Abstract
To understand the regulation of cap-dependent translation initiation mediated by specific 5′ untranslated region (UTR) RNA-protein interactions in mammalian cells, we have studied the selective translation of influenza virus mRNAs. Previous work has shown that the host cell mRNA binding protein guanine-rich sequence factor 1 (GRSF-1) bound specifically to conserved viral 5′ UTR sequences and stimulated translation of viral 5′ UTR-driven mRNAs in vitro. In the present study, we have characterized the functional domains of GRSF-1 and mapped the RNA binding activity of GRSF-1 to RRM 2 (amino acids 194 to 275) with amino-terminal deletion glutathione S-transferase (GST)-GRSF-1 proteins. When these mutants were assayed for functional activity in vitro, deletion of an Ala-rich region (Δ[2-94]) appeared to diminish translational stimulation, while deletion of the Ala-rich region in addition to RRM 1 (Δ[2-194]) resulted in a 4-fold increase in translational activation over wild-type GRSF-1 (an overall 20-fold increase in activity). We have also mapped the GRSF-1 RNA binding site on influenza virus NP and NS1 5′ UTRs, which was determined to be the sequence AGGGU. With polysome fractionation and cDNA microarray analysis, we have identified cellular and viral mRNAs containing putative GRSF-1 binding sites that were transcriptionally up-regulated and selectively recruited to polyribosomes following influenza virus infection. Taken together, these studies demonstrate that RRM 2 is critical for GRSF-1 RNA binding and translational activity. Further, our data suggest GRSF-1 functions by selectively recruiting cellular and viral mRNAs containing 5′ UTR GRSF-1 binding sites to polyribosomes, which is mediated through interactions with cellular proteins.
The ability of cells to respond to extracellular stimuli and intracellular cues, including mitogenic signals, is directly linked to the regulation of mRNA translation initiation. The control of initiation can be regulated by the specific interaction of RNA binding proteins and initiation factors (eIFs) with cis-acting elements contained in both the 5′ and 3′ untranslated regions (UTRs) of mature mRNAs (reviewed in reference 37). Accordingly, deregulation of protein synthesis is a key mechanism in both malignant transformation and viral replication. Influenza virus infection results in the selective translation of viral mRNAs, while host cell protein synthesis is markedly attenuated (reviewed in reference 38). The subversion of the host cell protein synthetic machinery to produce high levels of influenza virus proteins, which are required for infection and replication, is dependent on conserved sequences present in the 5′ UTRs of the influenza virus mRNAs (13).
Translation initiation relies upon the interactions of trans-acting factors with both ribosomal RNAs and cis-acting determinants in the mRNA. Influenza virus protein synthesis is a cap-dependent process mediated by highly conserved sequences contained in the 5′ UTRs of the viral mRNAs (15). As there is little destruction of cellular mRNA in the cytoplasm until late in infection, the selective initiation of translation on viral mRNAs must be dependent on the discrimination of viral and host cell mRNAs (4, 19). Interestingly, the 5′ UTRs of influenza virus mRNAs are relatively unremarkable, apart from the 5′ cap structure and 10 to 14 nucleotides obtained from host cell mRNAs by the “cap-stealing” mechanism of viral mRNA synthesis (20). Dependent upon the RNA segment, influenza virus 5′ UTRs are typically between 20 and 50 nucleotides in length, in addition to the host cell-derived nucleotides, and do not contain upstream AUGs (21). Although the influenza virus 5′ UTRs are predicted to contain little secondary structure, they result in profound stimulatory effects on viral protein synthesis in infected cells, while host cell protein synthesis is substantially reduced (reviewed in reference 38).
Our laboratory has demonstrated recently that the host cell mRNA binding protein GRSF-1 specifically interacts with the conserved sequences in the influenza virus 5′ UTR and that this interaction stimulated protein synthesis in in vitro translation-competent lysates prepared from infected HeLa cells. Moreover, we demonstrated that the stimulation of translation of mRNAs in vitro was dependent on the presence of the conserved viral 5′ UTR, as chimeric mRNAs containing a mutated viral 5′ UTR were translated at a lower efficiency and did not show GRSF-1 responsiveness (30). As further proof of the involvement of GRSF-1 in selective translation, the stimulation of protein synthesis could be abolished by the immunodepletion of GRSF-1 from the in vitro translation lysates (30). Augmented protein expression could be reconstituted in the immunodepleted extracts by the addition of exogenous GRSF-1. These experiments demonstrated that the interaction of GRSF-1 with the viral 5′ UTR was a critical determinant in the selective translation of viral 5′ UTR-containing mRNAs during infection.
GRSF-1 is a member of the RNP superfamily of RNA binding proteins (10), which have been implicated in a wide variety of cellular processes, including RNA processing, nuclear export, trafficking, mRNA stability, and mRNA translation (8, 12, 36). The members of the RNP superfamily are typified by the presence of single or multiple copies of an RNA binding domain called the RNA recognition motif (RRM) (8). The RRM is a conserved motif of approximately 80 amino acids, including two short, highly conserved regions, termed RNP1 and RNP2. The GRSF-1 protein contains three RRMs, in addition to two auxiliary regions comprising an N-terminal Ala-rich region and an acidic domain located between RRM 2 and RRM 3.
Our hypothesis is that GRSF-1 represents an alternative pathway for the selective translation of mRNAs utilized during cellular stress that has been subverted by influenza virus to preferentially synthesize viral proteins. To date, this is one of the few examples of a virus recruiting a host cell protein as a positive regulator of viral protein synthesis (11, 30, 39). To examine the role of GRSF-1 in selective, cap-dependent translation initiation, the regions of GRSF-1 responsible for mRNA binding and translational control were also identified by using a series of amino-terminal deletion mutants of GRSF-1. To further characterize the interactions of GRSF-1 with influenza virus mRNAs, we have determined the sequences in the viral 5′ UTR required for GRSF-1 binding. Moreover, we have used polysome fractionation and cDNA microarray analyses to identify mRNAs containing GRSF-1 5′ UTR binding sites that were selectively recruited to polyribosomes following infection, many of which are involved in viral replication, cell survival, and the inflammatory response. These studies help elucidate the role of GRSF-1 in selective translation during infection and provide a more detailed understanding of the selective control of protein synthesis in mammalian cells, as well as the replication of influenza virus.
MATERIALS AND METHODS
Constructs and preparation of RNAs.
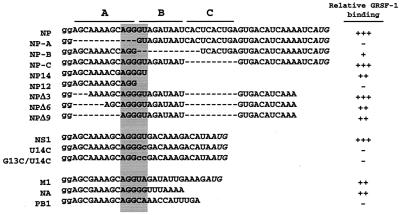
The HindIII-BstEII fragment containing the T7 promoter followed by influenza virus NP 5′ UTR was amplified by PCR from pBC/CMV/(NP)SEAP with the primers 5′-AAAAAGCTTAATACGACTCACTATAGGGAGCAAAAGCAGGGTAG-3′ and 5′-GTCGGTGACCATGATTTTGATGTCACTCAGT-3′. After being digested with HindIII and BstEII, fragments were subcloned between the HindIII and BstEII sites on pSP-luc+NF (Promega), yielding plasmid pSP-NP. Plasmid pSP-NP-A was constructed in the same way with primers 5′-AAAAAGCTTAATACGACTCACTATAGGGGTAGATAATCACTCACTGAG-3′ and 5′-GTCGGTGACCATGATTTTGATGTCACTCAGT-3′. For RNA gel shift and UV cross-linking analyses, unlabeled and radiolabeled RNAs were transcribed from linear double-stranded template DNAs prepared by annealing equal moles of complementary oligonucleotides containing the T7 RNA polymerase promoter (see Table 1 for RNA sequences).
