Abstract
Replication of the Carnation Italian ringspot virus genomic RNA in plant cells occurs in multivesicular bodies which develop from the mitochondrial outer membrane during infection. ORF1 in the viral genome encodes a 36-kDa protein, while ORF2 codes for the 95-kDa replicase by readthrough of the ORF1 stop codon. We have shown previously that the N-terminal part of ORF1 contains the information leading to vesiculation of mitochondria and that the 36-kDa protein localizes to mitochondria. Using infection, in vivo expression of green fluorescent protein fusions in plant and yeast cells, and in vitro mitochondrial integration assays, we demonstrate here that both the 36-kDa protein and the complete replicase are targeted to mitochondria and anchor to the outer membrane with the N terminus and C terminus on the cytosolic side. Analysis of deletion mutants indicated that the anchor sequence is likely to correspond approximately to amino acids 84 to 196, containing two transmembrane domains. No evidence for a matrix-targeting presequence was found, and the data suggest that membrane insertion of the viral proteins is mediated by an import receptor-independent signal-anchor mechanism relying on the two transmembrane segments and multiple recognition signals present in the N-terminal part of ORF1.
The genomes of positive-stranded RNA viruses from a number of supergroups are replicated in association with intracellular membranes (3). Depending on the virus, a variety of membrane systems can be concerned in infected cells, including the plasma membrane, endoplasmic reticulum, vacuole, chloroplasts, mitochondria, peroxisomes, and lysosomes (19, 22, 32, 38, 42, 47). Little is known about the molecular mechanisms supporting these virus-cell interactions, which are critical for the development of infection. Membrane interaction with both viral proteins and host-encoded factors (50) has been proposed. Viral RNA replication is often associated with infection-specific membrane proliferation and/or vesiculation.
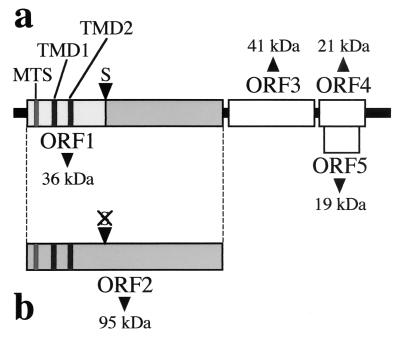
Plant infection with Carnation Italian ringspot virus (CIRV), a member of the genus Tombusvirus in the family Tombusviridae, triggers the development of conspicuous membrane bodies from modified mitochondria (11). The CIRV genome is composed of a monopartite, positive-sense RNA genome of 4,760 nucleotides (GenBank accession number X85215). It is not polyadenylated, lacks a 5′ cap structure, and contains five functional open reading frames (ORFs) (36) (Fig. 1). The 5′-most ORF (ORF1) in the CIRV genome encodes a 36-kDa protein (36K protein), while ORF2 codes for a 95-kDa protein (95K), which is expressed by readthrough of the amber stop codon of ORF1. Readthrough occurs at a frequency of about 10% in plant cells. Both pre- and complete readthrough products are essential for viral replication and were shown to contain the eight conserved motifs (PI to PVIII) of RNA-dependent RNA polymerases of supergroup II of the positive-stranded RNA viruses (16, 40). ORF3 encodes the coat protein of 41 kDa, and the two nested ORFs 4 and 5 code for 21-kDa and 19-kDa proteins which are involved in virus movement and symptom expression in infected plants, respectively (36).
FIG. 1.
Organization of CIRV genomic RNA. (a) Translation of ORF1 yields a 36-kDa polypeptide containing a predicted MTS and two putative transmembrane domains (TMD1 and TMD2). (b) Readthrough of the ORF1 stop codon (S) allows translation of ORF2, yielding the 95-kDa complete replicase.
Cytopathological studies of CIRV infections have identified vesiculated structures (multivesicular bodies [MVBs]), made up of a main body surrounded by many spherical to ovoid vesicles 80 to 150 nm in diameter, resulting from proliferation of the limiting mitochondrial membrane (11, 41). MVBs were suggested to be the replication site of the viral genome. Our previous studies demonstrated that the 36K protein encoded by the CIRV ORF1 is recovered in the mitochondrion- and MVB-containing subfractions of infected plants and is able to target the green fluorescent protein (GFP) reporter to mitochondria in vivo upon transient expression of a fusion protein in plant leaves and cell suspensions (39). The 36K protein was also shown to localize and target GFP to mitochondria upon transformation of yeast (Saccharomyces cerevisiae) cells (37). Both in plant and in yeast cells, the protein is in stable association with cell membranes, based on its resistance to extraction with carbonate, urea, or high salt (37, 38).
Mitochondrial targeting and import of nucleus-encoded proteins have been extensively characterized for animal, yeast, and plant cells (2, 13-15, 20, 21, 34, 46). Four distinct translocation machineries which mediate import and sorting of proteins into the mitochondrial subcompartments have been identified (14). Most proteins destined to the mitochondrial matrix are synthesized as extended precursors containing a cleavable, positively charged, amino-terminal presequence that is able to form an amphipathic α-helix. Proteins destined to the outer membrane, the intermembrane space, or the inner membrane are generally synthesized as noncleavable mature-size polypeptides and contain internal information for targeting and sorting.
Nucleus-encoded mitochondrial proteins usually cross the outer membrane through the TOM (translocase of the outer membrane) complex. Matrix-targeted polypeptides with an N-terminal presequence are subsequently recruited by the TIM23 (translocase of the inner membrane) complex and proceed through the inner membrane before the presequence is cleaved. Integral inner membrane proteins require the TIM22 complex to insert into their final location. Targeting and integration of nucleus-encoded proteins into the mitochondrial outer membrane is less documented (7, 25, 27, 35, 44). Several mechanisms have been described. Some proteins contain a unique transmembrane domain which functions both as a signal and as an anchor sequence specific for the mitochondrial outer membrane. Insertion leaves domains exposed to the cytosol and to the intermembrane space. The S. cerevisiae Tom70 protein relies on such an amino-proximal “signal-anchor sequence” (27), whereas mammalian Bcl-2 contains a carboxy-terminal signal-anchor (31) and yeast Tom6 harbors both a signal-anchor and additional targeting information (5).
Membrane insertion of polypeptides with an N-terminal signal-anchor is mostly mediated by the TOM machinery of the general protein import system, whereas many of the C-terminal signal-anchor polypeptides appear to be inserted into the outer membrane by unique pathways that are independent of the common protein import receptors (25). However, at least some C-terminally anchored proteins use components of the TOM complex (27, 48). Neurospora crassa Tom22 is representative of another targeting mechanism which relies on separate import and membrane anchor sequences: the transfer is driven by a matrix-type targeting signal but is stopped by the hydrophobic domain, which results in membrane anchoring (35). This mechanism also applies to proteins containing two transmembrane segments. The N-terminal domain of the rat liver carnitine palmitoyltransferase 1 (CPT1) contains a noncleavable matrix-targeting signal located immediately downstream of the second transmembrane domain (7).
Membrane proteins containing a matrix-targeting signal and a “stop transfer” sequence follow the same pathway as matrix-targeted preproteins and engage with the TOM machinery. The most abundant protein in the mitochondrial outer membrane, porin, also called the voltage-dependent anion channel, seems to be devoid of uniformly hydrophobic transmembrane domains and is imported without the aid of a presequence. It appears to contain important targeting and/or assembly information in its C-terminal region (8) and also uses the general protein import pathway (17). Although the precise nature of the targeting signals in β-barrel polytopic membrane proteins like porin or Tom40 remains unclear, it is believed that some structural elements and/or the folded state of the polypeptide contain the import information (8, 25, 33).
The aim of the present work is to determine whether the CIRV proteins participating in the replication complex use one of these mechanisms to target mitochondria. The N-terminal half of the 36K protein contains the determinants for the formation of MVBs from mitochondria rather than from peroxisomes, as for other tombusviruses like the Cymbidium ringspot virus (4, 38). A 14-amino-acid motif (FGSLPSSLERPVAK) corresponding to positions 32 to 45 at the N terminus of the 36K protein was predicted to be a putative mitochondrial targeting sequence (MTS) (Fig. 1). No such sequence was detected in the ORF1 of the other Tombusviruses, which target peroxisomes, whereas a possible MTS was highlighted in the ORF1 of carmoviruses, which develop MVBs from mitochondria (6).
To start evaluating the prediction of an MTS in the CIRV ORF1, the serine residue at position 34 was mutated to aspartic acid or arginine. The presence of Arg34 had no cytopathological consequence, but the Ser34 to Asp34 mutation resulted in multidirected targeting, MVBs being derived from mitochondria but also from some peroxisomes (38).
