Abstract
V-ATPases acidify multiple organelles, and yeast mutants lacking V-ATPase activity exhibit a distinctive set of growth defects. To better understand the requirements for organelle acidification and the basis of these growth phenotypes, ∼4700 yeast deletion mutants were screened for growth defects at pH 7.5 in 60 mm CaCl2. In addition to 13 of 16 mutants lacking known V-ATPase subunits or assembly factors, 50 additional mutants were identified. Sixteen of these also grew poorly in nonfermentable carbon sources, like the known V-ATPase mutants, and were analyzed further. The cwh36Δ mutant exhibited the strongest phenotype; this mutation proved to disrupt a previously uncharacterized V-ATPase subunit. A small subset of the mutations implicated in vacuolar protein sorting, vps34Δ, vps15Δ, vps45Δ, and vps16Δ, caused both Vma− growth phenotypes and lower V-ATPase activity in isolated vacuoles, as did the shp1Δ mutation, implicated in both protein sorting and regulation of the Glc7p protein phosphatase. These proteins may regulate V-ATPase targeting and/or activity. Eight mutants showed a Vma− growth phenotype but no apparent defect in vacuolar acidification. Like V-ATPase-deficient mutants, most of these mutants rely on calcineurin for growth, particularly at high pH. A requirement for constitutive calcineurin activation may be the predominant physiological basis of the Vma− growth phenotype.
VACUOLAR proton-translocating ATPases (V-ATPases) acidify organelles such as the lysosome or vacuole, Golgi apparatus, and endosomes in all eukaryotic cells (Nishi and Forgac 2002). Compartment acidification by V-ATPases is important for receptor-ligand uncoupling in the biosynthetic and endocytic pathways, zymogen activation, and cellular pH homeostasis. The pH gradient generated by the V-ATPases can energize uptake of Ca2+, amino acids, and other ions and metabolites (Nelson and Harvey 1999; Nishi and Forgac 2002).
V-ATPases are highly conserved from fungi to plants and animals. They consist of a complex of peripheral subunits containing the ATP-binding sites, called the V1 sector, attached to a complex of membrane subunits containing the proton pore, called the Vo sector (Stevens and Forgac 1997). The yeast V-ATPase has emerged as the predominant model system for eukaryotic V-ATPases because it closely resembles the enzyme in higher eukaryotes but is more easily manipulated genetically and biochemically (Nelson and Klionsky 1996; Kane and Smardon 2003). The yeast genes for the subunits that copurify with enzyme activity have been identified. Eight subunits (designated A–H) form the V1 sector, and six (designated a, c, c′, c′′, d, and e) form the Vo sector (Stevens and Forgac 1997; Sambade and Kane 2004). Thirteen of the 14 subunits are encoded by single-copy vacuolar membrane ATPase (VMA) genes; the last subunit, Vo subunit a, is present as two organelle-specific isoforms encoded by the VPH1 and STV1 genes (Manolson et al. 1994). V-ATPase subunit genes have been identified through a variety of biochemical approaches (Nelson et al. 1989, 1994, 1995; Beltran et al. 1992; Supekova et al. 1995) and genetic screens (Preston et al. 1989; Ohya et al. 1991; Ho et al. 1993).
Although loss of V-ATPase activity in higher eukaryotes is lethal, disruption of any of the VMA genes in Saccharomyces cerevisiae results in a characteristic conditional lethal phenotype, the Vma− phenotype (Nelson and Nelson 1990; Ohya et al. 1991). vma mutants are able to grow in medium buffered to pH 5, but do not grow at pH 7.5 and/or in elevated extracellular calcium concentrations, at typical concentrations of nonfermentable carbon sources, or in the presence of a variety of heavy metals. Vph1p was identified through a screen for vacuolar acidification defects (Preston et al. 1989; Manolson et al. 1992). Deletion of VPH1, which encodes the vacuole-specific a-subunit isoform, results in a partial Vma− phenotype, but when a vph1Δ mutation is combined with deletions in STV1, the a-subunit isoform found in the Golgi and late endosome/prevacuolar compartment, the double-mutant cells exhibit a Vma− phenotype indistinguishable from that of the other vma mutants (Manolson et al. 1994). This result suggests that the Vma− phenotype arises from disruption of V-ATPase activity in multiple compartments. Although its physiological basis is only partially understood (Ohya et al. 1991; Munn and Riezman 1994; Plant et al. 1999), the Vma− phenotype has proven to be extremely useful in characterization of V-ATPase structure and function. It has been used extensively to identify subunit genes, to assess the extent of in vivo function of V-ATPase complexes with mutations in known subunit genes, and to uncover new proteins that help regulate V-ATPase function in vivo (Ohya et al. 1991; Oluwatosin and Kane 1998; Curtis et al. 2002). For example, mutations in the CYS4 gene, which encodes an enzyme required for cysteine (and glutathione) biosynthesis, also result in a Vma− phenotype, arising at least in part from the sensitivity of the V-ATPase catalytic subunit to redox conditions in the cytosol (Oluwatosin and Kane 1997). These results emphasize that screening for mutants that exhibit a Vma− phenotype can reveal not only structural genes for the V-ATPase, but also proteins important for ATPase assembly and regulation. Significantly, no genetic screen for vma mutants appears to have been saturating, because in each screen, previously unidentified mutants have been uncovered along with a subset of the existing vma mutants.
The advent of genomic approaches has made it possible to assess the complete set of genes necessary for V-ATPase activity and organelle acidification. As a step toward this goal, we have screened a collection of haploid yeast mutants with individual deletions in most of the nonessential genes for growth characteristics resembling the vma mutants.
MATERIALS AND METHODS
Media and strains:
All strains were from the collection of haploid deletion mutants developed by the Saccharomyces Genome Deletion Project (Winzeler et al. 1999) and obtained from Research Genetics (Birmingham, AL). These strains are based on the BY4741 wild-type strain (MATa, his3-Δ1 leu2-Δ0 ura3-Δ0 met15-Δ0) and were constructed by replacing the open reading frames of all nonessential genes of at least 100 codons with the kanMX marker. The collection was arrayed in a 384-well format and grown on unbuffered yeast extract-peptone-dextrose (YEPD) plates. For the initial screening, mutants were transferred to YEPD plates buffered to pH 5 and YEPD plates buffered to pH 7.5 containing 60 mm CaCl2, using a Virtek Versarray robot (Bio-Rad, Hercules, CA). Mutants that grew substantially better at pH 5 than at higher pH and calcium concentrations after 24–48 hr at 30° were identified. The screen was performed in duplicate and repeated twice, and mutants common to both screens were characterized further. For dilution assays, cells were grown in YEPD, pH 5 to midlog phase and then serially diluted into the wells of a microtiter dish before transfer to the appropriate medium using a manual pinning tool.
YEPD and yeast extract-peptone, 3% ethanol, 2% glycerol (YPEG) media were prepared as described (Sherman et al. 1982). YEPD was buffered to pH 5 or 7.5 with 50 mm sodium phosphate and 50 mm sodium succinate. YEPD, pH 7.5 + CaCl2 plates were buffered with 50 mm 2-(N-morpholino)ethane sulfonic acid (MES) and 50 mm 4-morpholinepropanesulfonic acid (MOPS) and contained 60 mm CaCl2. Cyclosporin A sensitivity was tested by diluting logarithmically growing cells into YEPD buffered to pH 5 or YEPD buffered to pH 6 and adding cyclosporin A (Calbiochem, San Diego) from an ethanol stock to a final concentration of 150 μg/ml. Control cells had an equivalent concentration of ethanol added. The cultures were then grown for 24 hr at 30° and the OD600 was measured. To test for suppression of the growth phenotypes by iron and copper, 100 μm FeCl2 and 10 μm CuCl2 were added to YEPD, pH 7.5, YEPD, pH 7.5 + CaCl2, or YPEG (Serrano et al. 2004).
Fluorescence microscopy:
Yeast cells were stained with quinacrine as described (Roberts et al. 1991). After incubation, cells were visualized within 10 min using a Zeiss Axioskop II and fluorescein fluorescence optics. Images were captured using a Hamamatsu CCD camera and analyzed using Adobe Photoshop 4.0.
The VMA2 gene was tagged at the C terminus with GFP as described in Longtine et al. (1998), except that longer (∼200 bp) flanking regions were added to the GFP-kanMX cassette using fusion PCR (Wach 1996). The resulting fusion product was transformed into wild-type cells using a modified lithium acetate transformation protocol (Ito et al. 1983), and transformants were selected on YEPD containing 200 μg/ml G418 and tested for production of a larger fusion protein detectable with the anti-B-subunit monoclonal antibody. The Vma2p-GFP-containing wild-type strain is able to grow on YEPD, pH 7.5 + 60 mm CaCl2, suggesting that the V-ATPase is functional in the presence of the GFP fusion. To localize Vma2p-GFP in the deletion strains, which are already G418-resistant, the kanMX marker was switched to the natMX marker as described by Tong et al. (2001). The GFP-tagged B subunit was then visualized by fluorescence microscopy as described above.
