Abstract
The genetic trajectory leading to viral attenuation was studied in a canine parvovirus (CPV) strain grown on dog kidney cells for 115 transfers. Consensus sequences of viral populations at passages 0, 3, 30, 50, 80, and 115 were obtained from PCR products covering 86% of the genome; clones from each of the 80th and 115th passages were also sequenced, covering 69% of the genome. Sixteen changes were fixed in the 115th-passage virus sample. Levels of polymorphism were strikingly different over time, in part because of a plaque-cloning step at passage 112 that reduced variation: passage 80 had 19 variants common among the clones, but passage 115 had only a single common variant. Several mutations increased in the culture at the same time, with most reaching fixation only after the 80th passage. The pattern of evolution was consistent with recombination and not with separate selective sweeps of individual mutations. Thirteen of the changes observed were identical to or at the same positions as changes observed in other isolates of CPV or feline panleukopenia virus.
The most successful viral vaccines use live attenuated virus strains. For more than a century, attenuation has involved propagation of a virus under novel conditions so that it becomes less pathogenic to its original host as it evolves under the new conditions (9, 10). Despite the antiquity of this method and its remarkable success in generating many vaccines that are widely used in humans and other animals, little is known about the process by which the attenuating mutations arise and evolve. In addition, there are some drawbacks to this method: the outcome of an attempted attenuation is largely unpredictable, and a successfully attenuated virus may revert to virulence, depending on the nature of the attenuating mutation(s). Although newer approaches show promise, many contemporary vaccines are still being developed by these classic methods.
A better understanding of the classical attenuation process might thus suggest protocols to eliminate some disadvantages of these methods of attenuation. One of the obvious gaps in our understanding of attenuation is at the genetic level. Until recently, the molecular changes underlying the shift from virulence to attenuation were almost completely unknown for most attenuated vaccine viruses. Even now, the dynamics over an attenuation episode have yet to be described. To begin filling this gap, we examined the genetic evolution that occurred during the passaging of canine parvovirus (CPV) in tissue culture to give rise to a successfully attenuated vaccine that has been used effectively for 20 years.
CPV is a single-stranded DNA virus with a linear genome of ∼5.15 kb. CPV appears to have derived from a single ancestral cat virus that emerged in dogs during the mid-1970s (24) and spread worldwide in 1978 (30). The initial 1978 strain of the virus (designated CPV type 2) was replaced during 1979 and 1980 by a genetically and antigenically distinct variant termed CPV type 2a (25). Those viruses have continued to evolve slowly in nature, but some sequence changes have swept through the global CPV population (30).
The source of CPV genomes for this study were the frozen or lyophilized archive stocks of the CPV type 2 strain (78-0916) that had been passaged in dog or cat cells in culture at 33 and 35°C, during which time the virus became attenuated (2). Passage 0 was the original canine feces from which the virus was isolated, while passages 1 through 115 had been grown in primary dog kidney cells. A second line of virus passages was started with dog cell passage 103 and was continued for another 9 passages in feline renal cell line CRFK (ATCC CCL-94). Virus titers were approximately 106/ml at the end of each passage, whereupon they were diluted 1:50 or 1:100 to start the next passage. The virus was passaged from an endpoint dilution culture at the 64th and 73rd passages and plaque cloned once in A72 canine cells at the 112th passage. The virus stocks were tested for virulence at passages 30, 50, 80, and 115 by inoculation of dogs. They proved to be significantly attenuated by the 80th passage and formed larger plaques in tissue culture by the 94th passage, as detailed previously (2, 3).
Consensus PCR sequences.
In the first analysis, PCR products of archived viral suspensions were analyzed, thus representing a consensus of common changes in the viral population. Two overlapping PCR products were amplified with Taq DNA polymerase (between bases 480 and 2680 and between bases 2400 and 4889), spanning 86% of the 5,124-base genome. Those PCR products were completely sequenced for passages 1, 50, and 115 and for 103/9 virus (after 103 passages in dog cells and 9 passages in cat cells). Passages 3, 30, and 80 were sequenced only over the regions in which changes had been detected at passages 115 and 103/9. All sequencing reactions used ABI Big Dye chain-terminating reactions, with primers spaced every 300 to 400 bases. Reactions were read on an automated machine (ABI 377). ABI sequence data were then analyzed with DNA Star Lasergene (1998).
