Abstract
In this study, a zebrafish homologue of the coxsackievirus and adenovirus receptor (CAR) protein was identified. Although the extracellular domain of zebrafish CAR (zCAR) is less than 50% identical to that of human CAR (hCAR), zCAR mediated infection of transfected cells by both adenovirus type 5 and coxsackievirus B3. CAR residues interacting deep within the coxsackievirus canyon are highly conserved in zCAR and hCAR, which is consistent with the idea that receptor contacts within the canyon are responsible for coxsackievirus attachment. In contrast, CAR residues contacting the south edge of the canyon are not conserved, suggesting that receptor interaction with the viral “puff region” is not essential for attachment.
Coxsackie B viruses and many adenoviruses initiate infection by attaching to a 46-kDa transmembrane protein, the coxsackievirus and adenovirus receptor (CAR) (1, 16, 22). Mice are susceptible to infection by coxsackie B viruses, and a murine homologue of CAR has been shown to function as a receptor for both coxsackieviruses and adenoviruses (2, 22). CAR homologues identified in the rat, pig, dog (8), and cow (20) are nearly identical to human CAR (hCAR) and murine CAR, but these proteins have not been shown to function in virus infection. Several nonhuman viruses, including adenoviruses isolated from dogs (18), fowl (19), and chimpanzees (6), and a pig picornavirus, swine vesicular disease virus (11), have been shown to interact with hCAR. We wished to use interspecies CAR chimeras to map the site for virus attachment, and we isolated a CAR homologue from a nonmammalian vertebrate, the zebrafish (Danio rerio), in the expectation that it would not bind viruses. However, we found that zebrafish CAR (zCAR) functioned as a receptor when expressed in transfected cells.
Identification of a zCAR homologue.
A search of the expressed sequence tag database revealed several zebrafish cDNA clones with sequences similar to those of hCAR. When these cDNA clones were obtained and examined further, one clone (GenBank accession no. AI54913) was found to contain the entire coding sequence of a CAR homologue that showed 52% amino acid identity to hCAR overall, with 45% identity within the extracellular domain and 66% identity within the cytoplasmic domain (Fig. 1).
FIG. 1.
Predicted zCAR amino acid sequence. The hCAR sequence (h) and the sequence of a zebrafish homologue (z) (GenBank accession no. AF268197) are shown. The predicted hydrophobic leader (determined as described in reference 13) and transmembrane domains are in italics. Residues at the interface with adenovirus fiber (4) are underlined. (Residues at the interface with CB3 are shown in Fig. 5.)
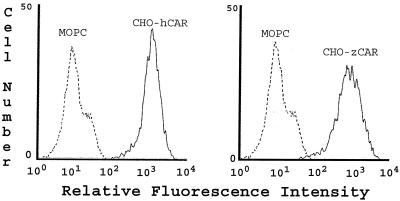
zCAR cDNA was inserted into a mammalian expression vector (pcDNA 3.1; Invitrogen, Carlsbad, Calif.) and introduced by electroporation into CHO dhfr cells as previously described for transfection with the integrin α2 subunit (3). Stably transfected cells were analyzed by flow cytometry with the monoclonal antibody RmcB, which recognizes hCAR (1) but not murine CAR (data not shown). Somewhat unexpectedly, RmcB detected zCAR on the surface of transfected CHO cells (Fig. 2); as was previously observed (1), mock-transfected CHO cells showed no staining with RmcB. To eliminate the possibility that we had inadvertently obtained CHO cells expressing hCAR, the zCAR cDNA was reisolated, resequenced, and used for an independent transfection; the same result was obtained.
FIG. 2.
Expression of zCAR on transfected CHO cells. CHO cells transfected with cDNA encoding hCAR or zCAR were incubated first with the anti-CAR monoclonal antibody RmcB or with a control antibody (MOPC 195) and then with fluorescein isothiocyanate-conjugated goat antibody to mouse immunoglobulin. Cells were examined by flow cytometry. As previously reported (1), no RmcB staining of mock-transfected CHO cells was observed (data not shown).
zCAR functions as a virus receptor.