TABLE 1.
RNAs used in analyses showing relative GRSF-1 bindinga
The GRSF-1 binding site is shaded gray, and the A-box sequences are indicated. Binding: +++, strong RNA-protein complex formation; −, no complex formation. The 5′-terminal guanosines result from transcription of the 3′ end of the T7 RNA polymerase promoter.
Unlabeled RNAs were prepared from linearized cDNA templates with the MAXIscript T7 kit (Ambion), while radiolabeled RNAs were similarly synthesized in the presence of 100 μCi of [α-32P]UTP (3,000 Ci/mmol; NEN). Radiolabeled RNAs used in gel mobility shift assays were further gel purified on an 8% polyacrylamide gel containing 6 M urea. Bands containing the full-length radiolabeled RNA were excised from the gel, and the RNAs were eluted by incubation in 2 M ammonium acetate (pH 5.4)-1% sodium dodecyl sulfate (SDS)-25 μg of tRNA per ml. For capped mRNA transcripts, plasmids were linearized with EcoRI and were transcribed with T7 RNA polymerase with a T7 mMESSAGE mMACHINE kit (Ambion). The resulting capped transcripts were further purified, and their concentration was determined from the A260. Size and integrity of all transcripts were checked by agarose gel electrophoresis and ethidium bromide staining prior to use.
Preparation of fusion proteins.
To prepare wild-type glutathione S-transferase (GST)-GRSF-1, Escherichia coli HB101 cells were transformed with the plasmid pGEX2TZQ-2.7 (31) and cultured in Luria-Bertani (LB)-ampicillin medium and induced with 0.3 mM isopropylthiogalactopyranoside (IPTG) at 30°C for 4 h. Cells were harvested and disrupted by sonication in buffer containing 1× phosphate-buffered saline (PBS), pH 7.4, 10% glycerol, 0.5 mM MgCl2, 1 mM EDTA, and protease inhibitors (EDTA-free complete protease inhibitor cocktail; Roche-BMB). Nested N-terminal deletions were constructed with PCR and cloning into the expression plasmid pGEX-2TZQ at the BamHI and KpnI sites. Four 5′ primers were run against the same 3′ primer. The sequences of the 5′ primers were Δ[2, 94], 5′-GGGCCAGGATCCGCCACCATGCTCATTCGAGCTCAA-3′; Δ[2, 194], 5′-GGGCCAGGATCCGCCACCATGGTGGTTCGTTTGAGA-3′; Δ[2, 275], 5′-GGGCCAGGATCCGCCACCATGCATGTCGGTTCTTAT-3′; Δ[2, 345], 5′-GGGCCAGGATCCGCCACCATGTTTGTCCACATGAGA-3′; and 5′-GGGCCAGGTACCTTATTTTCCTTTTGGACA-3′ for the 3′ primer.
The PCR products were digested with BamHI and KpnI, gel purified, and ligated into pGEX-2TZQ. The sequences were confirmed by automated sequencing. GST fusion proteins were attached on glutathione-Sepharose beads and were eluted with freshly prepared elution buffer (100 mM Tris-HCl [pH 8.0] and 10 mM reduced glutathione). Eluted protein, containing 10% glycerol and protease inhibitors, was stored at −80°C.
RNA binding analysis.
A binding reaction was a modification of that described by Meerovitch et al. (26). Briefly, GST-GRSF-1 (100 ng) was incubated with radiolabeled RNA (100,000 dpm) in buffer that contained 5 mM HEPES (pH 7.6), 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 100 mM NaCl, 0.02 mM dithiothreitol, 2 mM GTP, and 1.5 mM ATP at 30°C for 20 min. Samples were then electrophoresed on a 5% polyacrylamide gel at 4°C. The gels were dried and exposed to an X-ray film with an intensifying screen at −80°C. For UV-cross-linking experiments, equal moles of wild-type and mutant GST-GRSF-1 were incubated with 106 dpm of radiolabeled NP 5′ UTR RNA in buffer containing 5 mM HEPES (pH 7.6), 25 mM KCl, 2 mM MgCl2, 5% glycerol, 100 mM NaCl, 2 mM dithiothreitol, and 20 U of RNasin (Promega) at 30°C for 15 min. Samples were then exposed to UV light (1.0 J) in a Stratalinker (Stratagene), followed by incubation with 10 mg of RNase A per ml and 2 U of RNase T1 at 37°C for 30 min. Samples were suspended in 1× Laemeli buffer and separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE). The resulting gel was dried and visualized with a Storm 850 PhosphorImager (Molecular Dynamics).
Polyribosome fractionation.
HeLa cells were grown to ≈80% confluency and were infected with influenza virus type A/WSN/33 at a multiplicity of infection of 50 or with medium alone (mock infected). After 4 h at 37°C, medium was replaced with medium containing 100 μg of cycloheximide (Sigma) per ml and incubated for 10 min at 37°C. Cells were washed with 1× PBS, pH 7.5, plus 100 μg of cycloheximide per ml and released from the culture flask with a trypsin-EDTA solution containing 100 μg of cycloheximide per ml. Cells were then washed extensively in 1× PBS containing 100 μg of cycloheximide per ml and Complete EDTA-free protease inhibitors (BMB), pelleted by centrifugation (1,000 × g), and resuspended in 0.75 ml of buffer containing 200 mM Tris, pH 7.5, 100 mM NaCl, and 30 mM MgCl2 (LBS).
Following a 3-min incubation on ice, 0.25 ml of buffer containing 1.2% Triton N-101, 0.2 M sucrose in LBS was added and lysed in a Dounce homogenizer (8 strokes) and centrifuged briefly at 10,000 × g. Supernatants were transferred to fresh tubes, and 0.1 ml of solution containing 10 mg of heparin per ml and 1.5 M NaCl in LBS was added. Lysates were then layered on top of a 12-ml linear 15% to 50% sucrose gradient and centrifuged at 163,659 RCFave for 110 min at 4°C. Gradients were fractionated into 1.0-ml fractions (approximately 14 samples per gradient) with a syringe pump and fractionator (Brandel, Gaithersburg, Md.) with continuous monitoring based on the A254. Following polysome fractionation, poly(A)+ mRNA was isolated with an Oligotex mRNA purification kit (Qiagen, Valencia, Calif.) as previously described (17).
cDNA microarray analysis.
Briefly, fluorescently labeled cDNA probes were generated by reverse transcription, and unincorporated nucleotides were removed with 96-well multiscreen-FB filter plates (Millipore, Bedford, Mass.) followed by G-50 ProbeQuant columns (AP Biotech) (17). Human cDNA I.M.A.G.E. clones (18) were obtained from Research Genetics (Huntsville, Ala.) Homo sapiens 15K sequence verified set (UG Build 19, plates 1 to 44). cDNA inserts for I.M.A.G.E. clones and controls were PCR amplified and purified (see below). DNA pellets were suspended in a 50% solution of reagent D (AP Biotech) and deposited on coated glass microscope slides (75 mm by 25 mm; type VII; AP Biotech) with the use of a Molecular Dynamics (Sunnyvale, Calif.) Generation III microarray spotter. The appropriate indocarbocyanine (Cy3)- and indodicarbocyanine (Cy5)-labeled probes were combined, denatured by boiling, and applied to the slides under a glass coverslip. Microarrays were hybridized at 42°C in a humidified chamber for 16 to 20 h. Following hybridization, slides were washed extensively and scanned at 532 and 633 nm with an Avalanche dual laser confocal scanner (Molecular Dynamics).
