Abstract
In a recent vaccine trial, we showed efficient control of a virulent simian-human immunodeficiency virus SHIV-89.6P challenge by priming with a Gag-Pol-Env-expressing DNA and boosting with a Gag-Pol-Env- expressing recombinant-modified vaccinia virus Ankara. Here we show that long-term control has been associated with slowly declining levels of viral RNA and DNA. In the vaccinated animals both viral DNA and RNA underwent an initial rapid decay, which was followed by a lower decay rate. Between 12 and 70 weeks postchallenge, the low decay rates have had half-lives of about 20 weeks for viral RNA in plasma and viral DNA in peripheral blood mononuclear cells and lymph nodes. In vaccinated animals the viral DNA has been mostly unintegrated and has appeared to be largely nonfunctional as evidenced by a poor ability to recover infectious virus in cocultivation assays, even after CD8 depletion. In contrast, in control animals, which have died, viral DNA was mostly integrated and a larger proportion appeared to be functional as evidenced by the recovery of infectious virus. Thus, to date, control of the challenge infection has appeared to improve with time, with the decay rates for viral DNA being at the lower end of values reported for patients on highly active antiretroviral therapy.
Recent hope for vaccine-induced control of human immunodeficiency virus type 1 (HIV-1) has been raised by a series of trials in which vaccines designed to raise cellular immunity have shown good control of simian-human immunodeficiency virus SHIV-89.6P challenges in rhesus macaques (for a review, see reference 20). These vaccines, which include interleukin-2 (IL-2)-adjuvanted DNA and DNA combined with live viral vectors or live viral vectors alone (1, 1a, 3, 4, 24) have controlled both intravenous and mucosal challenges to the levels characteristic of those in HIV-1-infected humans who are long-term nonprogressors (≤1,000 copies of viral RNA per ml of plasma), have protected CD4 cells, and have prevented the onset of AIDS. With the exception of one escape that occurred within 6 months of challenge (2), the protected animals have successfully controlled their infections up to the present time (about 2 years) in the various trials.
The success of these recent vaccine trials has been largely attributed to the antiviral activities of CD8 T cells. The classical mechanism by which CD8 T cells control viral infections is by recognizing and lysing virus-infected cells (for reviews, see references 16 and 26). CD8 T cells also limit HIV-1 infections by producing chemokines such as MIP-1α, MIP-1β, or RANTES that bind to CCR-5 and block CCR-5 coreceptor activity for viral entry (9, 17). A third antiviral activity of CD8 T cells is the production of suppressive factors that inhibit viral replication (5, 22; for a review, see reference 15). These poorly defined factors are likely to have a number of activities, the currently best defined of which is the inhibition of the expression of proviral DNA (19, 25).
Control of HIV-1 infections in humans by highly active antiretroviral drugs is associated with long-lived reservoirs of viral DNA (for a review, see reference 18), with estimated half-lives ranging from 21 to 163 weeks (11, 12, 14). An important compartment in these reservoirs is integrated HIV-1 DNA in resting memory CD4 T cells; the half-life for this DNA has been estimated to be over 43 months (8). The stability of this latent reservoir is thought to reflect both a low level of ongoing viral replication in the presence of antiretroviral therapy and mechanisms that contribute to the homeostasis of memory CD4 T cells.
In this study, we further investigated the status of the challenge virus in our DNA/recombinant modified vaccinia virus Ankara (rMVA) vaccine trials (1) by testing for viral DNA in the peripheral blood mononuclear cells (PBMC) and lymph nodes of vaccinated and challenged animals. Our trial used two priming inoculations with a Gag-Pol-Env-expressing DNA at 0 and 8 weeks followed by one booster inoculation at 24 weeks with a Gag-Pol-Env-expressing rMVA. An intrarectal challenge was given 7 months following the booster immunization. Control of the challenge infection to the background level of detection (<500 copies of viral RNA per ml of plasma) was achieved in 23 out of 24 vaccinated animals. Here we show that vaccine-mediated control of the SHIV challenge is associated with slowly declining levels of viral DNA and RNA.
MATERIALS AND METHODS
Animals and vaccine trial.
Young adult rhesus macaques from the Yerkes breeding colony were cared for under guidelines established by the Animal Welfare Act and the National Institutes of Health Guide for the Care and Use of Laboratory Animals, with protocols approved by the Emory University Institutional Animal Care and Use Committee. Four groups of six rhesus macaques were primed with either 2.5 mg (high dose) or 250 μg (low dose) of a DNA vaccine that expressed SHIV-89.6 Gag-Pol and Env (DNA/89.6) by intradermal (i.d.) or intramuscular (i.m.) inoculations at 0 and 8 weeks and were boosted with 2 × 108 PFU of an rMVA vaccine that expressed SHIV-89.6 Gag-Pol and Env (MVA/89.6) at 24 weeks (1). An intrarectal challenge with SHIV-89.6P was administered 7 months later (1). The original designations for animals 1 to 28 are given in reference 1; new designations include unvaccinated control animals 29 (RPs-4) and 30 (RKj-5). Viral DNA in PBMC was quantified at weeks 5, 12, 16, 32, 40, 52, 58, and 70 postchallenge, and that in lymph nodes was quantified at weeks 2, 12, and 58 postchallenge. Integrated and unintegrated viral DNA was analyzed at weeks 2, 12, 16, and 58 for vaccinated animals and at weeks 5, 12, and 16 for control animals. Cocultivation assays were conducted at 2 and 52 weeks.
