2 through the 5′ end of gene 4.
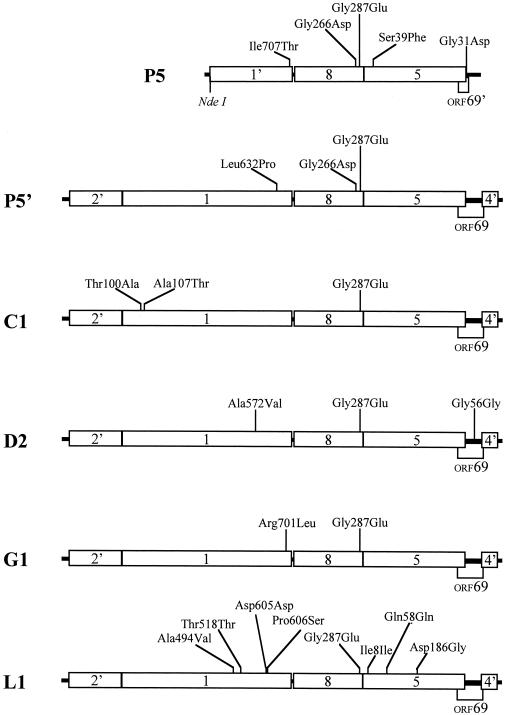
FIG. 2.
Escape mutations. The amino acid alterations that result from mutations present in cloned DNAs isolated from escape mutant phage are indicated. The wild-type amino acid precedes the amino acid number, which is followed by the mutant amino acid. The cloned segments conferred the escape phenotype upon marker rescue, and the phage from which they were derived were independently isolated (except for P5′, which was derived from P5). Note that each clone contains the same Gly287Glu mutation in the scaffold gene. The clone names are indicated on the left. The gene names are shown in the rectangles that approximate their relative sizes. A prime symbol indicates a truncated gene.
During the mapping of the escape mutations, it was discovered that bacteria harboring certain plasmid constructs grew poorly (e.g., if the EcoRI/EcoRI fragment was removed from the P5 or wild-type mapping clones). The cloning vector initially used for mapping (pCR-XL-TOPO) contained the ccdB gene, whose product is toxic to E. coli. We suspected that a low level of expression of this gene product in certain clones was responsible for the poor growth. To remedy this problem, the mapping plasmid was modified by PCR such that both the lac promoter that drove the expression of the insert and the ccdB gene were deleted. The modified plasmid, designated pTOPO-fix, was used for mapping all of the escape mutants isolated after P5; strains carrying constructs derived from this plasmid exhibited normal growth rates.
DNA sequencing.
The cloned mapping segments were sequenced from purified plasmid DNAs by using automated technology (Applied Biosystems sequencer; Lone Star Labs, Houston, Tex.). High-quality sequence data were obtained with 10 primers that primed from alternating strands approximately every 500 bases. The presence of the Gly287Glu mutation in the recombinant phage used for the biochemical studies was determined by sequencing from a PCR product generated from that phage stock. Genes 1 and 8 were sequenced in the parental phage strain without prior PCR amplification from total DNA that had been purified from phage pellets obtained from 25 ml of lysed culture by using a commercial DNA purification kit (Qiagen) (23).
Burst analysis.
β-Gal and portal protein hostcultures were grown to mid-log phase and induced with IPTG for 1.5 h at 37°C. Equal aliquots of the cultures at room temperature were infected with phage at an MOI of .05. After 15 min, the cultureswere diluted 1,000-fold in LB broth with IPTG. Samples were then withdrawn to determine the number of infected cells (total PFU minus non-chloroform-sensitive PFU), and the cultures were grown for 1 h at 20°C. The cultures were treated with chloroform, and the titers were determined. The burst was calculated by dividing the final titer by the number of infected cells.
Sucrose gradient analysis of lysates.
Bacteria were chilled on ice, harvested, and resuspended at 1/150 the original culture volume in ice-cold lysis buffer (100 mM K+ glutamate, 30 mM K+ morpholinepropanesulfonic acid [MOPS], 5 mM MgSO4, 0.02% NaN3 [pH 7.4]). To this mixture, lysozyme was added from a fresh 10-mg/ml stock to a final concentration of 0.05 mg/ml, and the cells were immediately frozen in 500-μl aliquots at −70°C. Lysates were prepared by thawing a sample, adding phenylmethylsulfonyl fluoride to a 1 mM final concentration, and then adding chloroform to 5% (vol/vol) to accelerate lysis. The samples were incubated at room temperature (∼20°C) for 20 to 30 min, during which time the initial gel-like viscosity of the lysates was greatly reduced. Lysates were again treated with 1 mM phenylmethylsulfonyl fluoride and centrifuged for 5 min at 14,000 × g in a microcentrifuge to remove large debris. The supernatant was removed and transferred to a clean tube.
Linear 5 to 45% sucrose gradients were prepared with Beckman SW-41 tubes by using a BioComp Gradient Master according to the manufacturer's instructions (sucrose in a mixture containing 100 mM NaCl, 50 mM Tris-Cl, 5 mM MgCl2, and 0.02% NaN3 [pH 7.4]). After the gradients were set, 300 μl was removed from the top of the gradients, and 150 μl of a CsCl underlay (55% CsCl in a mixture containing a 5% sucrose solution, 0.05% bromophenol blue, and 0.02% NaN3; d20, ∼1.7 g/cm3) was injected at the bottom of the gradients by using a pulled Pasteur pipette fitted to a syringe. A 250-μl quantity of lysate supernatant was then layered on the top of the gradients, and the gradients were centrifuged for 47 min at 35,000 rpm at 18°C. The gradients were fractionated into 850-μl aliquots by pumping from the bottom through thin plastic tubing with a peristaltic pump.
