Abstract
The subcellular localization of herpes simplex virus tegument proteins during infection is varied and complex. By using viruses expressing tegument proteins tagged with fluorescent proteins, we previously demonstrated that the major tegument protein VP22 exhibits a cytoplasmic localization, whereas the major tegument protein VP13/14 localizes to nuclear replication compartments and punctate domains. Here, we demonstrate the presence of a second minor population of VP22 in nuclear dots similar in appearance to those formed by VP13/14. We have constructed the first-described doubly fluorescence-tagged virus expressing VP22 and VP13/14 as fusion proteins with cyan fluorescent protein and yellow fluorescent protein, respectively. Visualization of both proteins within the same live infected cells has indicated that these two tegument proteins localize to the same nuclear dots but that VP22 appears there earlier than VP13/14. Further studies have shown that these tegument-specific dots are detectable as phase-dense bodies as early as 2 h after infection and that they are different from the previously described nuclear domains that contain capsid proteins. They are also different from the ICP0 domains formed at cellular nuclear domain 10 sites early in infection but, in almost all cases, are located in juxtaposition to these ICP0 domains. Hence, these tegument proteins join a growing number of proteins that are targeted to discrete nuclear domains in the herpesvirus-infected cell nucleus.
The herpes simplex virus (HSV) tegument, the region between the capsid and the envelope, comprises at least 15 virus-encoded proteins, including major structural proteins VP13/14, VP16, and VP22 (14, 41). The roles of many of these proteins in virus replication are not yet understood, but it is probable that they perform functions additional to their roles in virus assembly and maturation. An example of a protein with such dual functionality is tegument protein VP16, which acts as a transactivator of immediate-early gene expression at early stages of infection (28, 35) but is also essential for virus assembly at later stages (31, 44). While the roles played by, for instance, VP16 and the vhs protein (17, 20) are now well established, the functions of a number of other tegument proteins, including the major components VP22 and VP13/14, have yet to be defined.
The mechanism of tegument acquisition by the maturing virion is also poorly characterized. Because tegument must be incorporated into the virus prior to or at the same time as envelopment, the identification of the cellular site(s) of tegument protein assembly may help refine the current understanding of HSV maturation. While the details of virus assembly remain controversial, a collection of evidence in recent years has supported the theory that capsids in the nucleus undergo an envelopment-deenvelopment step at the nuclear membrane and then acquire their final envelope at a downstream location within the cytoplasm (1, 18, 29, 40, 42, 45). This model would allow for tegument proteins to be added in the nucleus, the cytoplasm, or both. The targeting of capsid proteins to the nucleus (33, 39) and envelope proteins to the secretory pathway (32, 34) is well documented, and the subcellular compartmentalization of these proteins correlates well with their putative sites of acquisition by the maturing virus. However, analysis of the targeting of several tegument proteins has revealed that the localization of this class of proteins is more varied and complex than that of the other structural components of the virion, making it difficult to correlate the subcellular localization of these proteins with potential sites of incorporation into the virus. For instance, the three proteins that make up the major part of the HSV tegument, namely, VP22, VP16, and VP13/14, have been shown to exhibit a range of localization patterns in infected cells. VP13/14 targets the nucleus (4, 30), VP16 localizes to nuclear replication compartments and the cytoplasm (7, 21), while conflicting reports on VP22 localization have shown it to be either cytoplasmic (11, 30) or nuclear (36). In addition, the homologues of VP22 from pseudorabies virus (PRV), Marek's disease virus, and bovine herpesvirus 1 have all been shown to localize to the nucleus of infected cells (2, 6, 23, 38).
Most studies on the subcellular localization of proteins in virus-infected cells have been carried out by immunofluorescence analysis of fixed cells. In contrast, our studies in recent years have involved the development of viruses expressing fluorescence-tagged structural proteins, enabling us to visualize the localization of these proteins in live cells and to monitor changes in their localization within the same cells as infection progresses. To this end, we have constructed HSV type 1 (HSV-1) recombinants in which individual major tegument proteins, namely, VP22 and VP13/14, have been tagged with green fluorescent protein (GFP) (4, 11). Both of these viruses replicate as efficiently as the parental virus and incorporate the fusion proteins into their structure at levels equivalent to those incorporated by the parental virus, suggesting that the addition of a GFP tag does not interfere with the behavior of these tegument proteins in infected cells. However, when these viruses were analyzed by live-cell microscopy, they produced quite different results with regard to the main cellular localization of the two proteins. VP22 was predominantly cytoplasmic throughout infection and exhibited localization and trafficking reminiscent of movement through the Golgi apparatus (11). In contrast, VP13/14 was efficiently targeted to the nucleus, localizing in a range of distinctive patterns as infection progressed, most notably in bright punctate dots (4). Although a small proportion of the protein could be seen in the cytoplasm, the exact behavior of this proportion of VP13/14 was difficult to discern. Considering that both of these proteins are packaged into the same compartment of the virion, we were intrigued by the contrasts in the subcellular targeting of the two tegument proteins.
To address the question of where in the cell VP22 and VP13/14 may colocalize, we have undertaken a detailed comparison of the localization and trafficking of these proteins in live infected cells. During these studies, we observed that while VP22 localized predominantly to the cytoplasm, it was also possible to detect a minor proportion of the protein in discrete nuclear dots. These dots were similar in appearance to the intense VP13/14-specific dots that have been described previously (4). We went on to construct a virus that expresses both cyan fluorescent protein (CFP)-VP22 and yellow fluorescent protein (YFP)-VP13/14 and showed that it is possible to simultaneously observe the two tegument proteins in live infected cells. The results obtained with this virus indicate that both VP22 and VP13/14 are targeted to the same nuclear dots, with VP22 localizing earlier and in smaller amounts than VP13/14. Furthermore, we showed that these tegument-specific domains are located adjacent to the nuclear dots formed early in infection at nuclear domain 10 (ND10) sites by the immediate-early protein ICP0. Hence, we have identified a novel tegument protein-containing compartment within the HSV-1-infected cell nucleus.
