Abstract
The V protein of the paramyxovirus simian virus 5 (SV5) is responsible for targeted degradation of STAT1 and the block in alpha/beta interferon (IFN-α/β) signaling that occurs after SV5 infection of human cells. We have analyzed the growth properties of a recombinant SV5 that was engineered to be defective in targeting STAT1 degradation. A recombinant SV5 (rSV5-P/V-CPI−) was engineered to contain six naturally occurring P/V protein mutations, three of which have been shown in previous transfection experiments to disrupt the V-mediated block in IFN-α/β signaling. In contrast to wild-type (WT) SV5, human cells infected with rSV5-P/V-CPI− had STAT1 levels similar to those in mock-infected cells. Cells infected with rSV5-P/V-CPI− were found to express higher-than-WT levels of viral proteins and mRNA, suggesting that the P/V mutations had disrupted the regulation of viral RNA synthesis. Despite the inability to target STAT1 for degradation, single-step growth assays showed that the rSV5-P/V-CPI− mutant virus grew better than WT SV5 in all cell lines tested. Unexpectedly, cells infected with rSV5-P/V-CPI− but not WT SV5 showed an activation of a reporter gene that was under control of the IFN-β promoter. The secretion of IFN from cells infected with rSV5-P/V-CPI− but not WT SV5 was confirmed by a bioassay for IFN. The rSV5-P/V-CPI− mutant grew to higher titers than did WT rSV5 at early times in multistep growth assays. However, rSV5-P/V-CPI− growth quickly reached a final plateau while WT rSV5 continued to grow and produced a final titer higher than that of rSV5-P/V-CPI− by late times postinfection. In contrast to WT rSV5, infection of a variety of cell lines with rSV5-P/V-CPI− induced cell death pathways with characteristics of apoptosis. Our results confirm a role for the SV5 V protein in blocking IFN signaling but also suggest new roles for the P/V gene products in controlling viral gene expression, the induction of IFN-α/β synthesis, and virus-induced apoptosis.
Alpha and beta interferons (IFN-α and IFN-β) are important cytokines that are produced during virus infection (reviewed in references 5 and 46). IFN-α/β signaling is initiated when secreted IFN binds to its receptor on the cell surface, resulting in the phosphorylation of a family of latent transcription factors called STAT1 and STAT2 (signal transducers and activators of transcription). IFN-activated STAT1 and STAT2 heterodimerize and associate with p48 (also known as IRF-9) to form the transcription factor ISGF3. This transcription factor binds to interferon-sensitive response elements (ISRE) located in the promoter region of IFN-inducible genes (reviewed in reference 24). Many of the identified IFN-induced gene products have potent antiviral activity and can contribute to the inhibition of host and viral protein synthesis, induction of apoptosis, and clearance of viral infections. As such, viruses have evolved mechanisms that counteract the induction of IFN, the activation of the IFN signaling pathways, or both of these processes (reviewed in references 15 and 18). Here we report that the paramyxovirus simian virus 5 (SV5) is a very poor inducer of IFN synthesis and that IFN signaling or cell death pathways are not activated in infected cells as shown previously (12). However, our results show that IFN synthesis and IFN signaling and apoptotic pathways are efficiently activated by infection with recombinant SV5 (rSV5) containing naturally occurring substitutions in the P/V gene.
Paramyxoviruses are a family of enveloped viruses whose genome is composed of a single strand of negative-sense RNA. SV5 is a prototypic member of the rubulaviruses, a paramyxovirus genus that includes mumps virus (MuV), human parainfluenza virus type 2 (HPIV2), and SV41 (29). The ∼15-kb SV5 genome contains seven tandemly linked genes that code for the nucleocapsid protein (NP), which binds the viral RNA genome; the phospho- (P) and large (L) proteins, which together constitute the viral polymerase; the matrix (M) protein, which is involved in virus assembly; and the membrane proteins HN and F, which are responsible for cell attachment and fusion, respectively. Two viral proteins regulate the host response to virus infection. The small hydrophobic (SH) protein serves an unknown function in the viral life cycle, but infection of MDBK cells with an rSV5 lacking the SH gene results in the induction of apoptosis (20). The V protein blocks IFN-α/β signaling by inducing the degradation of STAT1 (13).
Paramyxovirus gene expression is thought to occur by a “stop-start” mechanism in which the viral polymerase produces a decreasing gradient of monocistronic mRNAs for genes located further from the 3′-end promoter (29). Transcription of the paramyxovirus P/V gene differs from transcription of other viral genes, since the viral polymerase produces two mRNAs by a process called RNA editing (48). For SV5, accurate transcription of the P/V gene produces an mRNA that encodes the V protein, while the P mRNA contains two additional nontemplate G residues added cotranscriptionally at a precise location in the P/V transcript by the viral polymerase (48). Thus, the V and P proteins have the same amino-terminal segment (the P/V region) but different C-terminal regions. The phosphoprotein (P protein) plays multiple roles in the paramyxovirus growth cycle (6, 11, 22), including a function as a polymerase subunit with L and an assembly function with NP during genome replication. The V protein interacts with the 127-kDa subunit of the damage-specific DNA binding protein (DDB1) (31), acts to slow the cell cycle (32), and targets STAT1 for degradation (13).
Paramyxoviruses have evolved mechanisms to counteract the antiviral effects of IFN-α/β signaling (15, 18, 49). For example, infection with the paramyxoviruses Sendai virus (SeV) results in a block in IFN signaling (12, 49), and recent evidence suggests that the SeV C proteins are involved in inactivation of STAT1 (16, 17, 47). Remarkably, the rubulaviruses SV5, MuV, and HPIV2 block IFN signaling by a mechanism different from that of SeV or HPIV3 (26, 27, 36, 49). Previous results have shown that infection of human cells with SV5 results in the loss of STAT1 protein, an essential component of the IFN-α/β signaling pathway (12, 49). Transfection experiments and expression of V from heterologous vectors have shown that the SV5 V protein is sufficient to inhibit IFN signaling in human cells by targeting STAT1 for degradation by the proteasome (13).
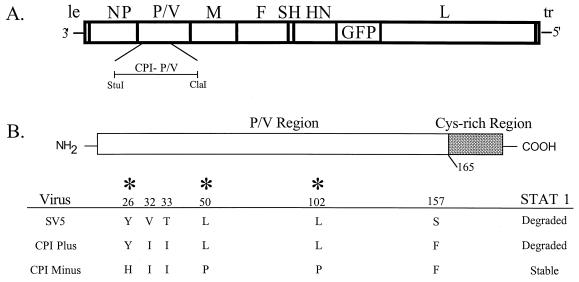
Naturally occurring strains of SV5 have been isolated that differ in their ability to block IFN signaling (7). Canine parainfluenza virus-plus (CPI+) is an SV5 strain isolated from a dog with neurological dysfunction (3). CPI− was subsequently isolated from a dog experimentally infected with CPI+ (4). Recent work has shown that while infection with CPI+ virus induces STAT1 degradation and blocks IFN-α/β signaling, the CPI− strain is defective in these two virus-induced alterations to the host cell (7). Sequence analysis has shown six amino acid differences in the amino-terminal region of the P/V genes of CPI−, which does not induce STAT1 degradation, and the W3 (wild-type [WT]) strain of SV5, which induces a loss of STAT1 (see Fig. 1B) (7, 45). The CPI+ and CPI− V proteins differ at only three residues in the P/V region, and all three of these amino acid differences contribute to their differential ability to block IFN signaling (7).
FIG. 1.
rSV5 genome structure and amino acid differences between the P/V region of the CPI+ and CPI− strains of SV5. (A) Structure of rSV5-GFP. The SV5 genome is depicted as a rectangle with vertical bars denoting the intergenic regions. The GFP open reading frame is found between the HN and L genes (19). The location of the StuI-ClaI restriction fragment used to introduce the CPI− P/V mutations is indicated. le, leader; tr, trailer. (B) Amino acid differences in the first 165 residues of the amino-terminal region of the P/V proteins are shown for WT, CPI+, and CPI− strains of SV5. Asterisks denote amino acid differences between CPI+ and CPI− (7, 45).
In this study, we have analyzed the growth properties of an rSV5 (rSV5-P/V-CPI−) that was engineered to encode the six substitutions in the N-terminal region of the CPI− P/V proteins that differ from WT rSV5, and these substitutions are sufficient to disrupt V-mediated STAT1 degradation (7). Our results show that rSV5-P/V-CPI− is altered in multiple phases of viral growth and virus-host interactions, including the control of viral gene expression, induction of IFN-α/β, and virus-induced cytopathic effect. Our results suggest new roles for the SV5 P/V gene products in the control of viral gene expression and the induction of IFN-α/β synthesis and paramyxovirus-mediated apoptosis.
MATERIALS AND METHODS
Cells, viruses, growth analysis, and plaque assays.
