Abstract
We studied a 15-year-old girl, patient X, who has maintained consistently low plasma loads of human immunodeficiency virus type 1 (HIV-1) RNA, as well as normal and stable CD4+ T-cell concentrations. She has presented no clinical manifestations of AIDS, despite having only received zidovudine monotherapy for a part of her life. Patient X's HIV-positive mother (patient Y) has also not progressed to AIDS and has never been treated with antiretroviral agents. HIV-1 isolated from patient X replicated poorly in human peripheral blood mononuclear cells (PBMC). In order to map the determinant of the poor growth of patient X's isolate, viral sequences from patient X were determined and examined for insertion or deletion mutations. These sequences contained a two-amino-acid insertion mutation in the Vif gene, which was also observed in uncultured PBMC acquired at different times. Furthermore, Vif sequences harbored by patient Y contained the identical mutation. These observations suggest that polymorphic HIV-1 was transmitted to patient X perinatally 15 years previously and has been maintained since that time. Recombinant HIV-1, engineered with Vif sequences from patient X, replicated in PBMC to levels approximately 20-fold lower than that of wild type. Removal of the insertion mutation from this recombinant restored replication efficiency to wild-type levels, while introduction of the insertion mutation into wild-type Vif sequences resulted in greatly decreased replication. Furthermore, Vif protein from patient X's HIV-1 was aberrantly cleaved, suggesting a mechanism for loss of Vif function. Since HIV-1 containing these sequences replicates poorly, the implication is that the two-amino-acid insertion mutation in Vif contributes significantly to the nonprogressor status of this mother and child. Further studies of these sequences might provide information regarding contributions of Vif structure and/or function to HIV-1 virulence.
The natural history of human immunodeficiency virus type 1 (HIV-1) infection and temporal progression to AIDS shows high variability. In those infected perinatally, disease may progress rapidly, often leading to death within 3 years of infection (30). In contrast, some HIV-1-infected individuals display no signs of progression for periods in excess of 10 years, even in the absence of antiretroviral therapy (3, 23, 27). The ability of these clinical nonprogressors (CNPs) to control HIV-1 replication is likely due to host and/or viral factors. For example, the CD4+ T cells of 25 to 30% of CNPs express mutant forms of CCR5 or CCR2, the most frequently used second receptors for HIV-1, rendering these cells less susceptible to infection (8, 18). Furthermore, many CNPs display more vigorous helper T-cell and cytotoxic T-lymphocyte responses to HIV-1 than typical progressors (13, 14). In addition to these host factors, some CNPs are infected with HIV-1 strains containing aberrant Nef, Rev, p17Gag, and Vpr sequences that may affect viral replication in vivo (5, 7, 19, 21, 36). In this report, we describe a CNP mother and child pair, the HIV-1 sequences they harbor, and the functions subserved by these sequences.
CNPs.
Patient Y (mother) is a 38-year-old female who was diagnosed as HIV-1 seropositive in June 1989. She is obese (present weight, 126 kg), has non-insulin-dependent diabetes mellitus and asthma, and has antibodies to hepatitis C virus. HIV-1 Vif sequences were amplified from her peripheral blood mononuclear cells (PBMC), which were obtained in July 2000, when her viral load was 62,800 copies of HIV-1 RNA/ml and her CD4+ T-lymphocyte count was 741 cells/μl. Since then, her viral load has ranged from 19,000 to 34,000 RNA copies/ml and her CD4+ T-lymphocyte count has ranged from 773 to 1,052 cells/μl. Patient Y has never taken any antiretroviral medication and has not had any HIV-associated illnesses. Her daughter, patient X, was born in January 1987. She developed lymphocytic interstitial pneumonitis in June 1989, at which time her positive HIV-1 serostatus was confirmed by enzyme-linked immunosorbent assay (ELISA) and Western immunoblotting. In April 1998, when infectious HIV-1 was isolated, her viral load was 4,800 RNA copies/ml and her CD4+ T-lymphocyte count was 990 cells/μl (Fig. 1). Patient X received zidovudine monotherapy between October 1989 and March 1997 but no subsequent antiretroviral therapy. She presently has no HIV-associated illnesses.