Sample collection and DNA extraction.
Blood samples were collected in EDTA, PBMC were prepared using Ficoll-Hypaque gradients, and pellets corresponding to 5 × 106 PBMC were stored at −70°C. Lymph node samples were mashed and filtered over a Medicons (BD Biosciences, San Jose, Calif.) prior to preparation of cells by using Ficoll-Hypaque gradients. DNA was extracted with QIAamp DNA minikit (Qiagen, Valencia, Calif.) and eluted in 100 μl of distilled water. The DNA concentration was measured by determining the optical density at 260 nm in triplicate and adjusted to 1 μg per 35 μl of distilled water.
Real-time PCR for viral DNA and RNA.
The PCR conditions for viral RNA detection have been described previously (1). The PCR primers and probe for DNA detection included the forward primer QC-F (5′-AGAAAGCCTGTTGGAIAACAAAGAAGG-3′), the reverse primer QC-R (5′-AGTGTGTTTCACTTTCTCTTCTGCGTG-3′), and the Taqman probe, QC-P (5′-FAM-CTGTCTGCGTCATTTGGTGC-TAMRA-3′). These generated and detected a 101-bp SIVmac239 gag fragment. For standardization of the real-time PCR detection assay, linearized plasmid pGEM/pSIV5′ was serially diluted from 106 to 101 copies, aliquoted, and stored at −20°C. Each aliquot was used only one time. For assays, splenocyte DNA from an uninfected rhesus macaque (1 μg/reaction mixture) was spiked with various levels of the plasmid standard.
Amplification was performed in a 50-μl reaction mixture containing 1 μg (35 μl) of test DNA; 200 nM primers QC-F and QC-R; 100 nM Taqman probe QC-P; 50 nM (each) dATP, dCTP, dTTP, and dGTP; 4 mM MgCl2; 1.25 U of AmpliTaq Gold polymerase; and 1× PCR Taqman buffer A (all reagents were from Perkin-Elmer Applied Biosystems, Foster City, Calif.). Amplification and detection were performed with a Perkin-Elmer Applied Biosystems 7700 sequence detector under the following conditions: 1 cycle at 95°C for 10 min followed by 45 two-step cycles of 30 s at 93°C and 60 s at 59.5°C. The slope of the standard curve was between −3.5 and −3.8, the correlation coefficient (r2) was >0.99, and the intra-assay coefficient of variation was <20% for >10 copies of viral DNA.
To avoid contamination, tissue culture hoods and PCR hoods located in separate rooms were used for cell collection, DNA extraction, pre-PCR mix preparation, and 96-well plate preparation. Following use, all equipment was cleaned with 10% bleach. All samples were run in duplicate, with the mean result reported. Viral DNA loads of from 6 to 54 copies per 106 PBMC (1 to 9 copies per μg of template DNA) are reported as <60 copies per 106 PBMC (<10 copies per μg of DNA). Viral DNA copies per milliliter of blood were calculated by multiplying the number of copies of viral DNA per picogram of PBMC DNA by the picograms of DNA per cell (assuming 6 pg of DNA per cell) by the absolute number of lymphocytes per milliliter of blood in the respective samples.
Hirt fractionation.
The Hirt fractionation procedure separates integrated from unintegrated DNA by using a high salt concentration in the presence of sodium dodecyl sulfate (SDS) to precipitate chromosomal DNA (10). Briefly, 5 × 106 fresh PBMC were washed with ice-cold PBS and lysed by adding 1 ml of lysis buffer (0.6% SDS, 10 mM EDTA, and 200 μg of proteinase K per ml in 10 mM Tris [pH 7.5]). The lysis mixture was incubated at 37°C for 2 h, and then NaCl was added to a final concentration of 1 M from a 5 M stock solution, followed by incubation at 4°C overnight. The mixture was then centrifuged at 14,000 rpm in a microcentrifuge (Eppendorf 5415C) for 30 min at 4°C, and the supernatant was carefully removed and diluted with an equal volume of distilled water to reduce the salt concentration. The SDS pellet was dissolved in 1 ml of 0.01 M EDTA. The DNA in the supernatant and pellet was extracted once each with STE-saturated phenol, phenol-CHCl3-isoamyl alcohol (25:24:1), and CHCl3-isoamyl alcohol, following which the DNA was precipitated by the addition of 10 μl of glycogen (20 μg/μl; Roche, Indianapolis, Ind.) and 2 volumes of absolute ethanol and incubation at −20°C. The DNA was then dissolved in 100 μl of distilled water and stored at −20°C. The lymphoblastoid cell line 8E5,which contains a single integrated genome of HIV-1-LAV per cell, was diluted into the parent cell line A301 and spiked with the same number of copies of linearized pGEMpSIV5′ to control for the ability of the Hirt method to separate integrated and unintegrated DNAs. Spiked samples were assayed for simian immunodeficiency virus (SIV) DNA by the PCR protocol described above. Assays for HIV-1 DNA used the primers HIV-F (5′-TGGCATGGGTACCAGCAC-3′) and HIV-R (5′-CTGGCTACTATTTCTTTTGCTA-3′) and the HIV Taqman probe (5′-TTTATCTACTTGTTCATTTCCTCCAATTCCTT-3′) to amplify and detect the subtype B HIV pol gene (6). The results from the Hirt fractionations are expressed as copies of viral DNA per 106 cells.