Data analysis and differentially expressed clone selection were performed as previously described (17). Briefly, each slide contained 4,608 cDNAs spotted in duplicate. Included in this number was a set of 384 selected cDNAs that were spotted on every slide. This set contained four influenza virus genes, nonhuman genes used as negative controls, and a variety of selected transcription factors, ligands, and receptors chosen from the Research Genetics 15K human gene set. For each of the polysome pooled fractions (I and II), duplicate slides were hybridized with the same RNAs but with the fluorescent labels reversed to control for dye-specific effects as described previously (17). Intensity values in Cy3 and Cy5 channels were extracted from each image, and the Cy3/Cy5 ratio was determined with Spot-on image software. Data for all replicates were combined and normalized with our software, Spot-on Unite. For each gene that was differentially expressed in at least two experiments, the mean intensity and standard deviation were extracted for all experiments (at each time point).
In vitro translation analysis.
To prepare HeLa extracts for cell-free translation, an S10 cytoplasmic lysate was prepared from an exponentially growing HeLa S3 suspension culture (4 × 109 cells), infected with influenza virus strain WSN (at a multiplicity of infection of ≈40 PFU per cell). Briefly, HeLa cells (2 × 109 cells) in log phase were harvested, washed three times with ice-cold PBS, and resuspended with 1.5× packed cell volume with hypotonic buffer (10 mM K-HEPES [pH 7.5], 10 mM potassium acetate, 1.5 mM magnesium acetate, 2 mM dithiothreitol). After incubation on ice for 10 min, cells were disrupted with a Wheaton Dounce homogenizer (type A) until approximately 95% of the cells were disrupted (about 20 strokes), as visualized by the trypan blue dye exclusion assay.
The cell lysate was centrifuged at 10,000 × g for 20 min. The resulting supernatant was dialyzed for 4 h against 1 liter of dialysis buffer (10 mM HEPES, pH 7.5, 90 mM potassium acetate, l0.5 mM magnesium acetate, 1.0 mM dithiothreitol, 5% glycerol) in the Slide-A-Lyzer dialysis cassette (10,000 molecular weight cutoff; Pierce). The S10 lysate was supplemented with 0.0156 mg of tRNA per ml, 0.62 mM ATP, 0.037 mM GTP, 6.22 mM creatine phosphate, 0.0156 mg of creatine kinase per ml, 11.8 mM HEPES (pH 7.6), 1.24 mM dithiothreitol, 15.6 μM complete amino acid mixture (Promega), and 0.156 mM spermidine. For in vitro translation, 200 ng of template mRNAs was incubated in the absence or presence of 0.2 μg of GST-GRSF-1 for 60 min at 30°C. The reactions were terminated by the addition of 9 volumes of PBS buffer. Samples (20 μl) were then analyzed for luciferase activity with a luciferase reporter assay system (Promega).
RESULTS
Determination of the regions of GRSF-1 responsible for mRNA binding.
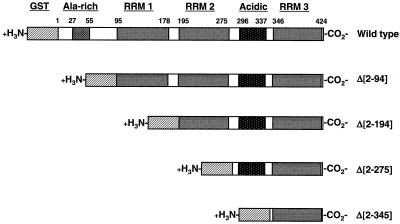
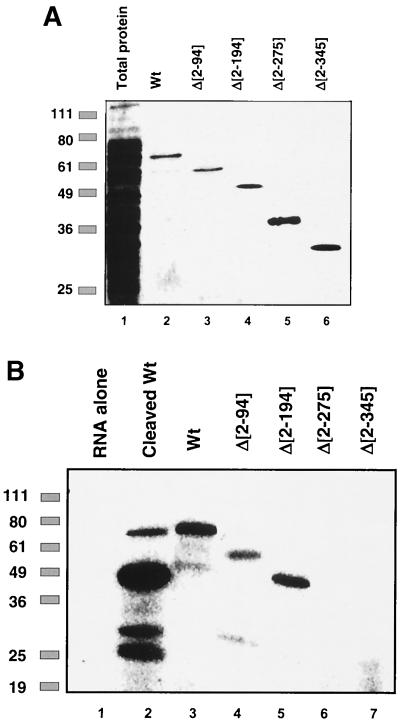
To identify the regions of the GRSF-1 protein which were required for the interaction with viral 5′ UTRs, a series of nested N-terminal deletion GRSF-1 mutant proteins were prepared, as shown in Fig. 1. These proteins were expressed in E. coli as GST fusion proteins and were incubated with radiolabeled NP 5′ UTR RNA and analyzed by UV cross-linking analysis. As shown in Fig. 2A, wild-type and mutant GST-GRSF-1 proteins were expressed in E. coli, purified, and eluted from glutathione-Sepharose, and analysis by SDS-PAGE demonstrated that all of the fusion proteins were expressed efficiently and purified to near homogeneity.
FIG. 1.
Diagram of wild-type and N-terminal deletion mutant GST-GRSF-1 proteins. Wild-type GRSF-1 is shown, containing the Ala-rich region (amino acids 27 to 55), RRM 1 (amino acids 95 to 178), RRM 2 (amino acids 195 to 275), acidic domain (amino acids 296 to 337), and RRM 3 (amino acids 346 to 424). Amino-terminal deletion mutants used are also shown.
FIG. 2.
Second RRM of GRSF-1 is required for mRNA binding in vitro. (A) Coomassie blue-stained denaturing 12% polyacrylamide gel of equal moles (1.25 × 10−11 mol) of wild-type and mutant GST-GRSF-1 fusion proteins purified by glutathione affinity chromatography from E. coli. (B) UV cross-linking analysis of thrombin-cleaved GRSF-1 (rGRSF-1) and intact wild-type (Wt) and mutant GST-GRSF-1 fusion proteins (1.25 × 10−11 mol) incubated with radiolabeled NP 5′ UTR RNA (106 dpm) in 5 mM HEPES (pH 7.6)-25 mM KCl-2 mM MgCl2-5% glycerol-100 mM NaCl-2 mM dithiothreitol-20 U of RNasin (Promega) at 30°C for 15 min. Samples were then exposed to UV light (1.0 J) in a Stratalinker (Stratagene), followed by incubation with 10 mg of RNase A per ml and 2 U of RNase T1 at 37°C for 30 min. Samples were suspended in 1× Laemmli buffer and separated by SDS-10% PAGE, and the gel was dried and visualized with a Storm 850 phosphorimager (Molecular Dynamics). These data are representative of two independent experiments.
For RNA binding analysis, our approach was to use the wild-type and deletion mutant GST-GRSF-1 proteins (diagrammed in Fig. 1) incubated with NP 5′ UTR RNA. Briefly, GST-GRSF-1 (1.25 × 10−11 mol) was incubated with radiolabeled NP 5′ UTR RNA (106 dpm) for 10 min at 30°C, followed by UV cross-linking, RNase A treatment, and SDS-10% PAGE. As shown in Fig. 2B, deletion of the N-terminal Ala-rich domain and RRM 1 of GRSF-1 did not affect NP RNA binding in vitro (Fig. 2B, compare lanes 3 to 5). However, deletion of RRM 2 resulted in the total loss of mRNA binding activity (lanes 6 to 7). Moreover, RRM 3 was unable to bind independently to NP 5′ UTR RNA (lane 6), although it is possible that RRM 3 may stabilize the RRM 2-mediated binding or may interact with other RNA ligands in vivo. These data demonstrated that, although GRSF-1 contains three RRMs, RRM 2 appeared to be required for binding of viral 5′ UTR RNA sequences, but we cannot exclude the possibility that RRM 2 and RRM 3 act in concert to bind RNA.