Monolayer cultures of cells were grown in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS). The W3A strain of SV5 and the Greer strain of HPIV2 were grown in MDBK and CV-1 cells, respectively. Single-step growth assays and plaque assays were carried out as described previously (37).
Recovery of rSV5 from cDNA.
The full-length infectious cDNA clone pBH311 (19) was kindly provided by B. He and R. A. Lamb (Northwestern University). To generate pRSV5-P/V-CPI−, a StuI-ClaI DNA fragment was excised from pEF.W3/CPI−/V, a plasmid encoding the CPI− V gene (7) and inserted into the corresponding region of pBH311. rSV5 was recovered using pRSV5-P/V-CPI− as described previously (19) with minor modifications (33, 37). Briefly, 3.5-cm dishes of A549 cells were infected at a multiplicity of infection (MOI) of ∼3 with vaccinia virus MVA and then transfected with plasmids carrying the NP (1.2 μg), P (0.15 μg), and L (1.5 μg) genes along with pRSV5-P/V-CPI− (5 μg). After overnight incubation, the medium was replaced with ∼105 uninfected Vero cells in DMEM containing 10% FBS. Medium was harvested 2 days later and used undiluted to infect fresh Vero cells. After 5 days, plaques were isolated on Vero cell monolayers and were used to generate virus stocks by growth in Vero cells. The presence of the CPI− P/V region was confirmed by sequencing reverse transcription-PCR products derived from infected-cell RNA.
Western blotting, isotopic labeling of polypeptides, and immunoprecipitation analysis.
Six-well dishes of cells were infected with WT rSV5-GFP or rSV5-P/V-CPI− as described in each figure legend. The cells were washed with phosphate-buffered saline (PBS) and incubated in DMEM containing 2% FBS. At each time point, cells were washed with PBS and lysed in 1% sodium dodecyl sulfate (SDS). Protein concentration of cell lysates was determined by the bicinchoninic acid assay (Pierce Chemicals). Equivalent amounts of protein were analyzed by Western blotting with rabbit antisera to the SV5 NP and P proteins (37) or the cellular STAT1 protein (clone 554; Santa Cruz Biotechnology) followed by horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence.
To detect radiolabeled proteins, MDBK cells that were mock infected or infected with rSV5-green fluorescent protein (GFP) or rSV5-P/V-CPI− were radiolabeled for 20 min at 8 h postinfection pi with 200 μCi of Tran 35S-label per ml. Cells were lysed in 1% SDS, and proteins were immunoprecipitated from cell extracts using polyclonal antibodies to NP, P, M, or GFP (Invitrogen) or a monoclonal V-specific antibody (clone 11); (39) before analysis by SDS-polyacrylamide gel electrophoresis (PAGE) as described previously (37).
Flow cytometry analysis of protein expression.
Dishes (diameter, 6 cm) of cells were mock infected or infected with rSV5-GFP or rSV5-P/V-CPI− as described in each figure legend. At each time p.i., medium was harvested and adherent cells were trypsinized. Media and trypsinized cells were centrifuged at 450 × g, and cells were resuspended in 600 μl of 3% paraformaldehyde (PFA) for fixation and stored at 4°C until ready for analysis by flow cytometry. The GFP fluorescence of individual cells was measured using a FACScalibur flow cytometer (Becton Dickinson Biosciences).
Analysis of viral transcription products.
The accumulation of viral RNA in infected MDBK cells was analyzed as described previously (37). At the indicated times p.i., poly(A)+ RNA was isolated from rSV5-GFP-infected, rSV5-P/V-CPI− mutant-infected or mock-infected cells using oligo(dT) cellulose. RNA samples were analyzed by Northern blotting with negative-sense 32P-labeled riboprobes specific for the SV5 NP gene (bases 1 to 180). NP RNA levels were determined by using a PhosphorImager instrument and software (Molecular Dynamics) and was normalized to glyceral dehyde-6-phosphate dehydrogerase GAPDH mRNA as described previously (37).
Transfection of reporter plasmids and IFN protection assays.
Induction of the IFN-β promoter was assayed using pβlux, a plasmid containing upstream bases 38 to 470 of the murine IFN-β regulatory region, including all four positive regulatory domains (PRDI to PRDIV) linked to a luciferase reporter gene (35). The pSV-βgal plasmid (Promega) consists of the β-galactosidase gene under the control of a constitutive SV40 promoter and was used to normalize for transfection efficiencies between samples. Six-well dishes of A549 cells were cotransfected with 1 μg of pSV-βgal and 5 μg of pβLux per well using 12 μl of FuGENE6 reagent (Roche Molecular Biochemicals) per well. At 24 h posttransfection, the cells were mock infected or infected with WT rSV5-GFP or rSV5-V-CPI−. Cells were harvested in reporter lysis buffer (Promega) at 20 h p.i., and luciferase and β-galactosidase activities were determined. Normalized luciferase activity was calculated as luciferase activity divided by β-galactosidase activity.
A pISRE-luc plasmid containing five copies of the ISG54 ISRE upstream of a TATA box and luciferase reporter gene (36) was used to measure the induction of ISRE transcription. Six-well dishes of A549 cells were transfected as described above using 1 μg of pISRE-luc, 1 μg of pSV-βgal, and 4 μl of FuGENE6. The results shown in Fig. 5A were obtained using Lipofectin transfection reagent, which consistently gave lower luciferase values than did FuGENE6. At 24 h posttransfection, the cells were mock infected or infected with WT rSV5-GFP or rSV5-V-CPI−. At 16 h later, the cell medium was supplemented for 6 h with or without 2,000 U of human universal IFN-α/β (αA/D; PBL Biomedical Laboratories), as described previously (49). Cells were harvested in reporter lysis buffer, and normalized luciferase activity was calculated.
FIG. 5.
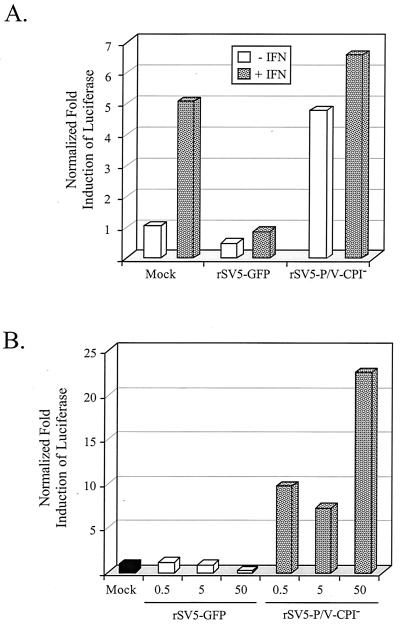
IFN signaling in cells infected with rSV5-V-CPI−. (A) Induction of ISRE. A549 cells were cotransfected with pSV-βgal and a plasmid containing the luciferase gene under the control of an ISRE. At 24 h later, the cells were mock infected or infected with rSV5-GFP or rSV5-P/V-CPI− at an MOI of 50. At 16 h p.i., the cells were incubated for 6 h with (hatched bars) or without (white bars) 2,000 U of IFN-α/β. Normalized luciferase activity was calculated by dividing luciferase activity by β-gal activity and is expressed as a fold induction over that seen in mock-infected cells. Data are representative of three independent experiments. (B) Effect of virus dose on the induction of ISRE. A549 cells were treated as described for panel A, except that the infections were at an MOI of 0.5, 5, or 50. At 20 h postinfection, normalized luciferase activity was determined as described for panel A. Data are representative of three independent experiments.
To quantitate the levels of IFN secretion, 3.5-cm dishes of A549 cells were mock infected or infected with individual viruses at an MOI of ∼5. At 24 h p.i., the medium was clarified, adjusted to pH ∼2 with HCl, and incubated at 4°C overnight to inactivate remaining virus. Samples were neutralized with NaOH, and serial dilutions in DMEM-2% FBS were added to fresh monolayers of HeLa cells in 96-well dishes. After overnight incubation, HeLa cell monolayers were infected with vesicular stomatitis virus (VSV; kindly provided by D. Lyles) at an MOI of ∼0.2. After overnight incubation, monolayers were stained with crystal violet and the optical density at 410 nm was determined. The number of IFN-equivalent units in the experimental samples that conferred 50% protection from the cytopathic effect of VSV was determined by comparison with samples analyzed in parallel using titrated units of human universal IFN-α/β.
Cell-killing assays.