Cocultivation assay.
Prior to cocultivations, uninfected rhesus macaques were screened to identify ones with PBMC suitable for the growth of SHIV-89.6P. PBMC from these macaques were then cryopreserved for future use as targets in cocultivation assays. For cocultivation assays, PBMC or lymph node cells (LNC) from test monkeys were thawed, stimulated overnight with concanavalin A, and cultured in triplicate microcultures of threefold serial dilutions with a constant number of prescreened target PBMC per well. Cultures were maintained with 10 U of IL-2 per ml. CD8 cell depletion of PBMC was done using magnetic beads coated with anti-CD8 monoclonal antibodies (ITI5C2; Dynal Corp, Lake Success, N.Y.). For 5-aza-2′-deoxycytidine (5-aza-dC) treatments, test cultures were set up for the first 3 days in the presence of 1 μM 5-aza-dC (Sigma, St. Louis, Mo.). Partial supernatant fluid collections were performed every 4 to 5 days with replenishment of fresh IL-2 medium. Supernatant fluids were tested for the presence of SIV p27 by an antigen capture assay (Coulter Immunotech, Hialeah, Fla.), and the frequency of cocultivation-positive cells per million PBMC was derived by using the Spearman-Karber equation.
Statistical analyses.
r2 correlation coefficients were calculated by using Excel (Microsoft Inc., Seattle, Wash.). To obtain an estimate for the rate of decline of viral DNA and RNA in peripheral blood, half-lives were determined for each test animal by calculating the best-fitting linear regression line for log-transformed data obtained between 12 and 72 weeks postchallenge (S-Plus 6 Statistical Package; Insightful Corp., Seattle, Wash.). These lines represented the predicted rate of decline for each monkey. The half-life was taken to be that time point at which the level predicted by each linear estimate reached one-half of the level of the starting point. Median half-lives were determined by calculating the median of the values for the individual animals. To determine the half-life for viral DNA in lymph nodes, data for 12 and 58 weeks were used to determine the half-life for each animal, following which the median was determined for the individual values. Results are presented as the median plus the range of half-life values. The significance of the half-life values, or the determination that the slopes for the decline of the levels of viral DNA or RNA were significantly different from zero, was determined by using a one-sample Wilcoxon signed rank test. P values of less than 0.05 were regarded as being statistically significant.
RESULTS
Postchallenge levels of viral DNA and RNA.
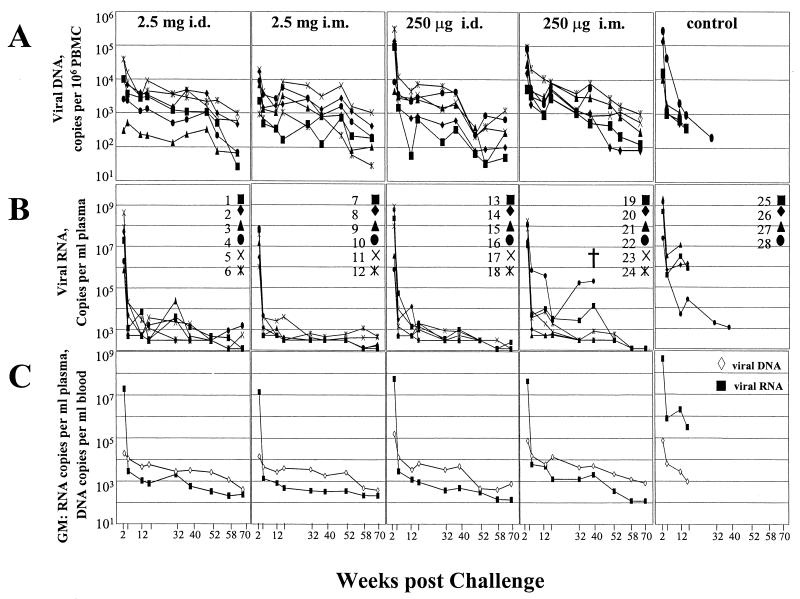
PBMC were analyzed at between 2 and 70 weeks postchallenge to determine the loads of viral DNA in the vaccinated and control animals over time. Real-time PCR analyses for a 101-bp gag fragment were done on samples from groups receiving both high (2.5-mg)- and low (250-μg)-dose i.m. and i.d. DNA priming and from the naive unvaccinated controls. Patterns of viral DNA and RNA for individual animals are presented as copies of viral DNA per 106 PBMC (Fig. 1A) and copies of viral RNA per milliliter of plasma (Fig. 1B). In Fig. 1C, the geometric means for the copies of viral RNA and DNA are presented. To facilitate comparisons, the levels of viral DNA have been normalized per milliliter of blood by determining the copies of DNA per picogram of PBMC DNA, multiplying by 6 to determine the number of copies per cell (each cell was considered to contain 6 pg of DNA), and then multiplying by the absolute number of lymphocytes per milliliter of blood in the test animal.
FIG. 1.
Levels of viral DNA and RNA in vaccinated and control animals over time postchallenge. (A) Copy numbers of viral DNA per 106 PBMC. (B) Copy numbers of viral RNA per milliliter of plasma. Data are taken in part from (1). (C) Geometric mean (GM) copies of viral DNA per milliliter of blood and of viral RNA per milliliter of plasma. Animal groups are indicated at the top of panel A. Designations for individual animals are indicated in panel B. †, died.