Downstream of the putative MTS, the CIRV 36K polypeptide contains two predicted hydrophobic domains sufficiently long to span the limiting outer membrane of mitochondria (Fig. 1). These domains can be considered putative transmembrane domains. They are separated by a possible hydrophilic “loop.” From the previous data and sequence analyses, the following model can be proposed. The 36K CIRV protein, as well as the complete 95K replicase (ORF2) which contains the same sequences, might be targeted to mitochondria by the 14-amino-acid motif and subsequently anchored to the organelle outer membrane by the two hydrophobic domains. The hydrophilic loop would protrude inside the intermembrane space, whereas the N- and C-terminal regions would be cytosolic (38). A stop-transfer mechanism would fit such a view of the process.
Both in vivo and in vitro experiments were developed to analyze the mitochondrial targeting and membrane integration of the 36K and 95K proteins. A set of deletion mutants were prepared to determine the relative importance of the different domains defined above in targeting and insertion into the mitochondrial membrane and to define the topology of insertion. No evidence was found of the presence of an efficient MTS in the N-terminal hydrophilic regions, either upstream or downstream of the hydrophobic domains, thus excluding the stop-transfer hypothesis. Altogether, the results are consistent with a predominant role of the two transmembrane domains and their immediately adjacent sequences in targeting of the pre- and complete readthrough viral replicase, which points to a signal-anchor pathway.
MATERIALS AND METHODS
Antiserum production.
To produce specific polyclonal antibodies potentially able to recognize the different domains of the polypeptide, the 36K protein was expressed in Escherichia coli from a cDNA (4) inserted into the vector pQE-60, purified according to the supplier (Qiagen), and used to generate a rabbit antiserum by conventional procedures.
Analysis of membrane integration in infected plants.
Nicotiana benthamiana plants were inoculated with CIRV and mitochondria were extracted as previously described (39). The fraction containing lightly vesiculated mitochondria (initial stages of transformation into MVBs; see Fig. 1 in Rubino et al. [39]) was subjected to proteinase K (100 μg/ml) digestion for 30 min on ice prior to electrophoresis in the discontinuous Tricine system of Schagger and von Jagow (43). The protease digestion product was detected by Western blotting as previously described (37).
Construction of recombinant plasmids.
The CIRV cDNA clones were all derived from the full-length infectious cDNA clone in pUC18 described previously (4). This was digested with EcoRV (cuts 46 nucleotides downstream of the stop codon of the 36K protein) and SmaI (cuts at the 3′ end of the viral genome) and religated. The resulting clone (pdES) contained the nontranslated leader and the 36K protein coding sequences fused to the T7 RNA polymerase promoter. Deletion mutants of plasmid pdES were obtained by PCR amplification of selected regions of the 36K protein coding sequence and reinsertion between the NcoI and EcoRI sites or by cleavage at appropriate restriction sites (present naturally or introduced by site-directed mutagenesis) within the 36K protein ORF and religation. Clone pdES and derivatives were used for in vitro transcription and protein synthesis.
For transient expression of the proteins in plants, deletion mutants of plasmid pRTL2-36K/GFP containing the 36K protein cDNA fused in-frame to the GFP gene under control of the 35S cauliflower mosaic virus (CaMV) constitutive promoter (39) were obtained by cleavage at appropriate restriction sites (present or introduced) and religation, or selected regions of the 36K protein-coding sequence were amplified by PCR and cloned into the NcoI and BamHI sites of plasmid pCK-GFP3 (24).
For expression of the viral proteins in S. cerevisiae, clones p36K, p36K-GFP, and pGFP-36K were used as previously described (37). They contained the native 36K protein cDNA or the 36K protein cDNA fused in-frame to the 5′ end or the 3′ end of the sequence coding for GFP, respectively, cloned in the yeast cell vector pYES2 (Invitrogen) under control of the galactose-activated GAL1 promoter. Deletion mutants of these clones were constructed by cleavage at appropriate restriction sites (present or introduced) and religation. Clones expressing the GFP fusion proteins were used to examine microscopically the localization of the 36K protein derivatives, whereas those expressing the unfused proteins were used mainly for biochemical analyses.
Similar constructs were prepared for in vitro transcription-translation and expression in plant or yeast cells of the complete CIRV replicase protein, i.e., the 95K polypeptide. For this purpose, the ORF2 sequence was cloned into the different vector systems described above using natural or introduced restriction sites, and the amber stop codon of ORF1 was mutated to an alanine codon.
Site-directed mutagenesis was carried out with a QuickChange site-directed mutagenesis kit (Stratagene). All constructs were routinely sequenced to avoid unwanted modifications.
In vitro protein synthesis and insertion tests with isolated mitochondria.
The Promega TNT-coupled transcription-translation kit was used for in vitro synthesis of [35S]methionine-labeled polypeptides following the instructions of the supplier. Mitochondria were isolated from potato (Solanum tuberosum) tubers mostly as described by Neuburger et al. (30) and from S. cerevisiae cells as described by Daum et al. (9). For the preparation of outer membrane fractions, plant mitochondria (5 to 10 mg of protein) were resuspended in 500 μl of 5 mM potassium phosphate buffer, pH 7.5, containing 1 mM phenylmethylsulfonyl fluoride (PMSF) and incubated on ice for 15 min with occasional resuspension with a pestle. After addition of another 500 μl of the same buffer and further incubation for 15 min, the suspensions were loaded on top of a three-layer discontinuous sucrose gradient (15, 38, and 60% [wt/vol] in 10 mM potassium phosphate [pH 7.5] and 1 mM PMSF) and centrifuged for 45 min at 290,000 × g. The outer membrane fraction was recovered at the interface between the 15% and 38% sucrose layers, diluted with 10 mM potassium phosphate (pH 7.5)-1 mM PMSF, and pelleted at 230,000 × g for 20 min.
Insertion tests with in vitro-synthesized viral polypeptides were performed using mitochondrial protein import procedures essentially according to Wischmann and Schuster (49) and Mayer and Neupert (23) for plant and yeast cell mitochondria, respectively. To characterize membrane-protected regions, incubation of the mitochondria with the labeled polypeptides was followed by treatment with proteinase K or trypsin (100 μg/ml) for 5 min at room temperature and 10 min on ice. Organelles were subsequently centrifuged through a 27% (wt/vol) sucrose cushion in 10 mM HEPES-KOH, pH 7.5. To test membrane integration of the different CIRV polypeptides, the mitochondrial samples were resuspended in 0.1 M Na2CO3 (pH 11.5)-1 mM PMSF, incubated for 30 min on ice, and centrifuged at 230,000 × g for 20 min. For some assays, mitochondria were pretreated with 50 μg of trypsin per ml for 15 min on ice. Following dilution and addition of 1 mg of soybean trypsin inhibitor per ml, mitochondria were reisolated and washed twice in the presence of 1 mg of soybean trypsin inhibitor per ml prior to use in insertion assays. Final samples were analyzed by classical sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (18) or with the Tricine system of Schagger and von Jagow (43).
Expression of gene constructs in plant and yeast cells.
For transient expression in plant cells, recombinant DNA was introduced into leaves of N. benthamiana using an Agrobacterium tumefaciens-based method as previously described (39). Briefly, a bacterial suspension was injected through the stomata into the intercellular space of N. benthamiana leaves. After 20 to 36 h, protoplasts were obtained from infiltrated leaf tissue (28, 29) and analyzed with an epifluorescence microscope (24). Analyses were run with or without previous incubation with the mitochondrion-specific dye MitoTracker (CMTMRos; Molecular Probes). For expression in yeast cells, transformation of S. cerevisiae, culture of transformants, and microscopy analyses were done as previously described (37).
RESULTS
Membrane integration of 36K protein in infected plants.
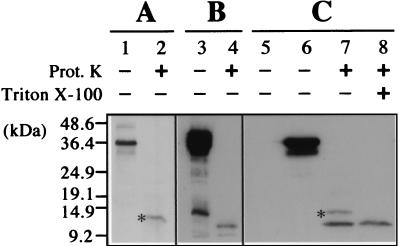
Subcellular fractions containing lightly vesiculated mitochondria were extracted from N. benthamiana plants infected with CIRV and digested with proteinase K prior to denaturing polyacrylamide gel electrophoresis. Western blotting with a polyclonal antibody raised against the purified CIRV 36K protein was used to detect a putative polypeptide which would be resistant to proteinase K due to embedding in the mitochondrial membrane. One band corresponding to a polypeptide with an apparent molecular mass of approximately 12 kDa resulted from proteinase K digestion (Fig. 2A). Since similar protease treatment of the 36K protein purified upon overexpression in bacteria resulted in complete degradation (Fig. 2B), it can be concluded that the 12-kDa polypeptide represents the portion of the 36K protein embedded in the membrane of MVBs, where it is inaccessible to proteolysis. This adds strength to the proposed model in which the protein anchors to the outer membrane of the mitochondrial envelope by the two hydrophobic transmembrane domains (38). However, in the proposed model of insertion, the embedded part was estimated to be only 8.7 kDa, consistently smaller than the resistant fragment found after protease digestion of purified multivesiculated mitochondria. The larger size of the protected fragment may indicate that hydrophilic regions flanking the hydrophobic domains at either the N-terminal or the C-terminal side also interact with the membrane and are protected from protease digestion. Further experimental evidence on this point from deletion mutants will be described below.