Samples of each fraction were mixed with loading buffer containing sodium dodecyl sulfate (SDS) and electrophoresed in 12.5% acrylamide-SDS gels. The gels were stained with 0.2 g of Coomassie blue R-250/liter (solvated in 10% methanol-10% acetic acid) and then destained in 10% acetic acid. The gels were digitally imaged with a charge-coupled device camera, and the protein band intensities were determined with Alpha Imager software (Alpha Innotech, San Leandro, Calif.) by drawing vertical rectangles around lanes and integrating the pixel intensities across the lanes. Baselines were then established, and the band peak intensities were determined by integration. Relative protein abundances were determined for each lane independently, and the protein ratios for a given particle were determined by averaging the three lanes that comprised the center of the particle peak in the gradient. Reimaging and recalculating the protein ratios gave values within 2 to 5% of each other.
Native agarose gel analysis.
Sucrose gradient fractions were mixed with a 1/3 volume of 4× native sample buffer (30% glycerol, 90 mM Tris, 90 mM borate, 2 mM MgCl2, 0.01% bromophenol blue, 0.02% NaN3), and 10-μl samples were electrophoresed in 1.2% agarose gels prepared in a buffer containing 45 mM Tris, 45 mM borate, and 2 mM MgCl2 at 8 V/cm and 4°C. The gels were then gently shaken in 20 mM EDTA for 30 min to chelate magnesium prior to staining with 0.15 g of Coomassie blue/liter (in a mixture containing 10% methanol-10% acetic acid). After several hours of staining, the gels were destained by being shaken in 10% acetic acid with several Kimwipes overnight. The gels were imaged on a white light box with a charge-coupled device camera (Alpha Innotech).
Electron microscopy.
Samples of gradient fractions were exchanged into a low-salt buffer by centrifugation in Bio-Spin 6 columns (Bio-Rad) preequilibrated with 10 mM Na2HPO4-10 mM NaCl-2 mM MgCl2-0.02% NaN3 [pH 7.4]. Ten-microliter samples were pipetted onto carbon-coated 300-mesh copper grids (EM Sciences), allowed to stand for 15 min, and blotted dry. After being stained for 20 s with 2% uranyl acetate, the grids were again blotted dry and then imaged in a Hitachi electron microscope with a 75-kV accelerating voltage.
RESULTS
Evolution of P22 mutants that escape excess portal protein.
P22 portal protein overexpressed from a plasmid strongly inhibits phage replication by causing the assembly of defective PLPs (23). As a result, wild-type P22 forms small plaques at 37°C on lawns of Salmonella overexpressing portal protein. At 20°C, no visible plaques are formed. This phenotype provided the basis for a screen to identify escape mutant phage that could form plaques in the presence of excess portal protein with the hope of identifying genes affecting the recruitment of portal protein into phage heads. The plating of wild-type phage at high densities on lawns of Salmonella overexpressing portal protein did not yield plaques. Therefore, to allow phage with rare or multiple escape mutations to accumulate, nonmutagenized phage were serially passaged in cultures overexpressing portal protein (see Materials and Methods).
After several rounds of passage in the presence of excess portal protein, the yield of phage increased, suggesting that escape mutants had accumulated (from initial titers of ∼1 × 1010 to titers of ∼9 × 1010 in round 6). The overall amplification during six rounds of selection was estimated to be approximately 1018 [(103)6]. Phage from round 5 were plated on lawns of Salmonella overexpressing either portal protein or control protein β-Gal. Phage that had been passaged on the β-Gal control host were unable to form plaques on lawns of the portal protein host. Phage that had been passaged on the portal protein host were able to form plaques on both the control host and the portal protein host, indicating that escape mutant phage had indeed evolved. Ten single-plaque isolates from round 5 were separately screened for the escape phenotype. Eight of the 10 isolates were portal escape mutants, indicating that the round 5 population was heterogeneous; moreover, these eight escape mutants were able to form plaques on the β-Gal host, indicating that they were not dependent on the presence of excess portal protein.
The escape mutations repeatedly mapped to genes 1, 8, and 5 and ORF69.
The escape mutants did not exhibit any cold- or temperature-sensitive growth (20, 37, or 42°C). Therefore, because there was no way to readily select against the input phage, it proved impossible to map the mutations by coinfection and recombination. Therefore, the mutations present in a representative isolate (termed P5) from round 5 were mapped by marker rescue. Segments of cloned DNA from the escape phage were inserted into plasmids, which were then tested for their ability to confer the escape phenotype on portal-sensitive phage by recombination (see Materials and Methods and Fig. 1) (7).
A cloned genomic segment from the escape mutant containing the latter half of gene 1 through the end of gene 5 conferred the escape phenotype during marker rescue, indicating that the escape mutations were present in that region. This cloned insert (approximately 3.3 kbp) was sequenced to identify the mutations (Fig. 2, clone P5).
Six mutations were present in the cloned segment from phage P5 (32). One of the mutations (Leu559Leu in gene 1) was also present in the parental phage stock used to start the evolution experiment and so was presumably silent and unrelated to the escape phenotype. The new mutations were in the portal (gene 1), scaffold (gene 8), and coat (gene 5) genes and ORF69. ORF69 has no assigned role in P22 morphogenesis. The large distance between the ORF69 mutation and the 1−csH137 marker suggested that it was not required for the escape phenotype, because the high frequency of the escape phenotype in the rescue recombinants suggested that the required escape mutations were nearer to the end of gene 1.
Multiple independent evolution experiments were subsequently performed to determine whether any particular gene region consistently was mutated for the escape phenotype. The evolution experiments were repeated in 12 parallel experiments (designated A through L). After five rounds of selection, escape mutant phage were present in each lineage. Eight plaque isolates from each lineage were plated on both the β-Gal and the portal protein hosts to isolate and characterize escape mutants. Interestingly, several of the escape mutants formed smaller plaques in the absence of excess portal protein, suggesting that they had become partially dependent on the presence of excess portal protein. In some instances, phage that formed large or small plaques were present in the same lineage, suggesting that phage present in a given lineage were heterogeneous in their ability to escape excess portal protein. In one instance (lineage L), all of the phage tested were dependent on the presence of excess portal protein for efficient growth.