MATERIALS AND METHODS
Cells and virus infections.
Vero cells and BHK-1 cells were maintained in Dulbecco's modified minimal essential medium containing 10% newborn calf serum. The parental virus used in this study was HSV-1 strain 17. Virions and infectious virus DNA were purified from extracellular virus released into the infected cell medium as described previously (11).
Fluorescence-tagged viruses.
The construction of the GFP-VP22-expressing virus (166v) and the YFP-VP13/14-expressing virus (179v) was described previously (4, 11). The HSV-1 recombinant expressing CFP-VP22 was constructed in the same manner as the GFP-VP22-expressing virus. Briefly, a CFP-UL49 cassette contained on a BamHI fragment was inserted into the BamHI site of plasmid pGE120 (11) to produce plasmid pGE180, which consisted of CFP-UL49 surrounded by the UL49 flanking sequences and hence driven by the UL49 promoter. Equal amounts (2 μg) of plasmid pGE180 and infectious HSV-1 strain 17 DNA were then transfected into 106 COS-1 cells grown in a 60-mm dish by the calcium phosphate precipitation technique modified with N,N-bis(2-hydroxyl)-2-aminoethanesulfonic acid-buffered saline in place of HEPES-buffered saline. About 3,000 PFU of progeny viruses were then plated on Vero cells and screened for possible recombinants by CFP fluorescence. The resulting virus that was chosen for analysis was called 180v. The HSV-1 recombinant expressing capsid protein VP26 fused at its C terminus to YFP (based on strain SC16) was constructed in the same manner as the VP26-GFP-expressing virus described previously (3).
To construct viruses expressing both CFP- and YFP-tagged structural proteins, Vero cells were coinfected with the two singly-tagged fluorescent viruses of interest. The progeny viruses from these coinfections were then screened for the presence of both fluorescent proteins in individual plaques, and the resulting doubly-tagged viruses were plaque purified four times prior to characterization. In this way, viruses 180v and 179v were used in coinfections to produce the CFP-VP22- and YFP-VP13/14-expressing virus 181v, while virus 180v and the VP26-YFP-expressing virus were used in coinfections to produce the CFP-VP22- and VP26-YFP-expressing virus 183v.
Virus genomic DNA screening.
Virus DNA for restriction digestion was purified from 5 × 107 infected Vero cells as described previously (11) and digested for 8 h with appropriate enzymes in the presence of RNase A. Electrophoresis was carried out overnight with 0.8% agarose, and the gel was transferred to a nylon membrane by standard procedures. Southern blots were then hybridized with a 32P-labeled DNA probe synthesized by random priming of fragments specific for UL49, UL47, UL35, or GFP.
One-step growth curves.
Vero cells grown in a six-well plate (106 cells per well) were infected at a multiplicity of 10 in 1 ml of medium per well. After 1 h (taken as 1 h postinfection), the inoculum was removed, the cells were washed with phosphate-buffered saline (PBS), and 2 ml of fresh medium was added to each well. At 1, 4, 8, 12, 16, and 24 h postinfection, one well of infected cells was harvested for both extracellular virus from the cell medium and intracellular virus from the cells scraped into 1 ml of PBS. Each virus sample was then titrated with Vero cells.
SDS-PAGE and Western blot analysis.
Solubilized proteins were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and the gels were either stained with Coomassie blue or transferred to nitrocellulose filters and reacted with the appropriate primary antibody. A horseradish peroxidase-linked secondary conjugate was used, and reactive bands were visualized with enhanced chemiluminescence detection reagents (Amersham).
Antibodies.
Polyclonal anti-VP13/14 antibody R220 was kindly provided by David Meredith. Polyclonal anti-VP22 antibody AGV30 was described previously (10). Antibodies against ICP0 (11060) and VP16 (LP1) were kindly provided by Roger Everett and Tony Minson, respectively. A monoclonal anti-GFP antibody was obtained from Clontech.
Live-cell microscopy.
Cells for short-term live analysis were plated in coverslip chambers and imaged in one of two ways. For confocal microscopy, the 488 laser on a Zeiss LSM 410 inverted confocal microscope was used to generate z sections through the cell. Alternatively, images were acquired by using a Photometrics Quantix digital camera mounted on a Zeiss Axiovert S100 TV inverted microscope with filter sets specific for CFP and YFP (Chroma Technology Corporation, set 86002), and images were processed by using Metamorph software. The resulting images from both systems were processed by using Adobe Photoshop software.
Immunofluorescence analysis.
Cells were either fixed for 20 min in 4% paraformaldehyde followed by permeabilization for 10 min with 0.5% Triton X-100 or fixed in 100% methanol for 10 min. The fixed cells were blocked by incubation for 30 min in PBS containing 10% newborn calf serum, primary antibody in the same solution was added, and the mixture was incubated for 30 min. Following extensive washing in PBS, Texas red-conjugated anti-mouse immunoglobulin G (Vector Labs) was added in the blocking solution described above, and the mixture was incubated for a further 30 min. The coverslips were then washed extensively in PBS and mounted in Vectashield (Vector Labs).
RESULTS
A minor population of VP22 localizes to discrete nuclear dots.