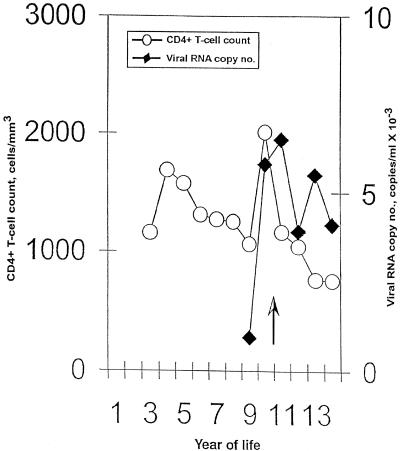
FIG. 1.
Plasma viremia and CD4+ T-cell counts for patient X. The black diamonds indicate the concentrations of HIV RNA in plasma and the white circles indicate the numbers of CD4+ T-cells/ml in PBMC from patient X. The vertical arrow indicates the time when zidovudine monotherapy was stopped.
Host-specific factors that have been associated with nonprogression.
To determine whether patient X and/or patient Y carried host-specific mutations that have been associated with nonprogression (e.g., CCR5:Δ32, CCR2:64I, or SDF-1:3′A) (8, 9, 18, 22, 24, 29, 37), cellular DNA was purified from 5 × 106 PBMC by using a saturated NaCl precipitation technique (2). For amplification of CCR5 sequences, the forward primer 5′CTGCCTCCGCTCTACTCACT3′ and the reverse primer 5′CCCTGTGCCTCTTCTTCTC3′ were used; for CCR2, the forward primer 5′ATGCTGTCCACATCTCGTTC3′ and the reverse primer 5′TGGGACAGAAGCAAACACAAG3′ were used; and for SDF-1, the forward primer 5′AGGCTTCTCTCTGTCCCATG3′ and the reverse primer 5′GATCAGGACTGCACTCAAAGG3′ were used. The resulting fragments were column purified, and the absence or presence of sequences that have been associated with nonprogression (i.e., CCR5:Δ32, CCR2:64I, and SDF-1:3′A) was determined (using the gene-specific forward primers described above) using an ABI 377 DNA sequencer (Perkin-Elmer Cetus, Norwalk, Conn.). These analyses revealed that neither patient X nor patient Y contained such mutations in these genes (data not shown).
Replication of HIV-1 isolated from patient X.
Since these individuals did not carry host-specific mutations associated with nonprogression, attention was focused on viral factors that could determine nonprogression. For this purpose, HIV-1 was isolated by cocultivation of PBMC (4). PBMC from patient X were incubated with phytohemagglutinin-stimulated PBMC from an HIV-seronegative donor in RPMI medium (Gibco-BRL, Grand Island, N.Y.) supplemented with 20% fetal bovine serum (FBS; Gibco-BRL) and 5% interleukin-2 (Advanced Biotechnologies). Fresh donor cells (∼5.0 × 107) were added once weekly for 3 weeks, and HIV-1 was detected by p24Gag ELISA (Coulter, Hialeah, Fla.). In these experiments, patient X's isolate replicated poorly in PBMC, only reaching a peak p24Gag concentration of 11 ng/ml in culture supernatants (data not shown), suggesting a defect in HIV-1 sequences.
HIV-1 sequences from an isolate from patient X.
In order to determine the viral sequences that conferred poor replication on the HIV-1 strain from patient X, cellular DNA was isolated from cultures infected with this isolate, which served as the template for PCR amplification of the entire HIV-1 genome. One microgram of cellular DNA was used as the template for PCR amplification of three overlapping HIV-1 fragments of approximately 3.5 kbp, using HIV-1-specific primers as previously described (3). The resulting fragments were column purified (Qiagen, Valencia, Calif.), and the entire HIV-1 sequence was determined using HIV-specific primers with an ABI 377 DNA sequencer.