Determination of the regions of GRSF-1 responsible for translational regulation.
We have previously established cell-free translation systems with influenza virus-infected HeLa cell lysates, which mimic the selective translation observed in intact influenza virus-infected cells (30). We have further employed these in vitro translation-competent lysates to report that GRSF-1 was required for the stimulation of viral 5′ UTR-driven luciferase protein.
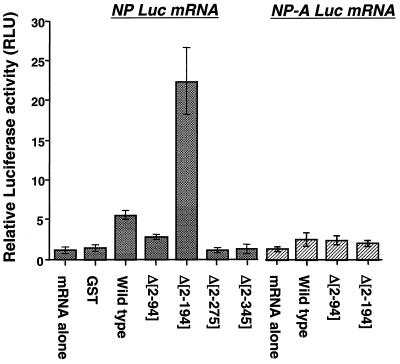
To determine the regions of GRSF-1 responsible for the stimulation of protein synthesis from influenza virus 5′ UTR-containing mRNAs, we added equal moles of wild-type and N-terminal deletion mutant GST-GRSF-1 proteins to in vitro translation-competent infected HeLa cell lysates. Chimeric luciferase mRNAs containing either the influenza virus NP or NP-A 5′ UTRs were added to the reactions, incubated for 60 min, and analyzed for luciferase activity. As shown in Fig. 3, GST protein was unable to stimulate luciferase translation, while an equal molar concentration of wild-type GST-GRSF-1 stimulated luciferase expression approximately 5-fold in an NP 5′ UTR A-box-dependent manner.
FIG. 3.
Second RRM of GRSF-1 is required for stimulation of translation in vitro. Equal moles (2.5 × 10−12 mol) of GST, wild-type, and deletion mutant GST-GRSF-1 proteins were incubated with in vitro translation-competent influenza virus-infected HeLa cell extracts containing 200 ng of capped NP-luciferase or NP-A-luciferase chimeric mRNAs in a 12-μl final volume, as described in Materials and Methods. Reactions were prepared in triplicate and incubated for 60 min at room temperature, and the reactions were terminated by the addition of 9 volumes of 1× PBS, pH 7.5. Luciferase activity in 25-μl aliquots of each reaction was determined with the luciferase assay kit (Promega). These data are representative of two independent experiments.
This selective 5-fold increase in luciferase protein synthesis driven by the wild-type NP 5′ UTR corresponds with our previous reported results (30). Deletion of the amino-terminal Ala-rich domain appeared to diminish the luciferase protein production, while deletion of the Ala-rich region and RRM 1 resulted in an approximately 20-fold increase in luciferase protein expression over NP 5′ UTR luciferase mRNA alone (4-fold higher than the wild-type GRSF-1 protein) which was still dependent on the presence of the wild-type NP 5′ UTR. This analysis demonstrated that the amino acid sequences N-terminal to RRM 2 contain a negative regulatory element and that RNA binding of the conserved viral 5′ UTR sequences (so-called A-box) was required for translational stimulation.
Determination of a minimal GRSF-1 RNA binding site.
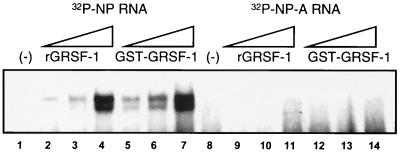
To clarify the role of GRSF-1 in the translational regulation of influenza virus mRNAs, we determined the GRSF-1 binding site on viral mRNA leaders. To resolve the minimal viral mRNA sequences required for GRSF-1 binding, we performed a series of RNA gel shift experiments with wild-type and mutant NP 5′ UTR RNAs. As shown in Fig. 4, increasing concentrations of either thrombin-cleaved GRSF-1 (rGRSF-1) or intact GST-GRSF-1 were incubated with radiolabeled NP or NP-A 5′ UTR RNA followed by native 5% PAGE. Incubation of NP RNA with both rGRSF-1 (lanes 2 to 4) and GST-GRSF-1 (lanes 5 to 7) resulted in a concentration-dependent RNP complex, demonstrating that the GST tag did not interfere with mRNA binding. In addition, these data demonstrated that deletion of the highly conserved A-box region of the NP 5′ UTR abolished binding by both rGRSF-1 and GST-GRSF-1. The sequences and relative GRSF-1 binding activity are shown in Table 1.
FIG. 4.
GRSF-1 binds the conserved A-box of the influenza virus NP 5′ UTR. Shown here is a representative autoradiogram of RNA gel shift analysis of radiolabeled influenza virus NP (lanes 1 to 7) or NP-A (lanes 8 to 14) 5′ UTR RNA incubated with increasing concentrations (50, 100, and 200 ng) of thrombin-cleaved (rGRSF-1) or uncleaved GST-GRSF-1. Proteins were incubated with radiolabeled RNA (100,000 dpm) in buffer that contained 5 mM HEPES (pH 7.6), 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 100 mM NaCl, 0.02 mM dithiothreitol, 2 mM GTP, and 1.5 mM ATP at 30°C for 20 min. Samples were then electrophoresed on a 5% polyacrylamide gel at 4°C and visualized by autoradiography. These data are representative of two independent experiments.
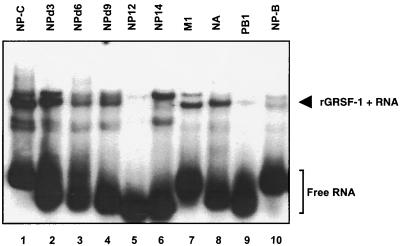
Next, we examined a series of mutant NP 5′ UTR RNA fragments with deletions in regions A and B by incubating GST-GRSF-1 (6.25 × 10−13 mol) with radiolabeled wild-type and mutant influenza virus 5′ UTR RNAs followed by 5% native PAGE. As shown in Fig. 5, GRSF-1 formed a complex with the first 14 nucleotides (AGCAAAAGCAGGGU; NP14) of the NP 5′ UTR, but not with the first 12 nucleotides (AGCAAAAGCAGG; NP12), indicating the last 2 nucleotides (GU) of NP14 were critical for GRSF-1 binding. In contrast, the RNA fragments that lacked 3 nucleotides (NP5Δ3), 6 nucleotides (NP5Δ6), or 9 nucleotides (NP5Δ9) from the 5′ end of the NP 5′ UTR showed binding activity with GRSF-1, although affinity for the last two RNAs was diminished (results of these in vitro binding assays are summarized in Table 1).
FIG. 5.
GRSF-1 interacts with specific sequences within the influenza virus mRNA 5′ UTR as detected by gel mobility shift analysis. Recombinant GRSF-1 (100 ng) was incubated with radiolabeled NP-c, NPD3, NPD6, NPD9, NP12, NP14, M1, NA, PB1, and NP-B RNAs (105 dpm) in buffer that contained 5 mM HEPES (pH 7.6), 25 mM KCl, 2 mM MgCl2, 3.8% glycerol, 100 mM NaCl, 0.02 mM dithiothreitol, 2 mM GTP, and 1.5 mM ATP at 30°C for 20 min. The resulting RNA-protein complexes were resolved by 5% native PAGE and visualized by autoradiography. These data are representative of two independent experiments.