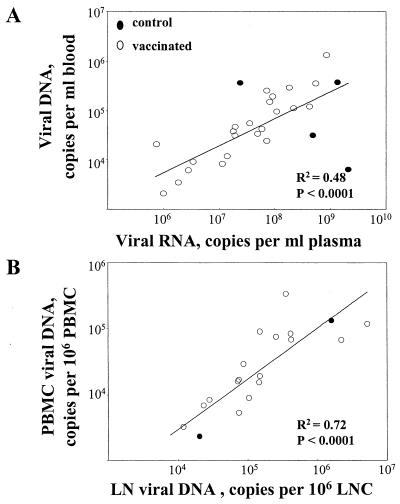
The data in Fig. 1 reveal a number of temporal differences between the levels of viral DNA and RNA in vaccinated and control groups. Peak copies of viral DNA and RNA occurred at 2 weeks postchallenge in both groups. In both groups the peaks of DNA were 1,000 to 10,000 times lower than the peaks of RNA and showed a reasonable correlation with the levels of RNA (Fig. 2A). However, by 5 weeks postchallenge, the correlation between viral DNA and RNA had been lost; vaccinated animals had more copies of viral DNA than viral RNA per ml of blood whereas the control animals had higher levels of viral RNA than DNA per milliliter of blood. At this time, the vaccinated animals had 100- to 1,000-times-lower levels of viral RNA, but overall similar levels of viral DNA, than the control animals. After 12 weeks, the low levels of RNA in the vaccinated animals showed occasional spikes (see animals 3, 19, and 22 in Fig. 1B). Concomitant increases were not seen in the peripheral blood lymphocyte-associated DNA except for animal 22 in the low-dose i.m. group, which progressed to disease (Fig. 1A). Animals within groups exhibited a relatively broad range of values for viral DNA (up to 50-fold differences at any time), which appeared to be broader than that observed for viral RNA (Fig. 1A and B). However, by 12 weeks postchallenge many animals had levels of viral RNA that were below the level of detection, limiting our ability to estimate the range of RNA values.
FIG. 2.
Correlations between levels of viral DNA and RNA in peripheral blood (A) or between levels of viral DNA in PBMC and lymph nodes (B).
Integrated versus unintegrated viral DNA.
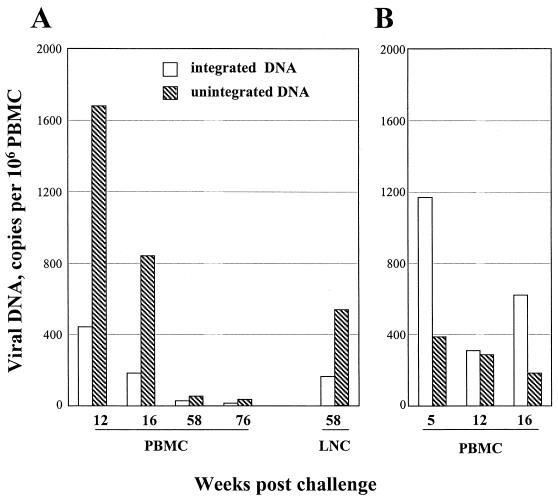
To learn more about the nature of the viral DNA at various times postchallenge, Hirt fractionations were undertaken to separate unintegrated from integrated viral DNA. These fractionations use a combination of high salt concentration and SDS to separate chromosomal (Hirt pellet) from nonchromosomal (Hirt supernatant) DNA. To determine how well Hirt fractionations would separate viral DNA that had, or had not, integrated into chromosomal DNA, the 8E5 cell line, which contains a single copy of HIV-1 DNA, and molecularly cloned SIV sequences were spiked at levels ranging from 10,000 to 0 into 5 × 106 A0.01 cells. These samples were then subjected to Hirt fractionation, and the supernatants and pellets were tested for copies of HIV-1 and SIV DNAs by using real-time PCR for HIV and SIV sequences (Table 1). Almost all of the integrated HIV DNA from 8E5 cells was observed in the pellet, whereas almost all of the unintegrated SIV plasmid was found in the supernatant. Furthermore, the dilution curves for integrated copies of HIV-1 DNA and unintegrated copies of SIV DNA were reasonably linear over the range of concentrations tested. These tests demonstrated that the Hirt method coupled with real-time PCR could be used to distinguish integrated and unintegrated viral DNAs. Unexpectedly, the PBMC of the vaccinated animals had accumulated predominantly unintegrated DNA, whereas the PBMC of the nonvaccinated animals had accumulated predominantly integrated viral DNA (Fig. 3). At all times postchallenge, more unintegrated than integrated DNA was present in the PBMC of vaccinated animals. In contrast, in the control animals, at all times tested, similar levels of integrated and unintegrated viral DNAs or even more integrated than unintegrated viral DNA was detected.
TABLE 1.
Use of the Hirt technique to distinguish integrated HIV DNA and unintegrated SIV DNAsa
| Sequence | Spiked copies | Copies in:
|
|
|---|---|---|---|
| Hirt supernatant | Hirt pellet | ||
| HIV | 10,000 | 23 | 1,200 |
| 1,000 | 23 | 180 | |
| 100 | 0 | 25 | |
| 10 | 1 | 13 | |
| 0 | 0 | 0 | |
| SIV | 10,000 | 4,200 | 6 |
| 1,000 | 314 | 0 | |
| 100 | 22 | 4 | |
| 0 | 0 | 0 | |
For details, see the text.