Isolation and biochemical characterization of vacuoles:
Vacuolar vesicles were isolated and ATPase activity in the vacuolar vesicles was assessed in the presence and absence of 100 nm concanamycin A (Wako Biochemicals, Richmond, VA) as described (Sambade and Kane 2004). Vacuolar vesicles and whole-cell lysates were prepared for immunoblotting as described (Kane et al. 1992) and V-ATPase subunits were detected with mouse monoclonal antibodies specific for the a, A, B, and C subunits (Kane et al. 1992) or with polyclonal antibody to the E subunit (a generous gift from Dan Klionsky, University of Michigan). Alkaline phosphatase (ALP) was detected by monoclonal antibody 1D3 obtained from Molecular Probes (Eugene, OR).
RESULTS
Growth of the haploid nonessential yeast deletion collection on YEPD plates buffered to pH 5.0 and on YEPD plates buffered to pH 7.5 containing 60 mm CaCl2 was compared. Strains from the library that grew well at pH 5.0 but failed to grow on pH 7.5, 60 mm CaCl2 plates in two independent platings were identified. Table 1 lists 64 strains that showed a growth defect at high pH and high calcium, but grew well at pH 5 in both of the duplicate screens. Where possible, the mutants were sorted by function on the basis of information from the Saccharomyces Genome Database.
TABLE 1.
Genes disrupted in mutants identified in initial library screen
| V-ATPase subunits and dedicated assembly factors |
VMA1*, VMA2*, VMA3*, VMA5*, VMA6*, VMA8*, VMA10*, VMA12*, VMA13*, VMA16*, VMA21*, VMA22*, VPH1* |
|---|---|
| Vacuolar protein sorting/vesicular transport | CLC1, SHP1, VPS15, VPS16, VPS34, VPS45, VPS54 |
| Amino acid biosynthesis/transport | ARO1, CYS4*, GLY1, ILV1, TAT1, TRP1, TRP2, TRP3, TRP4, TRP5 |
| Phosphate biosynthesis/metabolism | PHO2, PHO4, PHO81, PHO85 |
| Signal transduction | CAK1, CKB2, PTC1 |
| Transcription | ADA2, ADA3, CRZ1, RCS1, RPB4, SNF6, SNF5, SWI3, ZAP1 |
| Cell wall function | CWH36, KRE1, RMD7 |
| Other |
ANP1 (glycosylation), CTR1 (high-affinity copper transport), KEX2* (proteolytic processing), MAP1 (methionine aminopeptidase), RIB4 (vitamin B2 biosynthesis), RNR1 (purine and pyrimidine biosynthesis), TEF4 (translation) |
| Uncharacterized open reading frames |
YBL006c, YDR114c, YEL045c, YJL175w, YKL118w, YMR031w-a, YOR331c (dubious ORF opposite VMA4), YPL159c |
Mutant strains that grew on YEPD medium buffered to pH 5, but failed to grow on YEPD medium buffered to pH 7.5 containing 60 mM CaCl2, in two independent screens of the deletion mutant library are shown. The genes mutated in each strain are assigned a general function on the basis of information in the Saccharomyces Genome Database. Genes previously identified in screens for vma mutants are indicated by *.
The collection of mutants sensitive to high pH and high calcium concentrations included deletions in 13 of the 16 VMA genes identified previously as encoding subunits of the V-ATPase or dedicated assembly factors (Stevens and Forgac 1997). In addition, the kex2Δ and cys4Δ mutants, isolated in a previous screen for vma mutants (Oluwatosin and Kane 1997; Oluwatosin and Kane 1998), were again identified. Missing from our collection were mutants disrupted in the VMA4, VMA7, and VMA11 genes, which encode subunits of the V-ATPase. Another deletion mutant (yor331cΔ) that would remove most of the VMA4 open reading frame did show a strong Vma− phenotype. YOR331C is now designated as a dubious ORF and is on the strand opposite from VMA4.
Table 1 includes ∼50 mutants not previously associated with a Vma− defect. In addition to a severe growth defect at high pH and high calcium concentration, vma mutants are also sensitive to elevated pH without additional calcium and are unable to grow at typical concentrations of nonfermentable carbon sources (Ohya et al. 1991). To identify those mutants most similar to the known vma mutants, we tested the growth of this initial collection of mutant strains on medium buffered to pH 5, medium buffered pH 7.5 without added calcium (YEPD, pH 7.5), YEPD, pH 7.5 containing 60 mm CaCl2, and medium containing 3% ethanol and 2% glycerol (nonfermentable carbon sources) as the sole carbon sources. Growth of each strain on the different media was then scored, and the results are shown in Table 2. A number of the strains, for example, the entire collection of tryptophan biosynthesis (TRP) mutants and most of the phosphate metabolism (PHO) mutants, proved to have a calcium-specific growth defect; they failed to grow on pH 7.5 + CaCl2 medium, but were able to grow on pH 7.5 medium or on nonfermentable carbon sources. Because this suggests a weaker growth phenotype than that of the known vma mutants, we did not pursue them further. Sixteen new candidate vma mutants with a pattern of growth defects similar to that of the known vma mutants were selected. All of these strains showed little or no growth on pH 7.5 medium containing 60 mm CaCl2 and on glycerol/ethanol-containing medium in dilution assays (Table 2). Their growth at pH 5 and pH 7.5 is compared to that of the congenic wild-type strain and the vma3Δ mutant, which lacks Vo subunit c, in Figure 1.
TABLE 2.
Growth properties of potentialvma mutants
| Growth on
|
||||
|---|---|---|---|---|
| YEPD
| ||||
| Strain | pH 5 | pH 7.5 | pH 7.5 + 60 mm CaCl2 |
YPEG |
| Wild type (BY4741) | ++++ | +++ | + | +++ |
| vma3Δ | +++ | — | — | — |
| vma5Δ | +++ | — | — | — |
| clc1Δ | +++ | +/− | — | — |
| shp1Δ | +++ | + | — | — |
| vps34Δ | ++++ | + | — | — |
| vps45Δ | +++ | ++ | — | +/− |
| vps15Δ | ++++ | ++++ | — | + |
| vps16Δ | ++++ | ++++ | — | + |
| vps54Δ | ++++ | +++ | — | ++ |
| cys4Δ | ++++ | ++ | — | +++ |
| tat1Δ | ++++ | + | + | +++ |
| aro1Δ | ++++ | +++ | +/− | +++ |
| ilv1Δ | ++++ | ++++ | + | +++ |
| trp4Δ | ++++ | ++++ | — | +++ |
| trp5Δ | +++ | ++++ | — | +++ |
| trp1Δ | ++++ | ++++ | ++ | +++ |
| trp2Δ | ++++ | +++ | — | ++++ |
| trp3Δ | ++++ | +++ | + | ++++ |
| gly1Δ | ++++ | +++ | +/− | ++++ |
| pho85Δ | +++ | ++ | — | — |
| pho2Δ | ++++ | +++ | — | ++ |
| pho4Δ | ++++ | ++++ | — | +++ |
| pho81Δ | ++++ | ++++ | — | +++ |
| cak1Δ | ++++ | +++ | — | +++ |
| ptc1Δ | ++++ | +++ | — | +++ |
| ckb2Δ | ++++ | ++++ | — | ++++ |
| rcs1Δ | ++ | — | — | — |
| snf5Δ | ++++ | — | — | +/− |
| snf6Δ | +++ | — | — | + |
| rpb4Δ | +++ | + | — | ++ |
| ada3Δ | ++++ | +++ | — | ++ |
| ada2Δ | ++++ | ++++ | — | +++ |
| swi3Δ | ++++ | ++++ | — | +++ |
| crz1Δ | ++++ | ++++ | — | +++ |
| zap1Δ | ++++ | ++ | + | ++++ |
| cwh36Δ | +++ | — | — | — |
| rmd7Δ | +++ | ++ | — | — |
| kre1Δ | ++++ | +++ | — | +++ |
| kex2Δ | ++++ | — | — | — |
| ctr1Δ | +++ | ++ | — | — |
| anp1Δ | ++++ | ++++ | — | — |
| rib4Δ | +++ | — | — | +/− |
| rnr1Δ | +++ | +++ | — | +/− |
| ydr114cΔ | ++++ | ++++ | — | — |
| ybl006cΔ | ++++ | ++ | — | +++ |
| ypl159cΔ | ++++ | ++++ | +/− | +++ |
| yjl175wΔ | ++++ | ++++ | + | +++ |
| yel045cΔ | ++++ | ++ | + | ++++ |
| ymr031w-aΔ | ++++ | ++++ | + | ++++ |
All strains were grown overnight in liquid YEPD medium buffered to pH 5 and then diluted to an OD600 = 1.0 in YEPD, pH 5 medium. The diluted cultures were transferred to microtiter dishes and three serial 10-fold dilutions of each culture were made. Equal numbers of cells from each dilution were then transferred to plates containing the indicated media. Growth of the cells on the YEPD, pH 5 plates was scored after 48 hr at 30°, growth on the YEPD, pH 7.5 plates was scored after 72 hr at 30°, and growth on the other plates was scored after 96 hr. Strains are arranged by categories as shown in Table 1 and within categories, by the strength of the growth phenotype on YPEG (—, no growth to ++++, strong growth).