Analyses of these sequences revealed that HIV-1 isolated from patient X more than 1 year after cessation of zidovudine therapy contained two residues (67N and 219Q; data not shown) that confer resistance to this drug (16). The acquisition of zidovudine resistance is typical of individuals who fail monotherapy (16); at such times, the drug plays a negligible role in controlling viral replication. Zidovudine was first administered to patient X at the age of 33 months, an age at which many vertically infected children in the past had already developed AIDS-defining illnesses and when a low steady-state viral load was almost certainly established in this child (30). Although patient X has not received antiretroviral therapy for the past 5 years, there has been no substantial change in her viral load or CD4+ T-cell count (Fig. 1). These observations suggest that the nonprogressor status of patient X is likely not associated with her prior treatment with zidovudine.
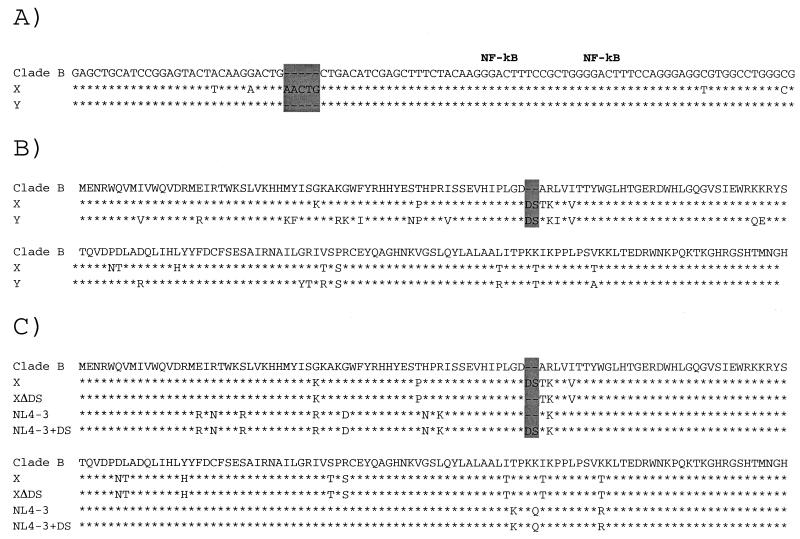
A 5-base insertion in the long terminal repeat (LTR) was observed in HIV-1 sequences from an isolate from patient X (Fig. 2A). HIV-1 sequences amplified directly from PBMC obtained from patient X 8 years earlier, in 1990, also contained these mutations (data not shown). However, this mutation, which was observed in all samples from patient X that were examined, was not present in the HIV-1 strain from patient Y (Fig. 2A).
FIG. 2.
(A) Nucleotide alignment of LTR sequences from strains from patients X and Y and clade B consensus LTR sequences. (B) Amino-acid alignment of HIV-1 Vif sequences from strains from patients X and Y and clade B consensus HIV-1 Vif sequences. (C) Amino-acid alignment of the sequence from the strain from patient X and Clade B consensus, NL4-3, and engineered HIV-1 Vif sequences derived from the strain from patient X or from NL4-3. The stars indicate conservation between the Clade B consensus sequence and the sequences from the strains from patient X. Dashes denote nucleotides or amino acids that are not contained in a particular sequence. The areas shaded in grey indicate the locations of the mutant sequences.
Sequence analysis revealed a two-amino-acid insertion in Vif sequences isolated from patient X (Fig. 2B). This mutation was also observed in sequences derived directly from PBMC as much as 8 years earlier (data not shown). Furthermore, this mutation was observed in sequences isolated from PBMC from patient Y (Fig. 2B). Thus, these sequences were almost certainly transmitted vertically from mother to child over 15 years ago. In fact, this mutation was observed in every clone, which in each case comprised two clones from each of two independent PCRs. The fact that this mutation was found in all samples obtained from these two individuals strongly suggests that the insertion mutation represents the sole Vif sequence in this mother and child.
Replication of recombinant HIV-1 containing polymorphic sequences isolated from patient X.