Compared to the unpassaged virus (passage 0), 16 changes appeared to be fixed in the consensus sequence of the final 115th passage in canine cells; one site appeared to be polymorphic. Of the fixed changes, there were 12 substitutions (11 transitions, one transversion) plus two deletions and two insertions (Fig. 1). The changes identified appeared to be fixed by virtue of showing a single peak in the sequence analysis, but as a PCR consensus sequence does not offer accurate information about frequencies, it is possible that other bases were also present but less common. The temporal pattern of evolution was that, of the 16 changes that ultimately appeared to be fixed by passage 115, 2 appeared to be fixed by passage 30 and only 1 more appeared to be fixed by passage 80, but at this time, 10 other changes were detectably polymorphic. Cat cell lineage virus 103/9 appeared to be fixed for all of the changes fixed in the 115 dog cell-passaged virus, and that virus stock also contained three additional substitutions, with one transition and two transversions (Fig. 1).
FIG. 1.
Sequence changes found in CPV during passage in dog cells between the wild-type virus (pass 0) and after 3, 30, 50, 80, and 115 passages in primary dog cells or after 103 passages in dog cells and 9 passages in cat cells (103/9). The left column (Changes detected) indicates the genomic nucleotide position(s) (24) and the gene affected. The next column gives the amino acid substitution(s) where those occurred (blank if the nucleotide change was noncoding). The residue number in the gene is given for each protein affected. The pass 0 column gives the ancestral base, and the new base is given in the 115 or 103/9 (cat) column. (Top) Twenty changes observed in PCR consensus sequences. Each of the variable sites that was detected in the PCR consensus sequence is shown as white (no change from pass 0), shaded (polymorphic signal in the PCR consensus sequence), or black (apparently fixed in that sequence). The number of variant nucleotides at each position in the nine clones of the 80th passage is shown as a ratio in parentheses for each position evaluated between bases 1099 and 4649 of that virus stock. The passage 115 data show the variation present at each position in eight clones of PCR products that were prepared as two segments and cloned into plasmids before sequencing. Virus isolate sequences (Virus isolates) were sequences obtained from GenBank or from other studies examining the sequences of natural isolates of CPV. (Bottom [Changes detected in p80 clones]) The lower seven changes were detected in only one to three of the nine clones prepared from the 80th-passage viruses, between nt 1099 and 4649 in the genome. The 115 column shows that only one of those passage 80 clone mutations was also seen in the eight clones from passage 115. Mutations found only in clones from passage 115 are not shown (all those not shown were present in just one of the eight clones). At the far right, those sequence changes observed in the passage 115 and 103/9 attenuation lineages are compared to the sequences of other viral isolates. Del or del, deletion; ins, insertion.
Analysis of cloned DNAs.
To examine the sequence variation in individual genomes, the viral replicative-form DNA was isolated from canine A72 cells infected with the 80th-passage virus. Low-molecular-weight DNA was recovered from the infected cells with a modified Hirt procedure (14). The viral genomic region between the EcoRI site at base 1099 and a PacI site at base 4649 was then cloned into plasmid pNEB193 (New England Biolabs, Beverly, Mass.), and the inserts were sequenced. The DNA of the 115th-passage virus was amplified between bases 480 and 2680 with XL Taq (Tth polymerase; Applied Biosystems, Forest City, Calif.) and between bases 2400 and 4889 with Deep Vent polymerase (New England Biolabs). The fragments were cloned into Topo plasmids (In Vitrogen, Carlsbad, Calif.) and sequenced.
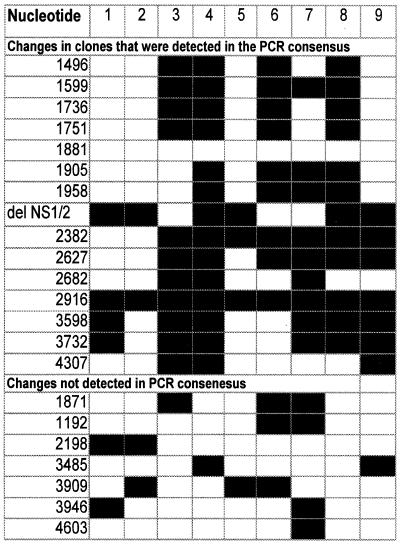
Nine DNA clones were prepared of the viral genome between nucleotides (nt) 1099 and 4649 of the passage 80 virus, which should each represent a single viral genome. Their sequences revealed polymorphisms at 20 sites (Fig. 2). The range of variation among these clones was impressive, as no two clones exhibited the same sequence. The clone sequences obtained for the 80th passage spanned 13 of the sites that appeared to have fixed changes by the 115th passage. One clone carried all 13 changes, and one clone carried only 2 of these changes. Seven of the 20 changes were never detected in the consensus sequences, and none of those was found in more than three isolates (Fig. 2).