The pilot evolution experiment that yielded mutant P5 had identified mutations in the portal, scaffold, and coat genes and ORF69. To determine whether the escape mutations derived from the 12 new lineages were also contained within these genes, the portal-scaffold-coat-ORF69 gene region (with additional flanking segments to allow for efficient homologous recombination) was cloned from representatives of each lineage and used in marker rescue experiments. When both excess portal protein-dependent and independent phage were present in the same lineage, a representative clone from each phenotype was included in the marker rescue experiment. A total of 18 clones were tested. All 18 clones conferred the escape phenotype, suggesting that the region from the portal gene through ORF69 carried the escape mutation in each one. Four clones were selected for DNA sequencing. Clones D2 and L1 were chosen because they were partially dependent on the presence of excess portal protein, whereas clones C1 and G1 were chosen because they were independent of excess portal protein (Fig. 2).
The cloned DNA from initial escape mutant P5 was used as a recombination template to construct a lysis-defective phage for biochemical analysis (data not shown). A large-plaque escape mutant arose during the recombination step and was later determined to be completely dependent on the presence of excess portal protein, suggesting that not all of the mutations present in clone P5 had been crossed back to the phage. Therefore, the portal-scaffold-coat-ORF69 gene region from this dependent phage (designated P5′) was also cloned and sequenced to determine how this phage differed from P5 (Fig. 2).
A particular scaffold mutation was present in each escape mutant.
Each of the sequenced clones had at least two mutations: one in the portal gene and one in the scaffold gene (Fig. 2). The mutations in the portal genes tended to cluster at the 3′ end. Surprisingly, all of the escape mutants contained the same nucleotide change in the scaffold gene. The mutation coded for a glycine-to glutamate substitution at residue 287, near the carboxyl-terminal end of the scaffold protein. This mutation was also present in the P5 escape mutant from the pilot experiment (Fig. 2). In addition to the common scaffold mutation, other mutations were present in each clone (Fig. 2).
To quantitatively assess the effect of the escape mutations on growth in the presence and absence of excess portal protein, the burst size of each mutant was determined (Table 1). As expected from its inability to form plaques in the absence of excess portal protein, the P5′ mutant had a very small burst size in the absence of excess portal protein (only 3.6 phage per cell). Each of the escape mutants grew significantly better than the wild type in the presence of excess portal protein. Additionally, each of the escape mutants was mildly dependent on the presence of excess portal protein.
TABLE 1.
Burst sizes of wild-type and escape mutant phage for control hosts and those overexpressing portal protein
| Phage strain | Burst size (PFU/cell) for the following host:
|
|
|---|---|---|
| β-Gal | Portal protein | |
| Wild type | 122 | 7.3 |
| P5 | 64 | 92 |
| P5′ | 3.6 | 70 |
| C1 | 61 | 102 |
| D2 | 24 | 72 |
| G1 | 74 | 112 |
| L1 | 26 | 48 |
| Gly287Glu | 85 | 53 |
The Gly287Glu mutation in the scaffold gene is sufficient for the escape phenotype.
The isolation of escape mutants with only a few coding changes allowed the importance of each to be addressed directly. A single EcoRI restriction site present in the scaffold gene allowed the portal gene mutations to be isolated from the scaffold and coat gene mutations in each clone. Ten new marker rescue plasmids were constructed by swapping the EcoRI/BglII restriction fragments (containing the scaffold gene, coat gene, and ORF69 mutations) from five escape mutant clones, P5′, C1, D2, G1, and L1, with a wild-type restriction fragment. The EcoRI/BlgII fragment from the wild-type clone was also inserted into the mutant clones to isolate the mutations in the portal genes. The EcoRI/BglII restriction fragment from clones C1 and G1 contained only the single Gly287Glu mutation in the scaffold gene. Each of the 10 clones was then tested for the ability to confer the escape phenotype by marker rescue.
For each clone tested, the recombination template containing mutations in the scaffold gene-coat gene-ORF69 mutant segment conferred the escape phenotype. Because subclones C1 and G1 contained only the Gly287Glu mutation in the scaffold gene, Gly287Glu was sufficient for the escape phenotype. Interestingly, the marker rescue plaques formed in the absence of the portal gene mutations in clones C1 and G1 were smaller than the escape mutant plaques generated from the complete clones, suggesting that the mutations in the portal genes were auxiliary to the Gly287Glu scaffold gene mutation.
The 1−csH137 marker that was used in the rescue experiments was very near to the portal gene mutations present in subclones P5′, D2, G1, and L1, which contained wild-type scaffold and coat genes and ORF69 (7). Few if any recombinants were observed in which the cs phenotype was rescued beyond the background level of reversion of the 1−csH137 marker when these subclones were used as recombination templates. This result suggests that the portal gene mutations by themselves were lethal to the phage. For subclone C1, approximately one-third of the recombinants formed very small fuzzy plaques, suggesting that when the portal gene mutations present in the N-terminal region of the C1 portal protein were recombined into the phage during marker rescue, the resulting phage were defective.
The Gly287Glu mutation in the scaffold protein reduces portal protein incorporation.