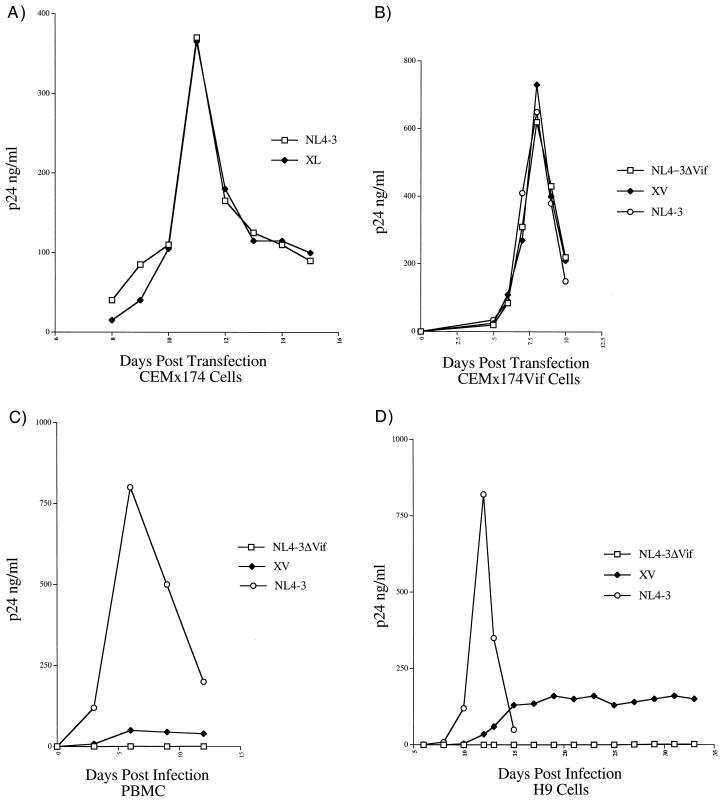
The 5-base insertion observed in LTR sequences from patient X (Fig. 2A) was introduced into the 5′ and 3′ LTR sequences of NL4-3 by overlap extension (17) to create strain XL. Plasmid DNA containing 5′ NL4-3, 3′ NL4-3, or derivative sequences was digested with EcoRI. The digested DNA was column purified (Qiagen), joined using T4 DNA ligase (New England Biolabs, Beverly, Mass.), and introduced into cells from the permissive cell line CEMx174 by DEAE-dextran transfection (26). The recombinant XL grew similarly to wild-type NL4-3 in these experiments (Fig. 3A). Since these data indicated that the LTR mutation was not a determinant of poor growth of HIV-1 from patient X in culture, strain XL was not subjected to further study.
FIG. 3.
(A) Replication in CEMx174 cells of HIV-1 strain NL4-3 and an NL4-3 recombinant (XL) that contains the 5-base insertion mutation in the LTR observed in HIV-1 isolated from patient X. (B) Replication in CEMx174 cells that express Vif (CEMx174Vif) of HIV-1 strain NL4-3, an NL4-3-based recombinant that contains a large deletion in Vif (ΔVif), and an NL4-3 recombinant that contains Vif sequences from the strain from patient X (XV). (C) Replication of the viral strains described in panel B in PBMC isolated from HIV-1-seronegative donors. (D) Replication in H9 cells of the viral strains described for panel B.
Vif sequences from the strain from patient X that contained a two-amino-acid insertion mutation (Fig. 2B) were introduced into the NL4-3 background to create the recombinant HIV-1 strain XV. In this experiment, the Vif gene sequences from the strain from patient X were exchanged for NL4-3 Vif sequences (1) in the plasmid p83-2, using NdeI and EcoRI restriction enzyme sites (New England Biolabs). XV, as well as wild-type NL4-3 and an NL4-3-based recombinant DNA containing a large deletion in Vif (HIVΔVif), were transfected into a Vif-complementing cell line (12) by DEAE-dextran transfection. All three strains replicated rapidly in these cells (Fig. 3B). Since the Vif protein is essential for efficient primate lentiviral replication in PBMC of primate origin (6, 10, 11, 32, 34, 35), the resulting stocks were assessed for replication efficiency in PBMC obtained from HIV-1-seronegative donors. In these experiments, Vif sequences diluted to contain 1 ng of p24Gag were used to infect 107 phytohemagglutinin-stimulated PBMC grown in RPMI medium supplemented with 20% FBS and 5% interleukin-2. The cell-free supernatant was harvested from infected cells, and levels of viral core protein were quantified by p24Gag ELISA. In this assay, wild-type HIV-1 replicated efficiently, the XV recombinant replicated poorly, and the HIVΔVif recombinant did not replicate to detectable levels (Fig. 3C). In addition to PBMC, Vif is required for efficient primate lentiviral replication in some T-cell lines (e.g., H9 and HUT78), monocyte-derived macrophages, and primary lymphocyte cultures (10, 11, 33, 34). To ensure that the poor XV growth was not cell-type dependent, stocks of XV, wild-type NL4-3, and HIVΔVif strains were used to infect H9 cells. For these experiments, 107 H9 cells that were grown in RPMI medium supplemented with 10% FBS were infected. Growth characteristics similar to those observed in PBMC (Fig. 3C) were observed in H9 cells (Fig. 3D).