FIG. 2.
Polymorphic sites in the nine clones prepared from the 80th-passage virus. The nucleotide positions are ordered along the vertical axis corresponding to changes observed in the PCR consensus sequence (upper 15) and those that were not observed in the consensus sequence (lower 7). Each of columns 1 to 9 represents a different clone corresponding to a different viral isolate. A black square denotes the evolved (nonancestral) base, which changed as indicated in Fig. 1. All of the clones have unique sequences, which is suggestive of recombination.
Eight clones were prepared from each of two overlapping regions of the 115th-passage virus after PCR amplification with high-fidelity polymerases. All mutations within the cloned region that had appeared to be fixed in the PCR consensus sequence were present in all eight clones (Fig. 1). The only base that appeared to be polymorphic in the 115th-passage PCR consensus sequence (base 2682) was mutant in five of eight clones. In addition, 23 mutations were observed singly (in one of eight clones, although different clones carried different mutations), and only one of these (3485T) had been seen in earlier passages.
There was moderate, but not perfect, agreement between the incidence of polymorphism in the clones versus the inference of polymorphism in the PCR consensus sequence based on multiple peaks. Bases seen in five or more of the clones were always detected in peaks of the consensus sequence, whereas bases seen in three or fewer of the clones were never detected in the consensus sequence; of bases seen in four clones, some were detected and some were not. Precise estimates of nucleotide frequencies are not possible with nine clones, but it is clear that consensus sequences did not detect even moderate frequencies of base substitutions. Thus, our classification of a base as “fixed” in a consensus sequence (or present in nine of nine or eight of eight clones) is compatible with a range of frequencies. Polymorphism may well have existed at many other sites in the genome in passages analyzed only with PCR consensus sequences but would have remained undetected because the wild-type base always dominated the consensus sequence.
Sequences of related feline panleukopenia virus (FPV), CPV, and mink enteritis virus (MEV) were obtained from GenBank (accession numbers: CPV-d, M38245; CPV-N, M19296; CPV-Y1, D26079; CPV-39, M74849; MEV, D00765; FPV-b, M38246; FPV-193, X55115), while the sequences of 4 CPV isolates between nt 180 and 4525 were determined from the cloned or PCR-amplified viral replicative-form DNA as described above.
Proteins affected.
Of the base substitutions that appeared to be fixed at passage 115, one was outside coding regions, six were silent within coding regions, and five were missense within NS1, VP1, or VP1 and VP2 (Fig. 1). Three additional missense changes were observed in the virus passaged in feline cells. The NS1 and NS2 proteins have partially overlapping open reading frames, with 87 residues at the N termini in common, with the last 90 residues of NS2 and NS1 being read from the same sequence in two different reading frames (27). There were no changes in the N-terminal sequence common to both NS1 and NS2, three missense mutations in the NS1 unique portion of the open reading frame, and a 15-nt deletion from the NS1-NS2 overlapping sequence that removed five amino acids from both NS1 (residues 591 to 595) and NS2 (residues 100 to 104) (Fig. 1). CPV encodes two capsid proteins, VP1 and VP2, and those proteins contained four changes in the virus passaged 115 times in dog cells (VP1 residue 109 and VP2 residues 44, 271, and 316) and two others in CPV 103/9. The two VP2 residue changes (residues 300 and 301) in CPV 103/9 are known to prevent infection of dogs or dog cells and therefore likely represent a host adaptation of the virus to growth in cat cells (18, 22).
Serial passage is a method that has been commonly applied to create vaccines currently in use, yet the genetic architecture of the process is largely unknown for any virus. Although many sequences of attenuated viruses relative to their wild-type ancestors are known, an understanding of how those changes accumulated and how much variation was present during the attenuation is lacking. Such information is vital if an attenuation process is to be repeated, extended, or truncated or if multiple isolates are to be drawn from a single attenuation series. Where substrains of classical human vaccine viruses have been examined, considerable diversity has been found in the strains that were passaged for different lengths of time or under different conditions, suggesting that the populations of those viruses were also highly diverse at various times during the process (11, 23). Comparison of rubella viruses attenuated independently shows them to have few or no changes in common (15).