To generate a conditional lysis-defective phage with the Gly287Glu mutation for biochemical characterization, a recombination template containing only the Gly287Glu mutation was used to rescue the 1−csH137 marker from 1−csH137 13−amH101 phage. Approximately two-thirds of the recombinant phage that were no longer cold sensitive were also portal escape mutants, suggesting that the Gly287Glu mutation was crossed into the rescued phage at a frequency consistent with the distance between the Gly287Glu mutation and 1−csH137. All of the recombinant phage required the amber suppressor for growth, demonstrating that the 13−am marker was still present in each recombinant. When phage carrying only the Gly287Glu mutation were grown in the β-Gal host, their burst size was slightly larger than that of phage G1 (85 versus 74), which carries a secondary mutation in the portal protein. This difference suggests that the Gly287Glu mutation alone was less detrimental to phage production than the combination of both mutations (Table 1). Unlike the double-mutant phage, the Gly287Glu mutant phage replicated less efficiently in the presence of excess portal protein, suggesting that the secondary mutation in the portal gene assisted in the escape mechanism (Table 1). An isogenic control phage was generated by rescue of the 1−csH137 13−am phage with a wild-type recombination template. The portal and scaffold genes in each 13−am phage were sequenced to verify the presence or absence of the Gly287Glu escape mutation.
To determine the effect of the Gly287Glu mutation on phage morphogenesis, liquid cultures of nonsuppressing control β-Gal and portal protein hosts were induced with IPTG to express protein for 1.5 h. The cultures were then evenly divided and infected with either the control 13−am phage or the Gly287Glu 13−am phage. After 2 h of infection, the cells were harvested and frozen (see Materials and Methods). Prepared lysates of each were layered on sucrose gradients, centrifuged to resolve the products of phage assembly from other cellular components, fractionated, and analyzed by SDS-polyacrylamide gel electrophoresis and native agarose gel electrophoresis (Fig. 3 and 4).
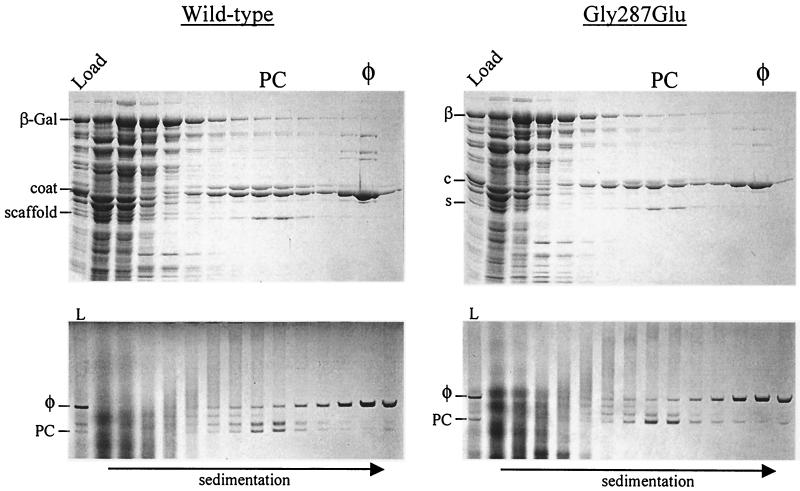
FIG. 3.
Wild-type and Gly287Glu phage infections of the β-Gal host. β-Gal-expressing cells that had been infected with either wild-type or mutant Gly287Glu phage were resolved in sucrose gradients and analyzed for protein content by SDS-PAGE (top panels) and for particle content by native agarose gel electrophoresis (bottom panels). Load (L), samples of the total lysate; PC, procapsid peak position; φ, phage peak position. Dominant proteins (or the first letter thereof) are labeled to the left of each SDS gel, and the positions of the procapsid and phage particles are indicated to the left of each native agarose gel. The direction of sedimentation is indicated at the bottom. In the SDS gels, one-quarter of the bottommost fraction was loaded to reduce distortions caused by excessive CsCl present in the sample.
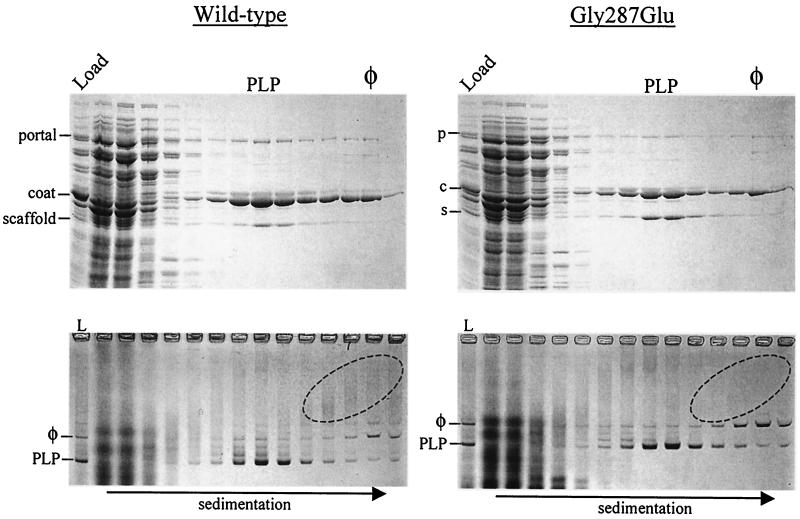
FIG. 4.
Wild-type and Gly287Glu phage infections of the portal protein host. P22 portal protein-overexpressing cells that had been infected with either wild-type or mutant Gly287Glu phage were resolved in sucrose gradients and analyzed for protein content by SDS-polyacrylamide gel electrophoresis (top panels) and for particle content by native agarose gel electrophoresis (bottom panels). Load (L), samples of the total lysate; PLP, procapsid-like particle peak position; φ, phage peak position. Dominant proteins (or the first letter thereof) are labeled to the left of each SDS gel, and the positions of the procapsid and phage particles are indicated to the left of each native agarose gel. The direction of sedimentation is indicated at the bottom. The ovals highlight regions in the native gels that contained aberrant, large, heterogeneous assemblies. Less sample was loaded from the bottommost fraction in the SDS gels to reduce CsCl distortion.