FIG. 3.
Relative levels of integrated and unintegrated viral DNA over time. (A) Samples from vaccinated animals. (B) Samples from unvaccinated controls. Samples from late times postchallenge are not available for the controls due to their development of AIDS.
Tests for functional DNA.
Cocultivation assays undertaken at 1 year postchallenge suggested that in our assay the viral DNA in the vaccinated animals was largely nonfunctional. Because CD8+ T cells can suppress proviral expression and methylation can inhibit the transcription of proviral DNA, CD8-depleted and 5-aza-dC-treated PBMC were tested in cocultivation assays (Table 2). The viral coculture titers per million unfractionated concanavalin A-activated PBMC were below detection in 17 out of the 17 vaccinated animals that were tested. After depletion of CD8 cells, 4 animals of 17 scored at 0.9 cocultivation-positive cells per million PBMC. In CD8-depleted and 5-aza-dC treated PBMC, 2 of 17 vaccinated animals scored at titers of 0.9 cocultivation-positive cells per million PBMC. In contrast, in cocultivation assays undertaken at 2 weeks postchallenge, both vaccinated and control animals readily scored (10 to >3,000 cocultivation-positive cells per million PBMC). Although the number of animals tested at 2 weeks postchallenge was limited, within this set of animals, the control animals scored with higher frequencies of cocultivation-positive cells than the vaccinated animals despite fairly similar levels of viral RNA and DNA (Table 2).
TABLE 2.
Cocultivation assays for frequencies of cells harboring replication-competent virusa
| Time postchallenge | Vaccine dose and animal no. | Viral RNA (copies/ml of plasma) | Viral DNA (copies/ml of blood) | Cocultivation-positive cells (TCID50b/million PBMC)
|
||
|---|---|---|---|---|---|---|
| PBMC | CD8-depleted cells | CD8-depleted and 5-aza-dC-treated cells | ||||
| 2 wk | 2.5 mg i.d. | |||||
| 1 | 1.9 × 107 | 3.1 × 104 | 2.7 × 102 | NTc | NT | |
| 3 | 7.3 × 105 | 8.1 × 102 | 1.0 × 101 | NT | NT | |
| 4 | 1.9 × 106 | 3.5 × 103 | 1.4 × 101 | NT | NT | |
| 6 | 8.3 × 107 | 1.5 × 105 | 2.7 × 102 | NT | NT | |
| Control | ||||||
| 29 | 2.1 × 109 | 6.4 × 103 | 2.5 × 103 | NT | NT | |
| 30 | 2.4 × 107 | 3.7 × 105 | >3 × 103 | NT | NT | |
| 1 yr | 2.5 mg i.d. | |||||
| 1 | 4.6 × 102 | 2.5 × 103 | <d | < | < | |
| 2 | <300 | 5.4 × 103 | < | < | < | |
| 3 | <300 | 6.8 × 102 | < | < | < | |
| 4 | <300 | 1.9 × 103 | < | < | 0.9 | |
| 5 | <300 | 3.0 × 102 | < | 0.9 | 1.5 | |
| 6 | <300 | 5.2 × 103 | < | < | < | |
| 2.5 mg i.m. | ||||||
| 7 | <300 | 1.3 × 103 | < | < | < | |
| 8 | <300 | 3.9 × 103 | < | 0.9 | < | |
| 9 | <300 | 1.9 × 103 | < | < | < | |
| 10 | <300 | 2.8 × 103 | < | < | < | |
| 11 | 3.8 × 102 | 1.6 × 104 | < | 0.9 | < | |
| 12 | 5.8 × 102 | 6.8 × 102 | < | < | < | |
| 250 μg i.m. | ||||||
| 19 | <300 | 1.1 × 103 | < | < | < | |
| 20 | <300 | 2.6 × 102 | < | < | < | |
| 21 | <300 | 6.2 × 103 | < | 0.9 | < | |
| 23 | <300 | 3.3 × 103 | < | < | < | |
| 24 | 6.0 × 102 | 8.2 × 103 | < | < | < | |
For details, see Materials and Methods.
TCID50, 50% tissue culture infective dose.
NT, not tested.
<, no replication-competent virus detected in 3 × 106 PBMC.
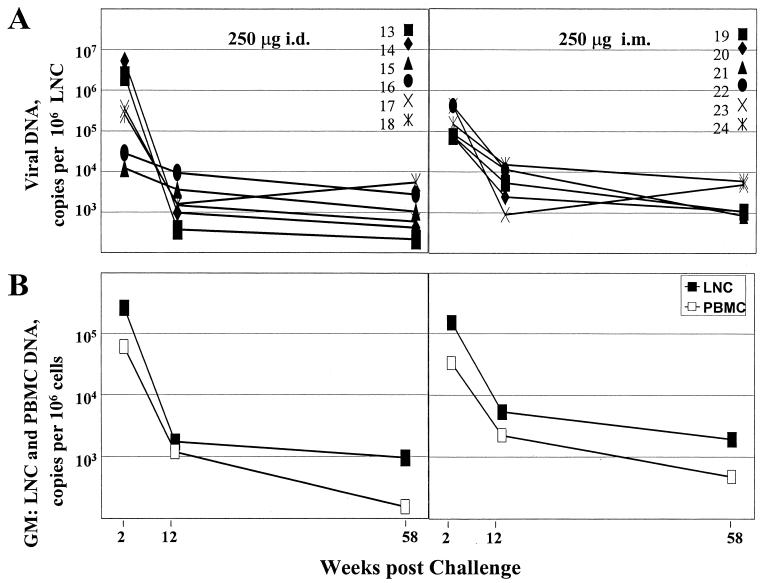
Viral DNA in lymph nodes.