Figure 1.—
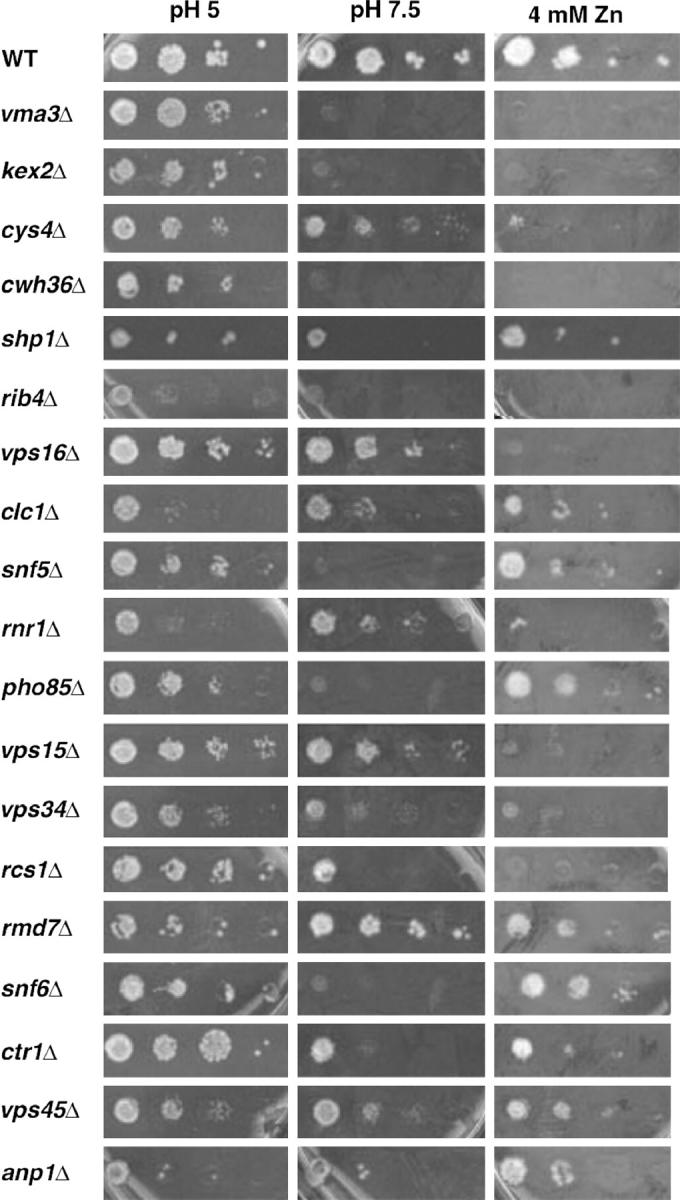
Growth of wild-type and mutant strains at pH 7.5 and in the presence of 4 mm ZnCl2. Logarithmically growing wild-type yeast strain BY4741 and the indicated kanMX deletion strains were transferred to YEPD plates buffered to pH 5, buffered to pH 7.5, or unbuffered but containing 4 mm ZnCl2. The undiluted culture is shown at the left along with 10-fold serial dilutions going left to right.
The growth of these strains on medium containing 4 mm ZnCl2 was also examined and the results are shown in Figure 1. Mutations that disrupt any of the “single-copy” VMA genes result in Zn2+ sensitivity, as shown in Figure 1 for the vma3Δ mutants. These genes encode subunits that are believed to be present in all V-ATPase complexes in the cell. Although disruption of both Vo a-subunit isoforms is necessary to obtain most Vma− phenotypes (Manolson et al. 1994), disruption of the vacuolar isoform (Vph1p) alone results in full sensitivity to 4 mm ZnCl2. This result has been attributed to the vacuole's major role in detoxification of heavy metals, and it has been argued that Zn2+ sensitivity may help to distinguish vma mutants with effects specific to the vacuole from those that affect V-ATPase function in multiple locations (Bachhawat et al. 1993). As shown in Figure 1, the candidate vma mutants selected after the second round of screening showed varied levels of sensitivity to 4 mm ZnCl2 that did not necessarily correlate with their sensitivity to growth at pH 7.5. The anp1Δ, rmd7Δ, pho85Δ, snf5Δ, snf6Δ, ctr1Δ, clc1Δ, vps45Δ, and shp1Δ mutants were relatively insensitive to added Zn2+, but the kex2Δ, cys4Δ, cwh36Δ, rib4Δ, vps16Δ, rnr1Δ, vps15Δ, vps34Δ, and rcs1Δ mutants were as sensitive as the vma3Δ mutants.
In a recent genomic screen for alkaline-dependent growth, Serrano et al. demonstrated that copper and iron become limiting factors for growth of wild-type cells at elevated pH and that a number of mutants that grew poorly at pH 7.2–7.5 could be rescued by either overexpressing iron and copper transporters or extracellular iron and/or copper supplementation (Serrano et al. 2004). We added 100 μm FeCl2 and 10 μm CuCl2 to YEPD, pH 7.5 medium and retested the mutant strains for growth. Consistent with the results of Serrano et al., the rcs1Δ mutant showed a pronounced improvement in growth. Only the vps34Δ and vps45Δ mutants also showed significant growth improvement, while ctr1Δ and ydr114cΔ mutants showed only a very slight improvement. The other strains in Figure 1 showed no change in growth in the presence of added iron and copper (data not shown). We also determined whether addition of the same concentrations of extracellular iron and copper would improve the growth of the strains on nonfermentable carbon sources (YPEG), because insufficient iron or copper might prevent respiration by preventing assembly of metal-containing complexes required for oxidative phosphorylation. Both the rcs1Δ and ctr1Δ strains showed greatly improved growth on YPEG supplemented with iron and copper, as did the shp1Δ strain. The vps15Δ, vps16Δ, and cys4Δ mutants also showed improved growth, but the improvement was not as pronounced, and the clc1Δ mutant showed only a very slight improvement. In those mutants that showed improved growth in the presence of added iron and copper, deficiency in these metals appears to be a major cause of their growth defect. Mutants that did not show improved growth may still have iron and copper deficiencies, but there may be other deficiencies as well, including an inability to take up, distribute, or utilize the additional extracellular metals.
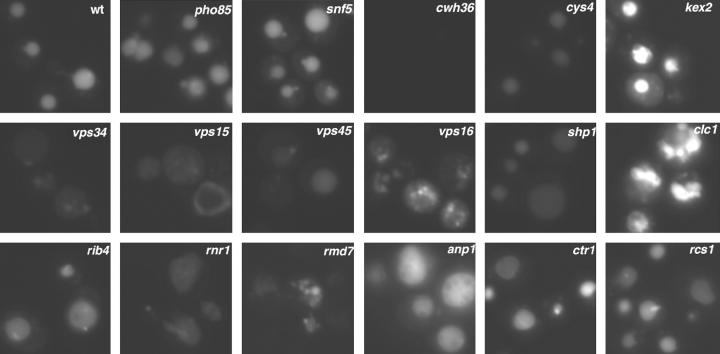
We next addressed whether the new candidate vma mutants had defects in vacuolar acidification, using a quinacrine uptake assay. Wild-type cells accumulate the fluorescent weak base quinacrine in the vacuole because it crosses membranes freely at near-neutral pH but becomes protonated and trapped in compartments of low pH. As shown in Figure 2, vacuoles of the wild-type strain were brightly stained with quinacrine. Mutant strains lacking any of the V-ATPase genes show no quinacrine staining (Yamashiro et al. 1990; data not shown). Only one of the candidate vma strains, the cwh36Δ mutant, completely lacked quinacrine staining, even after extended exposure times. There were, however, less dramatic staining defects among the other mutants. The vps45Δ, vps34Δ, vps15Δ, rnr1Δ, cys4Δ, and shp1Δ mutants all showed decreased quinacrine uptake relative to the wild-type strain. Staining in the rmd7Δ mutant was variable among different cells. The cys4Δ mutant was previously shown to be defective in quinacrine uptake (Oluwatosin and Kane 1997). The kex2Δ mutant was defective for quinacrine uptake in an earlier study (Oluwatosin and Kane 1998), but did not show a defect in this strain background. A number of the other strains showed aberrant cellular or vacuolar morphologies (shown in Figure 6). However, even in the vps16Δ and clc1Δ mutants, which have very highly fragmented vacuoles (Figure 2; Seeley et al. 2002), quinacrine uptake into membrane-bound compartments occurred. These experiments identify a subset of mutants with probable vacuolar acidification defects. They also suggest that quinacrine uptake occurs even under conditions where cellular or vacuolar morphology is disrupted and indicate that some of the mutants with a strong Vma− growth phenotype do not appear to have major defects in vacuolar acidification.
Figure 2.—
Quinacrine staining of wild-type and mutant strains. The indicated strains were stained with the lysosomotropic dye quinacrine and then visualized by fluorescence microscopy, as described in materials and methods. All images correspond to a 300-msec exposure. For the cwh36Δ mutant, there was no staining visible at any exposure, but cells containing vacuoles were first located under Nomarski optics and then visualized under the same conditions used for the other strains. Like the cwh36Δ mutant, mutants lacking V-ATPase subunits (vma3Δ and vma5Δ) were also visualized and showed no staining (data not shown).