FIG. 2.
Western blot analysis of mitochondrial fractions from CIRV-infected N. benthamiana plants and 36K protein-transformed yeast cells. (A) Mitochondria were purified from CIRV-infected N. benthamiana and analyzed directly (lane 1) or after proteinase K (Prot. K) treatment (lane 2). (B) A fraction of 36K protein overexpressed in E. coli and purified on an Ni-nitrilotriacetic acid-agarose column (Qiagen) was analyzed directly (lane 3) or after proteinase K digestion (lane 4). The extra band at the bottom of lane B3 corresponds to residual contamination by a low-molecular-weight E. coli polypeptide, as determined by N-terminal sequencing after large-scale purification of the overexpressed 36K protein and SDS-PAGE fractionation. (C) Mitochondria were purified from yeast cells transformed with a control plasmid (lane 5) or with a plasmid carrying the CIRV 36K protein cDNA (lanes 6 to 8) and analyzed directly (lanes 5 and 6) or after proteinase K treatment (lanes 7 and 8) in the absence (lane 7) or in the presence (lane 8) of 1% (vol/vol) Triton X-100. Blots were subjected to immunodetection with a polyclonal antiserum against the CIRV 36K protein. Binding of the primary antibodies was revealed by using peroxidase-conjugated secondary antibodies and enhanced chemoluminescence reagents (Amersham Biosciences). The full-length 36K protein and truncated forms expressed in yeast cells were used as size markers together with commercial proteins. The molecular size scale is indicated on the left. The specific polypeptides protected against proteinase K digestion in the mitochondrial fractions are indicated by an asterisk.
To test whether the viral polypeptide has the same mitochondrial membrane integration properties in a nonplant context, the protease protection assays were conducted on mitochondria isolated from S. cerevisiae cells transformed with the complete CIRV 36K protein cDNA (37). Mitochondria were isolated from wild-type and transformed yeast cells and subjected to proteinase K digestion as before. Again, a protease-protected fragment was detected on Western blots, with a somehow larger apparent molecular mass (approximately 13.5 kDa) than that obtained from mitochondria extracted from infected plant leaves (Fig. 2C). The nonionic detergent Triton X-100 solubilizes membranes and releases inserted proteins. As expected, when mitochondria isolated from yeast cells expressing the CIRV 36K protein were treated with Triton X-100 prior to proteinase K digestion, the 13.5-kDa fragment was no longer resistant (Fig. 2C), indicating that protease protection in the absence of detergent was likely to be due to insertion in the mitochondrial membrane.
In vitro insertion of CIRV 36K protein and complete replicase into the outer membrane of isolated plant and yeast cell mitochondria.
Mechanistic aspects of 36K protein and complete replicase membrane targeting were analyzed with in vitro assays involving either plant or yeast cell mitochondria and 35S-labeled viral polypeptides synthesized in a cell-free transcription-translation system. An ORF2 cDNA in which the amber stop codon of ORF1 has been mutagenized to an alanine codon was used to synthesize the complete 95-kDa replicase.
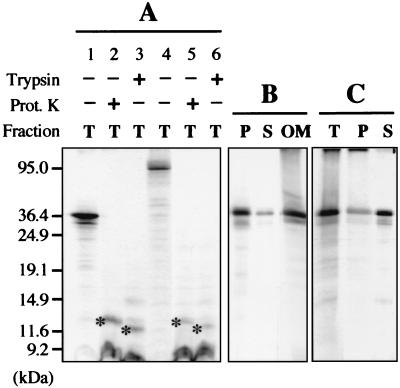
In standard in vitro protein import conditions (49), the 36K and 95K CIRV proteins stably associated with potato mitochondria (Fig. 3A). Previous sequence analysis highlighted a putative mitochondrial targeting sequence in the two polypeptides (see above), but no specific processing product of reduced size was observed upon incubation with isolated mitochondria, as would be the case for a matrix-targeted protein with a cleavable presequence. Moreover, association of the viral proteins with mitochondria did not seem to be sensitive to inhibitors of the MTS-driven protein import machinery (13).
FIG. 3.
In vitro protein insertion assays in the presence of isolated plant mitochondria. (A) After the insertion assay in the presence of potato mitochondria, incorporation of the in vitro-translated CIRV 36K (lanes 1 to 3) and 95K (lanes 4 to 6) polypeptides was analyzed in the total mitochondrial fraction (T); mitochondria were either untreated (lanes 1 and 4) or treated with proteinase K (lanes 2 and 5) or trypsin (lanes 3 and 6) after insertion. The specific polypeptides protected against proteinase K or trypsin digestion in the mitochondrial fractions are indicated by an asterisk. (B) After the insertion assay in the presence of potato mitochondria, incorporation of the CIRV 36K polypeptide was analyzed in the pellet (P) and supernatant (S) fractions following carbonate extraction of the mitochondria and in the mitochondrial outer membrane fraction (OM). (C) The insertion assay was run in the presence of a potato mitochondrial outer membrane preparation instead of intact mitochondria; incorporation of the CIRV 36K polypeptide was subsequently analyzed in the membrane fraction (T) and in the pellet (P) and supernatant (S) following carbonate extraction of the membrane fraction. Samples were analyzed by conventional SDS-PAGE. In vitro-translated full-length viral proteins and truncated forms were used as molecular size markers, and their positions are indicated on the left.
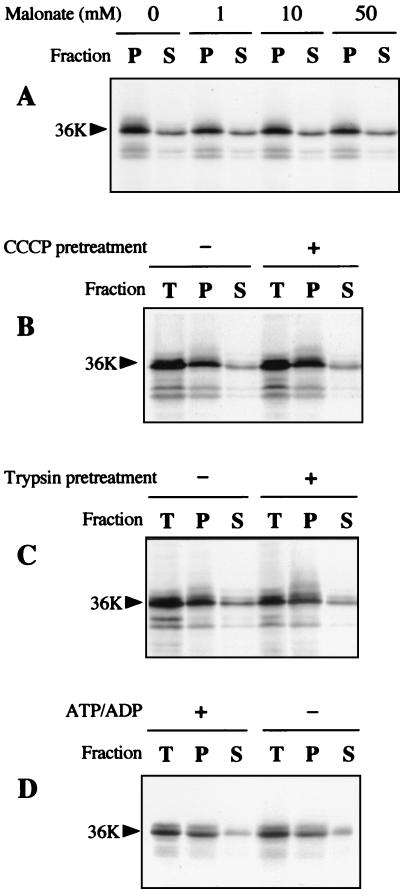
Import into the matrix through the TIM23 complex or insertion into the inner membrane mediated by the TIM22 complex requires the inner membrane electrochemical potential. Association of the viral 36K protein with mitochondria did not need the electrochemical potential because it was not impaired by treatment of the organelles with valinomycin or carbonyl cyanide m-chlorophenylhydrazone (CCCP) and could not be titrated out by increasing concentrations of malonate in the presence of succinate (Fig. 4A and B). The mobile carrier ionophore valinomycin permits diffusion of K+ and thus dissipates the existing membrane potential (ΔΨ) (12). The protonophore CCCP allows proton diffusion across the inner membrane and collapses the proton motive force (ΔpH, ΔΨ). Malonate inhibits the oxidation of succinate via complex II in a competitive manner, leading to a decrease in electron transport. Increasing amounts of malonate thus gradually decrease the ΔΨ membrane potential (26, 45). These observations indicated that the N-terminal hydrophilic domain of the CIRV 36K and 95K proteins does not drive import toward the matrix or the inner membrane and were consistent with the hypothesized outer membrane anchoring.
FIG. 4.
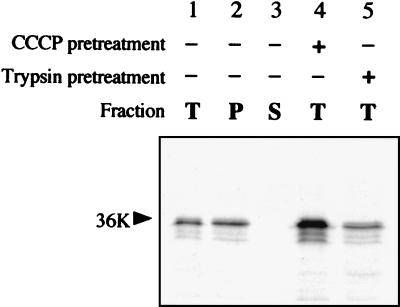
In vitro protein insertion assays in the presence of isolated plant mitochondria. (A) Insertion assay in the presence of potato mitochondria was run with 10 mM succinate and 0 mM, 1 mM, 10 mM, or 50 mM malonate; incorporation of the in vitro-translated CIRV 36K polypeptide was subsequently analyzed in the pellet (P) and supernatant (S) fractions following carbonate extraction of the mitochondria after insertion. (B) Potato mitochondria were untreated or treated with 1 μM CCCP prior to the insertion assay; incorporation of the CIRV 36K polypeptide was subsequently analyzed in the total mitochondrial fraction (T) and in the pellet (P) and supernatant (S) fractions following carbonate extraction. (C) Potato mitochondria were untreated or treated with 50 μg of trypsin/ml prior to the insertion assay; incorporation of the CIRV 36K polypeptide was subsequently analyzed in the total mitochondrial fraction (T) and in the pellet (P) and supernatant (S) fractions following carbonate extraction. (D) The insertion assay in the presence of potato mitochondria was run with and without added ATP and ADP in the medium; incorporation of the CIRV 36K polypeptide was subsequently analyzed in the total mitochondrial fraction (T) and in the pellet (P) and supernatant (S) fractions following carbonate extraction. Samples were analyzed by conventional SDS-PAGE. The position of the 36K protein is indicated by an arrow.