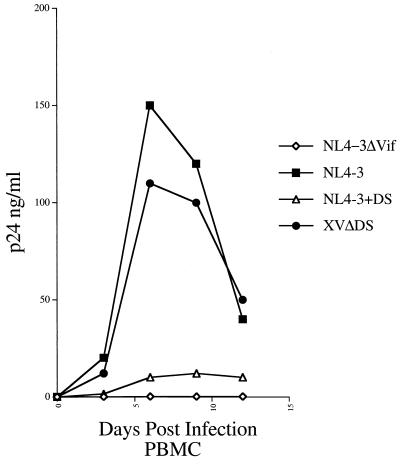
Next, we investigated whether the insertion mutation in the Vif sequences from the strain from patient X was the primary determinant of poor XV replication. For these experiments, two additional recombinants were engineered by overlap extension: XVΔDS, in which the two-amino-acid insertion mutation was deleted from XV, and NL4-3+DS, in which the insertion mutation was introduced into NL4-3 (Fig. 2C). The recombinant DNAs were transfected into the Vif-complementing cell line by using DEAE-dextran to produce viral stocks, which were then used to infect human PBMC isolated from HIV-1-seronegative donors. In these cultures, the XVΔDS strain replicated efficiently, whereas the NL4-3+DS strain replicated poorly (Fig. 4).
FIG. 4.
Replication in PBMC isolated from HIV-1-seronegative donors of HIV-1 strain NL4-3, an NL4-3-based recombinant that contains the insertion mutation (DS) observed in Vif from the strain from patient X (NL4-3+DS), and an XV-based recombinant with the insertion mutation removed (XVΔDS).
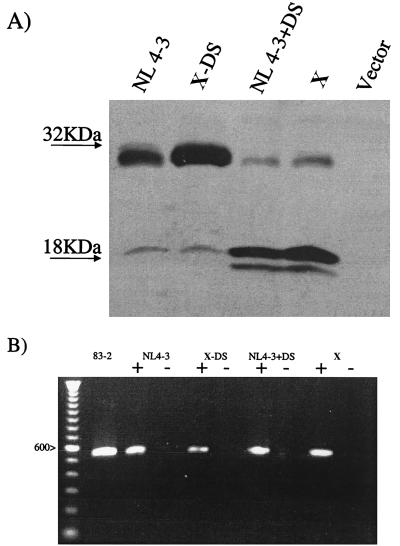
Since the Vif sequences from the strain from patient X had a marked inhibitory effect on HIV-1 replication, the nature of the Vif defect from the strain from patient X was further investigated by examining polypeptide expression. Vif sequences from the strain from patient X, as well as sequences from strains XΔDS, NL4-3, and NL4-3+DS, were fused at their carboxy termini to a Flag epitope by insertion into the pIRES-hrGFP-1a expression vector (Stratagene, La Jolla, Calif.), using BamHI and XhoI restriction sites. The resulting clones were introduced in 293T cells, using FuGene 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, Ind.). The cells were incubated for 48 h and lysed, and cellular protein was harvested (28). Cellular protein was electrophoresed through 10% polyacrylamide gel electrophoresis, transferred to a nitrocellulose membrane, and probed with a Flag monoclonal antibody (Sigma-Aldrich, St. Louis, Mo.) in a Western immunoblot. Vif protein was detected using an ECL detection kit (Amersham Pharmacia, Piscataway, N.J.). In this assay, the vast majority of NL4-3 and XΔDS Vif protein migrated as expected, whereas a small portion migrated markedly faster (Fig. 5A). Conversely, the vast majority of NL4-3+DS Vif protein and Vif protein from the strain from patient X migrated markedly faster than full-length Vif, whereas only a small portion comigrated with full-length Vif (Fig. 5A).