Figure 6.—
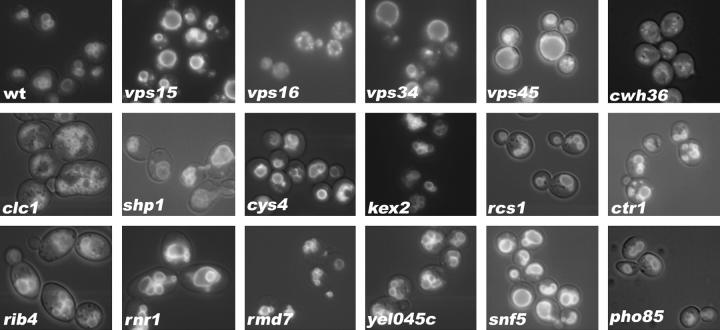
Distribution of Vma2p-GFP in wild-type and mutant strains. The V1 B subunit (Vma2p) of the V-ATPase was tagged at its C terminus with GFP and introduced into each strain as described in materials and methods. Distribution of the Vma2p-GFP fusion protein was visualized by fluorescence microscopy using an FITC filter set. To visualize the outline of the cells as well as the GFP tag, a low level of visible light was allowed to enter the microscope as the fields were photographed. Even though each strain contains a single GFP integrated at the VMA2 locus, there was considerable variability in the fluorescence intensities.
We next asked whether the mutants had another common feature that might lead to their common growth features with V-ATPase-deficient mutants despite their apparent differences in vacuolar acidification. The viability of the vma mutants depends on constitutive activation of calcineurin, the highly conserved Ca2+/calmodulin-dependent protein phosphatase that is activated by a number of conditions of ionic stress (Hemenway et al. 1995). As a result of this dependence, vma mutants are highly sensitive to cyclosporin A (CsA) and FK506, two inhibitors of calcineurin activity (Iida et al. 1990; Hemenway et al. 1995; Tanida et al. 1995), and mutations in the V-ATPase and calcineurin subunit genes are lethal when combined (Garrett-Engele et al. 1995; Parsons et al. 2004). Tanida et al. (1995) showed that the sensitivity of the vma mutants to CsA was dependent to some degree on the pH of the medium; at pH 5, some growth occurred at a CsA concentration that prevented growth at pH 6. We reasoned that the pattern of growth defects in the candidate vma mutants might be characteristic of mutants that are dependent on constitutive activation of calcineurin. Figure 3 compares the overnight growth of wild-type and candidate vma mutants in the presence and absence of 150 μg/ml CsA in medium buffered to pH 5 or pH 6. As reported previously, the growth of the wild-type cells was relatively insensitive to CsA at either pH, but the growth of the vma3Δ strain was highly sensitive to the drug, with slightly more inhibition at pH 6 than at pH 5 (Figure 3, bottom). A number of the candidate vma mutants, including kex2Δ, cys4Δ, cwh36Δ, shp1Δ, rib4Δ, vps16Δ, and clc1Δ, showed a similar pattern of sensitivity, although there was some variation in the absolute level of inhibition. Another set of mutants, including rnr1Δ, pho85Δ, vps15Δ, vps34Δ, rcs1Δ, and ydr114cΔ, was quite sensitive to CsA at pH 6, but relatively insensitive at pH 5. Similar behavior was observed in earlier studies for the vph1Δ mutant, which does not eliminate all cellular V-ATPase activity (Tanida et al. 1995). The remaining mutants (rmd7Δ, snf6Δ, ctr1Δ, vps45Δ, anp1Δ, and yel045cΔ) are largely insensitive to CsA inhibition. These results indicate that at least some of the mutants that have little if any defect in quinacrine uptake, such as kex2Δ and snf5Δ, still rely on constitutive activation of calcineurin for growth.
Figure 3.—
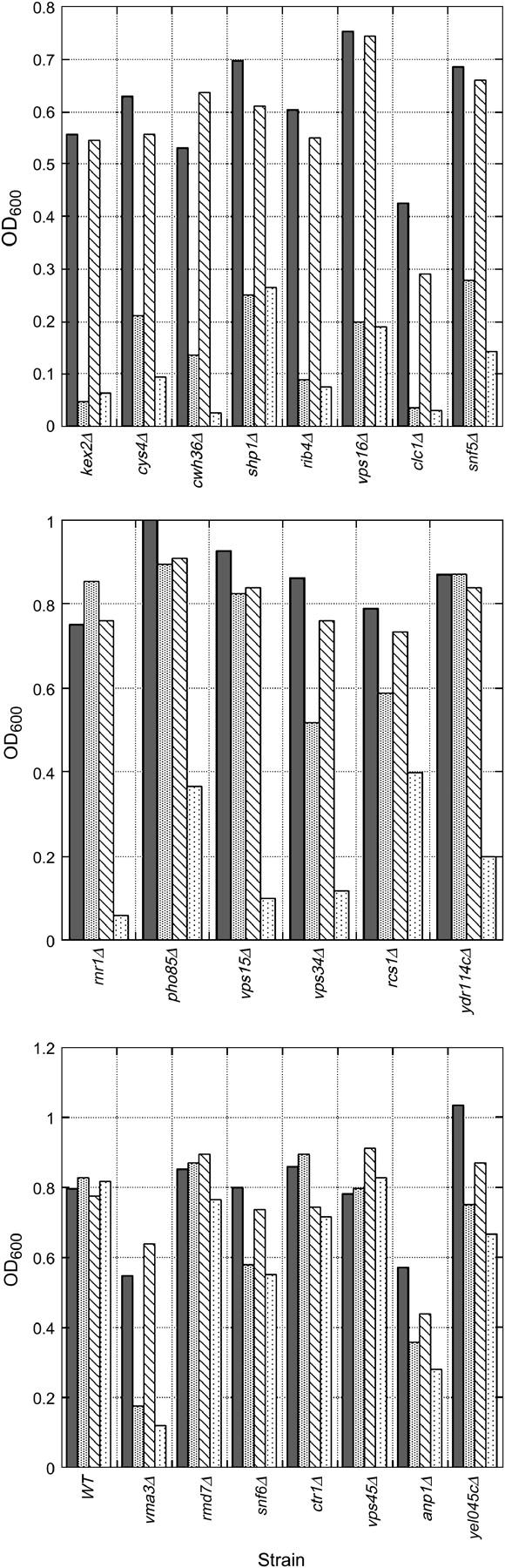
pH-dependent cyclosporin sensitivity in wild-type and mutant strains. The indicated strains were grown to midlog phase and then diluted into YEPD buffered to either pH 5 or pH 6. An ethanol stock of cyclosporin A (final concentration 150 μg/ml) was then added to half of the cells at each pH and an equivalent amount of ethanol was added to the other half as a control. The cells were incubated at 30° with shaking for 24 hr and then the culture density (OD600) was measured for each. For each strain: solid bar, YEPD, pH 5 without cyclosporin A; shaded bar, YEPD, pH 5 with cyclosporin A; hatched bar, YEPD, pH 6 without cyclosporin A; and stippled bar, YEPD, pH 6 with cyclosporin A. Top, strains showing acute sensitivity to cyclosporin A at both pH 5 and pH 6. Middle, strains showing greater sensitivity to cyclosporin A at pH 6 than at pH 5. Bottom, strains that are relatively insensitive to cyclosporin A at pH 5 or 6. The growth of congenic wild type and a strain lacking a known V-ATPase subunit (vma3Δ) are also included in the bottom for comparison.
We addressed whether several of the candidate mutants affected vacuolar H+-ATPase activity biochemically, by isolating vacuolar vesicles and assaying concanamycin A-sensitive ATPase activity. (Concanamycin A is a potent and specific inhibitor of V-ATPases from all eukaryotic species; Drose et al. 1993). Table 3 shows that vacuoles from the cwh36Δ and vps16Δ cells had the lowest levels of ATPase activity. The vps15Δ, vps34Δ, vps45Δ, shp1Δ, clc1Δ, and rmd7Δ mutant vacuoles also had <30% of the V-ATPase activity of the wild-type strain. Decreases seen in the rnr1Δ, yel045cΔ, and vps45Δ strains were smaller, and activities in vacuoles isolated from the pho85Δ, snf5Δ, snf6Δ, and anp1Δ mutants were >50% of wild type.
TABLE 3.