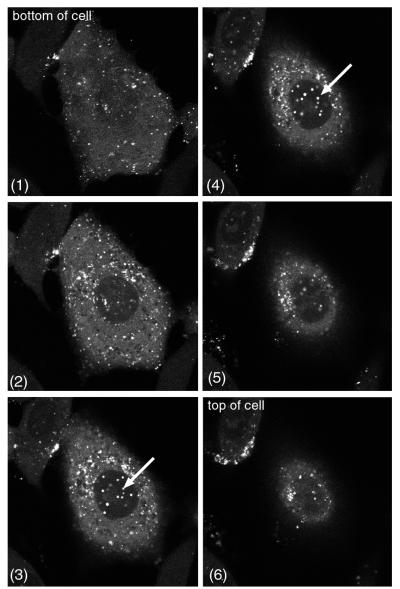
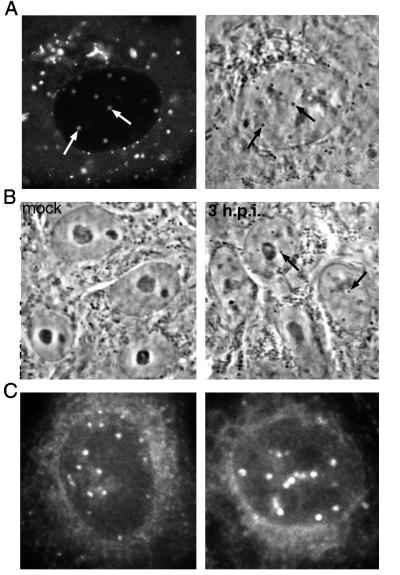
We previously examined the subcellular localization and trafficking of VP22 by using recombinant HSV-1 expressing GFP-VP22 and showed that the protein localizes predominantly to the cytoplasm of the cell. However, a more detailed examination of cells infected with this virus revealed that a small amount of GFP-VP22 was also present in discrete dots within the nucleus, a pattern that was often masked by the high concentration of punctate cytoplasmic GFP-VP22. To demonstrate the presence of these VP22-specific nuclear dots more clearly, Vero cells infected with the GFP-VP22-expressing virus (166v) were analyzed by confocal microscopy at about 8 h after infection, and z sections were collected through the depth of individual cells. Examples of such images are shown in Fig. 1, in which six z sections of a single cell are presented. As can be seen in slices 1 to 3, most of the GFP-VP22 fluorescence within the cell was contained within the cytoplasm. However, in slices 3 and 4, a number of fluorescent punctate dots were present within the nucleus (Fig. 1, arrows). When such cells were examined by bright-field microscopy, the GFP-VP22-specific fluorescent nuclear dots (Fig. 2A, left panel) were also visible as small phase-dense structures (Fig. 2A, right panel). Furthermore, these dense structures were apparent in infected cells as early as 2 h after infection and were easily detected by 3 h (Fig. 2B, compare 3 h.p.i. with mock). At 5 h, when GFP-VP22 localizes to these nuclear structures, bright-field microscopy showed that they were present in almost every nucleus (data not shown). To ensure that the localization of VP22 to these structures also occurred in the absence of a GFP tag, Vero cells infected with parental virus s17 were fixed 6 h after infection, and immunofluorescence analysis was carried out with anti-VP22 polyclonal antibody AGV30. As shown in Fig. 2C, similar VP22-specific dots were detected in a large number of infected cell nuclei, and it is likely that these immunoreactive dots are the same as those reported previously by Morrison and coworkers (30).
FIG. 1.
A minor population of GFP-VP22 localizes to nuclear dots in HSV-1-infected cells. Vero cells infected with GFP-VP22-expressing virus 166v at a multiplicity of 10 were examined by confocal microscopy at 8 h after infection. Six z sections through an individual live cell are shown. GFP-VP22-containing nuclear dots are indicated by arrows.
FIG. 2.
Nuclear dots containing GFP-VP22 are visible by bright-field microscopy. (A) The infected cells used in the experiment shown in Fig. 1 were examined by both fluorescent microscopy (left panel) and bright-field microscopy (right panel). Corresponding fluorescent and phase-dense dots are indicated by arrows. (B) Vero cells were infected with parental virus s17 at a multiplicity of 10 and examined live by bright-field microscopy. The newly formed phase-dense nuclear dots are indicated by arrows in the 3 h.p.i. image. h.p.i., hours postinfection. (C) Vero cells infected as decribed for panel B were fixed 6 h after infection, and immunofluorescence analysis was carried out with anti-VP22 polyclonal antibody AGV30.
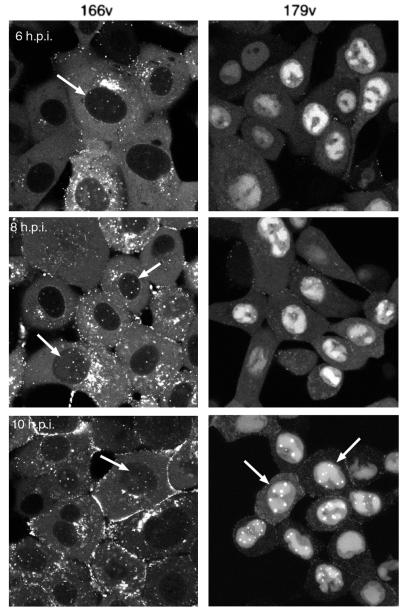
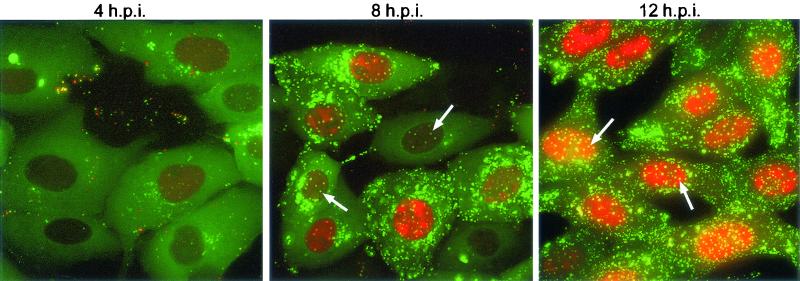
On making these observations, we were struck by the similarity between this nuclear VP22 pattern and the pattern previously observed for tegument protein VP13/14 (4). Hence, we decided to conduct a side-by-side comparison of cells infected with either the GFP-VP22-expressing virus (166v) or the YFP-VP13/14-expressing virus (179v). Confluent Vero cells were infected with 166v or 179v at a multiplicity of 10, and representative images were collected as single z sections by using confocal microscopy at 6, 8, and 10 h after infection. At 6 h, VP13/14 was accumulating in nuclear replication compartments (Fig. 3, 179v, 6 h.p.i.), whereas VP22 was present in cytoplasmic particles (Fig. 3, 166v, 6 h.p.i.). However, in addition to the intense cytoplasmic fluorescence of VP22, small nuclear dots were also present at this time in a number of 166v-infected cells (Fig. 3, 166v, 6 h.p.i., arrow) but were not apparent in 179v-infected cells (Fig. 3, 179v, 6 h.p.i.). Over the next 4 h, the VP22-specific dots became visible in a larger number of infected cells (Fig. 3, 166v, 8 and 10 h.p.i., arrows), with the number of dots per nucleus ranging from about 4 to 10. VP13/14-specific nuclear dots did not appear until 10 to 12 h after infection (Fig. 3, 179v, 10 h.p.i., arrows); hence, their appearance was delayed by up to 6 h in comparison with the appearance of the VP22-specific dots.