Cell viability assays were performed as described previously (38). Briefly, cells were mock infected or infected at an MOI of 50 with WT rSV5-GFP or rSV5-P/V-CPI−, washed with PBS, and replaced with DMEM containing 2% FBS. At 2 days p.i., cells remaining on the dish were trypsizined and viable cells were determined by trypan blue exclusion. Data are expressed as fold increase in cell number relative to the starting number of cells prior to infection (38). Time-lapse video microscopy was carried out as described previously (38) using 25-cm2 flasks of A549 or MDBK cells that were mock infected or infected with rSV5-GFP or rSV5-V-CPI−. Cells were cultured on a time-lapse video microscopy system (37°C at 5% CO2) using a Zeiss inverted Axiovert phase-contrast microscope with an attached view camera. Pictures were recorded at intervals on standard VHS tape and were viewed to monitor cytopathic changes in cell morphology. The accumulation of rounded cells that did not undergo cell division was determined and is expressed as a percentage of the total number of cells in the field (38).
Fluorescence microscopy assays.
Microscopy experiments were performed with a Nikon Eclipse fluorescence microscope using a 20× lens. Images were captured using a QImaging digital camera and processed using QCapture software. Exposure times were manually set to be constant between samples. For low-MOI experiments, 3.5-cm dishes of A549 cells were mock infected or infected with rSV5-GFP or rSV5-P/V-CPI− at an MOI of 0.05. At each time postinfection, medium was removed and the cells were washed with PBS. The cells were fixed with 2% paraformaldehyde (PFA) for 10 min in the dark and then analyzed by microscopy. For apoptosis assays, 3.5-cm dishes of A549 cells were mock infected or infected with rSV5 viruses at an MOI of 50. At 24 and 48 h p.i., the cells were washed in PBS and fixed with a 3:1 mix of ethanol-acetic acid for 10 min at room temperature. Terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining was performed as described by the In Situ death detection kit (Roche Molecular Biochemicals).
RESULTS
Recovery of rSV5 encoding the CPI− P/V amino-terminal region.
Infection with WT rSV5 or CPI+ results in the degradation of STAT1 in human cells, while infection with CPI− does not induce the degradation of STAT1 (7). The amino-terminal segment of the CPI− P/V proteins differs from the WT P/V sequence at six residues (shown in Fig. 1B at amino acids 26, 32, 33, 50, 102, and 157), but the V-specific Cys-rich segment is conserved (45). Three amino acid differences in the P/V region between CPI− and CPI+ (Y26H, L50P, and L102P) are thought to be important for V-mediated STAT1 degradation. To generate an rSV5 encoding the six substitutions in the CPI-minus common P/V region, the full-length SV5 infectious cDNA was modified to contain a StuI-ClaI restriction fragment encoding the amino-terminal region of the CPI− P/V gene (Fig. 1A) (7). Both the recombinant virus used in this study as a WT control (designated WT rSV5-GFP) and the CPI− P/V mutant contained an additional transcription unit encoding GFP between the HN and L genes (Fig. 1A) to facilitate the quantitation of infected cells. Previous work has shown that inserting the additional GFP gene into WT rSV5 does not affect virus growth properties (19). Virus encoding GFP and the CPI− P/V region was recovered from Vero cells that are defective in IFN production (14) by transfection with the resulting plasmid and was designated rSV5-P/V-CPI−.
Enhanced expression of viral proteins and mRNA in cells infected with the rSV5-P/V-CPI− mutant virus.
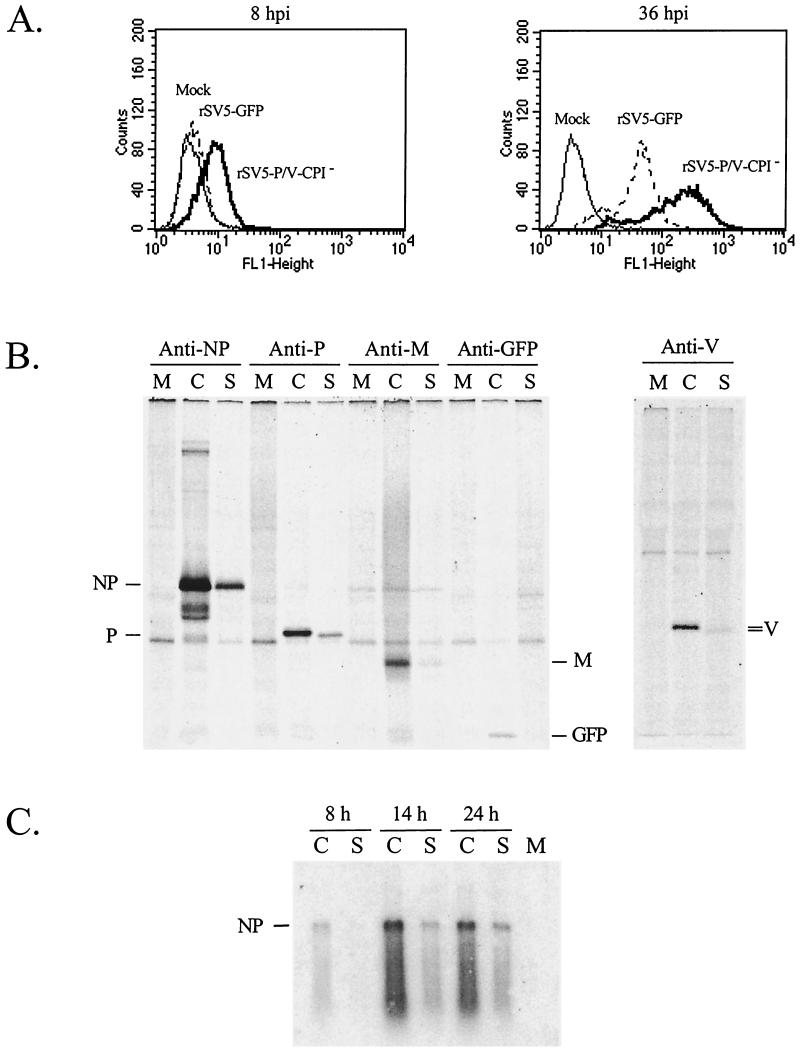
To determine if protein levels were affected by the P/V gene mutations, flow cytometry was performed for expression of GFP at 8 and 36 h p.i. Surprisingly, cells infected with rSV5-P/V-CPI− had higher fluorescence over time than did cells infected with WT rSV5-GFP. As shown in Fig. 2A, A549 cells infected with the rSV5-P/V-CPI− mutant expressed detectable levels of GFP by 8 h p.i. while the level of GFP in cells infected with WT rSV5-GFP was indistinguishable from that in the mock-infected control sample. By 36 h p.i., the level of GFP in rSV5-P/V-CPI− mutant-infected cells was ∼5- to 10-fold higher than in WT rSV5-GFP infected cells.
FIG. 2.
Elevated levels of viral protein and RNA in cells infected with rSV5-V-CPI−. (A) Elevated GFP expression in cells infected with rSV5-V-CPI−. A549 cells were mock infected or infected with rSV5-GFP or rSV5-V-CPI−. The cells were harvested at the indicated times p.i.and analyzed by flow cytometry for expression of GFP. (B) Elevated rate of protein synthesis in cells infected with rSV5-V-CPI−. MDBK cells were mock infected (M lanes) or infected with rSV5-GFP (S lanes) or rSV5-P/V-CPI− (C lanes) at an MOI of 50. At 8 h p.i., the cells were radiolabeled for 20 min using Tran 35S-label, and proteins were immunoprecipitated from cells extracts by using antibodies to NP, P, M, GFP, or V and then analyzed by SDS-PAGE. (C) Elevated mRNA accumulation in cells infected with rSV5-V-CPI−. MDBK cells were mock infected (M lane) or infected with rSV5-GFP (S lanes) or rSV5-P/V-CPI− (C lanes) at an MOI of 50. At the indicated times p.i., poly(A)+ RNA was isolated and analyzed by Northern blotting with a negative-sense NP-specific riboprobe.
Pulse-labeling was carried out to determine if the elevated accumulation of viral protein and mRNA for rSV5-P/V-CPI− correlated with a higher rate of viral protein synthesis. MDBK cells that were mock infected or infected with rSV5-P/V-CPI− mutant or WT rSV5-GFP were pulse-radiolabeled with 35S-labeled amino acids at 8 h p.i. Proteins were immunoprecipitated from cell extracts and analyzed by SDS-PAGE. The rate of protein synthesis in cells infected with rSV5-P/V-CPI− was higher than that seen for WT rSV5-GFP for each protein examined, including proteins derived from genes at the 3′ (e.g., NP) as well as 5′ (e.g., GFP) ends of the genome (Fig. 2B). In the experiment in Fig. 2B, the rate of NP, P, and M synthesis in rSV5-P/V-CPI− mutant-infected cells was increased by 16-, 14-, and 24-fold, respectively. Since P and V are produced from the same gene by RNA editing, the substitutions in the amino-terminal region of P/V gene could alter the ratio of P and V proteins. As shown in Fig. 2B, immunoprecipitation analysis with an antibody to the V-specific Cys-rich region showed that the rate of protein synthesis in cells infected with the rSV5-P/V-CPI− mutant was ∼12-fold higher than that seen for WT rSV5-GFP, and this was similar to the ∼15-fold increase in P-protein synthesis.