V-ATPase activity, cyclosporin A sensitivity, and quinacrine staining of candidatevma mutants
| Strain | % wild type ATPase activity |
CsA sensitive? | Quinacrine |
|---|---|---|---|
| Wild type | 100 | −pH 5/−pH 6 | ++ |
| vma3Δ | 5a | +pH 5/+pH 6 | − |
| kex2Δ | 103b | +pH 5/+pH 6 | ++ |
| rib4Δ | ND | +pH 5/+pH 6 | ++ |
| pho85Δ | 85 (1) | −pH 5/+pH 6 | ++ |
| snf5Δ | 80 (1) | +pH 5/+pH 6 | ++ |
| snf6Δ | 60 (1) | −pH 5/−pH 6 | ++ |
| rcs1Δ | ND | −pH 5/+pH 6 | ++ |
| anp1Δ | 90 (1) | −pH 5/−pH 6 | ++ |
| ctr1Δ | ND | −pH 5/−pH 6 | ++ |
| cwh36Δ | 5 ± 2 (2) | +pH 5/+pH 6 | − |
| vps34Δ | 17 ± 7 (2) | −pH 5/+pH 6 | + |
| vps15Δ | 15 ± 5 (2) | −pH 5/+pH 6 | + |
| vps45Δ | 32 ± 10 (2) | −pH 5/−pH 6 | + |
| vps16Δ | 1 ± 1 (2) | +pH 5/+pH 6 | ++?d |
| shp1Δ | 28 ± 9 (2) | +pH 5/+pH 6 | + |
| clc1Δ | 23 ± 12 (2) | +pH 5/+pH 6 | ++?d |
| rnr1Δ | 35 ± (2) | −pH 5/+pH 6 | + |
| rmd7Δ | 28 ± 3 (2) | −pH 5/−pH 6 | +?d |
| cys4Δ | 47c | +pH 5/ +pH 6 | + |
Comparison of V-ATPase activity is shown in isolated vacuolar membranes with quinacrine staining and cyclosporin A sensitivity. Vacuolar membranes were isolated from the BY4741 wild-type strain and from the indicated mutant strains, and ATPase activity sensitive to 100 nm concanamycin A was determined as described in materials and methods. The wild-type vacuolar vesicles had an average concanamycin-sensitive specific activity of 2.0 ± μmol Pi/min/mg protein when assayed at 37° (average of n = 7 independent vacuole isolations ±SEM). Activities from the mutant vacuole preparations are expressed as a percentage of the wild-type activity ± the range of duplicate assays where indicated. CsA sensitivity and quinacrine staining are derived from Figures 3 and 2, respectively.
From Umemoto et al. (1990).
These strains show either such perturbed vacuolar morphology (vps16Δ and clc1Δ) or a variability in staining among different cells (rmd7Δ) that assessment of quinacrine staining is difficult.
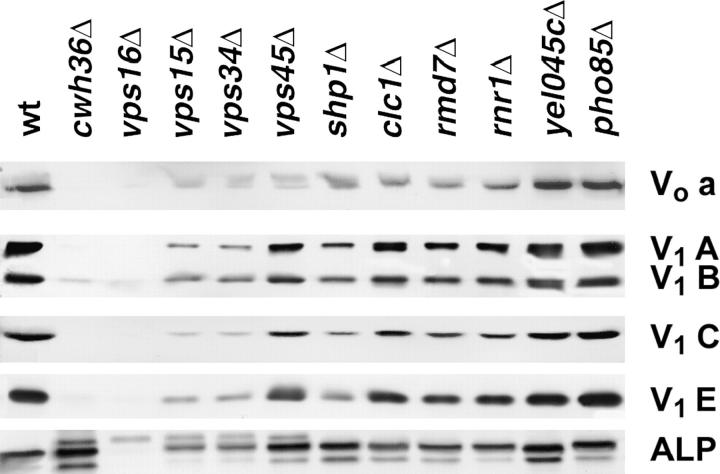
Levels of several V-ATPase subunits were assessed by Western blotting of vacuolar membranes using subunit-specific antibodies against V1 subunits A, B, C, and E and Vo subunit a. Figure 4 shows that there were almost no V-ATPase subunits in membranes isolated from the cwh36Δ and vps16Δ mutants, consistent with the very low V-ATPase activity. Levels of Vo subunit a were reduced in the vps15Δ, vps34Δ, and vps45Δ mutants, suggesting that the membrane sector of the V-ATPase was present at lower levels in these mutants. Loss of the membrane sector would remove sites for binding of the peripheral V1 sector, which contains sites for ATP hydrolysis and thus could account in part for the reduced ATPase activity. However, the vps15Δ and vps34Δ mutants have lower levels of all four V1 subunits than does vps45Δ, indicating that these two mutations may have an additional effect on binding of the V1 sector to Vo that does not occur from the vps45Δ mutation. The shp1Δ mutant has more of the Vo a subunit than the vps45Δ mutant vacuoles, but less of V1 subunits A, B, C, and E. This mutation may exert its primary effect on attachment of the peripheral V1 subunits to the membrane subunits. The clc1Δ, rmd7Δ, and rnr1Δ mutant vacuoles show some reduction in all of the subunits relative to wild-type vacuoles, but it is not clear whether this reduction is sufficient to account for the reduction in V-ATPase activity that is seen in these mutants. The pho85Δ and yel045cΔ mutant vacuoles both had near wild-type ATPase activity and approximately the same levels of all of the ATPase subunits as the wild-type strain.
Figure 4.—
Levels of V-ATPase subunits and alkaline phosphatase in vacuolar vesicles isolated from wild-type and mutant strains. Vacuolar vesicles were prepared from the indicated strains, and vesicles containing equivalent amounts of protein were solubilized and separated by SDS-polyacrylamide gel electrophoresis. Immunoblots were prepared and probed as described in materials and methods. V-ATPase subunit a (Vph1p) is part of the Vo (membrane) sector of the enzyme, and V-ATPase subunits A (Vma1p), B (Vma2p), C (Vma5p), and E (Vma4p) are part of the V1 (peripheral) sector. Alkaline phosphatase (ALP) is a vacuolar membrane protease.
Lower levels of V-ATPase subunits in the vacuolar membranes could arise from general vacuolar targeting defects or contamination of the vacuoles with other membranes. To help address this issue, we probed a Western blot of the vacuolar membranes with antibodies to another membrane-bound vacuolar enzyme, alkaline phosphatase (ALP; Klionsky and Emr 1989). Significantly, all of the mutant vacuolar membranes except those from vps16Δ contained near wild-type levels of ALP, although there was some variation in the amount of unprocessed ALP, which has a slightly lower mobility than the doublet corresponding to mature, proteolytically processed ALP (Klionsky and Emr 1989). From these results, it would appear that contamination of the vacuoles does not account for the differences in V-ATPase subunits or activity. This suggests that there is a relatively specific loss of the V-ATPase from the vacuolar membranes of all of the mutants except vps16Δ. The vps16Δ mutant was previously shown to have drastically disrupted vacuolar structure (Peterson and Emr 2001), and the fragmentation of the vacuoles in this mutant (Figure 2) may preclude vacuolar isolation by our typical protocol.
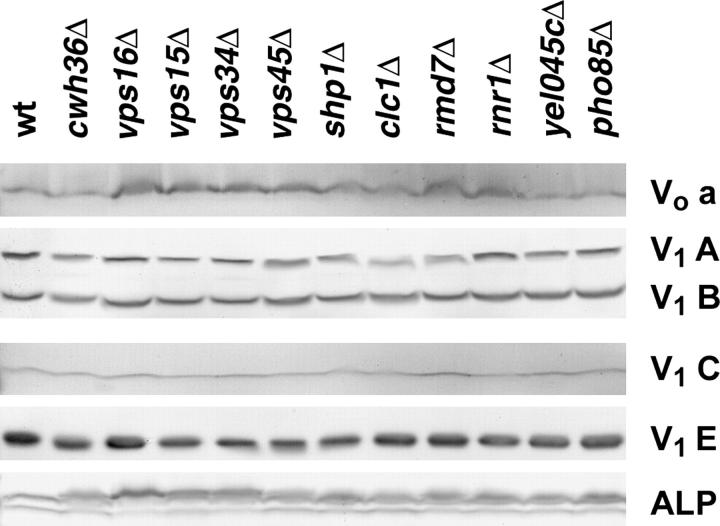
In most mutants tested to date, the cellular levels of V1 subunits were not altered even when they were not able to assemble with other subunits or the vacuolar membrane, but some of the Vo subunits became unstable in the absence of Vo assembly (Kane et al. 1992; Tomashek et al. 1997). We prepared whole-cell lysates from the mutant strains used for vacuole preparation and examined the cellular levels of the Vo a subunit and V1 A, B, C, and E subunits by Western blotting (Figure 5). In contrast to the vacuolar membranes, there was little if any difference in the cellular levels of any of the V-ATPase subunits or the control protein, ALP, even in the cwh36Δ and vma16Δ mutants, where none of the subunits was at the vacuole. This result indicates that the Vma− phenotype in these mutants does not arise from defects in synthesis or stability of these V-ATPase subunits.
Figure 5.—
Levels of V-ATPase subunits in whole-cell lysates isolated from wild-type and mutant yeast strains. Whole-cell lysates were prepared from the indicated strains as described in materials and methods and an equal amount of lysate protein was loaded in each lane. Whole-cell lysates were separated by SDS-polyacrylamide gel electrophoresis, the separated proteins were transferred to nitrocellulose, and immunoblots were probed as described in Figure 4.
As an alternate means for assessing the cellular distribution of the V-ATPase, we expressed a functional GFP-tagged version of the V-ATPase B subunit in each of the mutant strains (Figure 6). The B subunit is a peripheral membrane protein that is found in the cytosol under conditions that prevent V-ATPase assembly or stimulate disassembly (Kane and Stevens 1992; Kane 1995). In wild-type cells, the GFP signal is tightly localized around several vacuoles in most cells (Figure 6), and in a mutant lacking an intact Vo sector, the GFP signal is cytoplasmic (data not shown), consistent with results from immunofluorescence microscopy with anti-B-subunit antibody (Kane et al. 1992). As reported previously for a number of the strains (Raymond et al. 1992; Seeley et al. 2002), it was immediately clear that vacuolar morphology is perturbed in certain strains, including vps16Δ, clc1Δ, shp1Δ, and rnr1Δ. However, it is also notable that in most of the strains, with the possible exception of cwh36Δ, the GFP signal is predominantly membrane bound, indicating that some level of V-ATPase assembly was occurring in the mutants. The staining appeared to be concentrated in a spot adjacent to the vacuole in some cells of the vps34Δ mutant, and the ctr1Δ mutant showed evidence of a similar pattern, which could correspond to concentration of the V-ATPase in the prevacuolar compartment. In addition, even though the tagged subunits are all integrated into the genome and expressed from the endogenous promoter, there were variations in the intensity of staining, with some mutants, such as pho85Δ, cwh36Δ, and rcs1Δ, staining very poorly. This variability made it difficult to assess whether some of the mutants had higher levels of cytosolic signal than others.
