Abstract
Pregenomic RNA (pgRNA) plays two major roles in the hepadnavirus life cycle. It is the mRNA for two proteins required for DNA replication, C and P, and it is the template for reverse transcription. pgRNA is a terminally redundant transcript whose synthesis does not involve RNA splicing. For duck hepatitis B virus (DHBV), a spliced RNA is derived from pgRNA by removal of a single intron. The mechanism for the simultaneous cytoplasmic accumulation of unspliced (pgRNA) and spliced RNA was not known. We found that mutations within two regions of the DHBV genome reduced the level of pgRNA while increasing the level of spliced RNA. One region is near the 5′ end of pgRNA (region A), while the second is near the middle of pgRNA (region B). Inspection of the DHBV nucleotide sequence indicated that region A could base pair with region B. The 5′ and 3′ splice sites of the intron of the spliced RNA are within regions A and B, respectively. Substitutions that disrupted the predicted base pairing reduced the accumulation of pgRNA and increased the accumulation of spliced RNA. Restoration of base pairing, albeit mutant in sequence, resulted in restoration of pgRNA accumulation with a decrease in the level of spliced RNA. Our data are consistent with a model in which splicing of the pgRNA is suppressed by a secondary structure between regions A and B that occludes the splicing machinery from modifying pgRNA.
Reverse-transcribing viruses, such as retroviruses, caulimoviruses, and some hepadnaviruses, synthesize an unspliced “full-length” mRNA and one or more spliced derivatives (reviewed in reference 14). The unspliced full-length transcript, which is called pregenomic RNA (pgRNA) (or genomic RNA, in the case of retroviruses), is the template for reverse transcription, as well as the mRNA for proteins required for viral replication. In addition, members of these virus families can express one or more spliced derivatives of pgRNA. These spliced transcripts are mRNAs for other viral proteins. Therefore, these viruses must have a way to limit complete splicing and allow the simultaneous accumulation of unspliced and spliced mRNA from the same primary transcript. A complete understanding of these mechanisms for all reverse-transcribing viruses is not at hand. These processes are best understood for several retroviruses, in which several general themes have emerged (reviewed in reference 11). One theme is the use of suboptimal splice sites (13). A second theme is the use of cis-acting negative regulators of splicing that act at a distance to prevent efficient splicing (17). Both of these strategies prevent efficient splicing and allow the unspliced RNA an opportunity to transit out of the nucleus. Because splicing and nucleocytoplasmic transport are linked, unspliced RNAs must use alternative mechanisms to be transported out of the nucleus to the cytoplasm (reviewed in reference 3).
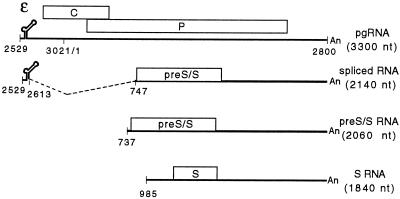
Hepadnaviruses are a family of reverse-transcribing DNA viruses that replicate in liver cells and cause diseases such as liver cirrhosis and hepatocellular carcinoma (reviewed in reference 10). The prototype member, hepatitis B virus (HBV), infects humans. Other family members, such as duck hepatitis B virus (DHBV), have been invaluable in understanding the molecular biology of viral replication. The DHBV virion contains a relaxed-circular 3.0-kb DNA genome (16). After the virus enters a susceptible cell, the genome is transported into the nucleus and converted to a covalently closed circular DNA (cccDNA) (23). Host RNA polymerase II transcribes the cccDNA to make the various viral RNAs. A detailed RNA transcription map of DHBV has been elucidated (4, 18) (Fig. 1). There is a single polyadenylation site on the genome; therefore, all transcripts have the same 3′ end. Two classes of RNA are synthesized: pregenomic (pg class) and subgenomic (sg class). The pg class of RNA is comprised of pgRNA and precore RNA. pgRNA is the reverse transcription template and the mRNA for the viral C and P proteins, which are required for reverse transcription. The C protein is the subunit of the nucleocapsid, while P is a multifunctional protein that has reverse transcriptase activity. The DHBV precore RNA is similar in structure to the pgRNA except for a 5′ extension of 82 nucleotides (nt). DHBV precore RNA, which is present at 1 to 5% of the level of pgRNA, is the mRNA for e-antigen, a secreted protein (20). The sg class of RNA contains three transcripts: two are 2.1 kb in size and the third is 1.8 kb (Fig. 1). The 1.8-kb transcript (S RNA) and one of the 2.1-kb transcripts (pre-S/S RNA) are transcribed from their own promoters and are synthesized without splicing (4). The other 2.1-kb RNA is a spliced variant of pgRNA (18). The subgenomic transcripts are the mRNAs for the two envelope proteins, L and S. The same open reading frame encodes the two envelope proteins. Initiation of translation from an upstream AUG (pre-S/S gene) gives rise to L protein, and initiation of translation from a downstream AUG (S gene) yields S protein. The 1.8-kb RNA only encodes the S protein because it does not contain the initiation codon of the pre-S/S gene. It is not clear whether one or both of the 2.1-kb transcripts is the mRNA for L protein.