The level of mRNA transcribed from the paramyxovirus genome is a major factor dictating viral protein levels (29). To determine if the higher levels of protein synthesis in cells infected with rSV5-P/V-CPI− correlated with higher mRNA levels, MDBK cells were infected with rSV5-P/V-CPI− or WT rSV5-GFP and the accumulation of viral poly(A)+ mRNA was measured by Northern blotting at 8, 14, and 24 h p.i. As shown in Fig. 2C, the level of NP mRNA in cells infected with the rSV5-P/V-CPI− mutant (C lanes) was higher than that seen for WT rSV5-GFP (S lanes) at all time points examined. Very similar results were obtained for P and M mRNAs (not shown). When NP mRNA levels in each sample were normalized to levels of cellular GAPDH, the rSV5-P/V-CPI− mutant was found to direct a ca. fivefold-higher accumulation of NP mRNA than that seen in cells infected with WT rSV5-GFP. Thus, naturally occurring substitutions in the amino-terminal P/V protein can lead to viruses with elevated transcription and translation properties.
rSV5-P/V-CPI− is not attenuated in single-step growth assays.
To determine how the P/V substitutions affected virus growth, a time course of virus production was analyzed in A549 cells infected at an MOI of 50 with WT rSV5-GFP or rSV5-V-CPI−. As shown in Fig. 3A, cells infected with rSV5-V-CPI− produced final virus yields that were very similar to those infected with WT rSV5-GFP, but the kinetics of virus growth for the mutant were slightly faster than for the WT strain. The rapid growth properties of rSV5-P/V-CPI− were not unique to A549 cells, since infection of a variety of cell lines with the rSV5-P/V-CPI− mutant always gave slightly higher yields by 16 h p.i. over that seen for WT rSV5-GFP (Fig. 3B), with the largest difference seen in MDBK cells. Thus, the mutant virus with P/V substitutions has the unexpected phenotypes of overexpressing viral protein and enhanced virus yield even in cells with intact IFN signaling pathways, as seen with CV-1, MDBK and A549 cells.
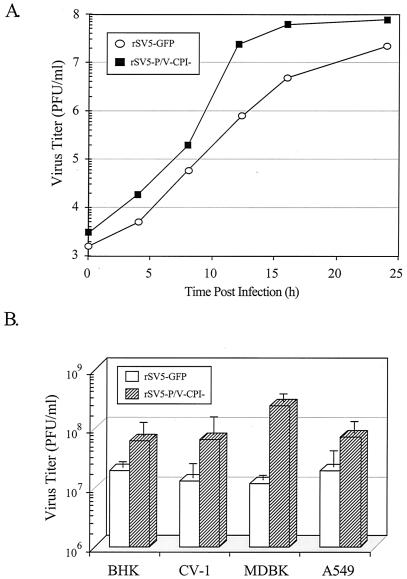
FIG. 3.
rSV5-P/V-CPI− grows to higher titers than rSV5-GFP. (A) Single-step growth assay. A549 cells were infected at an MOI of 50 with rSV5-GFP or rSV5-P/V-CPI−, and virus titers were determined at the indicated times p.i. by a plaque assay. The results shown are from one of three independent experiments. (B) Virus yield in different cell types. The indicated cell lines were infected at an MOI of 50 with rSV5-GFP or rSV5-P/V-CPI−. At 16 h p.i., medium was harvested and the virus titer was determined by a plaque assay. Data are the average of three determinations, and the standard deviation is shown by bars.
STAT1 levels remain constant during rSV5-P/V-CPI− infection.
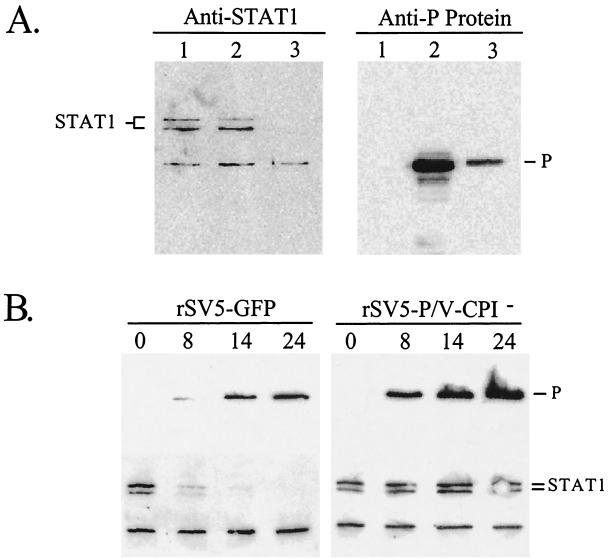
To determine if STAT1 levels were affected by infection with the rSV5-P/V-CPI− mutant, A549 cells were mock infected or infected with WT rSV5-GFP or rSV5-P/V-CPI− and cell lysates were analyzed by Western blotting with anti-STAT1 antibodies. As shown in Fig. 4A, STAT1 levels were greatly reduced in cells infected with WT rSV5-GFP (anti-STAT1 panel, lane 3) but there was no detectable difference in STAT1 levels between samples from control cells (lane 1) and cells infected with rSV5-P/V-CPI− (lane 2). To ensure that the lack of STAT1 degradation with rSV5-P/V-CPI− was not due to low levels of viral gene expression, samples were analyzed for P-protein levels by Western blotting. As shown in Fig 4A (anti-P panel), lysates from cells infected with rSV5-P/V-CPI− (lane 2) had higher levels of P protein than did lysates from cells infected with WT rSV5-GFP (lane 3). A time course experiment demonstrated that the STAT1 levels dropped dramatically by 8 h after infection of A549 cells with WT rSV5-GFP and were almost undetectable by 14 h (Fig. 4B). By contrast, the STAT1 levels remained relatively constant in cells infected with the rSV5-P/V-CPI− mutant (Fig. 4B). Analysis of viral protein expression showed higher levels of P (Fig. 4B) and NP (not shown) in cells infected with the rSV5-P/V-CPI− mutant than in cells infected with WT rSV5-GFP. These data support results from previous transfection experiments showing that the CPI− V mutations prevent V-mediated STAT1 degradation (7).
FIG. 4.
Protein expression and STAT1 levels in cells infected with rSV5-GFP or rSV5-P/V-CPI− virus. (A) Infection of human cells with rSV5-P/V-CPI− does not induce STAT1 degradation. A549 cells were mock infected (lanes 1) or infected with rSV5-P/V-CPI− (lanes 2) or rSV5-GFP (lanes 3) at an MOI of ∼50. At 24 h p.i., the cells were lysed and equivalent amounts of protein were analyzed by Western blotting with antisera specific for STAT1 (left) or SV5 P protein (right). (B) Time course of P-protein and STAT1 levels. A549 cells were infected with rSV5-GFP (left) or rSV5-P/V-CPI− (right) at an MOI of 50. At the indicated times p.i., cells were lysed and equivalent amounts of protein were analyzed by Western blotting with antisera specific for the SV5 P protein (top) or STAT1 (bottom).
Infection with the rSV5-P/V-CPI− mutant induces the ISRE promoter.
The above finding that rSV5 with P/V substitutions is defective in inducing STAT1 degradation suggested that the IFN signaling pathway would be functional in cells infected with the rSV5-P/V-CPI− mutant. To test this hypothesis, A549 cells were transfected with a ISRE-luciferase reporter plasmid and then 24 h later were mock infected or infected with WT rSV5-GFP or the rSV5-P/V-CPI− mutant. At 20 h p.i., media were supplemented with IFN-α/β for 6 h to induce the ISRE promoter or left unsupplemented, and normalized levels of luciferase were determined. Results of representative experiment are shown in Fig. 5A. Exogenously added IFN stimulated expression from the ISRE plasmid ca. fivefold in mock-infected cells, consistent with previous results using this reporter plasmid (36). In the case of cells infected with WT rSV5-GFP, addition of IFN did not induce ISRE-luciferase expression, consistent with previous published results that SV5 blocks the IFN signaling pathway (12, 13, 49). Surprisingly, cells infected with the rSV5-P/V-CPI− mutant showed luciferase levels matching that of the IFN-treated mock-infected sample, even in the absence of added IFN. The addition of IFN to the culture medium of rSV5-P/V-CPI− mutant-infected cells increased luciferase expression further. Infection of A549 cells with rSV5-P/V-CPI− induced the activation of the ISRE-luciferase plasmid at all MOIs tested (Fig. 5B), while cells infected with WT rSV5-GFP showed no induction. In the particular example shown in Fig. 5B, the sample from cells infected at an MOI of 0.5 with rSV5-P/V-CPI− is slightly higher than expected, but this was not routinely seen. These data showing that rSV5-P/V-CPI− induces the ISRE without exogenous IFN suggest that the rSV5-P/V-CPI− mutant-infected cells are secreting IFN that acts back on the infected cells to activate ISRE-luciferase expression.