In the β-Gal host culture infected with the 13−am phage, progeny phage were efficiently produced (most of the coat protein had migrated to the phage position) (Fig. 3, upper left panel). The amounts of coat protein at the procapsid (PC) and phage (φ) positions in the gradient were similar to those previously reported for P22-infected cells (23). Particles with native gel electrophoretic mobilities matching those of procapsids and phage were present in the center and bottom of the gradient, respectively (Fig. 3, lower left panel). Infections with the Gly287Glu mutant yielded similar distributions of coat protein in the gradient (Fig. 3, upper right panel). However, in a native agarose gel, the particles in the procapsid region of the Gly287Glu mutant gradient had distributions different from those of the wild type: the fast-migrating unexpanded procapsid form (Fig. 3, lower right panel, PC) was present in relative excess compared to the two slower-migrating intermediate bands.
The composition of the particles was characterized by densitometry of digital images of Coomassie blue-stained gels of procapsid- and phage-containing fractions (see Materials and Methods). In the control infection, the portal-to-coat protein ratio was the same for both the procapsid and the phage peaks. However, the procapsids produced in the Gly287Glu infection had only ∼80% as much portal protein as did the wild-type particles. The phage produced in this infection had a wild-type portal-to-coat protein ratio, suggesting that only particles with a full complement of portal protein had been selected to package DNA. The scaffold-to-coat protein ratios in the procapsid peak regions of both the control and the Gly287Glu gradients were, within densitometry error, the same. Thus, it appears that the Gly287Glu mutation results in a slight decrease in the amount of portal protein incorporated.
Parallel infections were carried out with host cells overexpressing portal protein, and the products were analyzed in a similar fashion. (Fig. 4). As expected, the wild-type phage infection produced significantly fewer mature phage in the presence of excess portal protein than in its absence (compare left panels of Fig. 3 and 4). The distribution of particles was similar to that previously reported for P22 infections in the presence of excess portal protein (23). PLPs (particles that resemble procapsids but that have not been demonstrated to be viable intermediates) that comigrated with procapsids were the most prevalent particles in the gradient (Fig. 4, lower left panel, lane L). Consistent with a previous report (23), densitometric analysis of the protein levels in the procapsid and phage peaks indicated that the portal-to-coat protein ratio was approximately twice that in the control infection (∼1.9- to ∼2.2-fold excess). Additionally, the amount of scaffold protein associated with the immature particles in the PLP region of the gradient was reduced relative to that in purified procapsids (by ∼45%; see reference 23 for comparisons).
In the presence of excess portal protein, the Gly287Glu mutant infection produced both PLPs and phage (Fig. 4, right panels). However, consistent with the increased titer, a higher fraction of the assembled particles had matured into phage (Fig. 4, lower right panel, lane L). Densitometry revealed that the amount of portal protein in the Gly287Glu PLPs was only slightly higher than that in normal procapsids (∼1.1-fold excess), suggesting that the Gly287Glu mutation in the scaffold protein had also reduced the efficiency of portal protein incorporation when portal protein was present in excess.
Multiple tails can bind to PLPs produced in the presence of excess portal protein.
In a previous report on the formation of PLPs with excess portal protein, it was suggested that PLPs contained excess portal protein as additional portal vertices (23). At that time, there was no direct evidence that this was the case (for instance, excess portal protein may have been nonspecifically trapped inside PLPs). In the present study, it was observed that 1 to 2% of the procapsids (rounded and thick walled) derived from the wild-type infection had tailspikes attached. Presumably, these structures prematurely acquired tailspikes prior to DNA packaging. Other tailed particles with a mature head morphology (angular and thin walled) were also present in the procapsid peak region; however, these mature particles had noticeable breaks in the coat lattice, suggesting that they were disrupted phage heads. Because portal rings are not readily detectable in coat lattices, the tailspikes bound to head structures serve as convenient markers to identify portal vertices.
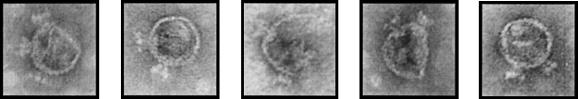
To determine if any of the PLPs with excess portal protein had also acquired tails, several micrographs of the PLP peak fractions from sucrose gradients were screened for the presence of tailed immature particles. Aberrant tailed immature particles were observed with the same frequency as in the control infection, and so tailed particles were rare (∼1 to 2%); however, out of 10 tailed immature particles that were clearly identifiable, 5 had more that one tail visible (Fig. 5). No multiple-tail particles were observed in micrographs of the procapsid peak fractions derived from β-Gal control hosts, and no multiple-tail particles were detected in fractions derived from any of the Gly287Glu mutant infections. These observations suggest that at least some of the PLPs with excess portal protein had the portal protein incorporated as properly assembled portal vertices that were competent to bind tail machines, and they support the observation that the Gly287Glu mutation rescues this phenotype.
FIG. 5.
Multiple-tail procapsids. Shown are electron micrographs of negatively stained particles from the procapsid peak fraction of the wild-type phage-infected portal protein host. Tailspikes were detected on approximately 1% of the total procapsids. Because tailspikes attach only to portal vertices, they serve as a marker for the presence of portals within procapsid shells. No multiple-tail procapsids were detectable in the wild-type phage-infected β-Gal host, and no multiple-tail procapsids were present in any samples derived from Gly287Glu infections.
The Gly287Glu mutation increases assembly fidelity in the presence of excess portal protein.
Aberrant head structures are formed during wild-type P22 infections as a small percentage of the total particles produced (12). In the absence of scaffold protein, the amount of spiral coat assemblies that are formed greatly increases, presumably because of unguided, off-pathway coat polymerization (8, 12, 19). When excess portal protein is present during wild-type phage infections, similar aberrant spirals are produced, suggesting that portal protein interacts with the growing procapsid (23). Spiral shells with sufficient mass comigrate with phage in sucrose gradients (observed by electron microscopy); however, the spiral populations tend to be distributed as slow-migrating smears in native agarose gels, presumably because the architecture of the spirals is varied (e.g., Fig. 4, lower left panel, and data not shown).