Upon separating the mitochondria into soluble and insoluble fractions after the in vitro assay, the 36K protein was associated with the total mitochondrial membrane fraction, and the incorporation was resistant to carbonate extraction, implying membrane integration (Fig. 3B). Moreover, when the insertion assay was scaled up so as to permit further submitochondrial fractionation, the viral protein was indeed recovered in the outer membrane fraction (Fig. 3B). Conversely, the 36K polypeptide was also able to associate with an isolated outer membrane fraction, but resistance to carbonate extraction was limited, suggesting less efficient integration (Fig. 3C). Proteinase K treatment of the mitochondria upon in vitro insertion of the 36K protein or the 95K protein yielded a protected polypeptide of the same apparent molecular mass (12 kDa) as that detected upon Western blot analysis of mitochondria from CIRV-infected plants (Fig. 3A), confirming that the viral polypeptides anchor to the outer membrane in vitro and indicating that the process is similar to the membrane integration occurring in vivo. A slightly smaller polypeptide was protected when trypsin was used instead of proteinase K (Fig. 3A).
Most mitochondrial outer membrane proteins, whatever targeting information they rely on (signal-anchor, stop-transfer, or other topogenic sequences), depend on surface-exposed import receptors for their membrane insertion (17, 25, 27, 35, 44). Pretreatment of the mitochondria with trypsin did not prevent membrane insertion of the CIRV 36K protein (Fig. 4C), suggesting that recognition by surface-accessible receptors is not a prerequisite. Finally, passage of some proteins such as porin to the outer membrane-embedded form requires elevated ATP, whereas signal-anchor protein insertion is not sensitive to ATP depletion (23, 27). The absence of added ATP and ADP in the incubation medium did not significantly influence in vitro membrane insertion of the CIRV 36K polypeptide (Fig. 4D).
The same observations were made when using a yeast cell in vitro mitochondrial import system. The viral protein integrated into the membrane fraction of isolated yeast cell mitochondria, and integration was resistant to carbonate extraction (Fig. 5). Again, the process was not prevented by treatment of the mitochondria with CCCP or trypsin.
FIG. 5.
In vitro protein insertion assays in the presence of isolated yeast cell mitochondria. The insertion assay was run with untreated (lanes 1 to 3), CCCP-treated (lane 4), or trypsin-treated (lane 5) yeast cell mitochondria; incorporation of the in vitro-translated CIRV 36K polypeptide was subsequently analyzed in the total mitochondrial fraction (T) and in the pellet (P) and supernatant (S) fractions following carbonate extraction of the mitochondria after insertion. Samples were analyzed by conventional SDS-PAGE. The position of the 36K protein is indicated by an arrow.
Expression of CIRV 36K protein, complete replicase, and deletion mutants in plant and yeast cells.
In vivo mitochondrial targeting of the CIRV 36K protein in plant and yeast cells was demonstrated previously (37, 39). To determine whether the complete 95K CIRV replicase has the same subcellular localization properties in vivo, the corresponding ORF2 cDNA with the amber stop codon of ORF1 mutagenized to an alanine codon was fused in frame with the GFP gene and placed under control of the 35S CaMV constitutive promoter or the galactose-activated GAL1 promoter for expression in plant leaves (39) and in yeast cells (37), respectively. In both plant and yeast cells, the 95K protein targeted the GFP to organelles which were easily identified as mitochondria by their size, their shape, and the positive reaction they exhibited with the specific dye MitoTracker, yielding fluorescence microscopy images similar to those obtained previously with the 36K protein (37, 39) (Fig. 6A and 7A). This indicates that the signals operating in the ORF1-derived protein also operate in the context of the complete replicase. In both types of cells, expression of the viral polypeptides promoted some aggregation of mitochondria, but this process was more pronounced in yeast cells than in plant cells and was accompanied by membrane proliferation (37).
FIG. 6.
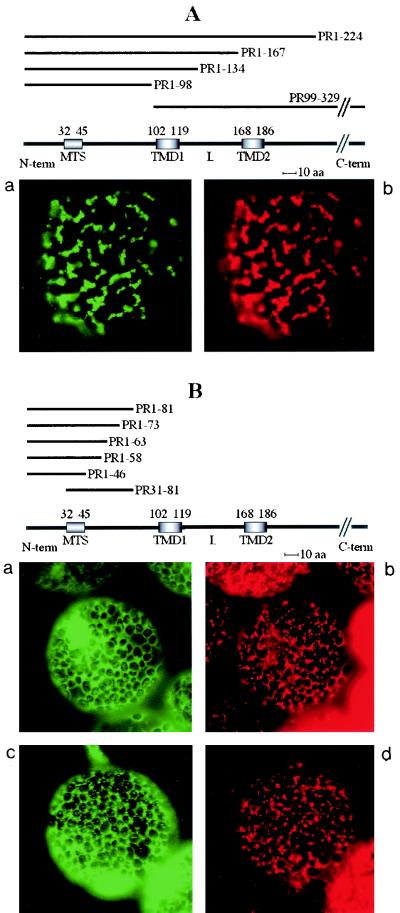
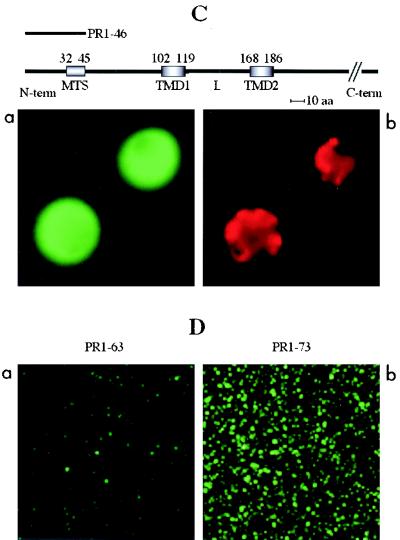
Transient expression of GFP fusions in N. benthamiana leaves. Constructs containing the unfused GFP gene, the GFP gene fused to the cDNA encoding the complete 36K or 95K protein, or the GFP gene fused to cDNAs encoding the different deletion variants (PR1-224, PR1-167, PR1-134, PR1-98, PR99-329, PR1-81, PR1-73, PR1-63, PR1-58, PR1-46, PR31-81, and PR186-225) were expressed. Fluorescence was observed at 40× or 100× magnification with a Nikon Eclipse E800 epifluorescence microscope. In each pair of panels, the image on the left shows the GFP fluorescence observed with a GFP band pass filter set (excitation, 460 to 500 nm; band pass emission, 510 to 560 nm), and the right-hand image shows the fluorescence of the mitochondrion-specific dye (MitoTracker) observed for the same cell by using a tetramethylrhodamine isothiocyanate filter set (excitation, 27.5 to 552.5 nm; emission, 577.5 to 632.5 nm). The structures of the deletion mutants yielding the different types of fluorescence pattern are presented above the image sets. The relative positions of the putative MTS (amino acids 32 to 45), the two putative transmembrane domains (TMD1, amino acids 102 to 119, and TMD2, amino acids 168 to 186), and the loop (L) in the ORF1 sequence are indicated. The sequences retained in the different deletion mutants (PRx-y) are indicated by heavy lines. Three different types of subcellular localization of the GFP fusion protein were observed and are illustrated by characteristic pictures. When fused to 95K protein, 36K protein, PR1-224, PR1-167, PR1-134, PR1-98, and PR99-329, the GFP was exclusively localized to mitochondria (A). When fused to PR1-81, PR1-73, PR1-63, PR1-58, PR1-46, and PR31-81, the GFP was diffuse in the cytosol (B, panel a), but additional localization to small structures poorly representative of the mitochondrial staining pattern occurred at a late stage of transient expression (B, panel c). GFP fused to PR186-225 was diffuse in the cytosol, like unfused GFP (C).
FIG. 7.