DISCUSSION
The major results of the screen can be summarized as follows and are discussed in more detail below.
We identified a new V-ATPase subunit, Vma9p, via the phenotype of the cwh36Δ mutant described here (Sambade and Kane 2004). The discovery of a new V-ATPase subunit gene was surprising because there was no previous biochemical evidence for this subunit in yeast, but the results support the specificity of this screen for V-ATPase defects and the power of the genomics approach.
Because the cwh36Δ mutant was the only mutant that exhibited such a strong phenotype, it is tempting to suggest that all of the yeast V-ATPase subunits have now been identified. (However, VMA9 itself was neither deleted in the library nor annotated originally as an ORF, so other small subunits could be present.)
A relatively small set of other gene products exhibits the characteristic Vma− phenotype when deleted (summarized in Table 3). Among these gene products is a small subset of the gene products implicated in vacuolar protein sorting (Bonangelino et al. 2002a). It was known that some of the vps mutants have defects in vacuolar acidification (Rothman et al. 1989; Raymond et al. 1992), but it is now clear that the vast majority of mutations that result in missorting of soluble vacuolar proteins do not sufficiently impair organelle acidification and/or V-ATPase activity to generate a Vma− phenotype. Conversely, the vps mutants that do have a relatively strong Vma− phenotype may have more direct effects on the V-ATPase.
Some mutants display a strong Vma− phenotype with no obvious acidification defect. These mutants have in common with the V-ATPase mutants a dependence on calcineurin for growth; this dependence may be the predominant physiological basis of the Vma− growth defects.
New mutants that affect V-ATPase activity:
Mutants lacking genes directly responsible for V-ATPase assembly and structure represent, as expected, a large fraction of the mutations that exhibit a strong Vma− phenotype. Thirteen of the 16 previously identified subunits and dedicated assembly factors are among the deletion mutants showing the strongest Vma− phenotype in Tables 1 and 2, and the other 3 known vma mutants were not represented in the deletion library (data not shown). Only the cwh36Δ mutant showed a phenotype identical to that of the V-ATPase subunit mutations. The predicted open reading frame for CWH36 is designated as a dubious ORF (Saccharomyces Genome Database), but we have now shown that the cwh36Δ mutation also deletes most of an unannotated V-ATPase subunit gene, VMA9, on the opposite strand, and deletion of VMA9 can account for the full range of phenotypes shown here (Davis-Kaplan et al. 2004; Sambade and Kane 2004).
Several other mutants mimic the growth phenotypes, pH-dependent sensitivity to calcineurin inhibitors, and quinacrine uptake defect of the vma mutants, but are not as severely affected in one or more of these characteristics as the vma mutants. These mutations may affect regulators of the V-ATPase. The majority have been implicated in protein trafficking. Although it might be argued that any vacuolar protein sorting (vps) mutant might affect V-ATPase activity by improperly targeting the enzyme to the vacuole, in fact, most of the vps mutants proved to have relatively little effect on vacuolar acidification or V-ATPase activity (Rothman et al. 1989; Preston et al. 1992; Raymond et al. 1992). Consistent with this, only 6 of the ∼150 mutants identified to date as having vacuolar protein sorting defects (Raymond et al. 1992; Bonangelino et al. 2002a) exhibited strong Vma− growth phenotypes in this genomic screen. Of the four vps mutants, the vps34Δ and vps15Δ mutants may be the most interesting. The vps15Δ, vps34Δ, and vps45Δ mutants were previously determined to have reduced vacuolar acidification (Rothman et al. 1989; Raymond et al. 1992). Vps34p is a phosphatidylinositol 3-kinase (PI 3-kinase) localized to the prevacuolar compartment by association with the serine/threonine protein kinase Vps15p (Schu et al. 1993; Stack et al. 1995). The product of Vps34p, the phosphatidyl inositol phospholipid PI3P, is enriched in endosomes, internal vesicles of multivesicular bodies, and the outer limiting membrane of the vacuole (Gillooly et al. 2000). At the vacuole, some of the PI3P in the limiting membrane is further processed by a PI-3P 5-kinase (Fab1p) (Gary et al. 1998). Interestingly, mutants lacking Fab1p, or its associated modulator, Vac14, have been reported to lack vacuolar acidification (Bonangelino et al. 2002b), but these mutants do not exhibit a Vma− growth phenotype (data not shown). Vacuolar protein sorting is also unaffected in a fab1Δ mutant (Wurmser et al. 1999). One possibility consistent with all of these results is that V-ATPase activity is influenced by certain inositol phospholipids, either by direct interaction or by recruitment of modulator proteins that affect V-ATPase activity. The vps34Δ and vps15Δ mutants would exhibit a Vma− phenotype because they compromise V-ATPase activity in both prevacuolar compartments and the vacuole itself, while the fab1Δ and vac14Δ mutants affect only the vacuole and thus allow function of the V-ATPase in earlier compartments. Alternatively, vps34Δ and vps15Δ may result in a stronger phenotype because they impact the V-ATPase at multiple levels: both in sorting to the vacuole and in the activity of the enzyme itself. Two other recent investigations have also implicated phospholipids in control of V-ATPase activity (Chung et al. 2003; Crider and Xie 2003), and another has suggested a link between PI3-kinase activity and glucose regulation of V-ATPase activity (Sautin et al. 2005). Vps45p binds to the PI3P-binding protein Vac1p and has been proposed to integrate signals coming from Vps34p/15p complex and Rab proteins at the Golgi-vacuole transport step (Peterson et al. 1999).
The vps16Δ mutant fails to form a coherent vacuolar structure and affects multiple steps of proton sorting (Peterson and Emr 2001). Even though we were not successful in isolating V-ATPase-containing vacuoles from the vps16Δ mutant, both the level of quinacrine staining and localization of Vma2-GFP to membrane structures argue that the V-ATPase is less affected in this mutant than in the other vps mutants isolated in the screen.
The shp1Δ and clc1Δ mutants may also affect protein trafficking (Bonangelino et al. 2002a). However, the effects of shp1Δ mutation on the V-ATPase are not easily dismissed as resulting solely from mistargeting. The shp1Δ vacuoles show a greater reduction of the peripheral V1 subunits than of the Vo subunit (Figure 4), suggesting that trafficking of the vacuolar membrane subunits is less affected than attachment of the V1 sector to the Vo sector. Shp1p is homologous to the mammalian p47 protein, which acts as an adaptor for the p97 ATPase in multiple membrane fusion events (Kondo et al. 1997), but in yeast it was initially shown to act as a regulator of the Glc7 protein phosphatase to affect glycogen accumulation and normal cell growth (Zhang et al. 1995). V-ATPase activity is regulated by glucose metabolism at the level of assembly of the V1 and Vo sectors (Kane 1995; Parra and Kane 1998), so the connection of Shp1p to both V-ATPase assembly and glucose storage is intriguing. In addition, it is interesting that Pho85p, also isolated in our screen, regulates Glc7p phosphatase activity as well (Tan et al. 2003). Glc7p, encoded by an essential gene, has been implicated in ion homeostasis (Williams-Hart et al. 2002) and vacuole-vacuole fusion (Peters et al. 1999). Although the target responsible for these effects is unknown, it is possible that either Glc7p has a direct effect on the V-ATPase or the general vacuolar features regulated by Glc7p affect V-ATPase activity. The clc1Δ mutant vacuoles do show a comparable reduction in the levels of all V-ATPase subunits, which might reflect a V-ATPase delivery defect.
Rnr1p, the large subunit of ribonucleotide reductase, is important for DNA replication and the DNA damage checkpoint (Nguyen et al. 1999; Enyenihi and Saunders 2003). It is not easy at this point to explain its association with reduced V-ATPase activity in isolated vacuoles. Rmd7p is implicated in meiotic division (Enyenihi and Saunders 2003), but, like the cwh36Δ mutant and certain known vma mutants (Lussier et al. 1997), also shows a calcofluor white sensitivity suggestive of cell wall defects (Hughes et al. 2000).
Among the mutants that were not detected in our screen that have been associated biochemically with regulation of V-ATPase activity are the rav1Δ, rav2Δ, fen1Δ, and sur4Δ mutants. Although all of these mutants have been shown to affect the V-ATPase in biochemical assays, only the sur4Δ mutant shows a Vma− growth phenotype at 30° (Seol et al. 2001; Chung et al. 2003). We detected sur4Δ in only one of our two duplicate screens of the library on YEPD, pH 7.5 + CaCl2 plates, so it was not pursued further. Taken together, these results indicate that we have identified most of the nonessential deletion mutants capable of exhibiting a Vma− phenotype.
What is the physiological basis of the Vma− phenotype?