FIG. 5.
(A) Western immunoblot of strains from patient X and strains with NL4-3 or derivative Vif sequences. Cells transfected with vector sequences without Vif sequences (Vector) served as the control. The migration of molecular size markers is indicated to the left. (B) RT-PCR of strains from patient X and strains with NL4-3 or derivative Vif sequences. A plus sign indicates that the isolated RNA was reverse transcribed prior to PCR amplification, and a minus sign indicates that the isolated RNA was not reverse transcribed prior to PCR amplification. A 100-nucleotide ladder is shown on the left, and the identity of the 600-nucleotide band is indicated.
To determine if the rapidly migrating forms of Vif protein resulted from aberrant cleavage of polypeptide or splicing of message, RNA from 293T cells transfected with DNA containing the various forms of Vif (i.e., those from the strain from patient X, XΔDS, NL4-3, and NL4-3+DS) was isolated and reverse transcription-PCR (RT-PCR) was performed. In these experiments, the cells were incubated for 48 h and lysed and cellular RNA was harvested using RNAwiz (Ambion, Austin, Tex.). The DNA in these preparations was eliminated using a RNAqueous-4PCR kit (Ambion). The Vif RNA was reverse transcribed with Moloney murine leukemia virus reverse transcriptase (Stratagene), using a primer that annealed to 3′-terminal Vif sequences (5′GTGTCCATTCATTGTGTGGCT3′). These products, as well as plasmid DNA containing the 5′ sequences of HIV-1 strain NL4-3 (83-2), were amplified using a primer that annealed to the 5′ terminal (5′ATGAAACAGATGGCAGGGTGA3′) and a primer that annealed to sequences adjacent to the 3′ terminal Vif sequences (5′CCCTCTGTGGCCCTTGGT3′). The amplified DNA fragments, along with a size marker, were electrophoresed through 0.8% agarose for size determination. NL4-3 (83-2) sequences, which were amplified by PCR with the same primers used to amplify the RT products, served as the control. In these experiments, the RT-PCR products derived from the RNA templates migrated indistinguishably from one another (Fig. 5B). Furthermore, analyses of these products revealed that they contained open Vif-specific sequences that had not undergone aberrant splicing (data not shown).
It is likely that a factor common to patient Y and patient X is critical to their nonprogressor status. One such factor is the insertion mutation observed in their Vif sequences (Fig. 2B), which was almost certainly transmitted vertically from mother to child over 15 years ago. We demonstrated that these sequences markedly reduce HIV-1 replication in PBMC from seronegative donors as well as in H9 cells (Fig. 3C and D). Vif is required for the maintenance of detectable SIV loads and progression to AIDS in experimentally infected rhesus macaques (12) and almost certainly for the maintenance of detectable HIV loads and progression to AIDS in infected humans. Thus, the sequences described here are likely to have affected replication efficiency in vivo, much as we have observed in culture. This strongly suggests that the aberrant Vif gene sequences are an important determinant of nonprogression of disease in this mother and child. Interestingly, a similar mutation (a D insertion), in the same locus of Vif as was observed in the HIV-1 strains from patient Y and patient X, was independently observed in another CNP (15); however, it was not determined whether these sequences affected HIV-1 replication. Our data suggest that this mutant could, indeed, have such an effect. Thus, insertion mutations in this region of Vif could determine nonprogressor status in some individuals.