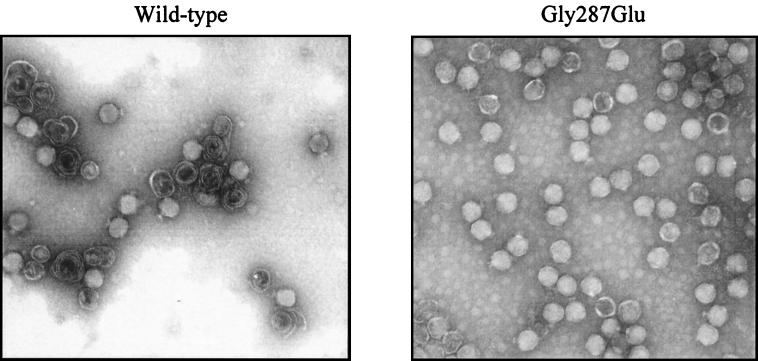
A notable difference between wild-type and Gly287Glu particle distributions in the native agarose gels in Fig. 4 was observed. The material that resolved as smears in the agarose gels toward the bottom of the wild-type gradient was not detectable in the Gly287Glu gradient (Fig. 4, lower panels, ovals). Because these fractions contained primarily coat protein, the smears were most likely aberrant assemblies. To visualize particles formed in both infections, selected fractions from sucrose gradients were analyzed by negative-stain electron microscopy. Figure 6 shows micrographs of the peak gradient fractions of wild-type and Gly287Glu phage infections in the presence of excess portal protein. The sample derived from the wild-type phage infection contained a high percentage of spiral aberrant heads (∼60% of the particles), consistent with the distribution of material in the native agarose gel (Fig. 4).
FIG. 6.
High-fidelity Gly287Glu mutation. Electron micrographs show negatively stained particles present in the phage peak fractions from the gradients shown in Fig. 4. The host in each example had overexpressed portal protein during the infection. Note that many of the particles in the sample derived from the wild-type phage infection are aberrant spirals and that practically all of the particles derived from the Gly287Glu infection have morphologies consistent with a T=7 capsid lattice.
The particles produced by the Gly287Glu mutant in the presence of excess portal protein had very homogeneous phage-like morphologies, and very few aberrant spirals were detected (<1%; Fig. 6, right panel). Some empty particles were also present, but because these had migrated in the sucrose gradient to the phage peak position, they had probably been disrupted after the gradient was fractionated. The absence of aberrant spirals in the Gly287Glu gradient indicates that this single mutation in the scaffold gene increased assembly fidelity. Because this mutation was present in every escape mutant that was sequenced, there appears to be a correlation between head assembly fidelity and portal vertex formation.
DISCUSSION
Escape mutants with synergistic mutations.
Mutant phage that escaped the presence of excess portal protein were isolated after multiple rounds of restricted growth. The need for multiple rounds was probably a consequence of the fact that wild-type phage were capable of replicating, albeit poorly, in liquid cultures. Thus, any mutation that conferred a growth advantage in the presence of excess portal protein would need several rounds to be present in a sufficient number to be detected. Some escape mutant phage were hypermutated, with as many as eight mutations in the portal-scaffold-coat gene region. Considering the extensive amplification of the phage populations during selection, it is probable that mutations arose that were unrelated to the escape phenotype. Such a scenario may explain why some phage had additional mutations beyond what was necessary for escape. Indeed, the two most fit mutants had the fewest mutations.
The escape phage contained mutations in the portal gene that appeared to be toxic on their own (they could not rescue nearby markers), suggesting that these mutations reduced the activity of the phage-encoded portal protein (Fig. 2). Because the inhibitory effect of excess portal protein is dose dependent (23), the acquisition of a mutation within the phage-encoded portal protein that reduced activity would confer a growth advantage. Interestingly, the mutant portal genes were functional in the context of the other mutations in the scaffold and coat genes and ORF69. One plausible explanation for this observation is that perhaps the escape mutations (such as Gly287Glu) slowed head assembly. Such a scenario could allow a partially defective portal protein sufficient time to assemble into the capsid, whereas in the context of an otherwise wild-type phage, it would be left out.
A surprising finding was that each of the sequenced escape mutants contained the Gly287Glu mutation in the scaffold gene. In each instance, the mutation was encoded by the same nucleotide change (GGA to GAA). If only single nucleotide changes are considered, codon 287 could have changed to code for glycine, alanine, valine, arginine, or glutamate. Of the 11 possible nonglycine codons that could arise from a single point mutation, only one (GAA) codes for glutamate, and none of the other possibilities is acidic. This finding suggests that glutamate (or at least an acidic residue) is probably required for the escape phenotype. A preliminary analysis of the activity of purified Gly287Glu scaffold protein in vitro has revealed that it polymerizes coat protein more slowly than wild-type scaffold protein (C. Fu, personal communication). Further experiments are under way to correlate the chemical identity of residue 287 to the escape mechanism.
The Serendipity of Gly287Glu.
Detailed structural information on the proteins of P22 is limited. Coincidentally, the Gly287Glu mutation in scaffold protein resides in a region for which an atomic resolution structure is known (residues 238 to 303) (28). This C-terminal portion of scaffold protein is required for coat protein binding (25, 28). In the nuclear magnetic resonance solution structure of this region, the Gly287 residue is positioned in the turn of a helix-turn-helix, and the side-group hydrogen of the glycine faces outward from the structure (28). A preliminary nuclear magnetic resonance analysis of a C-terminal scaffold fragment containing the Gly287Glu mutation suggests that most of the contacts present in the wild-type fragment are preserved in the mutant fragment, indicating that the Gly287Glu mutation does not dramatically alter the surrounding protein fold (data not shown). This finding suggests that the glutamate side chain probably projects away from scaffoldprotein and is capable of directly interacting with other phage proteins.
Excess portal protein and defective PLPs.