In this study, 16 genetic changes appeared to be fixed in the cultured population by the end of the passaging: 12 base substitutions, two insertions, and two deletions (one of the deletions lay in a region that encodes the overlapping NS1 and NS2 proteins). We have not tested the role of individual mutations or groups of mutations for their roles in viral virulence and cannot define which of these changes led to tissue culture adaptation or reduced virulence. The NS1 protein is a multifunctional protein that is required for many aspects of viral replication; it interacts with several cellular proteins and with viral DNA sequences and is regulated by phosphorylation (5, 7, 21). The changes found in the NS1 protein in these passaged viruses are all in a C-terminal region that, in the minute virus of mice (MVM), appears to affect transactivation of the viral p38 promoter and to affect the toxicity of the protein (16). The NS2 protein in MVM is required for capsid assembly and nuclear export within mouse cells (6, 19), although a similar role for NS2 of CPV was not detected (31). The changes in the capsid protein were all within the interior of the capsid structure, and no specific functions have been described for those residues (33). There were only four changes that were not within coding regions—single changes of nt 2382 within the small intron in the R3 message, insertion of 1 nt at position 4762, deletion of nt 4860, and an insertion of 63 bases in the 5′ noncoding region. In MVM, sequences within the small intron affect the splicing of that sequence (13), but replacement of nt 2384 is unlikely to affect that function in CPV, where only a single splice acceptor is used. We cannot predict any functions for the changes in the 5′ noncoding region, although functions in this region are likely required for DNA replication and possibly for DNA packaging into capsids (29). Insertions or deletions of sequences with unit lengths of about 60 nt have been seen in both wild-type and tissue culture-passaged viruses, suggesting that these are not, in themselves, associated with attenuation (27).
One surprising outcome was the high frequency (6 of 11) of fixed silent substitutions in coding regions, which are typically considered to be neutral (17). One model to explain the rapid ascent of neutral changes is hitchhiking (linkage) with a beneficial missense substitution. In the CPV data, however, this model is contraindicated because five of the silent substitutions evolved in vitro are identical to substitutions seen in other virus isolates. These independent, parallel evolutions suggest that the silent substitutions are themselves beneficial. Codon bias offers one possible advantage; direct interaction of the single-stranded genome with the capsid is another (4), as is a possible role of specific folded structures in the intracellular metabolism or packaging of these single-stranded DNA genomes. Parallel evolution of silent substitutions has also been seen in experimental populations of bacteriophage φX174 (1, 8, 32).
Dynamics of change.
A priori, there are various evolutionary processes that could underlie the evolution of these 16 fixations. At one extreme, the culture could have experienced 16 sequential selective sweeps, one at a time. At the other extreme, all changes could have ascended concurrently, so that the culture was simultaneously polymorphic for every one and thus contained a vast array of genotypes with different combinations of these changes. The latter picture is close to the data. At passage 80, 19 nucleotide changes were present as “common” polymorphisms, being observed in at least two clones, and hence present at a meaningful level and not simply quasispecies mutations. The simultaneous ascent of so many mutations suggests that recombination was a major factor in the evolution seen. We have not formally analyzed this conjecture, but to suppose that recombination was absent requires a highly specific and seemingly implausible combination of parallel mutation and epistasis of fitness effects. In the absence of recombination, beneficial mutations arising in different genomes remain genetically isolated and are doomed to compete with each other until all but one have been lost, resulting in a sequential evolution of different substitutions rather than a simultaneous evolution of them (12, 20). Regardless of whether recombination was the cause of this extensive polymorphism, however, the results illustrate the potential difficulty of obtaining two genetically identical or even similar isolates at certain passages in an attenuation process.
There was a strikingly higher level of polymorphism at passage 80 than at earlier and later passages, but it is unclear how much of the change in polymorphism was due to selection or to population bottlenecks. The decay of polymorphism between passages 80 and 115 can be attributed to the attempted plaque isolation at passage 112, for example, but the trend from passage 50 to passage 80 suggests that several fixations would have occurred in the absence of the bottleneck. As two passage-80 polymorphisms were retained at passage 115 (2682G and 3485T), three passages beyond the plaque isolation at passage 112, the bottleneck was no doubt larger than a single plaque. It would also be interesting to know why polymorphism increased so dramatically between passages 50 and 80; possibly, the mutations were ascending throughout the first 50 passages but had not increased to levels detectable by consensus PCR sequences. The biggest surprise, however, is the high levels of polymorphism at passage 80, since the culture was endpoint diluted at passages 64 and 73. If severe bottlenecks were indeed imposed at those two passages, it is puzzling that such high levels of variation were restored so quickly, especially given the rather slow evolution during the first 50 passages. It is thus likely that at least a moderate number of genomes survived these bottlenecks, a possibility further supported by the fact that no fixations occurred between passages 50 and 80. Despite these uncertainties about the causes underlying the dynamics of change during attenuation, our analysis illustrates precisely why it is important to use sequence data in conjunction with well-designed passaging protocols to achieve the desired goals and to achieve repeatability of the attenuation process.