Infection with rSV5-P/V-CPI− but not WT rSV5-GFP induces the IFN-β promoter.
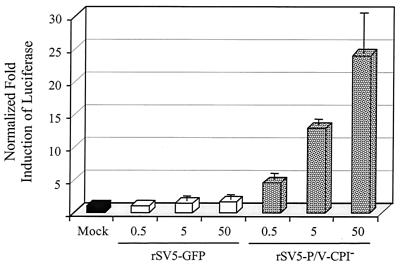
The experiments in Fig. 5 with the ISRE plasmid measured IFN signaling but did not measure IFN synthesis. To directly test if the IFN-β promoter is activated by rSV5-P/V-CPI− infection, A549 cells were transfected with a plasmid containing the luciferase gene under control of the IFN-β promoter (35) and then mock infected or infected with WT rSV5-GFP or rSV5-V-CPI−. As shown in Fig. 6, reporter gene activity from the IFN-β promoter in cells infected with rSV5-GFP was indistinguishable from that in mock-infected cells, even after infection at an MOI of ∼50. However, increased levels of expression from the transfected IFN-β-luciferase plasmid were detected when cells were infected with increasing amounts of the rSV5-P/V-CPI− mutant, with a ∼10-fold increase over the expression in mock-infected cells at an MOI of 50.
FIG. 6.
Induction of the IFN-β promoter by infection with rSV5-V-CPI− but not WT rSV5-GFP. A549 cells were transiently cotransfected with pSV-βgal and a plasmid encoding the luciferase gene under the control of the IFN-β promoter. At 24 h p.i., the cells were mock infected or infected with rSV5-GFP or rSV5-P/V-CPI− at the indicated MOI. Cells were lysed at 24 h p.i., and normalized luciferase activity was calculated as a fold induction over that seen in mock-infected cells. Data are the average of three determinations, and the standard deviation is shown by bars.
A bioassay was used to directly test if infection with the rSV5-P/V-CPI− mutant induced the secretion of IFN. A549 cells were mock infected or infected with WT rSV5-GFP, the rSV5-P/V-CPI− mutant, or HPIV-2, a known inducer of IFN-α/β secretion (26). At 24 h p.i., extracellular media were collected and virus was inactivated by low-pH treatment. Neutralized samples were then tested on HeLa cell monolayers for the ability to confer resistance to the cytopathic effect of VSV, a virus that is highly sensitive to the antiviral affects of IFN. The results of two independent experiments are shown in Table 1. In multiple experiments, no detectable protection from VSV challenge was seen in media from mock-infected or WT rSV5-GFP-infected A549 cells. By contrast, media from HPIV2-infected A549 cells conferred resistance to VSV challenge at a concentration that was equivalent to ∼5,100 to 20,000 U of IFN. Likewise, media from A549 cells infected with the rSV5-P/V-CPI− mutant conferred resistance to VSV challenge at a concentration that was equivalent to∼5,100 to 9,800 U of IFN. Taken together, the above data indicate that an rSV5 with substitutions in the P/V protein is defective in blocking STAT1-mediated IFN signaling. In addition, however, the mutant virus induces the activation of the β-IFN promoter and the secretion of an IFN-like cytokine, two properties which are not detected in cells infected with WT rSV5-GFP.
TABLE 1.
Antiviral activity in media from cells Infected with WT rSV5-GFP and rSV5-P/V-CPI−
| Samplea | Activity (U/ml)b in:
|
|
|---|---|---|
| Exp 1 | Exp 2 | |
| Mock infected | NDc | ND |
| rSV5-GFP | ND | ND |
| HPIV2 | 5,120 | 19,700 |
| rSV5-P/V-CPI− | 5,120 | 9,850 |
Monolayers of A549 cells were infected at an MOI of 5 with the indicated viruses, and medium was collected at 24 h p.i. Virus was inactivated by overnight incubation at pH ∼2.
Values are expressed as the number of units per milliliter which provide 50% inhibition of the VSV cytopathic effect on HeLa cell monolayers as described in Materials and Methods.
ND, none detected.
Growth of the rSV5-P/V-CPI− mutant after low-MOI infection.
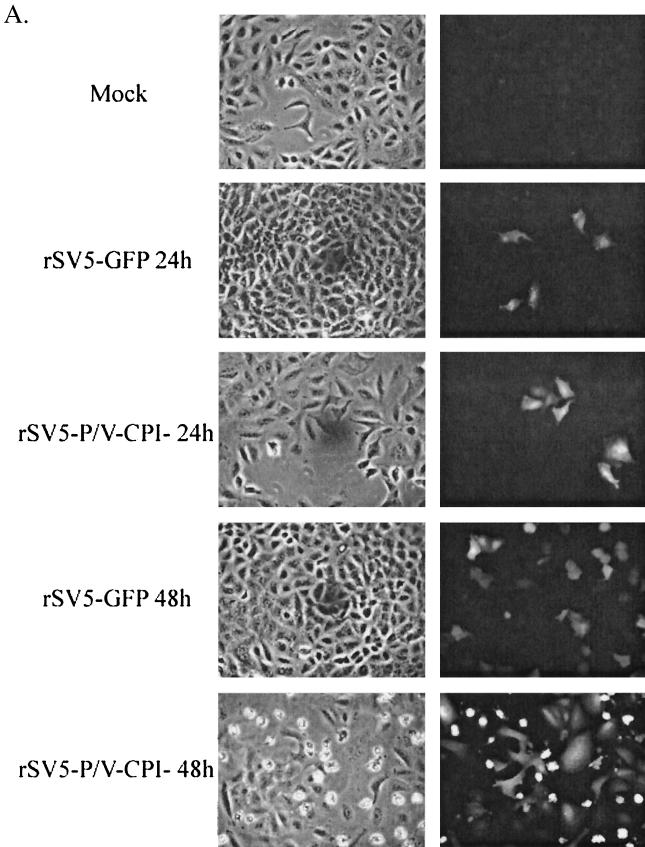
Because infection with rSV5-P/V-CPI− induces both IFN-α/β synthesis and signaling, we hypothesized that virus production and growth would be lower in multiple-cycle growth assays. To test this, A549 cells were infected at an MOI of 0.05 and at 24 and 48 h p.i. were monitored by microscopy for GFP expression. At 24 h p.i., approximately the same fraction of cells was expressing GFP in samples infected with either rSV5-GFP or rSV5-P/V-CPI− (Fig. 7A). Cells infected with rSV5-P/V-CPI− appear to have a greater fluorescence than cells infected with rSV5-GFP, consistent with the overexpression phenotype described above. At 48 h p.i., rSV5-P/V-CPI− had infected as many cells as had WT rSV5-GFP (Fig. 7A). The brightly fluorescing rounded cells in the rSV5-P/V-CPI− infection are indicative of dying cells that have rounded up on the dish, concentrating the GFP signal.
FIG. 7.
Multiple-step growth with WT rSV5-GFP and rSV5-P/V-CPI−. (A) Fluorescence microscopy assay for virus spread. A549 cells were mock infected or infected at an MOI of 0.05 with rSV5-GFP or rSV5-P/V-CPI−. At 24 or 48 h p.i., the cells were fixed with 2% PFA and then analyzed by microscopy for GFP fluorescence. Phase-contrast microscopy is shown in the left panels, and GFP fluorescence is shown in the right panels. (B) Multiple-step growth assay. A549 cells were infected with rSV5-GFP or rSV5-P/V-CPI− at an MOI of 0.05. Medium was harvested at the indicated times postinfection, and the viral titer was determined by a plaque assay on CV-1 cells.
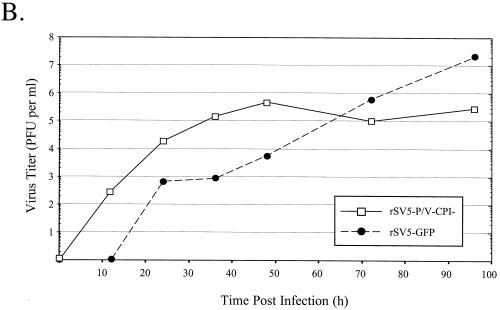
A multiple-step growth curve was generated to compare virus production in cells infected at low MOI with rSV5-GFP and rSV5-P/V-CPI−. A549 cells were infected at an MOI of 0.05, medium was harvested at various times postinfection, and plaque assays were performed to determine the viral titer. As shown in Fig. 7B, cells infected with rSV5-P/V-CPI− produced ∼100-fold more virus than did cells infected with WT rSV5-GFP up to 48 h p.i. However, 48 h p.i. was the peak of rSV5-P/V-CPI− virus production whereas WT rSV5-GFP production continued to increase and eventually reached final titers that were ∼100-fold higher than those of rSV5-P/V-CPI−. An added complication to interpreting these data is that cells infected with SV5 continued to grow and divide at a slightly lower rate than do mock-infected cells (32). By contrast, cells infected with the mutant virus were undergoing extensive cell death (see below). Thus, the ability of rSV5-P/V-CPI− infection to spread through the cell population may be underestimated due to loss of cells. Taken together, these data indicate that rSV5-P/V-CPI− grows to higher-than-WT levels at early times postinfection but that continuous spread of the mutant through the cell population is limited, perhaps due to the production of IFN.