We have demonstrated that the Vif protein with a DS insertion mutation displayed an electrophoretic migration pattern that is distinct from that of wild-type protein (Fig. 5A). Therefore, an abnormally low concentration of full-length Vif in cells infected with the HIV-1 strain from patient X, which may result from an altered protein structure, might affect documented activities of Vif such as the following, any of which could affect virulence: (i) packaging into the HIV-1 nucleoprotein complex through a direct interaction with viral RNA (20); (ii) virion association, which may reflect a role late in replication, such as in virion assembly (38); (iii) inhibition of a putative viral inhibitory factor (25, 31); or (iv) some other, novel Vif activity. Further studies of these sequences might provide information regarding contributions of Vif structure and/or function to HIV-1 virulence.
Nucleotide sequence accession number.
The Vif sequences from the strains from patients X and Y have been made available on GenBank under accession number AY064706.
Acknowledgments
We thank L. Denekamp for technical support, G. Miller for critical review of the manuscript, J. Simpson and S. Romano for collection of clinical samples, and patients X and Y for providing clinical samples.
This work was supported by NIH grant AI39015 and Elizabeth Glaser Pediatric AIDS Foundation grant PG-51179.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., Z. Du, A. Y. Howe, S. Czajak, and R. C. Desrosiers. 1999. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J. Virol. 73:5814-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O'Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aquino-De Jesus, M. J., C. Anders, G. Miller, J. W. Sleasman, M. M. Goodenow, and W. A. Andiman. 2000. Genetically and epidemiologically related “non-syncytium-inducing” isolates of HIV-1 display heterogeneous growth patterns in macrophages. J. Med. Virol. 61:171-180. [DOI] [PubMed] [Google Scholar]
- 5.Binley, J. M., X. Jin, Y. Huang, L. Zhang, Y. Cao, D. D. Ho, and J. P. Moore. 1998. Persistent antibody responses but declining cytotoxic T-lymphocyte responses to multiple human immunodeficiency virus type 1 antigens in a long-term nonprogressing individual with a defective p17 proviral sequence and no detectable viral RNA expression. J. Virol. 72:3472-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chowdhury, I. H., W. Chao, M. J. Potash, P. Sova, H. E. Gendelman, and D. J. Volsky. 1996. vif-negative human immunodeficiency virus type 1 persistently replicates in primary macrophages, producing attenuated progeny virus. J. Virol. 70:5336-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, et al. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 8.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, and S. J. O'Brien. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 9.Fauci, A. S. 1996. Host factors and the pathogenesis of HIV-induced disease. Nature 384:529-534. [DOI] [PubMed] [Google Scholar]
- 10.Fisher, A. G., B. Ensoli, L. Ivanoff, M. Chamberlain, S. Petteway, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1987. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science 237:888-893. [DOI] [PubMed] [Google Scholar]
- 11.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibbs, J. S., D. A. Regier, and R. C. Desrosiers. 1994. Construction and in vitro properties of HIV-1 mutants with deletions in “nonessential” genes. AIDS Res. Hum. Retrovir. 10:343-350. [DOI] [PubMed] [Google Scholar]
- 13.Greenough, T. C., D. B. Brettler, M. Somasundaran, D. L. Panicali, and J. L. Sullivan. 1997. Human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL), virus load, and CD4 T cell loss: evidence supporting a protective role for CTL in vivo. J. Infect. Dis. 176:118-125. [DOI] [PubMed] [Google Scholar]
- 14.Harrer, T., E. Harrer, S. A. Kalams, P. Barbosa, A. Trocha, R. P. Johnson, T. Elbeik, M. B. Feinberg, S. P. Buchbinder, and B. D. Walker. 1996. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 156:2616-2623. [PubMed] [Google Scholar]
- 15.Hassaine, G., I. Agostini, D. Candotti, G. Bessou, M. Caballero, H. Agut, B. Autran, Y. Barthalay, and R. Vigne. 2000. Characterization of human immunodeficiency virus type 1 vif gene in long-term asymptomatic individuals. Virology 276:169-180. [DOI] [PubMed] [Google Scholar]