Parallels with natural viral evolution.
The fact that many of the changes evolved in culture have also been seen in other virus isolates also constitutes an enigma. This recapitulation of natural evolution points to a selective environment common to the viruses adapted to cultured dog cells and the other isolates, although the causes of these parallel substitutions may be varied. For example, a change at nt 3685 (which evolved to A in the 103/9 virus stock but to G in CPV type 2a isolates) has been associated with changes in the antigenicity and host range of the virus (22, 26), so the capsid structure it affects may be sensitive to various factors of the host environment (18). Other parallels are two changes in a wild CPV isolate from Germany that had not been passaged in tissue culture, three changes in an MEV isolate from Japan, and seven changes each in two FPV isolates from the 1970s. We cannot predict what these substitutions might do, especially as five of the changes in coding regions are silent. Since the latter three viruses were passaged various numbers of times in feline tissue culture cells before sequence analysis, it is possible that some of the parallel changes between those viruses and ours evolved in response to the culture conditions. However, that possibility is somewhat diminished by the facts that the CPV-N isolate was an attenuated strain of CPV type 2 passaged in canine cells (28) and it had no changes in common with CPV 115.
Parallels with experimental systems.
The results of this study parallel results from experimental adaptations of bacteriophage φX174. The two viruses have several similarities. φX174 is a single-stranded DNA virus with a circular genome of 5.4 kb. Both viruses have proteinaceous icosahedral capsids. φX174 adaptation to different bacterial hosts and culture temperatures has been evaluated in a series of studies (1, 8, 32). The overall dynamics of both systems show many similarities. The 115 CPV passages likely represent a magnitude of population doublings similar to that which occurs during a standard 10-day chemostat experiment with φX174. The average number of genetic fixations in a φX174 chemostat experiment is around 12 but can range as high as 25 (1, 32), in the same range as seen for CPV (minimum population sizes in the φX174 cultures are at least hundreds of thousands, so bottlenecks have not been a factor in those fixations). The temporal pattern of substitutions in both systems was that more changes occurred late than early (32), although the pattern with CPV seems to be more pronounced in this regard. Other similarities between these two experimental systems include (i) the appearance of substitutions that do not fix (32) and (ii) evolution of identical changes in vitro and in nature (8). Rates of recombination may also be high in the φX174 chemostat systems, although the evidence in support of this possibility is indirect, as it is for our CPV passage (32). Taken together, these results suggest that broad generalities await to be found in experimental studies of viral evolution.
Acknowledgments
We thank Wendy Weichert for expert technical assistance.
This research was supported by grants GM 57756 (J.J.B.) and AI28385 (C.R.P.) from the National Institutes of Health.
REFERENCES
- 1.Bull, J. J., M. R. Badgett, H. A. Wichman, J. P. Huelsenbeck, D. M. Hillis, A. Gulati, C. Ho, and I. J. Molineux. 1997. Exceptional convergent evolution in a virus. Genetics 147:1497-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carmichael, L. E., J. C. Joubert, and R. V. Pollock. 1981. A modified live canine parvovirus strain with novel plaque characteristics. I. Viral attenuation and dog response. Cornell Vet. 71:408-427. [PubMed] [Google Scholar]
- 3.Carmichael, L. E., J. C. Joubert, and R. V. H. Pollock. 1983. A modified canine parvovirus vaccine. 2. Immune response. Cornell Vet. 73:13-29. [PubMed] [Google Scholar]
- 4.Chapman, M. S., and M. G. Rossmann. 1995. Single-stranded DNA-protein interactions in canine parvovirus. Structure 3:151-162. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, J., S. F. Cotmore, and P. Tattersall. 2001. Minute virus of mice initiator protein NS1 and a host KDWK family transcription factor must form a precise ternary complex with origin DNA for nicking to occur. J. Virol. 75:7009-7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore, S. F., and P. Tattersall. 1998. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J. Virol. 72:8477-8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crill, W. D., H. A. Wichman, and J. J. Bull. 2000. Evolutionary reversals during viral adaptation to alternating hosts. Genetics 154:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ebert, D. 1998. Experimental evolution of parasites. Science 282:1432-1435. [DOI] [PubMed] [Google Scholar]
- 10.Flint, S. J., L. W. Enquist, R. M. Krug, V. R. Racaniello, and A. M. Skalka. 2000. Principles of virology. ASM Press, Washington, D.C.