Infection with rSV5-P/V-CPI− but not WT rSV5-GFP induces cell death.
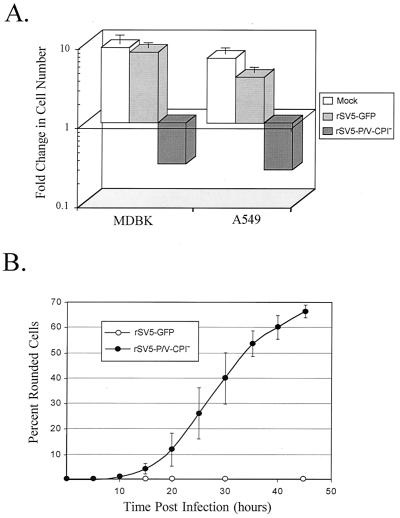
As observed during microscopy experiments (Fig. 7A), cells infected with the rSV5-P/V-CPI− mutant were found to show cytopathic effects not seen with WT rSV5-GFP infections. To quantitate the effect of rSV5-P/V-CPI− infection on cell viability, monolayers of A549 or MDBK cells were mock infected or infected with WT rSV5-GFP or the rSV5-P/V-CPI− mutant at high MOI. After 48 h, the cells were removed from the dishes and the number of viable cells was determined by trypan blue exclusion. A typical result is shown in Fig. 8A, where the number of viable cells remaining at 48 h p.i. is expressed as a fold change relative to the initial number of cells in the culture (38). Infection of MDBK cells with rSV5-GFP did not significantly affect the change in the number of viable cells relative to the mock-infected control (Fig. 8A), and the cell numbers increased by ca. seven- to eightfold. In A549 cells, the slight reduction in growth of infected (ca. fourfold change) relative to mock-infected (ca. sixfold change) samples is consistent with the report that SV5 infection slows the cell cycle (32). By contrast, infection of both MDBK and A549 cells with the rSV5-P/V-CPI− mutant resulted in ca. eightfold decrease in viable-cell numbers.
FIG. 8.
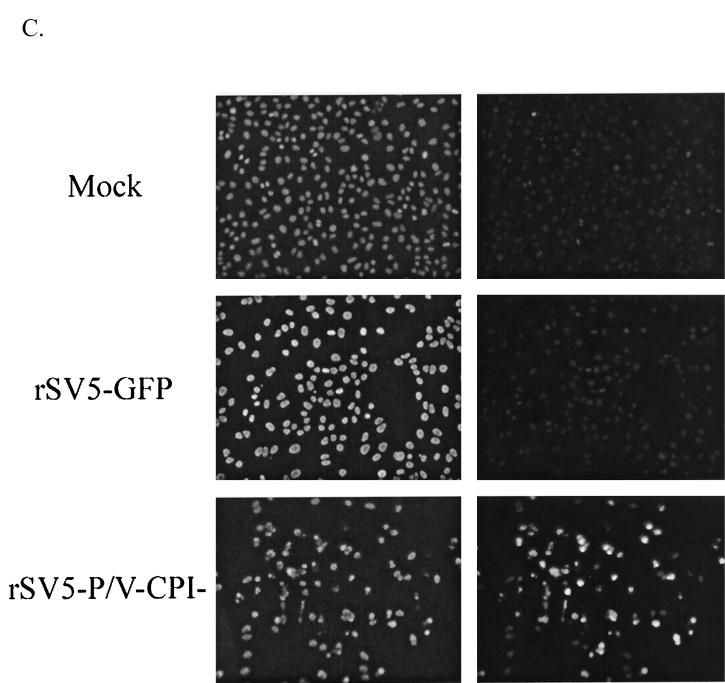
Infection with rSV5-P/V-CPI− but not WT rSV5-GFP results in cell death. (A) Cell viability during infection with rSV5-GFP or rSV5-V-CPI−. MDBK or A549 cells were mock infected or infected with rSV5-GFP or rSV5-P/V-CPI− at an MOI of 50. At 2 days p.i., viable-cell numbers were determined by trypan blue staining. Data are expressed as a fold change in cell number from the starting culture. (B) Time-lapse video microscopy of cytopathic effect. A549 cells were infected with rSV5-GFP or rSV5-P/V-CPI− at an MOI of ∼50, and cells were monitored by time-lapse video microscopy. At the indicated times p.i., the number of rounded cells in the field that did not later divide was counted as a percentage of the total number of cells in the field. Data are the mean of three determinations, and the standard deviation is shown by bars. (C) Fluorescence microscopy of TUNEL staining for DNA fragmentation. A549 cells were mock infected or infected at an MOI of 50. At 48 h p.i., TUNEL staining was performed as described in Materials and Methods. Photographs were taken at the same exposure for all samples.
Time-lapse video microscopy was carried out to more closely monitor cytopathic changes. Videotapes of the time course of infection were viewed for the number of rounded cells in a field that did not undergo cell division as a measure of a defined cytopathic effect. Figure 8B shows the results of three independent experiments in which the accumulation of rounded cells is expressed as a percentage of the total number of cells in the field. A549 cells infected with rSV5-GFP (Fig. 8B), were indistinguishable from mock-infected cells (data not shown), and by 45 h p.i., fewer than ∼2% of the cells showed cell rounding without undergoing cell division. By contrast, cytopathic effects were detected in rSV5-P/V-CPI− mutant-infected A549 cells starting at ∼15 h p.i., and by 45 h p.i., nearly 70% of the infected cells had rounded up.
Most of the rSV5-P/V-CPI− mutant-infected cells had additional characteristics of apoptosis including membrane blebbing and disintegration into cellular debris. To confirm that cell death was occurring by apoptosis, a TUNEL assay for DNA fragmentation was performed at 48 h p.i. and the results were observed by fluorescence microscopy. 4;6-Diamidino-2-pherylindole (DAPI) staining was used to detect all cells in the field, and rhodamine staining was used to display cells that are positive for DNA fragmentation. As seen in Fig. 8C, mock-infected A549 cells stain brightly with DAPI but very few cells were positive for TUNEL staining. The same result was seen in rSV5-GFP-infected A549 cells, where cells stained brightly with DAPI but TUNEL-positive cells were not readily detected. However, in rSV5-P/V-CPI− mutant-infected cells, a very large percentage of cells were found to be positive for TUNEL staining and also displayed a rounded phenotype, a morphological characteristic of apoptosis. To better quantitate the cells that were TUNEL positive, 10 randomly selected fields were chosen and the number of DAPI-stained vs. TUNEL-stained cells were counted. During rSV5-GFP infection, ∼2% of the cells were detected as TUNEL positive, while ∼68% of rSV5-P/V-CPI− infected cells stained positive for DNA fragmentation. These values are similar to those shown in Fig. 8B using time-lapse video microscopy to quantitate cytopathic effect. Collectively, these data indicate that mutations in the SV5 P/V gene convert a noncytopathic virus to one that kills cells by a mechanism with characteristics of apoptosis.
DISCUSSION
Paramyxoviruses have evolved mechanisms to counteract the host cell antiviral pathways induced by IFN. For example, infection with the rubulaviruses SV5 and HPIV2 induces the degradation of STAT1 and STAT2, respectively, both of which eliminate IFN-α/β signaling through the STAT-activated pathway (12, 13, 36, 49). In this study, we have examined the growth properties of an rSV5 that has lost the ability to target STAT1 for degradation. The rSV5-P/V-CPI− virus contains six substitutions in the P/V gene (Fig. 1B), three of which (Y26H, L50P, and L102P) are thought to be important for V-mediated STAT1 degradation (7). The effect of the other mutations on the growth properties of rSV5 are currently under investigation. While the rSV5-V-CPI− mutant had the expected phenotype of not targeting STAT1 for degradation, this recombinant virus also revealed three previously unknown roles for the SV5 P/V gene in the virus growth cycle and interaction with host cells. Our results show that an rSV5 strain with CPI− substitutions in the P/V gene expresses higher-than-WT levels of viral RNA and proteins, induces the synthesis of IFN-α/β, and induces cell killing.