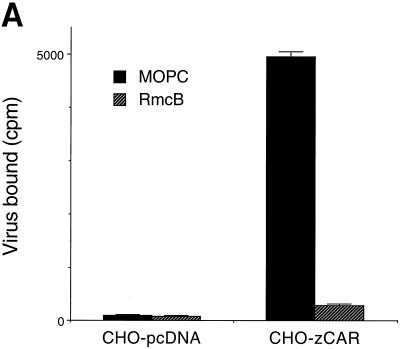
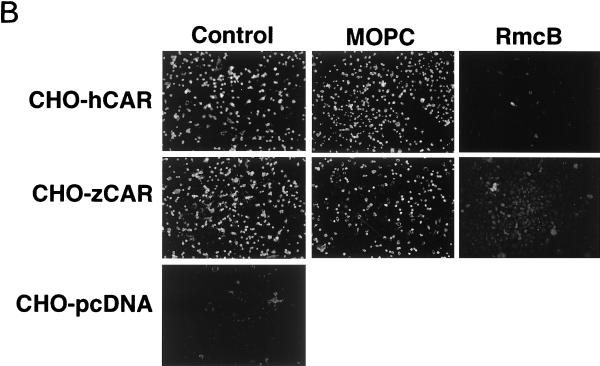
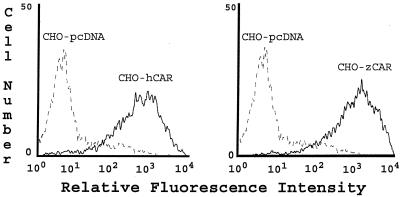
Wild-type CHO cells do not appear to express CAR (as detected by indirect immunofluorescence and immunoblotting with a polyclonal anti-CAR antibody that detects human, murine, and canine CAR [reference 5 and unpublished data]), and CHO cells do not bind radiolabeled coxsackievirus B3 (CB3) or adenovirus (1). However, CHO-zCAR cells, like CHO-hCAR cells, bound radiolabeled CB3 and binding was blocked by RmcB (Fig. 3A). CHO-zCAR cells became infected, as demonstrated by viral cytopathic changes (data not shown) and by expression of viral antigen after exposure to CB3 (Fig. 3B); CB3 infection was blocked by RmcB. CHO-zCAR cells, like CHO-hCAR cells, but not mock-transfected CHO cells, were efficiently transduced by an adenovirus type 5 vector carrying green fluorescent protein (Fig. 4); transduction of zCAR cells, like transduction of hCAR cells, was inhibited by a polyclonal anti-CAR antibody (5) (data not shown).
FIG. 3.
CB3 interaction with zCAR. (A) Virus attachment. In 24-well tissue culture plates, confluent CHO cells stably transfected with zCAR in pcDNA 3.1 (CHO-zCAR) or with expression vector alone (CHO-pcDNA) were exposed to monoclonal antibody RmcB (ascites fluid diluted 1/100) or to the control antibody MOPC 195 for 30 min at room temperature. Monolayers were rinsed and then incubated with 35S-labeled CB3-Nancy (prepared as described in reference 3) for 4 h at room temperature. Monolayers were washed four times, and then cell-bound radioactivity was determined. (B) Virus infection. CHO-zCAR, CHO-hCAR, or CHO-pcDNA cells grown on glass multiwell slides were preincubated with a control monoclonal antibody (MOPC 195), the anti-CAR monoclonal antibody RmcB, or no antibody before infection with CB3-Nancy (10 PFU per cell). After incubation for 24 h at 37°C, infected cells were visualized by staining for CB3 viral antigen with a polyclonal horse anti-CB3 serum, followed by a fluorescein isothiocyanate-conjugated goat antibody to horse immunoglobulin and examination by immunofluorescence microscopy.
FIG. 4.
zCAR mediates adenovirus type 5-mediated gene delivery. Cells were incubated with adenovirus type 5 carrying green fluorescent protein (1 PFU per cell) for 1 h at room temperature and then cultured for 48 h at 37°C. Green fluorescent protein expression was determined by flow cytometry.
CAR residues that make contact within the CB3 canyon are highly conserved.