FIG. 3.
Comparison of GFP-VP22- and YFP-VP13/14-specific nuclear dots in infected cells. Vero cells were infected with either a GFP-VP22-expressing virus (166v) or a YFP-VP13/140-expressing virus (179v) at a multiplicity of 10. Representative images of live cells were collected by confocal microscopy at 6, 8, and 10 h postinfection (h.p.i.) and are presented as single z sections. Nuclei containing fluorescent dots are indicated by arrows.
HSV-1 expressing two fluorescence-tagged tegument proteins.
To determine whether VP22 and VP13/14 were localizing to the same nuclear domains, we decided to construct a virus in which both tegument proteins were tagged with different fluorescent proteins. To this end, we first constructed an HSV-1 recombinant in which VP22 was tagged at its N terminus with the cyan variant of GFP (CFP). This virus, named 180v, was produced in a manner identical to that used for the previously constructed GFP-VP22-expressing virus (11) and exhibited similar characteristics (data not shown). To produce a doubly-tagged virus, the CFP-VP22-expressing virus was used to coinfect Vero cells with HSV-1 recombinant 179v (4), in which VP13/14 is tagged at its N terminus with YFP. Approximately 1,000 plaques of the progeny virus were then plated on Vero cells and screened 4 days later by using a filter set specific for CFP and YFP. Plaques that were fluorescent for both CFP and YFP were picked and purified four times, and one of these viruses, named 181v, was chosen for further characterization. The genomic DNA from 181v was examined by Southern blotting to ensure that recombination had occurred correctly (data not shown).
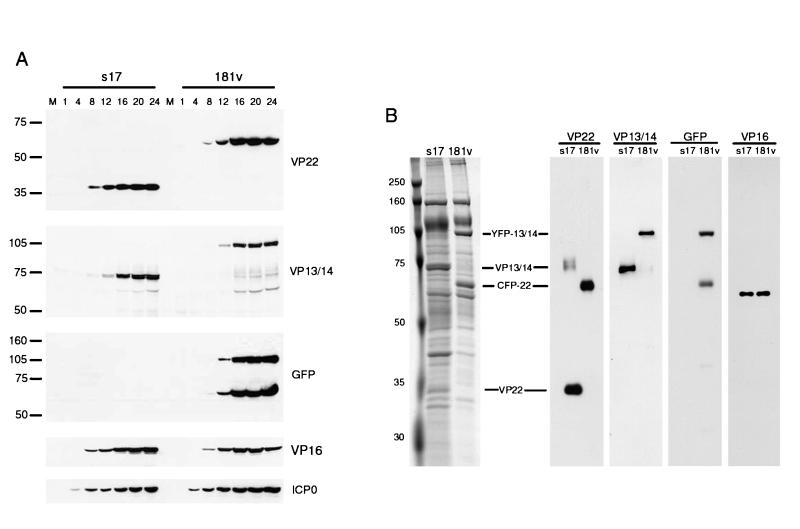
Although it was previously shown that the incorporation of a single GFP-tagged protein into the tegument of HSV-1 has little effect on virus replication or assembly and maturation, it was possible that the incorporation of two tagged proteins could affect the growth characteristics of the virus. Thus, to assess the replication efficiency of virus 181v, a high-multiplicity time course experiment was carried out. Individual monolayers of Vero cells were each infected at a multiplicity of 10 with either the wild-type parental virus (s17) or 181v, and at various times after infection, total cell lysates were prepared. The samples were analyzed by SDS-PAGE on 10% gels, followed by Western blotting with a range of antibodies (Fig. 4A). Blotting with antibodies against VP22 and VP13/14 demonstrated that the molecular masses of both proteins had increased by about 27 kDa in 181v-infected cells, confirming that both VP22 and VP13/14 were being expressed as fusion proteins (Fig. 4A, VP22 and VP13/14 panels). Furthermore, the new forms of VP22 and VP13/14 also reacted with an anti-GFP antibody (Fig. 4A, GFP panel). These fusion proteins were synthesized at similar rates in both s17- and 181v-infected cells, as were immediate-early protein ICP0 and another tegument protein, VP16 (Fig. 4A, ICP0 and VP16 panels). In addition, one-step growth curves for s17 and 181v demonstrated that the replication efficiencies of both viruses were similar (data not shown).
FIG. 4.