There are mutants in Table 3 that exhibited a Vma− growth phenotype that did not compromise quinacrine uptake into the vacuole, such as pho85Δ, snf5Δ, snf6Δ, rcs1Δ, and ctr1Δ. It is possible that some or all of these mutations do not prevent V-ATPase activity at the vacuole, but do affect activity or acidification in other organelles. One flaw in the quinacrine uptake assay and vacuole isolation as criteria for V-ATPase activity is that they fail to assess effects on other acidic compartments such as the prevacuolar compartment, endosomes, or late Golgi apparatus. Alternatively, these mutations may affect some aspect of pH and/or calcium homeostasis that allows them to mimic the growth defects of V-ATPase mutants and thus may provide important insights into the V-ATPase functions responsible for different aspects of the Vma− phenotype.
Several recent articles have highlighted the interconnections between alkaline pH stress and copper/iron deficiency (Davis-Kaplan et al. 2004; Serrano et al. 2004). This connection is likely to account for identification of the rcs1Δ mutant (also known as aft1Δ), which lacks the major transcription factor responsible for activation of the iron regulon in response to low iron, and the ctr1Δ mutant, which lacks the major plasma membrane copper transporter (Van Ho et al. 2002). Davis-Kaplan et al. (2004) demonstrated that failure to load the iron transporter Fet3p with copper was a major source of the iron deficiency in mutants lacking V-ATPase activity.
Another recent genomic screen also connected alkaline stress with calcium-mediated responses and calcineurin activation in particular (Viladevall et al. 2004). In this screen, alkaline pH stress was found to evoke a transient increase in cytosolic calcium capable of activating calcineurin, and substantial overlap was detected between mutations that confer sensitivity to FK506 and sensitivity to high pH. Significantly, most of the mutations in our screen show some level of CsA sensitivity even at moderately acidic pH, indicating that they are dependent on calcineurin activity for growth. Of the five that were not sensitive to CsA, rmd7Δ was shown elsewhere to be synthetically lethal with a calcineurin mutant (Parsons et al. 2004) and vps45Δ was found to be sensitive to another calcineurin inhibitor, FK506 (Serrano et al. 2004), indicating that these mutants are also calcineurin dependent even though they do not respond to CsA. Although a number of studies have demonstrated the strong calcineurin dependence of V-ATPase-deficient mutants (Garrett-Engele et al. 1995; Hemenway et al. 1995; Tanida et al. 1995; Forster and Kane 2000), the failure of these mutants to grow at elevated pH and calcium was not tightly linked to constitutive calcineurin dependence. Interestingly, none of these gene products, other than the V-ATPase, has been implicated directly in calcium homeostasis, so their deletions may cause calcineurin activation through one of the growing number of stresses that is translated into a calcium pulse resulting in calcineurin activation (Viladevall et al. 2004).
Acknowledgments
This work was supported by National Institutes of Health grant R01 GM50322 to P.M.K.
References
- Bachhawat, A. K., M. F. Manolson, D. G. Murdock, J. D. Garman and E. W. Jones, 1993. The VPH2 gene encodes a 25 kDa protein required for activity of the yeast vacuolar H(+)-ATPase. Yeast 9: 175–184. [DOI] [PubMed] [Google Scholar]
- Beltran, C., J. Kopecky, Y. C. Pan, H. Nelson and N. Nelson, 1992. Cloning and mutational analysis of the gene encoding subunit C of yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267: 774–779. [PubMed] [Google Scholar]
- Bonangelino, C. J., E. M. Chavez and J. S. Bonifacino, 2002. a Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol. Biol. Cell 13: 2486–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino, C. J., J. J. Nau, J. E. Duex, M. Brinkman, A. E. Wurmser et al., 2002. b Osmotic stress-induced increase of phosphatidylinositol 3,5-bisphosphate requires Vac14p, an activator of the lipid kinase Fab1p. J. Cell Biol. 156: 1015–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, J. H., R. L. Lester and R. C. Dickson, 2003. Sphingolipid requirement for generation of a functional v1 component of the vacuolar ATPase. J. Biol. Chem. 278: 28872–28881. [DOI] [PubMed] [Google Scholar]
- Crider, B. P., and X. S. Xie, 2003. Characterization of the functional coupling of bovine brain vacuolar-type H(+)-translocating ATPase. Effect of divalent cations, phospholipids, and subunit H (SFD). J. Biol. Chem. 278: 44281–44288. [DOI] [PubMed] [Google Scholar]
- Curtis, K. K., S. A. Francis, Y. Oluwatosin and P. M. Kane, 2002. Mutational analysis of the subunit C (Vma5p) of the yeast vacuolar H+-ATPase. J. Biol. Chem. 277: 8979–8988. [DOI] [PubMed] [Google Scholar]
- Davis-Kaplan, S. R., D. M. Ward, S. L. Shiflett and J. Kaplan, 2004. Genome-wide analysis of iron-dependent growth reveals a novel yeast gene required for vacuolar acidification. J. Biol. Chem. 279: 4322–4329. [DOI] [PubMed] [Google Scholar]
- Drose, S., K. U. Bindseil, E. J. Bowman, A. Siebers, A. Zeeck et al., 1993. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 32: 3902–3906. [DOI] [PubMed] [Google Scholar]
- Enyenihi, A. H., and W. S. Saunders, 2003. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163: 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, C., and P. M. Kane, 2000. Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 275: 38245–38253. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele, P., B. Moilanen and M. S. Cyert, 1995. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vacuolar H(+)-ATPase. Mol. Cell. Biol. 15: 4103–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary, J. D., A. E. Wurmser, C. J. Bonangelino, L. S. Weisman and S. D. Emr, 1998. Fab1p is essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J. Cell Biol. 143: 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly, D. J., I. C. Morrow, M. Lindsay, R. Gould, N. J. Bryant et al., 2000. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 19: 4577–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemenway, C. S., K. Dolinski, M. E. Cardenas, M. A. Hiller, E. W. Jones et al., 1995. vph6 mutants of Saccharomyces cerevisiae require calcineurin for growth and are defective in vacuolar H(+)-ATPase assembly. Genetics 141: 833–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, M. N., K. J. Hill, M. A. Lindorfer and T. H. Stevens, 1993. Isolation of vacuolar membrane H(+)-ATPase-deficient yeast mutants; the VMA5 and VMA4 genes are essential for assembly and activity of the vacuolar H(+)-ATPase. J. Biol. Chem. 268: 221–227. [PubMed] [Google Scholar]
- Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton et al., 2000. Functional discovery via a compendium of expression profiles. Cell 102: 109–126. [DOI] [PubMed] [Google Scholar]
- Iida, H., Y. Yagawa and Y. Anraku, 1990. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. A study of [Ca2+]i in single Saccharomyces cerevisiae cells with imaging of fura-2. J. Biol. Chem. 265: 13391–13399. [PubMed] [Google Scholar]
- Ito, H., Y. Fukuda, K. Murata and A. Kimura, 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, P. M., 1995. Disassembly and reassembly of the yeast vacuolar H(+)-ATPase in vivo. J. Biol. Chem. 270: 17025–17032. [PubMed] [Google Scholar]
- Kane, P. M., and A. M. Smardon, 2003. Assembly and regulation of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 35: 313–321. [DOI] [PubMed] [Google Scholar]
- Kane, P. M., and T. H. Stevens, 1992. Subunit composition, biosynthesis, and assembly of the yeast vacuolar proton-translocating ATPase. J. Bioenerg. Biomembr. 24: 383–393. [DOI] [PubMed] [Google Scholar]
- Kane, P. M., M. C. Kuehn, I. Howald-Stevenson and T. H. Stevens, 1992. Assembly and targeting of peripheral and integral membrane subunits of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267: 447–454. [PubMed] [Google Scholar]
- Klionsky, D. J., and S. D. Emr, 1989. Membrane protein sorting: biosynthesis, transport and processing of yeast vacuolar alkaline phosphatase. EMBO J. 8: 2241–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo, H., C. Rabouille, R. Newman, T. P. Levine, D. Pappin et al., 1997. p47 is a cofactor for p97-mediated membrane fusion. Nature 388: 75–78. [DOI] [PubMed] [Google Scholar]
- Longtine, M. S., A. McKenzie, III, D. J. Demarini, N. G. Shah, A. Wach et al., 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lussier, M., A. M. White, J. Sheraton, T. di Paolo, J. Treadwell et al., 1997. Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson, M. F., D. Proteau, R. A. Preston, A. Stenbit, B. T. Roberts et al., 1992. The VPH1 gene encodes a 95-kDa integral membrane polypeptide required for in vivo assembly and activity of the yeast vacuolar H(+)-ATPase. J. Biol. Chem. 267: 14294–14303. [PubMed] [Google Scholar]