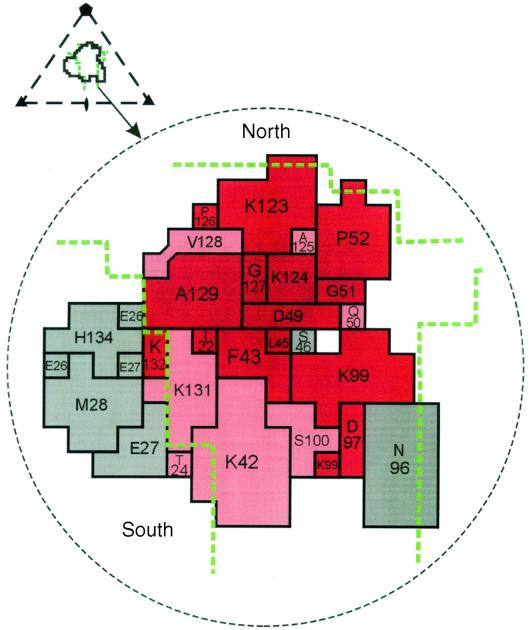
The structure of CB3 in complex with CAR was recently determined by cryo-electron microscopy (9). CAR's N-terminal immunoglobulin domain inserts into the canyon on the CB3 surface, with its distal end in contact with the north rim and floor of the canyon and the A and G beta strands in contact with the south rim, which is largely composed of residues from the “puff region,” a prominence formed by the EF loops of VP2. In zCAR, residues that make contact with virus deep within the canyon are highly conserved whereas those residues that contact the south rim of the canyon are not conserved (Fig. 5). This suggests that receptor interaction with the viral puff region—in poliovirus and rhinovirus, a recognition site for neutralizing antibodies (15, 17)—is not essential for virus attachment. In a previous study (21), mutation of hCAR K99, a conserved residue that contacts the north edge of the canyon, did not interfere with CB3 attachment to CAR. Thus, consistent with previous predictions (17), receptor contacts with viral residues deep within the canyon, but not with residues exposed on the virus surface, are likely to be essential for coxsackievirus attachment.
FIG. 5.
Receptor residues in contact with the virus surface. The CB3 footprint onto CAR is shown, with the boundaries of the canyon marked by dashed green lines. Residues in red are identical in zCAR and hCAR, and those in pink are similar in size and charge. Nonconserved residues are shown in grey. The triangular inset depicts the icosahedral asymmetric unit of the virus, the receptor footprint on the virus surface, and the boundaries of the canyon. CAR residues are labeled as in reference 1. This figure is adapted from He et al. (9).
Based on the crystal structure of a CAR-adenovirus fiber complex (4), 16 hCAR residues contact the fiber; 8 of these are identical in zCAR (Fig. 1), and many of the others have undergone conservative replacements.
The zCAR extracellular domain is less than 50% identical to that of hCAR, yet it is sufficiently similar to hCAR to permit recognition by monoclonal antibody RmcB and to permit attachment and infection by CB3 and adenovirus type 5. Although picornaviruses and adenoviruses have been isolated from fish (7), there is no evidence to suggest that zCAR functions naturally as a virus receptor. However, the observation that zCAR permits virus infection of transfected cells provides information about the structural basis of virus-receptor interaction and suggests that contacts within the canyon are primarily responsible for CB3 attachment.
Although CAR has been observed to mediate homotypic cell interactions (10), to contribute to the integrity of epithelial tight junctions (5), and to regulate the growth of tumor cells (14), its natural function is not well defined. In zebrafish, CAR is highly expressed in the embryonic nervous system (A. Chin, J. Bergelson, and M. Mullins, unpublished observations); techniques for gene targeting in zebrafish (12) may provide information about CAR function in these nonmammalian vertebrates. Search of the expressed sequence tag database reveals that there are CAR homologues in the frogs Silurana tropicalis (GenBank accession no. AL655801, AL594052, and AL630149) and Xenopus laevis (AW871784), but we have not identified CAR homologues in invertebrates.
Acknowledgments
J.P. and C.J.C. contributed equally to this work.
This work was supported by grants from the National Institutes of Health (HL35667 and AI50714), a Pediatric Infectious Diseases Society fellowship award sponsored by Glaxo-SmithKline, and an American Heart Association Beginning Grant-in-Aid.
We thank Jeffrey Faust for assistance with flow cytometry. We thank Michael Rossmann and Yongning He for comments on CAR structure and for providing Fig. 5.