Characterization of an HSV-1 recombinant expressing CFP-VP22 and YFP-VP13/14. (A) Vero cells infected with either wild-type virus (s17) or the doubly-tagged virus (181v) at 10 PFU per cell were harvested every 4 h after infection up to 24 h. Equal amounts of total cell lysates were analyzed by SDS-PAGE followed by Western blotting with antibodies against VP22, VP13/14, GFP, VP16, and ICP0. Lane M, markers (in kilodaltons). (B) Purified wild-type (s17) and recombinant (181v) virions were solubilized and analyzed by SDS-PAGE followed by either Coomassie blue staining (left panel) or Western blotting with antibodies against VP22, VP13/14, GFP, and VP16 (right panels).
To ensure that virus 181v incorporated CFP-tagged VP22 and YFP-tagged VP13/14 into its structure in the same proportions as wild-type virus, we examined the components of purified extracellular virions from both viruses. Equivalent amounts of purified virions were analyzed by SDS-PAGE, followed by either staining with Coomassie blue or Western blotting (Fig. 4B). The results indicated that the 35-kDa VP22 species and the 75-kDa VP13/14 species were both absent from the total protein profile of 181v virions, in which two novel species of 65 and 105 kDa were easily distinguished (Fig. 4B, left panel). Western blotting for VP22, VP13/14, or GFP demonstrated that these two new virion proteins represented the CFP-VP22 and YFP-VP13/14 fusions (Fig. 4B, VP22, VP13/14, and GFP panels). Moreover, both fusions were incorporated into 181v virions in amounts equivalent to those in s17 virions, as judged by comparison with a blot for tegument protein VP16, which indicated that the virions were loaded equally (Fig. 4B, VP16 panel). Taken together, these results suggest that the tagging of both VP22 and VP13/14 in the same virus had little effect on virus replication.
Simultaneous observation of VP22 and VP13/14 in live infected cells.
To determine whether we could examine the subcellular localization of both VP22 and VP13/14 in the same live cells, we infected cells with 181v at a multiplicity of 10 and took representative images at various times after infection. At 2 to 3 h after infection, before YFP-VP13/14 was detectable, CFP-VP22 localized in a diffuse pattern within the cytoplasm of the infected cells (data not shown). As YFP-VP13/14 became visible in a diffuse nuclear pattern, CFP-VP22 began to accumulate in fluorescent particles often located close to the nucleus (Fig. 5, 4 h.p.i., VP22 in green, VP13/14 in red). This particulate cytoplasmic CFP-VP22 material increased in intensity over the next few hours, while YFP-VP13/14 began to accumulate within the replication compartments of the nucleus (Fig. 5, 8 h.p.i.). Of note at this time, however, was the presence of CFP-VP22 in faint nuclear dots (Fig. 5, 8 h.p.i., arrows). This small proportion of nuclear CFP-VP22 was specifically located in these punctate domains, with little CFP fluorescence detectable in the nucleoplasm or replication compartments. As infection progressed, the cytoplasmic CFP-VP22 material increased in intensity, such that the entire cell became highly fluoresecent (Fig. 5, 12 h.p.i.). At the same time, YFP-VP13/14 began to localize in nuclear dots positioned around the replication compartments (Fig. 5, 12 h.p.i., arrows). Thus, CFP-VP22 and YFP-VP13/14 expressed by the doubly-tagged virus localized in the same way as those expressed by the singly-tagged viruses. This report is the first description of a recombinant HSV expressing two fluorescence-tagged structural proteins.
FIG. 5.
Live-cell analysis of Vero cells infected with recombinant virus 181v. Vero cells grown in a coverslip chamber were infected with virus 181v at a multiplicity of 10 and were examined live at various times after infection (h.p.i., hours postinfection). At each time point, images were acquired by using a digital camera and filters specific for CFP and YFP. CFP-VP22 fluorescence is shown in green; YFP-VP13/14 fluorescence is shown in red. Nuclear dots are indicated by arrows.
VP22 and VP13/14 colocalize in nuclear dots.
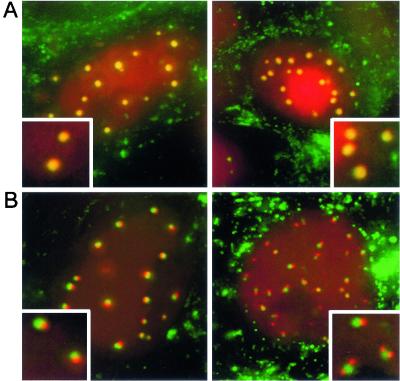
Our preliminary analysis of virus 181v suggested that VP22 and VP13/14 may colocalize in the same nuclear domains (Fig. 5, 12 h.p.i., arrows). To investigate this idea in more detail, we examined cells infected with virus 181v between 8 and 12 h after infection, the time when VP13/14 begins to localize in nuclear dots, and carried out detailed imaging of live cells. We found two slightly different relationships between the VP22-specific and the VP13/14-specific fluorescent dots. In the first of these, VP22 and VP13/14 indeed colocalized in nuclear domains (Fig. 6A). However, about half of the cells containing VP13/14-specific nuclear dots exhibited a slightly different localization, in which the VP13/14-specific dots were located adjacent to the VP22-specific dots, and in many cases two or three VP13/14-specific dots surrounded the VP22-specific dots (Fig. 6B, insets). We do not yet know the temporal relationship between these two patterns, but it is possible that the images shown in Fig. 6B are precursors to the images shown in Fig. 6A.
FIG. 6.
Relative localizations of VP22- and VP13/14-containing nuclear dots. Vero cells were infected with virus 181v expressing CFP-VP22 and YFP-VP13/14 at a multiplicity of 10. At 10 h after infection, the cell nuclei were examined for the presence of CFP-VP22- and YFP-VP13/14-containing dots. (A) In about half the nuclei, the CFP-VP22-specific dots colocalized with those of YFP-VP13/14. (B) In about half the nuclei, the YFP-VP13/14-specific dots were located beside the GFP-VP22-specific dots. Insets show magnified images of individual dots. CFP-VP22 is in green; YFP-VP13/14 is in red.
The tegument protein-containing nuclear domains are different from the VP26-containing nuclear domains.