- 11.Galler, R., P. R. Post, C. N. Santos, and I. I. Ferreira. 1998. Genetic variability among yellow fever virus 17D substrains. Vaccine 16:1024-1028. [DOI] [PubMed] [Google Scholar]
- 12.Gerrish, P. J., and R. E. Lenski. 1998. The fate of competing beneficial mutations in an asexual population. Genetica 102/103:127-144. [PubMed] [Google Scholar]
- 13.Haut, D. D., and D. J. Pintel. 1998. Intron definition is required for excision of the minute virus of mice small intron and definition of the upstream exon. J. Virol. 72:1834-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 15.Kakizawa, J., Y. Nitta, T. Yamashita, H. Ushijima, and S. Katow. 2001. Mutations of rubella virus vaccine TO-336 strain occurred in the attenuation process of wild progenitor virus. Vaccine 19:2793-2802. [DOI] [PubMed] [Google Scholar]
- 16.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, Mass.
- 18.Llamas-Saiz, A. L., M. Agbandje-McKenna, J. S. L. Parker, A. T. M. Wahid, C. R. Parrish, and M. G. Rossmann. 1996. Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range. Virology 225:65-71. [DOI] [PubMed] [Google Scholar]
- 19.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muller, H. J. 1932. Some genetic aspects of sex. Am. Nat. 66:118-138. [Google Scholar]
- 21.Nüesch, J. P. F., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated in vitro by the phosphorylation state of the polypeptide. J. Virol. 72:8002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parker, J. S. L., and C. R. Parrish. 1997. Canine parvovirus host range is determined by the specific conformation of an additional region of the capsid. J. Virol. 71:9214-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parks, C. L., R. A. Lerch, P. Walpita, H. P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Comparison of predicted amino acid sequences of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:910-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrish, C. R. 1990. Emergence, natural history, and variation of canine, mink, and feline parvoviruses. Adv. Virus Res. 38:403-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parrish, C. R. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. Virology 183:195-205. [DOI] [PubMed] [Google Scholar]
- 26.Parrish, C. R., C. Aquadro, M. L. Strassheim, J. F. Evermann, J.-Y. Sgro, and H. Mohammed. 1991. Rapid antigenic-type replacement and DNA sequence evolution of canine parvovirus. J. Virol. 65:6544-6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrish, C. R., C. F. Aquadro, and L. E. Carmichael. 1988. Canine host range and a specific epitope map along with variant sequences in the capsid protein gene of canine parvovirus and related feline, mink and raccoon parvoviruses. Virology 166:293-307. [DOI] [PubMed] [Google Scholar]
- 28.Reed, A. P., E. V. Jones, and T. J. Miller. 1988. Nucleotide sequence and genome organization of canine parvovirus. J. Virol. 62:266-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam, P., and C. R. Astell. 1993. Replication of minute virus of mice minigenomes: novel replication elements required for MVM DNA replication. Virology 193:812-824. [DOI] [PubMed] [Google Scholar]
- 30.Truyen, U., A. Gruenberg, S. F. Chang, B. Obermaier, P. Veijalainen, and C. R. Parrish. 1995. Evolution of the feline-subgroup parvoviruses and the control of canine host range in vivo. J. Virol. 69:4702-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, D., W. Yuan, I. Davis, and C. R. Parrish. 1998. Nonstructural protein-2 and the replication of canine parvovirus. Virology 240:273-281. [DOI] [PubMed] [Google Scholar]
- 32.Wichman, H. A., M. R. Badgett, L. A. Scott, C. M. Boulianne, and J. J. Bull. 1999. Different trajectories of parallel evolution during viral adaptation. Science 285:422-424. [DOI] [PubMed] [Google Scholar]
- 33.Xie, Q., and M. S. Chapman. 1996. Canine parvovirus capsid structure, analyzed at 2.9 Å resolution. J. Mol. Biol. 264:497-520. [DOI] [PubMed] [Google Scholar]