Cells infected with the rSV5-P/V-CPI− mutant express viral mRNA and protein earlier and to higher levels than do cells infected with WT rSV5-GFP. It is important to note that since the CPI− P/V substitutions are in a domain shared between P and V, we cannot assign this phenotype exclusively to P or V. One possibility is that activity of the P protein has been enhanced by the CPI− substitutions. In SeV, the P protein has multiple activities in viral gene expression (6, 10, 11, 22) and the enhanced gene expression could be due to gain-of-function substitutions attributed to one or more of these P functions. Alternatively, the CPI− P/V substitutions may have affected a V function in gene expression (10). It has been proposed that the SeV V protein interacts with NP to inhibit RNA replication by blocking NP encapsidation of the genome (23) and that the SV5 V protein binds to soluble but not polymeric NP (40). Thus, it is possible that the CPI− P/V substitutions decrease the ability of V to bind NP and effectively inhibit replication. In this hypothesis, the enhanced gene expression in cells infected with the rSV5-P/V-CPI− mutant would be due to an increase in genome replication, which in turn would increase the number of templates involved in transcription. An interesting possibility is that enhanced gene expression is due to a combination of loss of function for V protein and gain of function for P protein. Work is in progress to test these hypotheses using an in vivo minigenome system that reconstitutes SV5 RNA replication and transcription using cDNA-derived components (41, 42).
Previous transfection experiments have shown that the V protein is responsible for the block in IFN signaling in SV5-infected human cells (13) and that the CPI− V protein is defective in targeting STAT1 for degradation (7). Our results are consistent with these previous results, since the addition of exogenous IFN activated an ISRE reporter plasmid in cells infected with the rSV5-P/V-CPI− mutant. Surprisingly, however, the ISRE and IFN-β reporter plasmids were activated by infection with rSV5-P/V-CPI− but not with WT rSV5-GFP, even without added IFN. Importantly, we have shown that the medium in cultures of cells infected with rSV5-P/V-CPI− but not WT rSV5 contains an IFN-like activity that can confer resistance to VSV challenge. Together with the activation of the IFN-β and ISRE reporter plasmids, these results are consistent with the proposal that infection with rSV5-P/V-CPI− but not with WT rSV5-GFP results in the synthesis and secretion of IFN-α/β. These results suggest a previously unknown role for the SV5 P/V gene in controlling the induction of IFN synthesis.
Double-stranded RNA that is produced during viral infections can be a potent inducer of IFN, and viruses have evolved mechanisms to block the cellular response to or recognition of double-stranded RNA (5, 15, 18). It is possible that the rSV5-P/V-CPI− mutant induces IFN synthesis due to loss of a function that is normally active in the WT rSV5 virus. The P or V protein could normally function to sequester double-stranded RNA or directly inhibit a cellular factor that senses the virus infection, such as protein kinase R or a member of the family of IFN regulatory factors (IRF) (30, 44). Alternatively, the earlier and elevated levels of rSV5-P/V-CPI− gene expression could activate the IFN pathway before anti-IFN mechanisms are in place as would be the case for the WT rSV5 virus.
Minus-strand RNA viruses have evolved different approaches to counteract the host cell IFN responses (15, 18). For example, VSV blocks IFN production but is highly sensitive to the antiviral effects of IFN signaling. Conversely, Newcastle disease virus and HPIV2 do not block IFN production (Table 1) (15, 26) but can survive the effects of high levels of IFN by blocking IFN signaling and the expression of many IFN-induced antiviral genes. Respiratory syncytial virus (RSV) is different from these other two groups, since RSV infection induces IFN synthesis but there is no block IFN signaling and the virus can still replicate and spread through cells even in the presence of exogenously added IFN (1, 25, 49). It is therefore assumed that there must be additional mechanisms by which the effects of IFN are blocked in RSV-infected cells. Interestingly, RSV does not carry a V or C gene (9), which may explain why it does not block IFN-α/β signaling (25). Previous results from transfection studies have shown that compared to SeV, SV5 infection of human cells results in very poor induction of a reporter gene under the control of the IFN-β promoter (12). As shown in Fig. 6, there is no significant induction of the IFN-β promoter compared to mock-infected cells. Furthermore, our bioassays show no detectable antiviral activity in media from rSV5-WT-infected cells. Thus, our work reported here and the results of other studies (12, 49) show that WT SV5 is an unusual paramyxovirus which has evolved to block IFN signaling but is also a very poor inducer of IFN-α/β.
The enhanced gene expression in cells infected with the rSV5-P/V-CPI− mutant correlated with single-step growth that was higher than that for WT rSV5-GFP, even in cell lines with intact IFN signaling pathways. In multistep growth assays, the rSV5-P/V-CPI− mutant produced an initial wave of virus that reached a plateau earlier than WT rSV5. However, WT rSV5 continued to produce virus over a longer time frame than the rSV5-P/V-CPI− mutant and eventually reached higher final titers by late times postinfection. The high-level growth properties of the rSV5-P/V-CPI− mutant occur in the presence of activated pathways for IFN synthesis and signaling, raising the question of how an rSV5 can survive if IFN synthesis is induced and IFN signaling to ISRE promoters is activated.
The Cys-rich C-terminal region of the MuV and HPIV2 V proteins is important in preventing an IFN-induced antiviral state (27, 34), and recombinant HPIV2 with a V protein lacking the Cys-rich domain is severely attenuated for growth in IFN-competent cells (26). We have taken an alternative approach to disrupting V-mediated STAT1 degradation, which uses the exchange of genome segments between naturally occurring strains of SV5 that differ in their ability to induce STAT1 degradation. Thus, these mutations may result in P and V proteins that are only partially defective in counteracting the host antiviral response. For example, while the rSV5-P/V-CPI− mutant induces IFN synthesis and signaling, it is possible that the P or V proteins still retain other functions that can block antiviral pathways despite the activation of IRF-3 and/or IRF-7 (30, 44).
Infection of most cell types with SV5 results in little if any cell death (8, 20, 38). An unexpected result of our experiments was that infection with rSV5-P/V-CPI− resulted in extensive cell death in a number of cell types (Fig. 8). One explanation of this could be that the production of IFN-α/β induces apoptosis (2). Since STAT1 is not degraded in rSV5-P/V-CPI− mutant-infected cells, it is also possible that STAT1 is involved in the induction of cell death. For example, STAT1-null cell lines are resistant to apoptosis induced by tumor necrosis factor alpha (28), and it is possible that V-mediated STAT1 degradation by WT rSV5 is an important mechanism for avoiding virus-induced apoptosis. For SeV, activated IRF-3 is an important component of the virus-induced apoptotic pathway (21), and it is possible that WT rSV5 and the rSV5-P/V-CPI− differ in their ability to block IRF activities. We have shown that infection of cells with rSV5-P/V-CPI− results in both overexpression and premature expression of protein and RNA compared to infection with rSV5-GFP (Fig. 2). Another possibility is that during WT SV5 infection, the amount of viral protein or RNA produced is insufficient to trigger a response in the cell to initiate death. Consequently, when cells are infected with rSV5-P/V-CPI−, the increased protein exceeds a critical threshold and apoptotic pathways are activated. Future work is needed to determine the link between overexpression of virus proteins and activation of apoptotic pathways.
In summary, our results with an rSV5 strain containing CPI− P/V gene substitutions suggest previously unrecognized roles for the P/V gene in controlling the induction of IFN-α/β synthesis, the regulated expression of viral RNA and proteins, and virus-induced cell death. Recombinant viruses containing exchanges of genes between the SV5 and CPI viruses will be valuable tools for future studies to reveal additional P/V functions that are STAT1 dependent or independent.
Acknowledgments
We thank Mike Keller, Ginger Young, Doug Lyles, and David Ornelles for helpful comments on the manuscript. We are grateful to Barbara Sherry for the kind gift of pBlux plasmid and to Curt Horvath for supplying the ISRE-luciferase plasmid. We thank Rick Randall (University of St. Andrews, St. Andrews, Scotland) for the kind gift of CPI P/V plasmids and for essential insight during the initial phases of this work. We thank Nathan Iyer of the Comprehensive Cancer Center for excellent technical support.
E.K.W. is supported by NIH training grant AI-07401. This work was supported by NIH grant AI42023.