A previous analysis of capsid protein VP26 expressed as a GFP fusion in the context of virus infection had revealed the presence of VP26 in punctate nuclear spots (3), which have been proposed to represent the nuclear sites of capsid assembly (43). In order to determine whether the tegument proteins were localizing to capsid protein-containing domains, we constructed a virus that expressed both VP22 and VP26 as fluorescence-tagged proteins. HSV-1 expressing VP26 tagged at its C terminus with YFP was used to coinfect Vero cells with CFP-VP22-expressing virus 180v, and recombinant virus expressing CFP-VP22 and VP26-YFP was isolated as described above for virus 181v. This virus, named 183v, was fully characterized in the same way as 181v prior to use (data not shown).
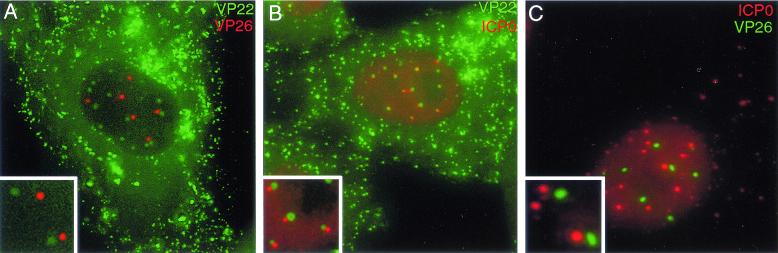
To examine the nuclear locations of both fluorescent proteins, Vero cells grown in a coverslip chamber were infected at a multiplicity of 10. At 6 to 8 h after infection, the cells were examined live, and representative images of infected cells were collected (Fig. 7A). At this stage of the infection, although the majority of VP22 was present in the cytoplasm, discrete nuclear dots of VP22 could also be seen (Fig. 7A, in green). The majority of VP26 expressed at this stage was also present in nuclear dots (Fig. 7A, in red), as observed previously (3). However, there was no colocalization or overlap between the VP26-specific dots and the VP22-specific dots, suggesting that the nuclear domains represented by these two proteins are different (Fig. 7A, inset).
FIG. 7.
Relative localizations of tegument protein-containing dots with other infected cell nuclear domains. (A) Vero cells were infected with virus 183v expressing CFP-VP22 and VP26-YFP at a multiplicity of 10. At 8 h after infection, the cells were examined live for the presence of CFP-VP22 (shown in green) and VP26-YFP (shown in red). (B) Vero cells were infected with virus 166v expressing GFP-VP22 at a multiplicity of 10. At 6 h after infection, the cells were fixed, and immunofluorescence analysis was carried out with an anti-ICP0 antibody. VP22 is shown in green; ICP0 is shown in red. (C) Vero cells were infected with an HSV-1 recombinant expressing VP26-YFP at a multiplicity of 10. At 6 h after infection, the cells were fixed, and immunofluorescence analysis was carried out with an anti-ICP0 antibody. VP26 is shown in green; ICP0 is shown in red. Insets show magnified images of individual nuclear domains.
The tegument protein-containing nuclear domains are located adjacent to the ICP0-containing domains.
We next compared the tegument protein-containing nuclear domains with those formed by immediate-early protein ICP0, which has been shown to localize to cellular ND10 sites early in infection (12). Vero cells infected with GFP-VP22-expressing virus 166v were fixed at 6 h after infection, and immunofluorescence analysis was carried out with a monoclonal antibody against ICP0. At this time, ICP0 was still nuclear in about half the cells, where it was present both in a diffuse nucleoplasmic background and in specific dots (Fig. 7B, ICP0 in red). These ICP0-specific domains did not colocalize with the VP22-specific domains. However, in many instances, the smaller, ICP0-specific dots were located exactly beside the larger, VP22-specific dots (Fig. 7B, inset), suggesting that there may be a distinct relationship between these two nuclear sites. Finally, we also examined the locations of VP26-specific dots and ICP0-specific dots. Cells infected with the VP26-YFP-expressing virus were fixed 6 h after infection and stained for ICP0. There was no colocalization between these two domains, although a number of the VP26-containing dots were located close to the ICP0-containing dots (Fig. 7C, inset). Hence, there are three distinct but closely associated punctate nuclear domains within HSV-1-infected cells—ICP0-containing dots, capsid protein-containing dots, and tegument protein-containing dots.
DISCUSSION
A number of herpesvirus-encoded proteins have been shown to localize to the infected cell nucleus. These include nonstructural proteins, such as ICP0 (12, 26) and ICP4 (19, 37), which are involved in gene expression, and structural proteins, such as ICP5 (33) and VP26 (3), which are assembled into the virus capsid within the nucleus. While such proteins have obvious and well-defined roles in the infected cell nucleus, the presence of tegument proteins in the nucleus is more difficult to interpret. Although tegument proteins are components of the virus particle and therefore must be assembled into the maturing virion, several if not all of them play dual roles in virus infection. For instance, major tegument protein VP16 has been shown to function as a transactivator of immediate-early gene expression in addition to its important structural role (25, 28, 32, 39). Hence, the previously demonstrated presence of newly synthesized VP16 in the nucleus during infection could be indicative of its role in gene expression, its site of assembly into the virion, or both.