The RNAs which were bound by GRSF-1 with high affinity (e.g., NP 5′ UTR, NP-C, NPΔ3, and NS 5′ UTR) all contain the AGGGU pentamer spanning regions A and B. These in vitro binding data suggested that the AGGGU pentamer might serve as a core element for GRSF-1 binding, whereas the flanking sequence of this core element may play a role in enhancing the binding affinity to GRSF-1, perhaps by stabilizing a necessary structural element.
We also performed RNA gel shifts with the 5′ UTRs from other influenza virus mRNAs, specifically the M1, NA, and PB1 mRNAs. Although not as strongly as AGGGU in NP, GRSF-1 appeared to bind to the AGGU and AGGGGU elements in M1 and NA, respectively. These data demonstrated that GRSF-1 was capable of binding influenza virus M1 and NA 5′ UTRs (Fig. 5, lanes 7 and 8), while in contrast, GRSF-1 did not show any binding activity to the 5′ UTR of influenza virus PB1 5′ UTR mRNA, which lacks any apparent GRSF-1 binding sites (Fig. 5, lane 9). In contrast to the M 5′ UTR, NP-B, which also contained region A and the AGGU, showed very weak binding to GRSF-1 (Fig. 5, lane 10). These data demonstrated that GRSF-1 displayed the highest affinity for the sequence AGGGU, although several permutations of this sequence, including AGGU and AGGGGU, still demonstrated a high affinity for GRSF-1.
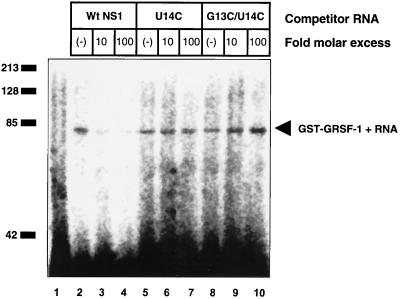
We have demonstrated that GRSF-1 can form stable complexes with the 5′ UTR of influenza virus NS1 mRNA, which contained the consensus A10GGGU14 binding site, and that GRSF-1 was able to discriminate between M1 and PB1 5′ UTRs. The principal difference between M1 and PB1 in the aligned GRSF-1 binding site (see Table 1) is AGGU in M1 and AGGC in PB1. Therefore, we next examined the importance of the G13 and U14 nucleotides by performing UV-cross-linking RNA binding competition analysis with wild-type NS1 5′ UTRs and two mutant NS1 5′ UTRs (U14C and G13C/U14C). Briefly, GST-GRSF-1 was incubated with radiolabeled wild-type NS1 RNA in the absence or presence of increasing concentrations of unlabeled wild-type, U14C, or G13C/U14C NS1 RNAs.
As shown in Fig. 6, incubation with a 10- to 100-fold molar excess of unlabeled wild-type NS1 RNA resulted in a competition for the binding of radiolabeled wild-type NS1 RNA. In contrast, incubation with a 100-fold molar excess of unlabeled U14C or G13C/U14C RNAs did not affect GRSF-1 binding of the radiolabeled wild-type NS1 RNA. As summarized in Table 1, these data demonstrated that the conserved A- and B-box regions of influenza virus mRNAs contained sequences required for GRSF-1 binding and that nucleotide U14 was critical for GRSF-1 binding. These data suggest that the interaction of GRSF-1 with these highly conserved 5′ UTR sequences might play a critical role in regulating viral protein synthesis during infection.
FIG. 6.
G13 and U14 in the NS1 5′ UTR are required for GRSF-1 binding. Wild-type (Wt) GST-GRSF-1 (1.25 × 10−11 mol) was incubated with 106 dpm of radiolabeled wild-type NS1 5′ UTR RNA in the absence or presence of increasing concentrations (10- or 100-fold molar excess) of unlabeled wild-type (lanes 2 to 4), U14C (lanes 5 to 7), or G13C/U14C (lanes 8 to 10) competitor RNAs in 5 mM HEPES (pH 7.6)-25 mM KCl-2 mM MgCl2-5% glycerol-100 mM NaCl-2 mM dithiothreitol-20 U of RNasin (Promega) at 30°C for 15 min. Samples were then exposed to UV light (1.0 J) in a Stratalinker (Stratagene), followed by incubation with 10 mg of RNase A per ml and 2 U of RNase T1 at 37°C for 30 min. Samples were suspended in 1× Laemmli buffer and separated by SDS-10% PAGE, and the gel was dried and visualized with a Storm 850 phosphorimager (Molecular Dynamics). These data are representative of three independent experiments.
In order to identify other mRNAs that were potential targets for GRSF-1-mediated translational regulation, we isolated, identified, and characterized cellular and viral mRNAs that were recruited to polyribosomes following influenza virus infection.
Analysis of polyribosome-associated mRNAs during infection.
We have previously used cDNA microarray analysis to report the changes in gene expression profiles following influenza virus infection (16). These analyses demonstrated that viral replication resulted in a marked overall decline in cellular gene expression. However, infection also resulted in the activation of many genes involved in the host cell antiviral response, cell proliferation, and the inflammatory response. To examine the role of translational activation during infection, we analyzed the distribution of cellular and viral mRNAs on polyribosomes with polysome fractionation and cDNA microarray analysis. Briefly, ribosomes were fractionated on linear 15% to 50% sucrose gradients following isolation from mock-infected or 8-h postinfection HeLa cells.
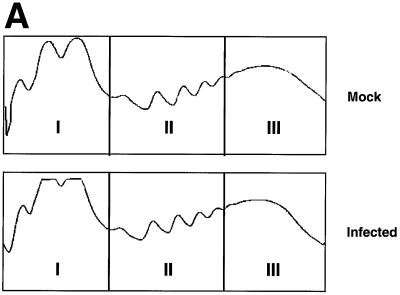
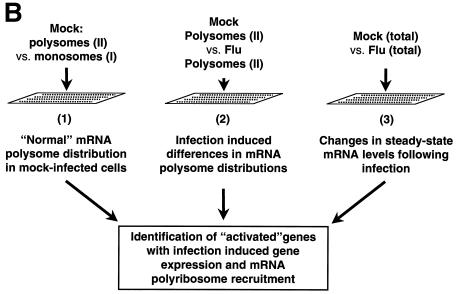
As shown in Fig. 7A, fractions were then pooled into three groups and mRNA was isolated by oligo(dT) selection. The first pooled group contained the 40S and 60S ribosome subunits in addition to the 80S monosome; the second pool consisted of the mRNAs associated with 2 to 6 ribosomes; and the third group contained the dense polysomal fraction. Pooled fractions I (representing monosomes) and pooled fractions II (representing polyribosomes) were used to interrogate cDNA microarrays. The association of the mRNAs with polyribosomes was verified by the addition of 25 mM EDTA to parallel samples that caused the release of these mRNAs via the dissociation of 80S ribosomes (data not shown). Pooled fraction III (containing the dense polyribosomes) was not included in the microarray analysis because previous work had suggested a block in elongation might occur during infection which would result in the accumulation of stalled ribosomes and produce anomalously high sedimentation coefficients (22).
FIG. 7.
Polyribosome and microarray analysis of infected cells. Polysome fractionation and microarray experiments were performed in duplicate with RNAs isolated from mock-infected and infected HeLa cells fractionated on linear 15 to 50% sucrose gradients. (A) Polysome profiles of mock-infected and infected HeLa cells as measured by real-time A254. Fractions pooled for microarray analysis are indicated. (B) Schematic representation of experiment design and classification of differentially expressed and polyribosome-associated mRNAs following influenza virus infection. (C) Diagram of the 492 genes present in all comparisons with a >1.5-fold change in steady-state concentration and polyribosome distribution. (D) Differentially polyribosome-associated mRNAs identified by microarray analysis containing consensus or putative variants of GRSF-1 binding sequences (GBS and vGBS, respectively). Sequences of 5′ UTR consensus and variant GRSF-1 binding sites identified by sequence analysis are indicated.