FIG. 1.
RNA transcription map of DHBV representing pgRNA and the three subgenomic transcripts. Coordinates of termini and locations of key features are indicated. The size of each RNA is indicated below its name. Indicated on pgRNA is the packaging signal, epsilon, and C and P genes. The initiation codons for the C and P genes are at nt 2547 and 170, respectively, and the termination codons for the C and P genes are at nt 412 and 2528, respectively. Epsilon is from nt 2560 to 2616. Indicated on spliced RNA is the position of the intron and the pre-S/S gene, which codes for the L protein. The start codons for the L and S proteins are at nt 800 and 1284, respectively, while their common termination codon is at nt 1785.
The mechanism by which the spliced RNA and the unspliced RNA (pgRNA) are simultaneously synthesized was not previously understood. We found that splicing is suppressed by a long-distance secondary structure that contains the 5′ and 3′ splice sites to permit the accumulation of pgRNA. This finding represents a previously undescribed mechanism for the regulation of RNA splicing.
MATERIALS AND METHODS
Molecular clones.
All molecular clones were derived from DHBV strain 3 (21). DHBV RNA was expressed from plasmids that contained approximately 1.5 copies of the DHBV genome starting from nt 1666 and ending at nt 3021. The wild-type reference used in our analyses did not express P protein due to a frameshift mutation at nt 424. All variants, except D2650/2671, contained this P gene mutation. The A2, A3, and A6 variants altered the coding capacity of the C gene. To prevent expression of variant forms of the C protein, all constructs containing these mutations were made null for C protein production by virtue of a frameshift mutation at nt 2845. All deletion and substitution mutations were created using oligonucleotides that contained an endogenous DHBV restriction site and the desired mutation. Restriction fragments containing the desired mutation were synthesized by PCR. These restriction fragments were initially cloned into a subgenomic molecular clone. DNA sequencing was performed to establish the presence of the desired mutation and the absence of secondary mutations within the PCR fragment. A restriction fragment was then transferred from the subgenomic clone into the 1.5-mer plasmid. Details of the construction of any individual variant will be provided upon request. The green fluorescent protein (GFP) expression plasmid, named 1929, was a kind gift from the laboratory of Bill Sugden, University of Wisconsin.
Cell cultures.
LMH (7), Huh7, and HepG2 cells were cultured in Dulbecco's modified Eagle's-F-12 medium (1:1) supplemented with 5, 10, and 10% fetal bovine serum, respectively. DNA transfections were accomplished with a calcium-phosphate precipitation protocol from Jesse Summers, University of New Mexico. Typically, 5 to 10 μg of the DHBV plasmid and 0.5 to 2 μg of GFP plasmid were transfected onto a 60-mm-diameter plate of cells. The precipitate was removed from the cultures after 16 h. Poly(A)+ RNA was harvested from the cultures 3 days later.
Isolation of poly(A)+ RNA.
Onto each 60-mm plate of cells, 1.0 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.5 M NaCl, 1% sodium dodecyl sulfate (SDS), and 200 μg of proteinase K/ml were added and incubated for at least 2 min at room temperature. The cell monolayer became a clear and gelatinous lysate. The lysate was sheared by centrifugation through a QIAshredder device (catalog no. 79654; Qiagen). Additional proteinase K was added to a final concentration of 300 μg/ml. The lysate, in a 1.5-ml microcentrifuge tube, was incubated at 37°C for 30 to 60 min. Using a 16-gauge needle, one scoop of oligo(dT) cellulose resin (catalog no. 808229; Roche) was added to each tube. Poly(A)+ RNA was annealed to the oligo(dT) cellulose resin by placing the tube on a rotating wheel at room temperature for 1 h. Each tube was spun briefly in a microcentrifuge for 10 to 15 s to pellet the oligo(dT) resin and the annealed poly(A)+ RNA. The supernatant was discarded. The poly(A)+ RNA-oligo(dT) resin was washed three times with 1.0 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.5 M NaCl, and 1% SDS, and then washed once with 1.0 ml of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 0.1 M NaCl, and 1% SDS. Poly(A)+ RNA was eluted from the resin with two washes of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA. Ethanol and sodium acetate were added to the RNA prep and stored at −20°C.