In our laboratory, using a virus expressing major tegument protein VP13/14 fused to YFP, it was recently shown that VP13/14 is another virus-encoded protein that is efficiently targeted to the nucleus throughout infection (4). When in the nucleus, VP13/14 localizes initially to replication compartments and at later times to punctate domains usually located at the edges of these replication compartments. The significance of this characteristic intranuclear localization of VP13/14 is as yet unclear because, like VP16, the nuclear population of VP13/14 could reflect either the site of VP13/14 incorporation into the virion or an additional functional role of VP13/14 unrelated to virus assembly. In this report, we show that the characteristic VP13/14-containing punctate nuclear domains are also a cellular localization site for the tegument protein VP22 in HSV-1-infected cells. In earlier studies on cells infected with a virus expressing GFP-VP22, such nuclear VP22 was not initially obvious (11). This was primarily because the GFP-VP22-specific nuclear population represented only a very small fraction of the overall GFP-VP22 fluorescence within the cell and was easily masked by the high levels of cytoplasmic punctate material. However, here we used confocal microscopy to examine infected cells in more detail and show that while the majority of GFP-VP22 is located in the cytoplasm of infected cells, a small proportion of it localizes specifically to nuclear dots. Furthermore, we also constructed the first-described virus expressing two fluorescent proteins, namely, CFP-VP22 and YFP-VP13/14, to show that both tegument proteins localize to the same small compartments within the infected cell nucleus. Live-cell fluorescence indicated that while VP22 could be detected in these structures at about 5 h after a high-multiplicity infection, VP13/14 could not be detected in them until 10 to 12 h after infection, suggesting that they may play roles within these domains at different times. Nonetheless, it is clear that VP22 and VP13/14 share a common physical target within the nucleus. These nuclear dots may be the same as those defined previously by Morrison and coworkers, when they carried out immunofluorescence analysis of HSV-1-infected cells with an antibody specific for VP22 (30). Moreover, it was recently demonstrated that the PRV homologue of VP22 localizes to multiple punctate dots within the infected cell nucleus early in infection (2). While these nuclear dots represent the predominant localization of PRV VP22 and are more numerous than HSV-1 VP22-specific nuclear dots, the spatial organization of PRV VP22-specific dots outside of virus replication compartments may imply that they represent the same virus-induced domains (2).
It was previously shown that VP13/14 contains an efficient nuclear localization signal at its amino terminus that targets the protein to the nucleus even in the absence of other virus proteins (5). VP22 does not contain a recognizable nuclear localization signal and, when expressed in isolation from other virus proteins, tends to concentrate initially in the cytoplasm of expressing cells (8-10). Thus, when expressed during virus infection, it is likely that a small proportion of VP22 enters the nucleus by an indirect mechanism, such as interaction with a second protein, or by a virus-induced modification required for its nuclear import. It was also previously shown that VP13/14 has the ability to shuttle between the nucleus and the cytoplasm (5). More recently, a domain in the C terminus of VP22 that causes a heterologous protein to accumulate in the cytoplasm has been characterized in our laboratory, a feature that would also be consistent with a nuclear export signal in this region of VP22 (25). Hence, it is possible that both tegument proteins shuttle between the cytoplasm and nuclear dots in infected cells, with VP13/14 exhibiting a nuclear steady-state localization and VP22 exhibiting a cytoplasmic steady-state localization.
A number of characterized nuclear compartments, of both cellular and viral origins, exhibit a punctate appearance similar to that of VP22 and VP13/14. The tegument-containing nuclear dots described here appeared in the nucleus as new phase-dense structures at about 2 to 3 h after infection, several hours before GFP-VP22 was detectable in them, suggesting that they had been induced by other virus-encoded proteins. Infection with HSV-1 has been shown to result in a number of different nuclear punctate domains, including those formed early in infection by ICP0 when it localizes to cellular ND10 sites (12); dense bodies described previously as the nuclear sites of localization for virus proteins ICP22, UL3, and UL4 (16, 22, 24); and domains termed assemblons that are formed slightly later by capsid proteins (3, 43). Our studies with a doubly-tagged virus expressing CFP-VP22 and capsid protein VP26 tagged with YFP showed that the tegument protein and the capsid protein compartments were not related. However, immunofluorescence analysis of ICP0 in cells infected with the GFP-VP22-expressing virus indicated that while the tegument-containing domains did not colocalize with the ICP0-containing domains, in almost all instances the GFP-VP22-specific dots were located exactly beside the ICP0-containing domains, suggesting a relationship between these two sites. It was previously shown that incoming virus genomes are deposited adjacent to the ND10 sites to which ICP0 localizes (15). Furthermore, it has also been suggested that both virus transcription and DNA replication are initiated from these original deposition sites next to ND10 sites (27). Thus, it is possible that these sites of genome deposition are the precursors of the phase-dense domains that we observed in infected nuclei and that the targeting of both VP22 and VP13/14 to these sites may be related to their as-yet-undefined roles in virus replication. Interestingly, members of our laboratory have previously suggested that VP13/14 may play a role in the trafficking of viral RNA, based on both its ability to shuttle between the nucleus and the cytoplasm and the similarity of its nuclear targeting signal to that of the HIV-1 RNA binding protein Rev (5).
The presence of two tegument proteins in the same discrete domains within the nucleus raises the alternative possibility that these domains are involved in tegument assembly into the virion. While we cannot rule out an involvement of these nuclear domains in tegument assembly, the low levels of VP22 in these sites compared to the high levels of VP13/14 do not reflect the relative concentrations of the two proteins within the assembled virus, in which they are almost equivalent. In addition, for PRV-infected cells, it has been demonstrated that there is no detectable VP22 on virions located in the perinuclear space, suggesting that in this virus at least, VP22 is assembled into the tegument at a site downstream in the assembly pathway (13). With the recent characterization of a range of VP22 and VP13/14 mutants in transient expression assays (5, 25), it should now be possible to transfer the mutations to the HSV-1 genome and investigate the relationship between targeting to nuclear domains and incorporation into the virus tegument.
Acknowledgments
We thank Tony Minson for antibody LP1, Roger Everett for antibody 11060, and David Meredith for antibody R220.
I.H. and A.W. were funded by the Community Fund (previously the National Lotteries Charity Board). G.E. was funded by Marie Curie Cancer Care. H.B. was funded by the Welcome Trust, UK, and an MRC Cooperative Group award.