- 16.Hirsch, M. S., B. Conway, R. T. D'Aquila, V. A. Johnson, F. Brun-Vezinet, B. Clotet, L. M. Demeter, S. M. Hammer, D. M. Jacobsen, D. R. Kuritzkes, C. Loveday, J. W. Mellors, S. Vella, D. D. Richman, and International AIDS Society—USA Panel. 1998. Antiretroviral drug resistance testing in adults with HIV infection: implications for clinical management. JAMA 279:1984-1991. [DOI] [PubMed] [Google Scholar]
- 17.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 18.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 19.Iversen, A. K., E. G. Shpaer, A. G. Rodrigo, M. S. Hirsch, B. D. Walker, H. W. Sheppard, T. C. Merigan, and J. I. Mullins. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J. Virol. 69:5743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khan, M. A., C. Aberham, S. Kao, H. Akari, R. Gorelick, S. Bour, and K. Strebel. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252-7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 22.Kostrikis, L. G., Y. Huang, J. P. Moore, S. M. Wolinsky, L. Zhang, Y. Guo, L. Deutsch, J. Phair, A. U. Neumann, and D. D. Ho. 1998. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat. Med. 4:350-353. [DOI] [PubMed] [Google Scholar]
- 23.Learmont, J. C., A. F. Geczy, J. Mills, L. J. Ashton, C. H. Raynes-Greenow, R. J. Garsia, W. B. Dyer, L. McIntyre, R. B. Oelrichs, D. I. Rhodes, N. J. Deacon, and J. S. Sullivan. 1999. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N. Engl. J. Med. 340:1715-1722. [DOI] [PubMed] [Google Scholar]
- 24.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 25.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naidu, Y. M., H. W. Kestler III, Y. Li, C. V. Butler, D. P. Silva, D. K. Schmidt, C. D. Troup, P. K. Sehgal, P. Sonigo, M. D. Daniel, et al. 1988. Characterization of infectious molecular clones of simian immunodeficiency virus (SIVmac) and human immunodeficiency virus type 2: persistent infection of rhesus monkeys with molecularly cloned SIVmac. J. Virol. 62:4691-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pantaleo, G., S. Menzo, M. Vaccarezza, C. Graziosi, O. J. Cohen, J. F. Demarest, D. Montefiori, J. M. Orenstein, C. Fox, L. K. Schrager, et al. 1995. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N. Engl. J. Med. 332:209-216. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 30.Scott, G. B., C. Hutto, R. W. Makuch, M. T. Mastrucci, T. O'Connor, C. D. Mitchell, E. J. Trapido, and W. P. Parks. 1989. Survival in children with perinatally acquired human immunodeficiency virus type 1 infection. N. Engl. J. Med. 321:1791-1796. [DOI] [PubMed] [Google Scholar]
- 31.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 32.Sodroski, J., W. C. Goh, C. Rosen, A. Tartar, D. Portetelle, A. Burny, and W. Haseltine. 1986. Replicative and cytopathic potential of HTLV-III/LAV with sor gene deletions. Science 231:1549-1553. [DOI] [PubMed] [Google Scholar]
- 33.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strebel, K., D. Daugherty, K. Clouse, D. Cohen, T. Folks, and M. A. Martin. 1987. The HIV 'A' (sor) gene product is essential for virus infectivity. Nature 328:728-730. [DOI] [PubMed] [Google Scholar]
- 35.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, B., Y. C. Ge, P. Palasanthiran, S. H. Xiang, J. Ziegler, D. E. Dwyer, C. Randle, D. Dowton, A. Cunningham, and N. K. Saksena. 1996. Gene defects clustered at the C terminus of the vpr gene of HIV-1 in long-term nonprogressing mother and child pair: in vivo evolution of vpr quasispecies in blood and plasma. Virology 223:224-232. [DOI] [PubMed] [Google Scholar]
- 37.Winkler, C., W. Modi, M. W. Smith, G. W. Nelson, X. Wu, M. Carrington, M. Dean, T. Honjo, K. Tashiro, D. Yabe, S. Buchbinder, E. Vittinghoff, J. J. Goedert, T. R. O'Brien, L. P. Jacobson, R. Detels, S. Donfield, A. Willoughby, E. Gomperts, D. Vlahov, J. Phair, and S. J. O'Brien. 1998. Genetic restriction of AIDS pathogenesis by an SDF-1 chemokine gene variant. ALIVE Study, Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort (SFCC). Science 279:389-393. [DOI] [PubMed] [Google Scholar]
- 38.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]