REFERENCES
- 1.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 2.Bergelson, J. M., A. Krithivas, L. Celi, G. Droguett, M. S. Horwitz, T. Wickham, R. L. Crowell, and R. W. Finberg. 1998. The murine CAR homologue (mCAR) is a receptor for coxsackie B viruses and adenoviruses. J. Virol. 72:415-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergelson, J. M., N. St. John, S. Kawaguchi, M. Chan, H. Stubdal, J. Modlin, and R. W. Finberg. 1993. Infection by echoviruses 1 and 8 depends on the α2 subunit of human VLA-2. J. Virol. 67:6847-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bewley, M. C., K. Springer, Y.-B. Zhang, P. Freimuth, and J. M. Flanagan. 1999. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science 286:1579-1583. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, C. J., J. T.-C. Shieh, R. J. Pickles, T. Okegawa, J.-T. Hsieh, and J. M. Bergelson. 2001. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc. Natl. Acad. Sci. USA 98:15191-15196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, C. J., Z. Q. Xiang, G.-P. Gao, H. C. J. Ertl, J. M. Wilson, and J. M. Bergelson. 2002. Chimpanzee adenovirus CV-68 adapted as a gene delivery vector interacts with the coxsackievirus and adenovirus receptor (CAR). J. Gen. Virol. 83:151-155. [DOI] [PubMed] [Google Scholar]
- 7.Essbauer, S., and W. Ahne. 2001. Viruses of lower vertebrates. J. Vet. Med. B. 48:403-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fechner, H., A. Haack, H. Wang, X. Wang, K. Eizema, M. Pauschinger, R. G. Schoemaker, R. V. Veghel, A. B. Houtsmuller, H.-P. Schultheiss, J. M. J. Lamers, and W. Poller. 1999. Expression of coxsackie adenovirus receptor and alpha v-integrin does not correlate with adenovector targeting in vivo indicating anatomical vector barriers. Gene Ther. 6:1520-1535. [DOI] [PubMed] [Google Scholar]
- 9.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda, T., H. Saitoh, M. Masuko, T. Katagiri-Abe, K. Tominaga, I. Kozakai, K. Kobayashi, T. Kominishi, Y. G. Watanabe, S. Odani, and R. Kuwano. 2000. The coxsackievirus adenovirus receptor as a cell adhesion molecule in the developing mouse brain. Mol. Brain Res. 77:19-28. [DOI] [PubMed] [Google Scholar]
- 11.Martino, T. A., M. Petric, H. Weingartl, J. M. Bergelson, M. A. Opavsky, C. D. Richardson, J. F. Modlin, R. W. Finberg, K. C. Kain, N. Willis, C. J. Gauntt, and P. P. Liu. 2000. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B, and by swine vesicular disease virus. Virology 271:99-108. [DOI] [PubMed] [Google Scholar]
- 12.Nasevicius, A., and S. C. Ekker. 2000. Effective targeted gene “knockdown” in zebrafish. Nat. Genet. 26:216-220. [DOI] [PubMed] [Google Scholar]
- 13.Nielson, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 14.Okegawa, T., R.-C. Pong, Y. Li, J. M. Bergelson, A. I. Sagalowsy, and J.-T. Hsieh. 2001. The mechanism of the growth inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of CAR protein structure. Cancer Res. 61:6592-6600. [PubMed] [Google Scholar]
- 15.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roelvink, P. W., A. Lizonova, J. G. M. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. Brough, E, I. Kovesdi, and T. J. Wickham. 1998. The coxsackie-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H.-J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Moser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 18.Soudais, C., S. Boutin, S. S. Hong, M. Chillon, O. Danos, J. M. Bergelson, P. Boulanger, and E. J. Kremer. 2000. Canine adenovirus type 2 attachment and internalization: coxsackievirus-adenovirus receptor, alternative receptors, and an RGD-independent pathway. J. Virol. 74:10639-10649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan, P. K., A.-I. Michou, J. M. Bergelson, and M. Cotten. 2001. Defining CAR as a cellular receptor for the avian adenovirus CELO using a genetic analysis of the two viral fiber proteins. J. Gen. Virol. 82:1465-1472. [DOI] [PubMed] [Google Scholar]
- 20.Thoelen, I., E. Keyaerts, M. Lindberg, and M. van Ranst. 2001. Characterization of a cDNA encoding the bovine coxsackie and adenovirus receptor. Biochem. Biophys. Res. Commun. 288:805-808. [DOI] [PubMed] [Google Scholar]
- 21.Tomko, R. P., C. B. Johansson, M. Totrov, R. Abagyan, J. Frisen, and L. Philipson. 2000. Expression of the adenovirus receptor and its interaction with the fiber knob. Exp. Cell Res. 255:47-55. [DOI] [PubMed] [Google Scholar]
- 22.Tomko, R. P., R. Xu, and L. Philipson. 1997. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc. Natl. Acad. Sci. USA 94:3352-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]