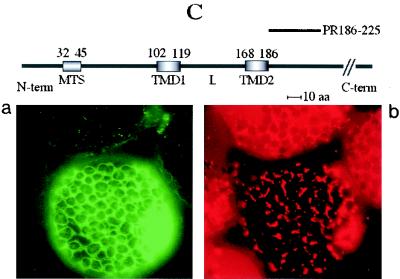
Expression of GFP fusions in transgenic yeast cells. Constructs containing the unfused GFP gene, the GFP gene fused to the cDNA encoding the complete 36K or 95K protein, or the GFP gene fused to cDNAs encoding the different deletion variants (PR1-224, PR1-167, PR1-98, PR99-329, PR1-83, PR1-73, PR1-63, and PR1-46) were expressed. Fluorescence was observed at 40× or 100× magnification with a Nikon Eclipse E800 epifluorescence microscope. In panels A, B, and C, the left-hand image shows the GFP fluorescence observed using a GFP band pass filter set (excitation, 460 to 500 nm; BP emission, 510 to 560 nm), and the right-hand image shows the fluorescence of the mitochondrion-specific dye (MitoTracker) observed for the same cells using a tetramethylrhodamine isothiocyanate filter set. Panels A, B, and C also include the structures of the deletion mutants yielding the type of fluorescence pattern presented in the panel. The relative positions of the putative MTS (amino acids 32 to 45), the two putative transmembrane domains (TMD1, amino acids 102 to 119, and TMD2, amino acids 168 to 186), and the loop (L) in the ORF1 sequence are indicated. The sequences retained in the different deletion mutants (PRx-y) are indicated by heavy lines. Three different types of subcellular localization of the GFP fusion protein were observed and are illustrated by characteristic pictures. When fused to complete 95K protein, complete 36K protein, PR1-224, PR1-167, PR1-98, and PR99-329, the GFP was exclusively localized to mitochondria (A). Expression of these fusion proteins promoted aggregation of the organelles. When fused to PR1-83, PR1-73, and PR1-63, the GFP was localized to structures poorly representative of the mitochondrial staining pattern (B). GFP fused to PR1-46 was diffuse in the cytosol, like unfused GFP (C). (D) Similar amounts of mitochondria extracted from yeast cells expressing the PR1-63 (left) or the PR1-73 (right) GFP fusion were spread on a microscope slide, and fluorescence was observed with the GFP band pass filter set.
To evaluate the relative contributions of the different domains in the N-terminal part of the viral proteins to mitochondrial targeting, a set of deletion mutants of CIRV ORF1 was prepared. They are referred to as PRx-y, where PR stands for pre-readthrough protein and x and y indicate the positions of the first and last amino acids of the mutant in the complete ORF1 sequence, respectively. The deleted ORF1 variant sequences were also fused in-frame with the GFP reporter gene and placed under control of the 35S CaMV or the galactose-activated GAL1 promoter. Deletion mutant PR1-224 contained the ORF1 sequence down to amino acid 224, after the second predicted transmembrane domain (TMD2), and included all potentially remarkable regions: the N-terminal hydrophilic region (amino acids 1 to 101), including the putative MTS (amino acids 32 to 45) and the two putative transmembrane domains TMD1 (amino acids 102 to 119) and TMD2 (amino acids 168 to 186) connected by the potential loop (L, amino acids 120 to 167). This mutant was prepared to confirm previous studies showing that the coding sequence downstream of the SacI restriction site (positions +667 to +672 in the ORF1 cDNA sequence) did not contribute to mitochondrial targeting (4). Plant cells expressing construct PR1-224 indeed showed the GFP confined to mitochondria, giving rise to targeting pictures which were indistinguishable from those obtained with the wild-type 36K and 95K proteins (Fig. 6A).
When ORF1 was progressively deleted farther from the C-terminal side up to amino acid 98, successively excluding the second hydrophobic domain (mutant PR1-167), most of the putative loop (mutant PR1-134), and even the first hydrophobic domain (mutant PR1-98), expression of GFP fusions in plant cells still yielded the same patterns, with the fluorescence clearly restricted to mitochondria as for the wild-type and PR1-224 fusions (Fig. 6A). By contrast, when fusions between the GFP and partial ORF1 sequences from the N-terminal hydrophilic domain down to amino acid 81 (mutants PR1-46, PR1-58, PR1-63, PR1-73, PR1-81, and PR31-81), all containing the predicted MTS at amino acids 32 to 45 (38), were transiently expressed in plant leaves, the fluorescence was diffuse in the cytosol around the chloroplasts (Fig. 6B, panel a). Partial and progressive localization to small spots, which were not or only to a limited extent representative of the MitoTracker staining pattern, appeared at a later stage of transient expression (30 to 32 h) (Fig. 6B, panel c). These images are completely different from those generated with the wild-type proteins or the PR1-224, PR1-167, PR1-134, and PR1-98 deletion mutants (Fig. 6A and B).
The foregoing cytological observations indicate that the region of ORF1 between amino acids 1 and 98 contains sufficient information for mitochondrial targeting in plant cells, but amino acids 1 to 81 alone do not possess efficient targeting properties. We conclude that the motif highlighted by computer prediction (amino acids 32 to 45) and previous point mutation (38) is not an efficient MTS, even when placed at the very N terminus of the fusion protein (mutant 31-81). Instead, amino acids 81 to 98 likely contain important targeting signals. However, these conclusions only partially reflect the properties of the ORF1 sequence, because a GFP fusion protein (mutant PR99-329) totally excluding amino acids 1 to 98 was also targeted to mitochondria as efficiently as the wild-type and PR1-224 fusions in plant cells (Fig. 6A).
It thus appears that the signals present in amino acids 81 to 98 are not indispensable for mitochondrial recognition and that other domains of the N-terminal part of ORF1 downstream of position 98 also contain organelle-targeting signals. These are likely to be included in the hydrophobic domains and/or the connecting loop, as GFP fused to the ORF1 region downstream of TMD2 (mutant PR186-225) was not targeted at all in plant cells (Fig. 6C). Note that the behavior of the PR186-225 deletion mutant is in contrast to the situation with the rat liver carnitine palmitoyltransferase 1 (CPT1) (7). This polypeptide contains two transmembrane domains in its N-terminal part and an MTS immediately downstream of the second transmembrane domain. Owing to the absence of subcellular targeting of the PR186-225/GFP fusion, there does not seem to be any unpredicted MTS downstream of the two transmembrane domains in the CIRV ORF1.
Expression of the GFP-fused PR1-224, PR1-167, PR1-98, and PR99-329 deletion mutants in transformed yeast cells led to the same observations and conclusions as with infiltrated plants, the fluorescence being clearly localized to mitochondria in all cases (Fig. 7A). The PR1-46/GFP fusion yielded diffuse fluorescence patterns, similar to those observed with unfused GFP, when expressed in yeast cells (Fig. 7C), confirming that amino acids 32 to 45 do not behave like an MTS. Upon expression in yeast cells of GFP fusions involving sequences from the ORF1 N-terminal hydrophilic domain down to amino acid 83 (mutants PR1-63, PR1-73, and PR1-83), the fluorescence was entirely restricted to defined structures, which, however, showed only very partial colocalization with the MitoTracker staining pattern (Fig. 7B). This differs somehow from the behavior of the equivalent constructs in plant cells, in which, even at late stages of transient expression, most of the fluorescence remained diffuse in the cytosol (Fig. 6B, panel c).
Altogether, the cytological observations upon expression of GFP fusions strongly suggest that multiple recognition signals for mitochondria are spread within amino acids 81 to 186 of the CIRV ORF1. These signals are likely to compensate for each other in the corresponding deletion mutants, leading to identical mitochondrial targeting patterns for mutants having no sequence in common (PR1-98 and PR99-329). The in vitro analyses developed below with plant mitochondria show that, although a wide range of deletion mutants are still able to promote association of the GFP with mitochondria, as measured by cytological observations, this reflects very different situations at the level of membrane integration.
Mitochondrial membrane integration of CIRV 36K protein deletion mutants in yeast cells.
Concurrent with microscopic observations, mitochondria were extracted from yeast cell transformants expressing the GFP fusions. Mitochondria isolated from yeast cells transformed with mutants PR1-224, PR1-167, and PR99-329 fluoresced identically as in vivo. When subjected to carbonate treatment and separated into pellet and supernatant fractions, the bulk of the fusion proteins was found in the pellet fraction, as expected for integral proteins. As for the mitochondria extracted from yeast cells transformed with mutant PR1-98, most of them were fluorescent, but the protein was equally distributed between the pellet and supernatant after carbonate treatment, indicating that the interaction was weaker than for the variants containing at least one of the predicted transmembrane domains.
By contrast, very few of the mitochondria from yeast cells transformed with mutants PR1-46 and PR1-63 were fluorescent (Fig. 7D). In the case of PR1-46, only a small portion of the protein was found in the pellet after carbonate treatment, most of it being in the supernatant, whereas for PR1-63 the distribution was more balanced. Finally, despite the ambiguous patterns obtained with these deletion variants upon microscopic analysis of whole cells, a large proportion of the mitochondria extracted from yeast cells transformed with mutants PR1-73 and PR1-83 was also fluorescent (Fig. 7D), and the protein was about equally distributed between the pellet and supernatant fractions resulting from carbonate extraction. These results indicate that the proteins expressed from constructs PR1-83 and PR1-73 retained some capacity to interact with yeast cell mitochondrial membranes and suggest that, in the case of yeast cells, the mitochondrial anchor domain, which indeed seemed to be a little larger (see above), extends farther upstream in the ORF1 sequence than the region defined in anchoring assays with isolated plant mitochondria (see below).