Analysis of viral DNA in lymph nodes revealed similar temporal patterns but 5- to 10-times-higher copy numbers of viral DNA per 106 LNC than per 106 PBMC (Fig. 4). The copies of viral DNA in the LNC underwent the steepest decline (10- to 100-fold) between 2 and 12 weeks postchallenge, following which the levels continued to slowly decline in most animals (9 of 11 animals). At 2 weeks postchallenge there was a good correlation between the levels of viral DNA in LNC and PBMC (Fig. 2B). By 12 weeks postchallenge, this correlation had been lost (data not shown).
FIG. 4.
Levels of viral DNA in lymph nodes over time. (A) Levels for individual animals. (B) Geometric mean (GM) copy numbers of viral DNA in LNC (closed symbols) compared to geometric mean copy numbers of viral DNA in PBMC (open symbols).
Half-lives of viral DNA and RNA.
The geometric means for the copies of viral DNA and RNA in peripheral blood and the geometric mean for the copies of viral DNA in lymph nodes showed a slow decline over time during the chronic phase of the postchallenge infection (Fig. 1C). To obtain an estimate for the decline, half-lives were calculated for each animal for viral RNA per milliliter of plasma, viral DNA per 106 PBMC, and viral DNA per 106 LNC (Table 3). For the peripheral blood, 22 out of 23 animals showed a decline in viral RNA and 22 out of 23 showed a decline in viral DNA. The one animal that did not show a decline in RNA was animal 12 in the high-dose i.m. group (Fig. 1B), and the one animal that did not show a decline in DNA was animal 13 in the low-dose i.d. group (Fig. 1A). Animal 13 distinguished itself by having a potentially spuriously low point for viral DNA at 12 weeks postchallenge. The median half-life for RNA was 21 weeks, with a range of negative values from 9 weeks (animal 19) to 417 weeks (animal 4). The median half-life for viral DNA in PBMC was 16 weeks, with negative values ranging from 8 weeks (animal 18) to 780 weeks (animal 7). Lymph nodes from 11 animals were available for analysis. Of these, nine underwent a decline over time. The median half-life of viral DNA in LNC was 26 weeks, with a range of negative values from 12 to 57 weeks. Thus, each of the measured parameters suggested a slow decline in levels of viral RNA and DNA in the vast majority of the vaccinated animals. The median rate of decline in viral DNA and RNA in PBMC differed significantly from zero (P < 0.001), clearly indicating that this typical pattern among the animals was for steady declines in these measures. Although 9 of 11 monkeys showed declines in viral DNA in LNC, this trend did not reach statistical significance.
TABLE 3.
Half-lives of viral RNA and DNA in the chronic phase of the postvaccine infectiona
| Assay | Time (wk) | No. of animals (declined/ tested)b | Half-life (wk)
|
P | |
|---|---|---|---|---|---|
| Median | Range | ||||
| Viral DNA in: | |||||
| PBMC | 12-70 | 22/23 | 16 | 8-780 | <0.0001 |
| LNC | 12-58 | 9/11 | 26 | 12-57 | 0.1 |
| Viral RNA in plasma | 12-70 | 22/23 | 21 | 9-417 | <0.0004 |
For details, see Materials and Methods.
Declined/tested, number of animals with negative trends/number of animals tested.
DISCUSSION
Our study reveals striking differences in the postchallenge dynamics of viral DNA and RNA in DNA/rMVA-vaccinated and control macaques. The vaccinated, but not the nonvaccinated, animals controlled their levels of plasma viral RNA, while both groups maintained overall similar levels of viral DNA in their PBMC. The viral DNA in the vaccinated animals distinguished itself from that in the unvaccinated animals by consisting predominantly of unintegrated species (Fig. 3). In the vaccinated animals, the levels of viral DNA and RNA underwent an initial contraction. This was followed by a steady slow decline in the levels of viral RNA and DNA, with a half-life of about 20 weeks. Below we discuss these phenomena.
Different relative levels of viral RNA and DNA in vaccinated and control animals.
By 12 weeks postchallenge, the vaccinated and control groups had similar levels of viral DNA but very different levels of viral RNA (Fig. 1). The vaccine reduced the peak titers of plasma viral RNA by only 20- to 40-fold over those in control animals (Fig. 1B). However, the vaccinated, but not the nonvaccinated, animals rapidly controlled their levels of plasma viral RNA. This control did not extend to the levels of viral DNA (Fig. 1). At 12 weeks postchallenge, the four vaccinated groups had geometric mean levels of DNA ranging from 1.8 × 103 to 6.2 × 103 copies per ml of blood, while the unvaccinated animals had a geometric mean of 2.7 × 103 copies.