We previously reported that the maturation of PLPs containing excess portal protein was blocked at the DNA packaging stage even though the phage DNA had replicated efficiently (23). This finding suggested that DNA packaging may have been inhibited either because excess portal protein was competing with procapsids for terminase binding or because the PLPs were somehow defective in progressing through the maturation process that normally accompanies packaging (for example, scaffold protein exit and/or capsid expansion). The amount of scaffold protein associated with PLPs is reduced in the presence of active terminase, suggesting that terminase is capable of interacting with PLPs containing excess portal protein (23). Because no escape mutations were found in the terminase genes, it seems likely that the maturation defect of the PLPs was due to a defective capsid architecture. Additionally, this screen did not identify any uncharacterized genes responsible for the process of portal vertex formation.
The accumulation of aberrant heads in the presence of excess portal protein is reminiscent of the effect of the absence of scaffold protein, suggesting that excess portal protein may have sequestered scaffold protein away from the assembly processes in the cell. However, this mechanism cannot account for the observations that the PLPs with excess portal protein contained the bulk of scaffold protein present in the lysates (indicating that scaffold protein was sufficiently available to bind and assemble PLPs) and that the PLPs contained both portal and minor proteins (a feature that requires functional scaffold protein). Also, if scaffold protein depletion were the cause of the aberrant assembly, then one might expect to find escape mutants that simply up-regulate the level of wild-type scaffold protein expression. The recovery of a common escape mutation in a region of scaffold protein required for coat protein binding suggests that a scaffold-coat protein interaction is involved in portal vertex formation.
In the absence of excess portal protein, the Gly287Glu mutation reduced phage yield (Table 1). The morphology and electrophoretic mobilities of both phage and procapsids produced with the Gly287Glu scaffold protein were indistinguishable from those of wild-type particles (Fig. 3 to 5 and data not shown). This finding suggests that the reduced phage production was due either to a reduced rate of assembly or to the production of defective particles. The Gly287Glu lysate contained relatively more procapsids than the wild-type lysate, suggesting that maturation of the Gly287Glu procapsids may have been partially impeded (Fig. 3). The slight reduction in the level of portal protein relative to those of the other head proteins in the Gly287Glu procapsids may have been the cause.
As anticipated from the ability to form plaques on lawns of bacteria overexpressing portal protein, the Gly287Glu escape mutation allowed more phage to be produced in the presence of excess portal protein (Table 1). Quite unexpectedly, the Gly287Glu mutation increased the fidelity of head assembly, such that nearly all of the particles produced in the presence of excess portal protein appeared to be T=7 capsid lattices. In addition, the level of portal protein in these procapsids was reduced to nearly normal, indicating that the escape mechanism provided by the Gly287Glu mutation involved reducing portal vertex formation and not, for example, allowing the maturation of capsids with more than one portal.
P22 coat protein expressed alone is capable of forming ordered lattice arrays with proper local contacts; however, the correct relative assembly of pentamers and hexamers is compromised. As a result, a large proportion of the structures formed in the absence of scaffold protein are not T=7 (12, 13, 30). An increase in the fidelity of assembly arising from the single Gly287Glu mutation in scaffold protein suggests a link between the fidelity of coat protein polymerization and portal vertex formation. This idea lends support for a model of portal vertex formation in which the portals are defined after the initiation of capsid assembly, during the growth of the coat lattice.
This model is consistent with the relationships between portal incorporation and the fidelity of head assembly among other dsDNA phages as well. For example, during the assembly of the prolate phage φ29, the absence of portal protein leads to the formation of isometric particles and spiral “monsters” (6, 21). This dependence on portal protein has also been observed for phages T3, λ, and SPP1 (11, 17, 31). In the absence of the phage T4 portal protein, long, open-ended tubes of coat protein form that contain a scaffold core (20). Moreover, a mutation in the major scaffold protein of T4 was identified (22tsA74) that caused aberrant heads to form when the phage was grown semipermissively (18). These aberrant heads had improperly spaced pentameric vertices, and many of the aberrant heads had multiple tails, suggesting that multiple portals had been incorporated (18). Interestingly, the tails were located at adjacent vertices, suggesting that the portal vertices had formed at similar times during head assembly.
At present, we do not know whether the increase in assembly fidelity is a side effect of reducing portal incorporation or if assembly fidelity is directly coupled to forming portal vertices. It is possible that portals are incorporated (and symmetry is interrupted) during difficult stages of capsid growth that are prone to driving assembly off the pathway. Image analysis of P22 procapsids by cryoelectron microscopy has revealed that scaffold protein primarily binds under the hexameric lattice positions (29). If portal vertex formation requires flexibility of the surrounding coat lattice, then the Gly287Glu mutant may bind to coat protein more tightly and constrain the curvature of the growing lattice such that pentameric vertices form in lieu of portal vertices. This model predicts that a mutation that slightly weakens the scaffold-coat protein interaction may allow for the formation of multiple portals when portal protein is not present in gross excess.
Acknowledgments
We thank Jonathan King, Miriam Susskind, and Sherwood Casjens for providing bacterial and phage strains and Sherwood Casjens for guidance throughout this project.
S.D. Moore was supported, in part, by an NIH training fellowship (AI07150). This work was supported by NIH grant GM47980.