In vitro insertion of CIRV 36K protein deletion mutants in the membranes of isolated plant mitochondria.
Isolation of relevant amounts of mitochondria from N. benthamiana plants agroinfiltrated with the different gene constructs was not feasible. Therefore, membrane integration of the different 36K protein deletion mutants in a plant context was studied in vitro after transcription-translation of the corresponding [35S]methionine-labeled polypeptides and incubation with isolated plant mitochondria in protein import conditions (49). For these assays, the deleted ORF1 variant sequences were not fused to a reporter gene.
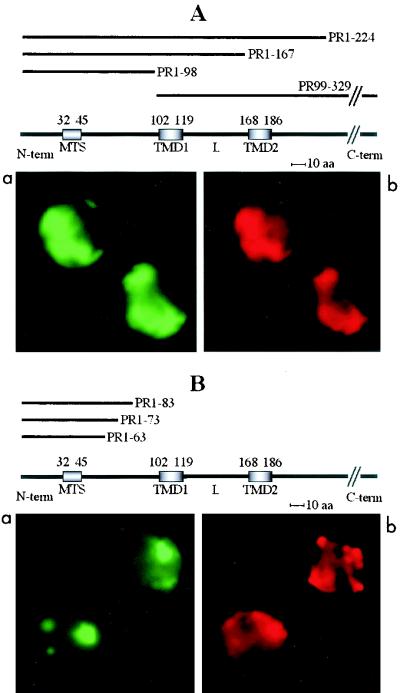
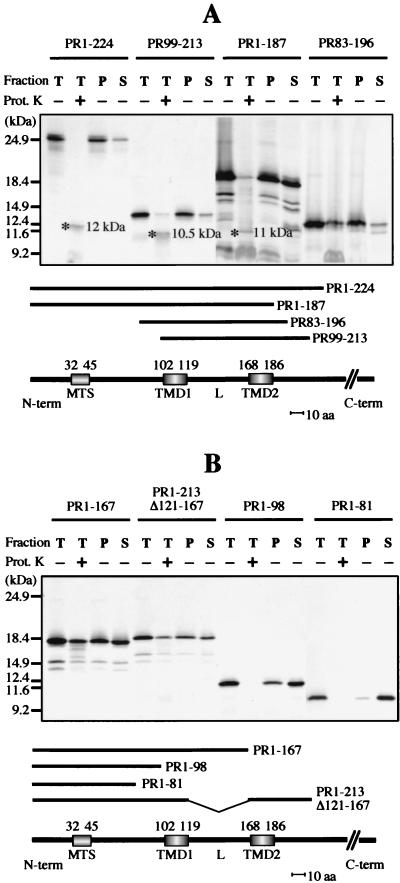
The PR1-224 deletion mutant polypeptide, which contains all the potential targeting domains, associated stably with isolated potato mitochondria (Fig. 8A). Proteinase K treatment of the mitochondria upon insertion yielded a protected polypeptide of the same apparent molecular mass (12 kDa) as that detected when the complete 36K protein or 95K protein was used in the assay (Fig. 3A). When the mitochondria were subsequently subjected to carbonate treatment and separated into pellet and supernatant fractions, the PR1-224 polypeptide was almost completely retained in the membrane fraction, as was the case for the complete 36K and 95K proteins (Fig. 3, 4, and 8). The same behavior was observed with the PR99-213 deletion mutant lacking the whole N-terminal hydrophilic region upstream of the first transmembrane domain (TMD1) (Fig. 8A). In that case, the polypeptide protected against proteinase K digestion was slightly smaller (10.5 kDa), implying that the full-length mitochondrial anchor domain of the 36K and 95K proteins begins in the hydrophilic region upstream of amino acid 99.
FIG. 8.
In vitro protein insertion assays in the presence of isolated plant mitochondria. Insertion assays in the presence of potato mitochondria were run with deletion mutants PR1-224, PR99-213, PR1-187, and PR83-196 (A) and PR1-167, PR1-213Δ121-167, PR1-98, and PR1-81 (B). Each panel includes the structures of the relevant deletion mutants. The relative positions of the putative MTS (amino acids 32 to 45), the two putative transmembrane domains (TMD1, amino acids 102 to 119, and TMD2, amino acids 168 to 186), and the loop (L) in the ORF1 sequence are indicated. The sequences retained in the different deletion mutants (PRx-y) are indicated by heavy lines. For each assay, incorporation of the in vitro-translated deletion mutant polypeptide was analyzed in the total mitochondrial fraction (T) and in the pellet (P) and supernatant (S) fractions following carbonate extraction of the mitochondria after insertion. Mitochondria were either untreated ordigested with proteinase K (Prot. K) after insertion, as indicated; the specific polypeptides protected against proteinase K digestion in the mitochondrial fractions are indicated by an asterisk, and the estimated apparent molecular mass is shown (A). Samples were analyzed by conventional SDS-PAGE. In vitro-translated full-length 36K protein and truncated forms were used as size markers, and their positions are indicated on the left.
In turn, a smaller proteinase K-protected fragment (11 kDa) was also observed following mitochondrial association of the mutant PR1-187 polypeptide, which lacks all sequences downstream of the second transmembrane domain (TMD2) (Fig. 8A). This finding indicates that the full-length mitochondrial anchor domain of the 36K and 95K proteins ends in the hydrophilic region downstream of amino acid 187. Deletion of this short downstream sequence obviously perturbs membrane integration, because the PR1-187 polypeptide was about equally distributed between the pellet and the supernatant upon carbonate extraction.
Taken together, the foregoing observations imply that the anchor includes the two transmembrane domains and the connecting region. Considering the ORF1 amino acid sequence, the apparent molecular masses of the full-length (12 kDa), N-terminally truncated (10.5 kDa), and C-terminally truncated (11 kDa) proteinase K-protected fragments obtained with the PR1-224, PR99-213, and PR1-187 mutants, respectively, suggest that the 36K and 95K protein mitochondrial anchor domain extends approximately from amino acid 84 to amino acid 196.
Taking into account that sizing of polypeptides with large hydrophobic domains by SDS-PAGE might be unreliable, we constructed mutant PR83-196 to synthesize in vitro the polypeptide corresponding to the estimated anchor region (the choice of amino acid 83 instead of 84 at the N terminus was dictated by cloning constraints). The PR83-196 polypeptide comigrated on SDS gels with the full-length proteinase K-protected fragment observed in mitochondrial insertion assays with PR1-224 and the complete 36K or 95K protein. Furthermore, it was apparently fully protected against proteinase K upon association with mitochondria and was almost completely resistant to carbonate extraction when anchored (Fig. 8A). Thus, the PR83-196 polypeptide indeed shows the properties of the 36K/95K protein mitochondrial anchor, and its sequence should be very close to that of the viral membrane insertion domain. As a consequence, the targeting information highlighted in amino acids 81 to 98 by the expression of GFP fusions in plant cells (see above) is not a cryptic MTS; rather, most of this region is likely to be part of the authentic anchor domain.
Further deletion mutant analyses in vitro revealed that some sequences of the ORF1 which are able to target the GFP to mitochondria in vivo in fact have poor anchoring properties. The polypeptide synthesized from mutant PR1-167, lacking the second transmembrane domain, associated with isolated plant mitochondria, but its membrane integration was impaired, as it was about equally distributed between the pellet and supernatant after carbonate treatment (Fig. 8B). The translation product of mutant PR1-134, lacking the second transmembrane domain plus most of the connecting loop, also associated with mitochondria, but membrane integration was not very efficient, as there was more material in the supernatant than in the pellet upon carbonate treatment. Deletion of only the hydrophilic connecting loop in mutant PR1-213Δ121-167 led to in vitro insertion properties similar to those of the PR1-167 polypeptide (Fig. 8B).
A significant portion of the complete PR1-167 and PR1-213Δ121-167 polypeptides was apparently resistant to proteinase K treatment of the mitochondria after the insertion assay. Control tests run in the same conditions demonstrated that resistance to proteinase K did not occur in the absence of mitochondria (not shown). A possible explanation for these observations is that the PR1-167 and PR1-213Δ121-167 polypeptides tend to aggregate in the presence of mitochondria. The PR1-98 deletion mutant polypeptide, excluding both hydrophobic domains but still retaining a small part of the anchor sequence based on the above results, bound to mitochondria but showed poor membrane integration, as only a small portion was found in the pellet after carbonate treatment (Fig. 8B). Finally, the PR1-81 polypeptide, which ends just before the anchor sequence, associated significantly with plant mitochondria but was indeed unable to integrate into the membrane, as it was completely extracted with carbonate (Fig. 8B).