Surprisingly, the polysome profiles obtained from mock-infected and infected cells were indistinguishable, which suggested that the translational apparatus remained largely unmodified following infection. This is in contrast to the polysome profile of poliovirus-infected cells that were shown to be dramatically affected by infection, resulting in the displacement of cellular mRNAs from polyribosomes (5, 6).
To verify the integrity of various translation apparatus components, we performed Western blot analysis for many of the canonical initiation factors, including eIF2α, IF3, eIF4A, eIF4B, eIF4E, Paip1, PABP, eIF4GI, and eIF4GII. We did not observe any changes in the steady-state protein levels of any of these initiation factors (data not shown). These data indicated the integrity and activity of the cap-dependent translational apparatus in the infected cells.
The experimental design of the array experiments is shown in Fig. 7B. Microarray analysis was performed on mRNA in pooled groups I and II prepared from mock-infected and infected cells. Comparisons were performed with mock-infected II versus mock-infected I to determine the “normal” polysome distribution of host cell mRNAs in an uninfected cell and influenza virus-infected II versus mock-infected II to determine relative polysome distribution following infection. The polysome fractionation data were then combined with our previous expression array data that revealed significant decreases in cellular gene expression following influenza virus infection of HeLa cells (16).
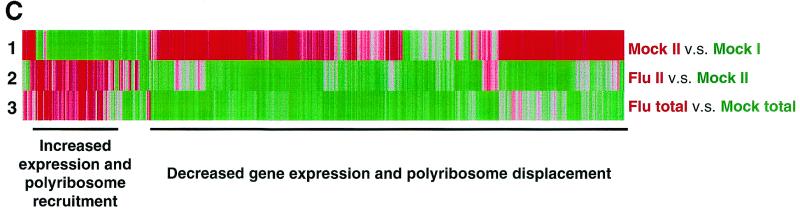
Out of 4,600 randomly selected human and influenza virus genes present in duplicate on the array, 1,045 genes were differentially regulated more than 1.5-fold, with red indicating a relative increase in hybridization signal and green indicating a relative decrease (data not shown). However, for consideration in this study, genes had to present in all comparisons, i.e., both this study and our earlier published work (16), and be regulated greater than 1.5-fold in at least two comparisons. Following application of these filters, the polysome profiles of 492 cellular and viral genes were compiled. As shown in Fig. 7C, row 1, comparison of mock-infected II versus mock-infected I demonstrated that the majority of cellular mRNAs were polysome associated (shown as red), while a minority of genes were associated with monosomes (shown as green).
Following infection with influenza virus, the comparison of influenza virus-infected II with mock-infected II showed that only a small minority of mRNAs, including both viral and host mRNAs, were polyribosome associated (shown as red in row 2). Analysis of the steady-state mRNA levels present in total RNA isolated from mock-infected and infected HeLa cells demonstrated that most cellular gene expression was down-regulated following infection (compare the number of up-regulated red genes and down-regulated green genes shown in Fig. 7C, row 3). Comparison of the normal mRNA polysome distribution (mock-infected II versus mock-infected I) with the distribution of mRNAs following infection (influenza virus-infected II versus mock-infected II) documented that there was a dramatic displacement of a majority (>85%) of the mock-infected polysome-associated mRNAs (compare the red genes in row 1 with the green genes in row 2), while there was a distinct increase in the polyribosome association of a subset of mRNAs (compare the red genes in row 2 with the green genes in row 1).
This analysis showed that from a pool of 492 genes, approximately 10% were induced in expression and mobilized to polyribosomes following infection. This group of “activated” genes that were transcriptionally induced and recruited to polyribosomes included the influenza virus genes present on the slide, in addition to a handful of cellular genes. One clear indication of the active translational state of the mRNAs in pooled polyribosomal fraction II was the presence of the influenza virus mRNAs, which are heavily translated during infection. Our hypothesis that GRSF-1 stimulated translation initiation via sequence-specific RNA-protein interactions predicted that a subset of these polyribosome-recruited mRNAs should contain a recognizable 5′ UTR GRSF-1 binding site. For 5′ UTR sequence analysis, we concentrated on genes with the following characteristics: increased steady-state mRNA levels following infection, localization to monosomes in mockinfected cells, and recruitment to polyribosomes following infection.
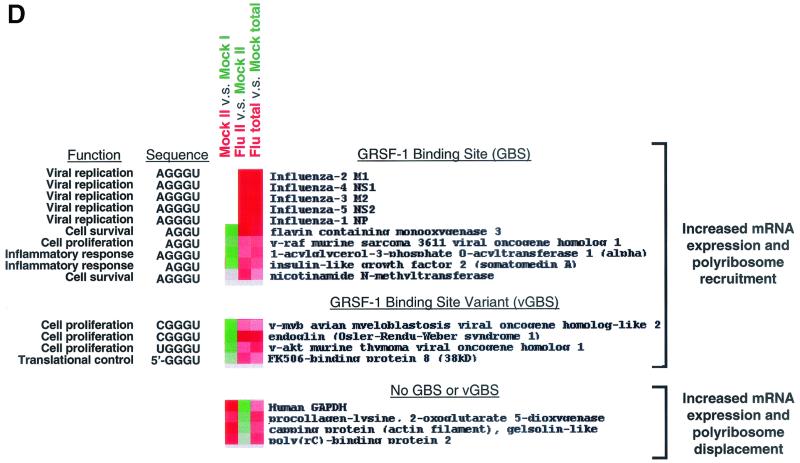
As shown in Fig. 7D, sequence analysis performed on the 5′ UTRs of the activated genes shown in Fig. 7C revealed that approximately 25% contained matches for a GRSF-1 binding site, including the influenza virus genes on the array, while a further 10% contained a variant GRSF-1 binding site. Interestingly, of the transcriptionally and translationally activated mRNAs that did not contain a GRSF-1 binding site or variant GRSF-1 binding site, 25% contained putative 5′-terminal oligopyrimidine (TOP) sequences, while the remaining 40% did not have reported 5′ UTR sequences. For contrast, we examined the sequences of the four mRNAs (out of six total) with reported 5′ UTRs whose expression was induced by infection but were displaced from polyribosomes and did not observe any sequences resembling a GRSF-1 binding site or a putative 5′-TOP element (see Fig. 7D).
One concern was that some of the polyribosome-mobilized mRNAs identified in our analyses might result from the accumulation of stalled ribosomes. At present, it is unknown if an inhibition of elongation was a primary consequence of infection (i.e., direct, targeted inhibition of an elongation factor by a viral protein) or a secondary effect arising from the infection-induced translation shutoff and how this affected the localization of mRNAs in the sucrose gradient. However, our hypothesis is that GRSF-1 contributed to protein expression by stimulating initiation on the 5′ UTR of mRNAs containing a GRSF-1 binding site and would not likely be involved in control of the downstream process of elongation.
Given the induction of mRNA expression and for the genes identified in Fig. 7D, their sheer presence on polyribosomes suggested they were able to bypass the infection-induced block in initiation. Moreover, it was clear that while a significant majority of cellular mRNAs were displaced from polyribosomes after infection, a distinct subset of genes are moving from monosomes to polysomes. We believe polyribosome recruitment was due to the presence of a GRSF-1 binding site, irrespective of the presence or absence of an elongation block.