REFERENCES
- 1.Browne, H., S. Bell, T. Minson, and D. W. Wilson. 1996. An endoplasmic reticulum-retained herpes simplex virus glycoprotein H is absent from secreted virions: evidence for reenvelopement during egress. J. Virol. 70:4311-4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Desai, P., and S. Person. 1998. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J. Virol. 72:7563-7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnelly, M., and G. Elliott. 2001. Fluorescent tagging of herpes simplex virus tegument protein VP13/14 in virus infection. J. Virol. 75:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donnelly, M., and G. Elliott. 2001. Nuclear localization and shuttling of herpes simplex virus tegument protein VP13/14. J. Virol. 75:2566-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorange, F., S. El Mehdaoui, C. Pichon, P. Coursaget, and J. F. Vautherot. 2000. Marek's disease virus (MDV) homologues of herpes simplex virus type 1 UL49 (VP22) and UL48 (VP16) genes: high-level expression and characterization of MDV-1 VP22 and VP16. J. Gen. Virol. 81:2219-2230. [DOI] [PubMed] [Google Scholar]
- 7.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott, G., and P. O'Hare. 2000. Cytoplasm-to-nucleus translocation of a herpesvirus tegument protein during cell division. J. Virol. 74:2131-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., and P. O'Hare. 1998. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J. Virol. 72:6448-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, W., and B. Roizman. 1974. Proteins specified by herpes simplex virus. X. Staining and radiolabeling properties of B capsid and virion proteins in polyacrylamide gels. J. Virol. 13:155-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishov, A. M., and G. G. Maul. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahedi, S., N. S. Markovitz, F. Filatov, and B. Roizman. 1999. Colocalization of the herpes simplex virus type 1 UL4 protein with infected cell protein 22 in small, dense nuclear structures formed prior to onset of DNA synthesis. J. Virol. 73:5132-5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, F. E., C. A. Smibert, and J. R. Smiley. 1995. Mutational analysis of the herpes simplex virus virion host shutoff protein: evidence that vhs functions in the absence of other viral proteins. J. Virol. 69:4863-4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61:276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBoissiere, S., and P. O'Hare. 2000. Analysis of HCF, the cellular cofactor of VP16, in herpes simplex virus-infected cells. J. Virol. 74:99-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liang, X., B. Chow, Y. Li, C. Raggo, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markovitz, N. S., and B. Roizman. 2000. Small dense nuclear bodies are the site of localization of herpes simplex virus 1 U(L)3 and U(L)4 proteins and of ICP22 only when the latter protein is present. J. Virol. 74:523-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, A., P. O'Hare, J. McLauchlan, and G. Elliott. 2002. The herpes simplex virus tegument protein VP22 contains overlapping domains for cytoplasmic localization, microtubule interaction, and chromatin binding. J. Virol. 76:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 27.Maul, G. G., A. M. Ishov, and R. D. Everett. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67-75. [DOI] [PubMed] [Google Scholar]
- 28.McKnight, J. L., T. M. Kristie, and B. Roizman. 1987. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc. Natl. Acad. Sci. USA 84:7061-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison, E. E., A. J. Stevenson, Y. F. Wang, and D. M. Meredith. 1998. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J. Gen. Virol. 79:2517-2528. [DOI] [PubMed] [Google Scholar]
- 31.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro, D., I. Qadri, and L. Pereira. 1991. A mutation in the ectodomain of herpes simplex virus 1 glycoprotein B causes defective processing and retention in the endoplasmic reticulum. Virology 184:253-264. [DOI] [PubMed] [Google Scholar]
- 33.Nicholson, P., C. Addison, A. M. Cross, J. Kennard, V. G. Preston, and F. J. Rixon. 1994. Localization of the herpes simplex virus type 1 major capsid protein VP5 to the cell nucleus requires the abundant scaffolding protein VP22a. J. Gen. Virol. 75:1091-1099. [DOI] [PubMed] [Google Scholar]
- 34.Norrild, B., I. Virtanen, B. Pedersen, and L. Pereira. 1983. Requirements for transport of HSV-1 glycoproteins to the cell surface membrane of human fibroblasts and Vero cells. Arch. Virol. 77:155-166. [DOI] [PubMed] [Google Scholar]
- 35.Pellett, P. E., J. L. McKnight, F. J. Jenkins, and B. Roizman. 1985. Nucleotide sequence and predicted amino acid sequence of a protein encoded in a small herpes simplex virus DNA fragment capable of trans-inducing alpha genes. Proc. Natl. Acad. Sci. USA 82:5870-5874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pomeranz, L. E., and J. A. Blaho. 1999. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J. Virol. 73:6769-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 67:2163-2177. [DOI] [PubMed] [Google Scholar]
- 38.Ren, X., J. S. Harms, and G. A. Splitter. 2001. Bovine herpesvirus 1 tegument protein vp22 interacts with histones, and the carboxyl terminus of vp22 is required for nuclear localization. J. Virol. 75:8251-8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rixon, F. J., C. Addison, A. McGregor, S. J. MacNab, P. Nicholson, V. G. Preston, and J. D. Tatman. 1996. Multiple interactions control the intracellular localisation of the herpes simplex virus type 1 capsid proteins. J. Gen. Virol. 77:2251-2260. [DOI] [PubMed] [Google Scholar]
- 40.Skepper, J. N., A. Whiteley, H. Browne, and A. Minson. 2001. Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75:5697-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Genderen, I. L., R. Brandimarti, M. R. Torrisi, G. Campadelli, and G. van Meer. 1994. The phospholipid composition of extracellular herpes simplex virions differs from that of host cell nuclei. Virology 200:831-836. [DOI] [PubMed] [Google Scholar]
- 43.Ward, P. L., W. O. Ogle, and B. Roizman. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus type 1. J. Virol. 70:4623-4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle II. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. Effects of targeting herpes simplex virus type 1 glycoprotein D to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]