The last observation contrasts with the outcome of the analyses made with mitochondria extracted from yeast cells expressing the different CIRV ORF1 deletion mutants, where the transition in the resistance of the mitochondrially bound polypeptides to carbonate extraction occurred between the PR1-63 and PR1-73 mutants (see above). No obvious protected fragment of reduced size was detected after proteinase K treatment in the assays conducted with mutants PR1-167, PR1-134, PR1-213Δ121-167, PR1-98, and PR1-81, although the presence of the bulk of the mitochondrial material would not allow us to properly resolve very small polypeptides on either type of gel. Also, it cannot be excluded that a protected fragment contains no methionine and is therefore not labeled.
DISCUSSION
Virus survival relies on efficient multiplication and spread. Multiplication is primarily dependent on replication of the viral genome. Considering the in vivo data presented here, it seems that the CIRV replicase has developed a very potent molecular mechanism for recognition of the viral subcellular replication site. Thus, mitochondrial targeting in vivo was lost only after considerable sequence deletion, up to amino acid 81. However, in vitro experiments showed that, as judged from resistance to alkaline extraction, almost all the deletions which do not hinder the recognition of mitochondria in fact more or less severely impair the proper anchoring of the corresponding polypeptide into the mitochondrial membrane. This is the case not only for mutants lacking, for instance, one of the transmembrane domains, but also for PR1-187, which presumably lacks only a short sequence upstream of the second transmembrane domain.
Strikingly, amino acids 81 to 98, most of which are likely to be part of the anchor, bring a decisive contribution to mitochondrial targeting when only amino acids 1 to 98 in the N-terminal hydrophilic region are included in the constructs but are dispensable for targeting and efficient membrane anchoring when the rest of the remarkable regions are present. This suggests that the ORF1 sequence contains redundant information for mitochondrial targeting. Analysis of mitochondria from yeast cells expressing the various deletion mutants indicates that membrane integration of the plant virus polypeptides in this nonplant context is actually less stringent, as there was no obvious loss in efficiency after deletion of one of the transmembrane domains or the connecting loop.
Although the results obtained do not confirm the presence of an MTS, they do support anchoring of the CIRV ORF1 and ORF2 polypeptides to the mitochondrial outer membrane with an insertion topology following the initially proposed model (38). In particular, in vitro analyses of the interactions between the PR99-213 and PR1-187 variants and isolated mitochondria demonstrate that mitochondrial membrane anchoring of the CIRV replicase is indeed a property of the region predicted to contain two transmembrane domains. Protease resistance analyses both in vivo with infected plants and transformed yeast cells and in vitro with isolated mitochondria established that the membrane-embedded region spans the predicted transmembrane domains plus short upstream and downstream sequences.
Considering all the data, it can be proposed that in a plant context, the anchor, including the presumed hinge protruding in the intermembrane space, corresponds approximately to amino acids 84 to 196. The fact that, in the complete 36K and 95K proteins, the mitochondrially protected fragment, as judged from its size, corresponds to the anchor domain defined by the deletion analyses confirmed the hypothesized topology of membrane insertion, with the very N-terminal part and the C-terminal part (in the case of the 36K polypeptide) or the bulk of the protein (in the case of the 95K polypeptide) remaining on the cytosolic side of the outer membrane (Ncyto-Ccyto insertion topology). The CIRV replicase therefore appears to be anchored but accessible in the interior of the cytosol-connected vesicles which develop from the mitochondrial outer membrane during infection (11, 41), excluding the need for a putative transmembrane transport of the genomic RNA to access the viral replicase.
The main concern in this work was to evaluate to what extent the CIRV proteins use the targeting and insertion mechanism of authentic mitochondrial outer membrane proteins (see the introduction). Our starting hypothesis was that the viral polypeptides follow some kind of stop-transfer pathway driven by the putative MTS (amino acids 32 to 45) and stopped by the first hydrophobic domain (amino acids 102 to 119). According to the experimental evidence presented here, amino acids 32 to 45 are not an efficient MTS, and the targeting and insertion properties do not depend on the N-terminal hydrophilic domain (amino acids 1 to 98).
The CIRV ORF1 protein and replicase appear to anchor to the mitochondrial outer membrane through two transmembrane domains, leaving both their N terminus and their C terminus on the cytosolic side (Ncyto-Ccyto insertion). Rat liver CPT1 also anchors to the mitochondrial outer membrane with an Ncyto-Ccyto insertion topology involving two transmembrane domains. The stop-transfer integration of CPT1 is driven by an MTS located downstream of the second transmembrane domain (7). Our results show that there is also no MTS downstream of the second transmembrane domain in the CIRV polypeptides, which excludes that the viral replicase follows a stop-transfer pathway similar to that of CPT1. Finally, membrane proteins containing stop-transfer sequences initially engage with the same pathway as matrix-targeted proteins and therefore rely mostly on the regular, surface-exposed protein import receptors. This does not seem to be the case for the CIRV polypeptides.
Altogether, the experimental evidence rejects the possibility that the 36K and 95K proteins are inserted into the mitochondrial outer membrane following a stop-transfer mechanism. On the other hand, the viral polypeptides contain a linear targeting-insertion sequence including two uniformly hydrophobic stretches. It is thus unlikely that they use a mitochondrial anchoring pathway similar to that of the β-barrel-type proteins, such as Tom40 and porin, in which noncontinuous structural elements and folding features contain the targeting-insertion information. That membrane insertion of the 36K protein in vitro is not dependent on elevated ATP and surface import receptors also argues against an insertion mechanism resembling that of porin (17, 27).
The Ncyto-Ccyto outer membrane insertion topology of the CIRV ORF1 protein and replicase is not typical of the common signal-anchor model, which relies on one transmembrane domain and leads to Ncyto-Cin or Nin-Ccyto insertion. Considering all data, a signal-anchor mechanism nevertheless remains the most plausible candidate to account for mitochondrial targeting and membrane insertion of the CIRV polypeptides. Both in vivo and in vitro deletion mutant data indicate that the N-terminal part of the 36K and 95K proteins contains multiple recognition-insertion signals for the mitochondrial outer membrane within the two hydrophobic domains and their flanking regions, which might together constitute an efficient signal-anchor sequence. That membrane targeting and insertion of the CIRV proteins lack obvious sensitivity to trypsin treatment of the mitochondria is not necessarily a counterargument in this case, as dependence of the membrane integration on surface-accessible import receptors varies among signal-anchored proteins.
On the other hand, there are atypical pathways accounting for targeting and insertion of some mitochondrial proteins with an N-terminal or C-terminal membrane anchor, and these are still poorly documented (25). Multiple determinants for association with another specific membrane system, the endoplasmic reticulum, were revealed in brome mosaic virus protein 1a, a multifunctional protein with an N-terminal RNA capping domain and a C-terminal helicase domain (10). Sequences in the N-terminal RNA capping module of 1a are necessary and sufficient for high-affinity endoplasmic reticulum membrane association, but additional sequences contribute to the normal subcellular distribution of the protein.
Two domains have also been implicated in the endosomal and lysosomal membrane association of protein NSP1, carrying the capping activity of Semliki Forest virus, and one of these membrane association determinants was mapped in a position similar to that in brome mosaic virus protein 1a (1, 10). Membrane binding of NSP1 was shown to be due to affinity for specific lipids (1), another option which remains possible for a contribution to membrane targeting and insertion of the CIRV polypeptides. For both brome mosaic virus and Semliki Forest virus, enzymatic activities of the replication complex seem to be sensitive to the proper lipid composition of the partner membrane (1, 19).
In conclusion, one may note that insertion of the CIRV 36K protein does not by itself trigger membrane vesiculation (37, 39), so that the mechanism and the molecular partners enabling the CIRV components to promote mitochondrial outer membrane proliferation and MVB formation still remain to be defined.
Acknowledgments
This work was supported by funding from the Centre National de la Recherche Scientifique (CNRS), the Université Louis Pasteur (ULP), the Consiglio Nazionale delle Ricerche (CNR), and the Italian-French Galileo cooperation program.
We thank A. Antonacci and A. Cosset for skillful technical assistance, K. Richards for critical reading of the manuscript, and P. Hamman for sequencing of gene constructs.