REFERENCES
- 1.Atreya, P. L., and S. Kulkarni. 1999. Respiratory syncytial virus A2 is resistant to the antiviral effects of type I interferons and human MxA. Virology 261:227-241. [DOI] [PubMed] [Google Scholar]
- 2.Balachandran, S. P. C. Roberts, T. Kipperman, K. N. Bhalla, R. W. Compans, D. R. Archer, and G. N. Barber. 2000. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/caspase-8 death signaling pathway. J. Virol. 74:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgartner, W. K., A. E. Metzler, S. Krakowka, and A. Koestner. 1981. In vitro identification and characterization of a virus isolated from a dog with neurological dysfunction. Infect. Immun. 31:1177-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumgartner, W. K., S. Krakowka, and B. Durchfeld. 1991. In vitro cytopathogenicity and in vivo virulence of two strains of canine parainfluenza virus. Vet. Pathol. 28:324-331. [DOI] [PubMed] [Google Scholar]
- 5.Biron, C. A., and G. C. Sen. 2001. Interferons and other cytokines, p. 321-349. In D. Knipe, P. Howley, D. Griffin, et al. (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 6.Bowman, M. C., S. Smallwood, and S. A. Moyer. 1999. Dissection of individual functions of the Sendai virus phosphoprotein in transcription. J. Virol. 73:6474-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatziandreou, N., D. Young, J. Andrejeva, S. Goodbourn, and R. E. Randall. 2002. Differences in interferon sensitivity and biological properties of two related isolates of simian virus 5: a model for virus persistence. Virology 293:234-242. [DOI] [PubMed] [Google Scholar]
- 8.Choppin, P. W. 1964. Multiplication of a myxovirus (SV5) with minimal cytopathic effects and without interference. Virology 23:224-233. [DOI] [PubMed] [Google Scholar]
- 9.Collins, P. 1996. Respiratory syncytial virus, p. 1313-1351. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 10.Curran, J., R. Boeck, and D. Kolakofsky. 1991. The SeV P gene expresses both an essential protein and an inhibitor of RNA synthesis by shuffling modules via mRNA editing. EMBO J. 10:3079-3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Curran, J., T. Pelet, and D. Kolakofsky. 1994. An acidic activation-like domain of the SeV P protein is required for RNA synthesis and encapsidation. Virology 202:875-884. [DOI] [PubMed] [Google Scholar]
- 12.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. Sendai virus and SV5 block activation of IFN-responsive genes: importance of virus pathogenesis. J. Virol. 73:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didcock, L., D. F. Young, S. Goodbourn, and R. E. Randall. 1999. The V protein of SV5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 16.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons:cell signaling, immune modulation, antiviral responses and virus countermeasures. J. Gen. Virol. 81:2341-2364 [DOI] [PubMed] [Google Scholar]
- 19.He, B., R. G. Paterson, C. D. Ward, and R. A. Lamb. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249-260. [DOI] [PubMed] [Google Scholar]
- 20.He, B., G. Y. Lin, J. E. Durbin, R. K. Durbin, and R. A. Lamb. 2001. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J. Virol. 75:4068-4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heylbroeck, C., S. Balachandran, M. J. Servant, C. DeLuca, G. N. Barber, R. Lin, and J. Hiscott. 2000. IRF-3 transcription factor mediates Sendai virus-induced apoptosis. J. Virol. 74:3781.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horikami, S., M., J. Curran, D. Kolakofsky, and S. A. Moyer. 1992. Complexes of Sendai virus NP-P and P-L proteins are required for defective interfering particle genome replication in vitro. J. Virol. 66:4901.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horikami, S. M., S. Smallwood, and S. A. Moyer. 1996. The Sendai virus V protein interacts with the NP protein to regulate viral genome RNA replication. Virology 222:383.. [DOI] [PubMed] [Google Scholar]
- 24.Horvath, C. M. 2000. STAT proteins and transcriptional responses to extracellular signals. Trends Biochem. Sci. 25:496-502. [DOI] [PubMed] [Google Scholar]
- 25.Jamaluddin, M., S. Wang, R. P. Garofalo, T. Elliott, A. Casola, S. Baron, and A. R. Brasier. 2001. IFN-beta mediates coordinate expression of antigen-processing genes in RSV-infected pulmonary epithelial cells. Am. J. Physiol. Lung Cell. Mol. Physiol. 280:L248-L257. [DOI] [PubMed] [Google Scholar]
- 26.Kawano, M., M. Kaito, Y. Kozuka, H. Komada, N. Noda, K. Nanba, M. Tsurudome, M. Ito, M. Nishio, and Y. Ito. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99-112. [DOI] [PubMed] [Google Scholar]
- 27.Kubota, T., N. Yokosawa, S. Yokota, and N. Fujii. 2001. C-terminal CYS-rich region of mumps virus structural V protein correlates with block of interferon α and γ signal transduction pathway through decrease of STAT 1. Biochem. Biophys. Res. Commun. 283:255-259. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A., M. Commane, R. W. Flickinger, C. M. Horvath, and G. R. Stark. 1997. Defective TNF-alpha-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science 278:1630-1632. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A., and D. Kolakofsky. 1996. Paramyxoviridae: the viruses and their replication, p. 1177-1204. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields Virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 30.Levy, D. E., I. Marie, E. Smith, and A. Prakash. 2002. Enhancement and diversification of IFN induction by IRF-7 mediated positive feedback. J. Interferon Cytokine Res. 22:87-93. [DOI] [PubMed] [Google Scholar]
- 31.Lin, G. Y., R. G. Paterson, C. D. Richardson, and R. A. Lamb. 1998. The V protein of SV5 interacts with damage-specific DNA binding protein. Virology 249:189-200. [DOI] [PubMed] [Google Scholar]
- 32.Lin, G. Y., and R. A. Lamb. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152-9166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy, S. K., and G. D. Parks. 1997. Genome nucleotide lengths that are divisible by six are not essential but enhance replication of defective interfering RNAs of the paramyxovirus SV5. Virology 232:145-157. [DOI] [PubMed] [Google Scholar]
- 34.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, H. Komada, and Y. Ito. 2001. High resistance of human parainfluenza type 2 virus protein-expressing cells to the antiviral and anti-cell proliferate activities of alpha/beta interferons: cysteine-rich V-specific domain is required for high resistance to the interferons. J. Virol. 75:9165-9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noah, D. L., M. A. Blum, and B. Sherry. 1999. Interferon regulatory factor 3 is required for viral induction of beta interferon in primary cardiac myocyte cultures. J. Virol. 73:10208-10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parisien, J. P., J. F. Lau, J. J. Rodriguez, B. M. Sullivan, A. Moscona, G. D. Parks, R. A. Lamb, and C. M. Horvath. 2001. The V protein of human parainfluenza virus 2 antagonizes type I interferon responses by destabilizing signal transducer and activator of transcription 2. Virology 283:230-239. [DOI] [PubMed] [Google Scholar]
- 37.Parks, G. D., K. R. Ward, and J. C. Rassa. 2001. Increased readthrough transcription across the simian virus 5 M-F gene junction leads to growth defects and a global inhibition of viral mRNA synthesis. J. Virol. 75:2213-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parks, G. D., V. A. Young, C. Koumenis, E. K. Wansley, J. L. Layer, and K. M. Cooke. 2002. Controlled cell killing by a recombinant nonsegmented negative strand RNA virus. Virology 193:203.. [DOI] [PubMed] [Google Scholar]
- 39.Paterson, R. G., G. P. Leser, M. A. Shaughnessy, and R. A. Lamb. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121-131. [DOI] [PubMed] [Google Scholar]
- 40.Randall, R. E., and A. Bermingham. 1996. NP:P and NP:V interactions of the paramyxovirus simian virus 5 examined using a novel protein:protein capture assay. Virology 224:121-138. [DOI] [PubMed] [Google Scholar]
- 41.Rassa, J. C., and G. D. Parks. 1998. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology 247:274-286. [DOI] [PubMed] [Google Scholar]
- 42.Rassa, J. C., and G. D. Parks. 1999. Highly diverse intergenic regions of the paramyxovirus simian virus 5 cooperate with the gene end U tract in viral transcription termination and can influence reinitiation at a downstream gene. J. Virol. 73:3904-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlender, J., B. Bossert, U. Buchholz, and K. K. Conzelmann. 2000. Bovine respiratory syncytial virus nonstructural proteins NS1 and NS2 cooperatively antagonize alpha/beta interferon-induced antiviral response. J. Virol. 74:8234-8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Servant, M. J., B. Tenoever, and R. Lin. 2002. Overlapping and distinct mechanisms regulating IRF-3 and IRF-7 function. J. Interferon Cytokine Res. 22:49-58. [DOI] [PubMed] [Google Scholar]
- 45.Southern, J. A., D. F. Young, F. Heaney, W. K. Baumgartner, and R. E. Randall. 1991. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 72:1551-1557. [DOI] [PubMed] [Google Scholar]
- 46.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi, K., T. Komatsu, J. Yokoo, A. Kato, T. Shioda, Y. Nagai, and B. Gotah. 2001. Sendai virus C protein physically associates with STAT1. Genes Cells 6:545-557. [DOI] [PubMed] [Google Scholar]
- 48.Thomas, S. M., R. A. Lamb, and R. G. Paterson. 1988. Two mRNAs that differ by two non templated nucleotides encode the amino co-terminal proteins P and V of the paramyxovirus SV5. Cell 54:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young, D. F., L. Didcock, S. Goodbourn, and R. E. Randall. 2000. Paramyxoviruses use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383-390. [DOI] [PubMed] [Google Scholar]