Taken together, these data provide new evidence for our hypothesis that the selective translation of influenza virus mRNAs results from the selection of mRNAs at the level of 5′ UTR sequence discrimination mediated in part by GRSF-1.
DISCUSSION
In the present study, we have determined the key functional domains of GRSF-1 required for mRNA binding and translational stimulation. This analysis demonstrated that RRM 2 is required for mRNA binding and that the region carboxy-terminal to RRM 1 was sufficient for translational stimulation. Moreover, our data suggested that the regions N-terminal to RRM 2 might contain a negative regulatory element that attenuated GRSF-1 function. Analysis of the sequence requirements for interaction of the host cell mRNA binding protein GRSF-1 with influenza virus 5′ UTRs demonstrated that the sequence AGGGU was a GRSF-1 binding consensus sequence (GRSF-1 binding site). We have further demonstrated that the nucleotides G and U in positions 4 and 5 of the virally encoded 5′ UTR of NS1 were important determinants of GRSF-1 mRNA binding and likely made critical contacts with residues in RRM 2. Moreover, with translation state array analysis, we have identified consensus GRSF-1 binding sites in the 5′ UTRs of mRNAs translationally activated by influenza virus infection.
GRSF-1 functional domain mapping.
With GRSF-1 N-terminal deletion proteins, we demonstrated that RRM 2 was primarily responsible for NP and NS1 5′ UTR RNA binding. However, given this set of mutant proteins, we have not been able to determine the role RRM 3 might play in stabilizing the binding of RNA by RRM 2, and studies are under way to better characterize the interplay between these two domains. Moreover, we cannot exclude the possibility that RRMs 1 and 3 bind other RNA ligands that may aid in the formation of a 48S preinitiation complex on viral mRNAs.
Deletion of the N-terminal and Ala-rich regions (Δ[2-94]) inhibited translation activation in vitro, while deletion of the Ala-rich region and RRM 1 (Δ[2-194]) yielded a hyperactive mutant with a 4-fold increase in activity over wild-type GRSF-1. One explanation of these data is the region N-terminal to RRM 2 contains a negative regulatory element that represses the extent of translational stimulation. A second possible explanation is the Δ[2-194]) GRSF-1 protein was not correctly folded and was unable to interact efficiently with other molecules, presumably regulatory proteins. Once RRM 2 is deleted, GRSF-1 was no longer able to bind mRNA and was unable to stimulate protein synthesis. Studies are currently under way to identify the proteins that interact with GRSF-1, particularly those that interact with RRM 2.
The role of host cell proteins, including La, polypyrimidine tract binding protein, and the poly(rC) RNA binding protein 2 in the stimulation of cap-independent initiation has been demonstrated in many picornavirus systems (6). Although studies in numerous other viral systems have shown how a viral protein can act as a positive regulator of viral protein synthesis, to date there are very few examples of a cellular protein required for the translational stimulation of capped viral mRNAs.
Viral 5′ UTR RNA recognition site.
The influenza virus 5′ UTRs are thought to have little stable secondary structure as predicted by Zuker's M-fold program (data not shown), suggesting that GRSF-1 might be binding to single-stranded regions. Perhaps an important function of GRSF-1 is to maintain the viral 5′ UTR in an unfolded state, allowing easier access of the RNA sequences to translation initiation factors. Of the AGGGU consensus GRSF-1 binding site, the 3′ nucleotides G and U were shown to be important determinants for GRSF-1 binding. The observation that one or two nucleotides in a particular RNA binding site can determine specificity is shown in the example of the RRM proteins U1A and U2B′. The human U1A protein has been shown to bind stem-loop II of human U1 small nuclear RNA by recognizing the loop sequence AUUGCACUCC, while the related protein U2A′ binds the homologous sequence (U)AUUGCAGUAC(CU) in the loop of stem-loop IV of the human U2 small nuclear RNA (28, 34). Thus, although the RNA binding sites of these two proteins share common structural and sequence elements, the sequences can be discriminated by only two nucleotides within the AUUGCA sequence.
We have demonstrated the interaction of GRSF-1 with sequences AGGU, AGGGU, and AGGGGU with only small changes in affinity (as summarized in Table 1). We have further shown that binding of the sequence AGGGU required the 3′ GU nucleotides. It is important to note that two influenza virus mRNAs, PB1 and HA, do not contain a recognizable GRSF-1 binding site (AGGC and AGGGGA, respectively), and our analysis has demonstrated that GRSF-1 does not bind the PB1 5′ UTR. At present, we do not understand this discrepancy, but we believe it may reflect differences in GRSF-1 RNA binding activity in vitro and in vivo, differences in the requirements for temporal regulation and accumulation of viral proteins early or late in infection, or the presence of another factor that specifically stimulates PB1 and HA protein synthesis. Moreover, GRSF-1 may recognize and bind in vivo to alternative G-rich sequences when present in different contexts or in the presence of other mRNA binding proteins. However, it is important to note that viral proteins that have very high expression levels, including NS1 and NP, contain GRSF-1 binding sites. We believe the GRSF-1-stimulated translation of these viral mRNAs, NS1 in particular for its role in PKR repression, would help the virus to circumvent the host cell antiviral response early in infection.
Analysis of polysome-recruited mRNAs containing GRSF-1 binding sites.
Our goal for these experiments was to identify cellular and viral mRNAs that were targets of GRSF-1-mediated translational regulation in vivo with cDNA microarray analysis of sucrose density gradient-separated mRNAs. Of particular interest were mRNAs that might be involved in either the host cell antiviral response or recruited by influenza virus to facilitate viral replication. Our prediction was that genes implicated in these two processes would have several criteria. First, gene expression would be transcriptionally induced by infection, and second, the corresponding mRNA would be recruited to polyribosomes.
Microarray analysis demonstrated that following influenza virus infection, more than 85% of the cellular mRNAs present on the slide were displaced from polyribosomes. Analysis of the 5′ UTRs of the mRNAs that were recruited to polyribosomes following infection demonstrated that approximately 25% of the transcriptionally and translationally induced genes contained 5′ UTR sequences containing variations of the AGGGU GRSF-1 consensus binding sequence. The mRNAs shown to be recruited to polyribosomes and containing a putative GRSF-1 binding site are involved in the control of cell proliferation and survival, mitogenesis, the inflammatory response, or viral replication. This was in marked contrast to the absence of a GRSF-1 binding site on genes that were transcriptionally induced but whose mRNAs were displaced from the polyribosomes during infection.
At present we are performing additional experiments to characterize the in vivo association of GRSF-1 with these 5′ UTRs and the biological consequence of this interaction. While we clearly need to verify the infection-induced synthesis of these proteins by metabolic labeling, the data presented in this report support our hypothesis that GRSF-1 represents an alternative stress-induced translation initiation pathway that has been adopted by influenza virus to stimulate viral protein synthesis early in infection. Furthermore, we believe that the genomic analysis of polyribosome-fractionated mRNA will provide important new information about the global changes in protein synthesis during viral infection and will additionally reveal functional relationships between 5′ UTR sequences and the regulation of polyribosome distribution.
Of the genes examined by microarray, approximately 25% of the polyribosome-recruited mRNAs contain a putative 5′-terminal oligopyrimidine tract (TOP). This is a known translational control element present in ribosomal and other cellular protein 5′ UTRs (reviewed in reference 37). Fractionation experiments have demonstrated that 5′-TOP mRNAs alternate between repressed (mRNP associated) and active (polysome associated) states and that in an active state are translated to maximum efficiency (1, 18, 25, 27, 35).