- Manolson, M. F., B. Wu, D. Proteau, B. E. Taillon, B. T. Roberts et al., 1994. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J. Biol. Chem. 269: 14064–14074. [PubMed] [Google Scholar]
- Munn, A. L., and H. Riezman, 1994. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J. Cell Biol. 127: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, H., and N. Nelson, 1990. Disruption of genes encoding subunits of yeast vacuolar H(+)-ATPase causes conditional lethality. Proc. Natl. Acad. Sci. USA 87: 3503–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, H., S. Mandiyan and N. Nelson, 1989. A conserved gene encoding the 57-kDa subunit of the yeast vacuolar H+-ATPase. J. Biol. Chem. 264: 1775–1778 (erratum: J. Biol. Chem. 264 (9): 5313). [PubMed] [Google Scholar]
- Nelson, H., S. Mandiyan and N. Nelson, 1994. The Saccharomyces cerevisiae VMA7 gene encodes a 14-kDa subunit of the vacuolar H(+)-ATPase catalytic sector. J. Biol. Chem. 269: 24150–24155. [PubMed] [Google Scholar]
- Nelson, H., S. Mandiyan and N. Nelson, 1995. A bovine cDNA and a yeast gene (VMA8) encoding the subunit D of the vacuolar H(+)-ATPase. Proc. Natl. Acad. Sci. USA 92: 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, N., and W. R. Harvey, 1999. Vacuolar and plasma membrane proton-adenosinetriphosphatases. Physiol. Rev. 79: 361–385. [DOI] [PubMed] [Google Scholar]
- Nelson, N., and D. J. Klionsky, 1996. Vacuolar H(+)-ATPase: from mammals to yeast and back. Experientia 52: 1101–1110. [DOI] [PubMed] [Google Scholar]
- Nguyen, H. H., J. Ge, D. L. Perlstein and J. Stubbe, 1999. Purification of ribonucleotide reductase subunits Y1, Y2, Y3, and Y4 from yeast: Y4 plays a key role in diiron cluster assembly. Proc. Natl. Acad. Sci. USA 96: 12339–12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi, T., and M. Forgac, 2002. The vacuolar (h+)-ATPases—nature's most versatile proton pumps. Nat. Rev. Mol. Cell. Biol. 3: 94–103. [DOI] [PubMed] [Google Scholar]
- Ohya, Y., N. Umemoto, I. Tanida, A. Ohta, H. Iida et al., 1991. Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet− phenotype are ascribable to defects of vacuolar membrane H(+)-ATPase activity. J. Biol. Chem. 266: 13971–13977. [PubMed] [Google Scholar]
- Oluwatosin, Y. E., and P. M. Kane, 1997. Mutations in the CYS4 gene provide evidence for regulation of the yeast vacuolar H+-ATPase by oxidation and reduction in vivo. J. Biol. Chem. 272: 28149–28157. [DOI] [PubMed] [Google Scholar]
- Oluwatosin, Y. E., and P. M. Kane, 1998. Mutations in the yeast KEX2 gene cause a Vma(−)-like phenotype: a possible role for the Kex2 endoprotease in vacuolar acidification. Mol. Cell. Biol. 18: 1534–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra, K. J., and P. M. Kane, 1998. Reversible association between the V1 and V0 domains of yeast vacuolar H+-ATPase is an unconventional glucose-induced effect. Mol. Cell. Biol. 18: 7064–7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, A. B., R. L. Brost, H. Ding, Z. Li, C. Zhang et al., 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22: 62–69. [DOI] [PubMed] [Google Scholar]
- Peters, C., P. D. Andrews, M. J. Stark, S. Cesaro-Tadic, A. Glatz et al., 1999. Control of the terminal step of intracellular membrane fusion by protein phosphatase 1. Science 285: 1084–1087. [DOI] [PubMed] [Google Scholar]
- Peterson, M. R., and S. D. Emr, 2001. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic 2: 476–486. [DOI] [PubMed] [Google Scholar]
- Peterson, M. R., C. G. Burd, S. D. Emr, M. Peterson and C. R. Cowles, 1999. Vac1p coordinates Rab and phosphatidylinositol 3-kinase signaling in Vps45p-dependent vesicle docking/fusion at the endosome. A novel Sec18p/NSF-dependent complex required for Golgi-to-endosome transport in yeast. Curr. Biol. 9: 159–162. [DOI] [PubMed] [Google Scholar]
- Plant, P. J., M. F. Manolson, S. Grinstein and N. Demaurex, 1999. Alternative mechanisms of vacuolar acidification in H(+)-ATPase-deficient yeast. J. Biol. Chem. 274: 37270–37279. [DOI] [PubMed] [Google Scholar]
- Preston, R. A., R. F. Murphy and E. W. Jones, 1989. Assay of vacuolar pH in yeast and identification of acidification-defective mutants. Proc. Natl. Acad. Sci. USA 86: 7027–7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, R. A., P. S. Reinagel and E. W. Jones, 1992. Genes required for vacuolar acidity in Saccharomyces cerevisiae. Genetics 131: 551–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond, C. K., I. Howald-Stevenson, C. A. Vater and T. H. Stevens, 1992. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell 3: 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, C. J., C. K. Raymond, C. T. Yamashiro and T. H. Stevens, 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194: 644–661. [DOI] [PubMed] [Google Scholar]
- Rothman, J. H., C. T. Yamashiro, C. K. Raymond, P. M. Kane and T. H. Stevens, 1989. Acidification of the lysosome-like vacuole and the vacuolar H+-ATPase are deficient in two yeast mutants that fail to sort vacuolar proteins. J. Cell Biol. 109: 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambade, M., and P. M. Kane, 2004. The yeast vacuolar proton-translocating ATPase contains a subunit homologous to the Manduca sexta and bovine e subunits that is essential for function. J. Biol. Chem. 279: 17361–17365. [DOI] [PubMed] [Google Scholar]
- Sautin, Y. Y., M. Lu, A. Gaugler, L. Zhang and S. L. Gluck, 2005. Phosphatidylinositol 3-kinase-mediated effects of glucose on vacuolar H+-ATPase assembly, translocation, and acidification of intracellular compartments in renal epithelial cells. Mol. Cell. Biol. 25: 575–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schu, P. V., K. Takegawa, M. J. Fry, J. H. Stack, M. D. Waterfield et al., 1993. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 260: 88–91. [DOI] [PubMed] [Google Scholar]
- Seeley, E. S., M. Kato, N. Margolis, W. Wickner and G. Eitzen, 2002. Genomic analysis of homotypic vacuole fusion. Mol. Biol. Cell 13: 782–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seol, J. H., A. Shevchenko and R. J. Deshaies, 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3: 384–391. [DOI] [PubMed] [Google Scholar]
- Serrano, R., D. Bernal, E. Simon and J. Arino, 2004. Copper and iron are the limiting factors for growth of the yeast Saccharomyces cerevisiae in an alkaline environment. J. Biol. Chem. 279: 19698–19704. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1982 Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Stack, J. H., D. B. DeWald, K. Takegawa and S. D. Emr, 1995. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J. Cell Biol. 129: 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, T. H., and M. Forgac, 1997. Structure, function and regulation of the vacuolar (H+)-ATPase. Annu. Rev. Cell Dev. Biol. 13: 779–808. [DOI] [PubMed] [Google Scholar]
- Supekova, L., F. Supek and N. Nelson, 1995. The Saccharomyces cerevisiae VMA10 is an intron-containing gene encoding a novel 13-kDa subunit of vacuolar H(+)-ATPase. J. Biol. Chem. 270: 13726–13732. [DOI] [PubMed] [Google Scholar]
- Tan, Y. S., P. A. Morcos and J. F. Cannon, 2003. Pho85 phosphorylates the Glc7 protein phosphatase regulator Glc8 in vivo. J. Biol. Chem. 278: 147–153. [DOI] [PubMed] [Google Scholar]
- Tanida, I., A. Hasegawa, H. Iida, Y. Ohya and Y. Anraku, 1995. Cooperation of calcineurin and vacuolar H(+)-ATPase in intracellular Ca2+ homeostasis of yeast cells. J. Biol. Chem. 270: 10113–10119. [DOI] [PubMed] [Google Scholar]
- Tomashek, J. J., L. A. Graham, M. U. Hutchins, T. H. Stevens and D. J. Klionsky, 1997. V1-situated stalk subunits of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 272: 26787–26793. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Van Ho, A., D. M. Ward and J. Kaplan, 2002. Transition metal transport in yeast. Annu. Rev. Microbiol. 56: 237–261. [DOI] [PubMed] [Google Scholar]
- Viladevall, L., R. Serrano, A. Ruiz, G. Domenech, J. Giraldo et al., 2004. Characterization of the calcium-mediated response to alkaline stress in Saccharomyces cerevisiae. J. Biol. Chem. 279: 43614–43624. [DOI] [PubMed] [Google Scholar]
- Wach, A., 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12: 259–265. [DOI] [PubMed] [Google Scholar]
- Williams-Hart, T., X. Wu and K. Tatchell, 2002. Protein phosphatase type 1 regulates ion homeostasis in Saccharomyces cerevisiae. Genetics 160: 1423–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906. [DOI] [PubMed] [Google Scholar]
- Wurmser, A. E., J. D. Gary and S. D. Emr, 1999. Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J. Biol. Chem. 274: 9129–9132. [DOI] [PubMed] [Google Scholar]
- Yamashiro, C. T., P. M. Kane, D. F. Wolczyk, R. A. Preston and T. H. Stevens, 1990. Role of vacuolar acidification in protein sorting and zymogen activation: a genetic analysis of the yeast vacuolar proton-translocating ATPase. Mol. Cell. Biol. 10: 3737–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, S., S. Guha and F. C. Volkert, 1995. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol. 15: 2037–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]