REFERENCES
- 1.Bazinet, C., and J. King. 1985. The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol. 39:109-129. [DOI] [PubMed] [Google Scholar]
- 2.Bazinet, C., and J. King. 1988. Initiation of P22 procapsid assembly in vivo. J. Mol. Biol. 202:77-86. [DOI] [PubMed] [Google Scholar]
- 3.Bazinet, C., R. Villafane, and J. King. 1990. Novel second-site suppression of a cold-sensitive defect in phage P22 procapsid assembly. J. Mol. Biol. 216:701-716. [DOI] [PubMed] [Google Scholar]
- 4.Botstein, D., and M. Levine. 1968. Intermediates in the synthesis of phage P22 DNA. Cold Spring Harbor Symp. Quant. Biol. 33:659-667. [DOI] [PubMed] [Google Scholar]
- 5.Botstein, D., C. H. Waddell, and J. King. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage p22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 80:669-695. [DOI] [PubMed] [Google Scholar]
- 6.Camacho, A., F. Jimenez, J. De La Torre, J. L. Carrascosa, R. P. Mellado, C. Vasquez, E. Vinuela, and M. Salas. 1977. Assembly of Bacillus subtilis phage φ29. 1. Mutants in the cistrons coding for the structural proteins. Eur. J. Biochem. 73:39-55. [DOI] [PubMed] [Google Scholar]
- 7.Casjens, S., K. Eppler, L. Sampson, R. Parr, and E. Wyckoff. 1991. Fine structure genetic and physical map of the gene 3 to 10 region of the bacteriophage P22 chromosome. Genetics 127:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S., and J. King. 1974. P22 morphogenesis. I. Catalytic scaffolding protein in capsid assembly. J. Supramol. Struct. 2:202-224. [DOI] [PubMed] [Google Scholar]
- 9.Caspar, D. L. D., and A. Klug. 1962. Physical principles in the construction of regular viruses. Cold Spring Harbor Symp. Quant. Biol. 27:1-40. [DOI] [PubMed] [Google Scholar]
- 10.Dokland, T. 1999. Scaffolding proteins and their role in viral assembly. Cell Mol. Life Sci. 56:580-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Droge, A., M. A. Santos, A. C. Stiege, J. C. Alonso, R. Lurz, T. A. Trautner, and P. Tavares. 2000. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J. Mol. Biol. 296:117-132. [DOI] [PubMed] [Google Scholar]
- 12.Earnshaw, W., and J. King. 1978. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 126:721-747. [DOI] [PubMed] [Google Scholar]
- 13.Greene, B., and J. King. 1996. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology 225:82-96. [DOI] [PubMed] [Google Scholar]
- 14.Guo, P. X., S. Erickson, W. Xu, N. Olson, T. S. Baker, and D. Anderson. 1991. Regulation of the phage phi 29 prohead shape and size by the portal vertex. Virology 183:366-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hendrix, R. W. 1978. Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA 75:4779-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvik, J., and D. Botstein. 1975. Conditional-lethal mutations that suppress genetic defects in morphogenesis by altering structural proteins. Proc. Natl. Acad. Sci. USA 72:2738-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellenberger, E. 1990. Form determination of the heads of bacteriophages. Eur. J. Biochem. 190:233-248. [DOI] [PubMed] [Google Scholar]
- 18.Keller, B., J. Dubochet, M. Adrian, M. Maeder, M. Wurtz, and E. Kellenberger. 1988. Length and shape variants of the bacteriophage T4 head: mutations in the scaffolding core genes 68 and 22. J. Virol. 62:2960-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, J., E. V. Lenk, and D. Botstein. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. II. Morphogenetic pathway. J. Mol. Biol. 80:697-731. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K., E. Molbert, M. Showe, and E. Kellenberger. 1970. Form-determining function of the genes required for the assembly of the head of bacteriophage T4. J. Mol. Biol. 49:99-113. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. S., and P. Guo. 1995. Sequential interactions of structural proteins in phage φ29 procapsid assembly. J. Virol. 69:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lurz, R., E. V. Orlova, D. Gunther, P. Dube, A. Droge, M. F. Weise, van Heel, and P. Tavares. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310:1027-1037. [DOI] [PubMed] [Google Scholar]
- 23.Moore, S. D., and P. E. Prevelige, Jr. 2002. Bacteriophage p22 portal vertex formation in vivo. J. Mol. Biol. 315:975-994. [DOI] [PubMed] [Google Scholar]
- 24.Murialdo, H., and A. Becker. 1978. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol. Rev. 42:529-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parker, M. H., M. Jablonsky, S. Casjens, L. Sampson, N. R. Krishna, and P. E. Prevelige, Jr. 1997. Cloning, purification, and preliminary characterization by circular dichroism and NMR of a carboxyl-terminal domain of the bacteriophage P22 scaffolding protein. Protein Sci. 6:1583-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossmann, M. G. 1984. Constraints on the assembly of spherical virus particles. Virology 134:1-11. [DOI] [PubMed] [Google Scholar]
- 27.Simpson, A. A., Y. Tao, P. G. Leiman, M. O. Badasso, Y. He, P. J. Jardine, N. H. Olson, M. C. Morais, S. Grimes, D. L. Anderson, T. S. Baker, and M. G. Rossmann. 2000. Structure of the bacteriophage phi29 DNA packaging motor. Nature 408:745-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun, Y., M. H. Parker, P. Weigele, S. Casjens, P. E. Prevelige, Jr., and N. R. Krishna. 2000. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 297:1195-1202. [DOI] [PubMed] [Google Scholar]
- 29.Thuman-Commike, P. A., B. Greene, J. Jakana, B. V. Prasad, J. King, P. E. Prevelige, Jr., and W. Chiu. 1996. Three-dimensional structure of scaffolding-containing phage p22 procapsids by electron cryo-microscopy. J. Mol. Biol. 260:85-98. [DOI] [PubMed] [Google Scholar]
- 30.Thuman-Commike, P. A., B. Greene, J. A. Malinski, J. King, and W. Chiu. 1998. Role of the scaffolding protein in P22 procapsid size determination suggested by T=4 and T=7 procapsid structures. Biophys. J. 74:559-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valpuesta, J. M., and J. L. Carrascosa. 1994. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q. Rev. Biophys. 27:107-155. [DOI] [PubMed] [Google Scholar]
- 32.Vander Byl, C., and A. M. Kropinski. 2000. Sequence of the genome of Salmonella bacteriophage P22. J. Bacteriol. 182:6472-6481. [DOI] [PMC free article] [PubMed] [Google Scholar]