Intriguingly, ribosomal protein mRNA 5′-TOPs have been demonstrated to escape the host cell shutoff induced by poliovirus (9) and herpes simplex virus type I (17). The translation of 5′-TOP-containing ribosomal proteins in poliovirus-infected cells is surprising because these mRNAs lack a bona fide internal ribosome entry site. Regardless of the role for 5′-TOP translation in other infection systems, the absence of a putative 5′-TOP element in the influenza virus mRNAs, which are purine rich, suggests that the regulation of translation of viral and cellular 5′-TOP-containing mRNAs arises from distinct pathways.
Our work suggests that at early times in infection, when the levels of viral proteins are low, the 5′ UTR of the viral mRNAs plays a critical role in stimulating translation through the interaction of GRSF-1, while at later times this specificity may be relaxed (10). This model further suggests that by adapting a cellular stress response pathway to help augment viral protein synthesis, the virus is able to get a jump on the host cell antiviral response. This may be particularly important for the production of the NS1 protein that has been demonstrated to perform a critical function for viral replication by inhibiting the double-stranded RNA-inducible protein kinase PKR (7, 14, 24). Other studies have demonstrated that ectopic NS1 protein expression inhibited host cell pre-mRNA splicing (32), polyadenylation (29), and nuclear export (2, 3, 33). Therefore, once sufficient levels of the NS1 protein are synthesized to accomplish these functions, the need for selective translation may be reduced.
Taken together, these data provide new evidence for our hypothesis that the selective translation of influenza virus mRNAs results from the adaptive discrimination of 5′ UTR sequences mediated in part by GRSF-1.
Acknowledgments
We thank the members of the Katze lab, past and present, for their contributions to our work.
Work in the Katze lab is supported by NIH grants AI-22646, RR-00166, and AI-41629. J. Wilusz is supported by NIH grants GM-63832 and CA-80062. J. C. Kash is supported by postdoctoral NIH grant GM-20785.
REFERENCES
- 1.Agrawal, M. G., and L. H. Bowman. 1987. Transcriptional and translational regulation of ribosomal protein formation during mouse myoblast differentiation. J. Biol. Chem. 262:4868-4875. [PubMed] [Google Scholar]
- 2.Alonso-Caplen, F. V., and R. M. Krug. 1991. Regulation of the extent of splicing of influenza virus NS1 mRNA: role of the rates of splicing and of the nucleocytoplasmic transport of NS1 mRNA. Mol. Cell. Biol. 11:1092-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alonso-Caplen, F. V., et al. 1992. Nucleocytoplasmic transport: the influenza virus NS1 protein regulates the transport of spliced NS2 mRNA and its precursor NS1 mRNA. Genes Dev. 6:255-267. [DOI] [PubMed] [Google Scholar]
- 4.Beloso, A., et al. 1992. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J. Gen. Virol. 73:575-581. [DOI] [PubMed] [Google Scholar]
- 5.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Bergmann, M., et al. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burd, C. G., and G. Dreyfuss. 1994. Conserved structures and diversity of functions of RNA-binding proteins. Science 265:615-621. [DOI] [PubMed] [Google Scholar]
- 9.Cardinali, B., et al. 1999. Resistance of ribosomal protein mRNA translation to protein synthesis shutoff induced by poliovirus. J. Virol. 73:7070-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cassetti, M. C., et al. 2001. Efficient translation of mRNAs in influenza virus A virus-infected cells is independent of the viral 5′ untranslated region. Virology 289:180-185. [DOI] [PubMed] [Google Scholar]
- 11.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dreyfuss, G., et al. 1993. hnRNP proteins and the biogenesis of mRNA. Annu. Rev. Biochem. 62:289-321. [DOI] [PubMed] [Google Scholar]
- 13.Gale, M., S. L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza virus A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 15.Garfinkel, M. S., and M. G. Katze. 1993. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J. Biol. Chem. 268:22223-22226. [PubMed] [Google Scholar]
- 16.Geiss, G. K., et al. 2001. Global impact of influenza virus on cellular pathways is mediated by both replication-dependent and -independent events. J. Virol. 75:4321-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greco, A., A. M. Laurent, and J. J. Madjar. 1997. Repression of beta-actin synthesis and persistence of ribosomal protein synthesis after infection of HeLa cells by herpes simplex virus type 1 infection are under translational control. Mol. Gen. Genet. 256:320-327. [DOI] [PubMed] [Google Scholar]
- 18.Hornstein, E., et al. 1999. The expression of poly(A)-binding protein gene is translationally regulated in a growth-dependent fashion through a 5′-terminal oligopyrimidine tract motif. J. Biol. Chem. 274:1708-1714. [DOI] [PubMed] [Google Scholar]
- 19.Inglis, S. C. 1982. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza virus and herpes simplex virus. Mol. Cell. Biol. 2:1644-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katze, M. G., and R. M. Krug. 1984. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol. Cell. Biol. 4:2198-2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katze, M. G., and R. M. Krug. 1990. Translational control in influenza virus-infected cells. Enzyme 44:265-277. [DOI] [PubMed] [Google Scholar]
- 22.Katze, M. G., D. DeCorato, and R. M. Krug. 1986. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J. Virol. 60:1027-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lennon, G., et al. 1996. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics 33:151-152. [DOI] [PubMed] [Google Scholar]
- 24.Levy, D. E., and A. Garcia-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 25.Loreni, F., and F. Amaldi. 1997. Translational control of terminal oligopyrimidine mRNAs requires a specific regulator. FEBS Lett. 416:239-242. [DOI] [PubMed] [Google Scholar]
- 26.Meerovitch, K., J. Pelletier, and N. Sonenberg. 1989. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 3:1026-1034. [DOI] [PubMed] [Google Scholar]
- 27.Meyuhas, O., E. A. Thompson, Jr., and R. P. Perry. 1987. Glucocorticoids selectively inhibit translation of ribosomal protein mRNAs in P1798 lymphosarcoma cells. Mol. Cell. Biol. 7:2691-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagai, K., et al. 1990. Crystal structure of the RNA-binding domain of the U1 small ribonucleoprotein A. Science 348:515-520. [DOI] [PubMed] [Google Scholar]
- 29.Nemeroff, M. E., et al. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 30.Park, Y. W., J. Wilusz, and M. G. Katze. 1999. Regulation of eukaryotic protein synthesis: selective influenza virus mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA 96:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian, Z., and J. Wilusz. 1994. GRSF-1: a poly(A)+ mRNA binding protein which interacts with a conserved G-rich element. Nucleic Acids Res. 22:2334-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 small nuclear RNA and inhibits U6-U2 and U6-U4 small nuclear RNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 33.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scherly, D., et al. 1990. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B' and their cognate RNAs. Science 345:506-507. [DOI] [PubMed] [Google Scholar]
- 35.Shima, H., et al. 1998. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siomi, H., and G. Dreyfuss. 1997. RNA-binding proteins as regulators of gene expression. Curr. Opin. Genet. Dev. 7:345-353. [DOI] [PubMed] [Google Scholar]
- 37.Sonenberg, N., J. W. B. Hershey, and M. B. Mathews (ed.). 2000. Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Tan, S. L., M. Gale, and M. G. Katze. 2000. Translational reprogramming during influenza virus infection, p. 933-950. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Wells, D. R., et al. 1998. HSP101 functions as a specific translational regulatory protein whose activity is regulated by nutrient status. Genes Dev. 12:3236-3251. [DOI] [PMC free article] [PubMed] [Google Scholar]