The very low levels of viral RNA associated with the viral DNA in the vaccinated animals suggested that this DNA was either suppressed for expression or defective. The suppression of provirus expression is a documented function of CD8 cells (15). DNA/rMVA vaccines raise very high levels of CD8 T cells (21, 23) that rapidly mobilize in response to the infection (1). Thus, the low levels of viral RNA in the presence of good levels of viral DNA could have been due to CD8 cell suppressive activities (for a review, see reference 15). However, depletion of CD8 cells prior to cocultivation assays to block CD8 suppressive activities changed the frequency of virus recovery from 0 of 17 to only 4 of 17 animals (Table 2). This suggests that the low levels of virus in plasma did not appear to be accounted for solely by CD8 suppressive activities, at least those that we could relieve by CD8 cell depletion in our cocultivation assays. Treatment of the cocultivation cultures with CD8 depletion and 5-aza-dC to release proviral DNA from methylation-induced transcriptional silencing also failed to provide significant recovery of virus from the DNA-containing PBMC of the vaccinated macaques (Table 2). The poor ability to recover infectious virus could have reflected our assay not effectively scoring the virus that had been selected in vivo.
Further analysis of the viral DNA in the vaccinated macaques suggested that the majority of the DNA at all times postchallenge was unintegrated (Fig. 3). This was in contrast to the case for the nonvaccinated animals, where as much or more integrated than unintegrated DNA was found in peripheral blood. For the control animals, which remained highly viremic, the predominance of integrated viral DNA could reflect ongoing viral replication in activated T cells or monocytes. For the vaccinated animals which controlled their infections, the predominance of unintegrated DNA could reflect host effector responses selecting for cells with nonfunctional viral DNA as well as ongoing infections in resting CD4 cells where integration does not occur (18). The preservation of CD4 cells in the vaccinated groups would have resulted in these groups having plentiful resting CD4 cells as targets for infection and the concomitant generation of unintegrated DNA. In contrast, the loss of CD4 cells in the control animals would have resulted in essentially no target cells in the blood, which in turn could have limited the levels of unintegrated DNA. A novel explanation might be that a highly active immune response favors the accumulation of unintegrated DNA. In vaccinated animals, postchallenge viral replication is likely to preferentially take place in virus-specific CD4 cells that are actively responding to the infection (7). Hypermutation, or the change of G to A in the dinucleotide context GA or GG, occurs in viral DNA that is undergoing replication in activated T cells (13). This could have resulted in the generation of hypermutated DNA that was defective for integration and formed a graveyard of slowly decaying nonfunctional DNA. Further studies will be needed to resolve the origin and genetic integrity of the unintegrated DNA.
Continuing decline in the level of the postchallenge infection.
Encouragingly, after an initial rapid contraction between 2 and 12 weeks postchallenge, the levels of both viral RNA and DNA continued to slowly decrease in the vast majority of the vaccinated animals. Levels of viral RNA were measured in the peripheral blood, whereas levels of viral DNA were measured in the peripheral blood and the lymph nodes (Fig. 2 and 4). The medians of the half-lives for individual animals indicated that overall, each of these parameters had a half-life of about 20 weeks (Table 3). The patterns of control as well as the half-lives of viral RNA and DNA were remarkably similar to those seen for patients on successful highly active antiretroviral therapy (18). Encouragingly, the half-lives for vaccine-mediated controls were at the lower end of the range of half-lives (21 to 40 weeks) reported for patients successfully treated with antiretroviral drugs (11, 14). These shorter half-lives could reflect differences in the kinetics of the highly virulent SHIV-89.6P infection, which causes AIDS within months, as opposed to HIV-1 infections, which require years for the onset of AIDS. However, they also could indicate that effective immune responses have the potential to provide at least as good, if not more stringent, control of immunodeficiency virus infections than antiretroviral drugs.
Acknowledgments
We thank Natalia Kozyr for assistance with developing the assay for viral DNA and Helen Drake-Perrow for expert administrative assistance.
This work was supported by an Integrated Preclinical/Clinical AIDS Vaccine Development program project (P01 AI43045), the Emory/Atlanta Center for AIDS Research (P30 DA 12121), and a Yerkes Regional Primate Research Center base grant (P51 RR0165).