REFERENCES
- 1.Ahola, T., A. Lampio, P. Auvinen, and L. Kaariainen. 1999. Semliki Forest virus mRNA capping enzyme requires association with anionic membrane phospholipids for activity. EMBO J. 18:3164-3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, H. P., and U. K. Schmitz. 1999. The protein-import apparatus of plant mitochondria. Planta 209:267-274. [DOI] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgyan, J., L. Rubino, and M. Russo. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967-1974. [DOI] [PubMed] [Google Scholar]
- 5.Cao, W., and M. G. Douglas. 1996. Specific targeting of ISP6 to mitochondria is mediated by sequences other than its amino terminus. Biochem. Biophys. Res. Commun. 224:457-461. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffreda, P., L. Rubino, and M. Russo. 1998. Molecular cloning and complete nucleotide sequence of galinsoga mosaic virus genomic RNA. Arch. Virol. 143:173-180. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, I., F. Guillerault, J. Girard, and C. Prip-Buus. 2001. The N-terminal domain of rat liver carnitine palmitoyltransferase 1 contains an internal mitochondrial import signal and residues essential for folding of its C-terminal catalytic domain. J. Biol. Chem. 276:5403-5411. [DOI] [PubMed] [Google Scholar]
- 8.Court, D. A., R. Kleene, W. Neupert, and R. Lill. 1996. Role of the N- and C-termini of porin in import into the outer membrane of Neurospora mitochondria. FEBS Lett. 390:73-77. [DOI] [PubMed] [Google Scholar]
- 9.Daum, G., S. M. Gasser, and G. Schatz. 1982. Import of proteins into mitochondria. Energy-dependent, two-step processing of the intermembrane space enzyme cytochrome b2 by isolated yeast cells mitochondria. J. Biol. Chem. 257:13075-13080. [PubMed] [Google Scholar]
- 10.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Franco, A., M. Russo, and G. P. Martelli. 1984. Ultrastructure and origin of multivesicular bodies induced by carnation Italian ringspot virus. J. Gen. Virol. 65:1233-1237. [Google Scholar]
- 12.Douce, R. 1985. Mitochondria in higher plants: structure, function and biogenesis. Academic Press, Orlando, Fla.
- 13.Glaser, E., S. Sjoling, M. Tanudji, and J. Whelan. 1998. Mitochondrial protein import in plants. Signals, sorting, targeting, processing and regulation. Plant Mol. Biol. 38:311-338. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann, J. M., and W. Neupert. 2000. Protein transport into mitochondria. Curr. Opin. Microbiol. 3:210-214. [DOI] [PubMed] [Google Scholar]
- 15.Koehler, C. M. 2000. Protein translocation pathways of the mitochondrion. FEBS Lett. 476:27-31. [DOI] [PubMed] [Google Scholar]
- 16.Koonin, E. V. 1991. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J. Gen. Virol. 72:2197-2206. [DOI] [PubMed] [Google Scholar]
- 17.Krimmer, T., D. Rapaport, M. T. Ryan, C. Meisinger, C. K. Kassenbrock, E. Blachly-Dyson, M. Forte, M. G. Douglas, W. Neupert, F. E. Nargang, and N. Pfanner. 2001. Biogenesis of porin of the outer mitochondrial membrane involves an import pathway via receptors and the general import pore of the TOM complex. J. Cell Biol. 152:289-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lee, W. M., M. Ishikawa, and P. Ahlquist. 2001. Mutation of host delta9 fatty acid desaturase inhibits brome mosaic virus RNA replication between template recognition and RNA synthesis. J. Virol. 75:2097-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lithgow, T. 2000. Targeting of proteins to mitochondria. FEBS Lett. 476:22-26. [DOI] [PubMed] [Google Scholar]
- 21.Macasev, D., E. Newbigin, J. Whelan, and T. Lithgow. 2000. How do plant mitochondria avoid importing chloroplast proteins? Components of the import apparatus Tom20 and Tom22 from Arabidopsis differ from their fungal counterparts. Plant Physiol. 123:811-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayer, A., R. Lill, and W. Neupert. 1993. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J. Cell Biol. 121:1233-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Menand, B., L. Maréchal-Drouard, W. Sakamoto, A. Dietrich, and H. Wintz. 1998. A single gene of chloroplast origin codes for mitochondrial and chloroplastic methionyl-tRNA synthetase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95:11014-11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mihara, K. 2000. Targeting and insertion of nucleus-encoded preproteins into the mitochondrial outer membrane. Bioessays 22:364-371. [DOI] [PubMed] [Google Scholar]
- 26.Millar, A. H., O. K. Atkin, H. Lambers, J. T. Wiskich, and D. A. Day. 1995. A critique of the use of inhibitors to estimate partitioning of electrons between mitochondrial respiratory pathways in plants. Physiol. Plant. 95:523-532. [Google Scholar]
- 27.Millar, D. G., and G. C. Shore. 1996. Signal anchor sequence insertion into the outer mitochondrial membrane. Comparison with porin and the matrix protein targeting pathway. J. Biol. Chem. 271:25823-25829. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, J. I., and P. Maliga. 1976. Callus induction and plant regeneration from mesophyll protoplasts of Nicotiana sylvestris. Z. Pflanzenphysiol. 78:453-455. [Google Scholar]
- 29.Negrutiu, I., R. D. Shillito, I. Potrykus, G. Biasini, and F. Sala. 1987. Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8:363-373. [DOI] [PubMed] [Google Scholar]
- 30.Neuburger, M., E. P. Journet, R. Bligny, J. P. Carde, and R. Douce. 1982. Purification of plant mitochondria by isopycnic centrifugation in density gradients of Percoll. Arch. Biochem. Biophys. 217:312-323. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen, M., D. G. Millar, V. W. Yong, S. J. Korsmeyer, and G. C. Shore. 1993. Targeting of Bcl-2 to the mitochondrial outer membrane by a COOH-terminal signal anchor sequence. J. Biol. Chem. 268:25265-25268. [PubMed] [Google Scholar]
- 32.Prod'homme, D., S. Le Panse, G. Drugeon, and I. Jupin. 2001. Detection and subcellular localization of the turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281:88-101. [DOI] [PubMed] [Google Scholar]
- 33.Rapaport, D., and W. Neupert. 1999. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 146:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rassow, J., and N. Pfanner. 2000. The protein import machinery of the mitochondrial membranes. Traffic 1:457-464. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Cousino, N., F. E. Nargang, R. Baardman, W. Neupert, R. Lill, and D. A. Court. 1998. An import signal in the cytosolic domain of the Neurospora mitochondrial outer membrane protein TOM22. J. Biol. Chem. 273:11527-11532. [DOI] [PubMed] [Google Scholar]
- 36.Rubino, L., J. Burgyan, and M. Russo. 1995. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch. Virol. 140:2027-2039. [DOI] [PubMed] [Google Scholar]
- 37.Rubino, L., A. Di Franco, and M. Russo. 2000. Expression of a plant virus non-structural protein in Saccharomyces cerevisiae causes membrane proliferation and altered mitochondrial morphology. J. Gen. Virol. 81:279-286. [DOI] [PubMed] [Google Scholar]
- 38.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 39.Rubino, L., F. Weber-Lotfi, A. Dietrich, C. Stussi-Garaud, and M. Russo. 2001. The open reading frame 1-encoded (′36K protein') protein of carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 82:29-34. [DOI] [PubMed] [Google Scholar]
- 40.Russo, M., J. Burgyan, and G. P. Martelli. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381-428. [DOI] [PubMed] [Google Scholar]
- 41.Russo, M., A. Di Franco, and G. P. Martelli. 1983. The fine structure of Cymbidium ringspot virus infections in host tissues. III. Role of peroxisomes in the genesis of multivesicular bodies. J. Ultrastruct. Res. 82:52-63. [DOI] [PubMed] [Google Scholar]
- 42.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schagger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 44.Shore, G. C., H. M. McBride, D. G. Millar, N. A. Steenaart, and M. Nguyen. 1995. Import and insertion of proteins into the mitochondrial outer membrane. Eur. J. Biochem. 227:9-18. [DOI] [PubMed] [Google Scholar]
- 45.Tanudji, M., P. Dessi, M. Murcha, and J. Whelan. 2001. Protein import into plant mitochondria: precursor proteins differ in ATP and membrane potential requirements. Plant Mol. Biol. 45:317-325. [DOI] [PubMed] [Google Scholar]
- 46.Tokatlidis, K., S. Vial, P. Luciano, M. Vergnolle, and S. Clemence. 2000. Membrane protein import in yeast cells mitochondria. Biochem. Soc. Trans. 28:495-499. [PubMed] [Google Scholar]
- 47.Van Der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wattenberg, B., and T. Lithgow. 2001. Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2:66-71. [DOI] [PubMed] [Google Scholar]
- 49.Wischmann, C., and W. Schuster. 1995. Transfer of rps10 from the mitochondrion to the nucleus in Arabidopsis thaliana: evidence for RNA-mediated transfer and exon shuffling at the integration site. FEBS Lett. 374:152-156. [DOI] [PubMed] [Google Scholar]
- 50.Yamanaka, T., T. Ohta, M. Takahashi, T. Meshi, R. Schmidt, C. Dean, S. Naito, and M. Ishikawa. 2000. TOM1, an Arabidopsis gene required for efficient multiplication of a tobamovirus, encodes a putative transmembrane protein. Proc. Natl. Acad. Sci. USA 97:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]