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H.-L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson.2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 1a.Amara, R. R., F. Villinger, S. Staprans, J. D. Altman, D. Montefiori, N. L. Kozyr, Y. Xu, L. Wyatt, P. L. Earl, J. G. Herndon, H. M. McClure, B. Moss, and H. L. Robinson. 2002. Different patterns of immune responses but similar control of a mucusal immunodeficiency virus challenge by MVA and DNA/MVA vaccines. J. Virol. 76:7625-7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barouch, D. H., J. Kuntsman, M. J. Kuruda, J. E. Schmitz, S. Santra, F. W. Peyeri, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gargonne, D. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., S. Santra, M. J. Kuroda, J. E. Schmitz, R. Plishka, A. Buckler-White, A. E. Gaitan, R. Zin, J. H. Nam, L. S. Wyatt, M. A. Lifton, C. E. Nickerson, B. Moss, D. C. Montefiori, V. M. Hirsch, and N. L. Letvin. 2001. Reduction of simian-human immunodeficiency virus 89.6P viremia in rhesus monkeys by recombinant modified vaccinia virus Ankara vaccination. J. Virol. 75:5151-5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Chun, T. W., J. S. Justement, S. Moir, C. W. Hallahan, L. A. Ehler, S. Liu, M. McLaughlin, M. Dybul, J. M. Mican, and A. S. Fauci. 2001. Suppression of HIV replication in the resting CD4+ T cell reservoir by autologous CD8+ T cells: implications for the development of therapeutic strategies. Proc. Natl. Acad. Sci. USA 98:253-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desire, N., A. Dehee, V. Schneider, C. Jacomet, C. Goujon, P. M. Girard, W. Rozenbaum, and J. C. Nicolas. 2001. Quantification of human immunodeficiency virus type 1 proviral load by a TaqMan real-time PCR assay. J. Clin. Microbiol. 39:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douek, D. C., J. M. Brenchley, M. R. Betts, D. R. Ambrozak, B. J. Hill, Y. Okamoto, J. P. Casazza, J. Kuruppu, K. Kunstman, S. Wolinsky, Z. Grossman, M. Dybul, A. Oxenius, D. A. Price, M. Connors, and R. A. Koup. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95-98. [DOI] [PubMed] [Google Scholar]
- 8.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 9.Garzino-Demo, A., A. L. DeVico, F. Cocchi, and R. C. Gallo. 1998. Beta-chemokines and protection from HIV type 1 disease. AIDS Res. Hum. Retroviruses 14(Suppl. 2):S177-S184. [PubMed] [Google Scholar]
- 10.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 11.Izopet, J., G. Salama, C. Pasquier, K. Sandres, B. Marchou, P. Massip, and J. Puel. 1998. Decay of HIV-1 DNA in patients receiving suppressive antiretroviral therapy. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 19:478-483. [DOI] [PubMed] [Google Scholar]
- 12.Izopet, J., C. Tamalet, C. Pasquier, K. Sandres, B. Marchou, P. Massip, and J. Puel. 1998. Quantification of HIV-1 proviral DNA by a standardized colorimetric PCR-based assay. J. Med. Virol. 54:54-59. [DOI] [PubMed] [Google Scholar]
- 13.Janini, M., M. Rogers, D. R. Birx, and F. E. McCutchan. 2001. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4+ T cells. J. Virol. 75:7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson, A. C., M. Birk, S. Lindback, H. Gaines, J. E. Mittler, and A. Sonnerborg. 2001. Initiation of therapy during primary HIV type 1 infection results in a continuous decay of proviral DNA and a highly restricted viral evolution. AIDS Res. Hum. Retroviruses 17:409-416. [DOI] [PubMed] [Google Scholar]
- 15.Levy, J. A., C. E. Mackewicz, and E. Barker. 1996. Controlling HIV pathogenesis: the role of the noncytotoxic anti-HIV response of CD8+ T cells. Immunol. Today 17:217-224. [DOI] [PubMed] [Google Scholar]
- 16.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 17.Pal, R., A. Garzino-Demo, P. D. Markham, J. Burns, M. Brown, R. C. Gallo, and A. L. DeVico. 1997. Inhibition of HIV-1 infection by the beta-chemokine MDC. Science 278:695-698. [DOI] [PubMed] [Google Scholar]
- 18.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 19.Powell, J. D., D. P. Bednarik, T. M. Folks, T. Jehuda-Cohen, F. Villinger, K. W. Sell, and A. A. Ansari. 1993. Inhibition of cellular activation of retroviral replication by CD8+ T cells derived from non-human primates. Clin. Exp. Immunol. 91:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson, H. L. 2002. New hope for an AIDS vaccine. Nat. Rev. Immunol. 2:239-250. [DOI] [PubMed]
- 21.Robinson, H. L., J. M. Smith, and R. R. Amara. 2000. AIDS vaccines: heterologous prime/boost strategies for raising protective T cell responses. AIDS Rev. 2:105-110. [Google Scholar]
- 22.Rubbert, A., D. Weissman, C. Combadiere, K. A. Pettrone, J. A. Daucher, P. M. Murphy, and A. S. Fauci. 1997. Multifactorial nature of noncytolytic CD8+ T cell-mediated suppression of HIV replication: beta-chemokine-dependent and -independent effects. AIDS Res. Hum. Retroviruses 13:63-69. [DOI] [PubMed] [Google Scholar]
- 23.Schneider, J., S. C. Gilbert, C. M. Hannan, P. Degano, E. G. Sheu, M. Plebanski, and A. V. S. Hill. 1999. Induction of CD8+ T cells using heterologous prime-boost immunisation strategies. Immunol. Rev. 170:29-38. [DOI] [PubMed] [Google Scholar]
- 24.Shiver, J. W., T. Fu, L. Chen, D. Casimiro, M. E. Davies, R. K. Evans, Z.-Q. Zhang, A. Simon, W. L. Trigona, S. Dubey, L. Huang, V. A. Harris, R. S. Long, L. Xiaoping, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Iospi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 25.Tomaras, G. D., S. F. Lacey, C. B. McDanal, G. Ferrari, K. J. Weinhold, and M. L. Greenberg. 2000. CD8+ T cell-mediated suppressive activity inhibits HIV-1 after virus entry with kinetics indicating effects on virus gene expression. Proc. Natl. Acad. Sci. USA 97:3503-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, O. O., and B. D. Walker. 1997. CD8+ cells in human immunodeficiency virus type I pathogenesis: cytolytic and noncytolytic inhibition of viral replication. Adv. Immunol. 66:273-311. [DOI] [PubMed] [Google Scholar]