Northern blotting.
Typically, 1/10 of the RNA isolated from a single 60-mm plate was analyzed. RNA samples were denatured in a formamide-formaldehyde solution at 65°C for 5 min immediately prior to electrophoresis. RNA samples were electrophoresed in a 1% agarose, 0.66 M formaldehyde, 1× running buffer (20 mM morpholinepropanesulfonic acid, 8 mM sodium acetate, 1 mM EDTA, pH 7.0). After electrophoresis, RNA was transferred to a Hybond N (Amersham) membrane. Hybridization was carried out according to the protocol of Church and Gilbert (6). The membrane was hybridized with two plus-strand-specific probes, one that detects DHBV nt 988 to 1665 and another that detects GFP mRNA. Quantification of autoradiographic images was done with a Molecular Dynamics PhosphorImager. Included on each Northern blot were two amounts (1× and 2×) of wild-type DHBV RNA to determine whether the hybridization signal was proportional to the amount of RNA loaded onto the gel.
RNase protection analyses.
Typically, 1/10 of the RNA isolated from a single 60-mm plate was analyzed. A 750-pg aliquot of each of the in vitro-transcribed DHBV standards was analyzed in each experiment. Radiolabeled DHBV and GFP probes were synthesized using standard methods. One nanogram of DHBV probe and 0.5 ng of GFP probe were added to each reaction mixture. To demonstrate that the procedure was quantitative, we always analyzed two different amounts (1× and 2×) of wild-type DHBV RNA, with the expectation that the signals for each RNA in these samples would be twofold different. If this expectation was not met, we repeated the analysis. Sample and probe RNA were coprecipitated and resuspended in 10 μl of hybridization buffer (80% formamide, 40 mM PIPES [pH 6.8], 0.4 M NaCl, 1 mM EDTA) and incubated at 42°C overnight. Next, 150 μl of digestion buffer (10 mM Tris [pH 8.0], 0.3 M NaCl, 5 mM EDTA, 7.67 μg of RNase A/ml, 7.67 U of RNase T1/ml) was added to each reaction mixture and incubated at 27 to 30°C for 30 min. The digestion reaction was terminated by the addition of 160 μl of solution D (4 M guanidinium thiocyanate, 25 mM sodium citrate [pH 7.0], 0.5% [wt/vol] sodium N-lauroylsarcosine, 0.1 M β-mercaptoethanol). RNA was precipitated by the addition of 75 μl of ethanol and 325 μl of isopropanol. Samples were resuspended in a formamide loading buffer and electrophoresed in a 5% acrylamide, 7.6 M urea, 1× Tris-borate-EDTA, 40-cm gel. Quantification of autoradiographic images was done with a Molecular Dynamics PhosphorImager.
RESULTS
Two different deletions unexpectedly result in lack of accumulation of DHBV pgRNA.
While studying DHBV DNA synthesis, we observed that deletions in two different regions of the genome resulted in a large reduction in the level of pgRNA (Fig. 2). One deletion was from nt 2650 to 2671. This variant, named D2650/2671, was expected to express a pgRNA with a 22-nt deletion near its 5′ end (Fig. 2, lane 3). The second deletion was from nt 686 to 717 (named D686/717) (Fig. 2, lane 2) and was expected to express a pgRNA with a 32-nt deletion approximately 1,200 nt from the 5′ end. Previously published reports did not offer an explanation for the low level of pgRNA synthesized by the two variants.
FIG. 2.
Northern blotting indicates that deletion variants do not accumulate pgRNA. Deletion of nt 2650 to 2671 or nt 686 to 717 results in accumulation of less pgRNA. Extending the nt 686-to-717 deletion to nt 840 restores accumulation of pgRNA. LMH cells were transfected with plasmids to express DHBV and GFP RNA, respectively. Poly(A)+ RNA was isolated from cells and analyzed by Northern blotting. Lane 1, wild-type RNA standard; lane 2, D686-717 RNA; lane 3, D2650-2671 RNA; lane 4, D686-840 RNA. Blots were hybridized with two probes, one that detects DHBV plus-strand sequence between nt 988 and 1665 and one that detects GFP mRNA. Positions of pregenomic class (PG), subgenomic class (SG), and GFP RNA are indicated on the left side.
In our analysis LMH cells were transfected with plasmids containing 1.5 tandem copies of the DHBV genome to express the full complement of DHBV RNAs. Synthesis of the individual DHBV RNAs was under the control of their respective endogenous regulatory elements. Three days after the transfection, poly(A)+ RNA was isolated from the cell culture. Included in each transfection was an expression plasmid for GFP, whose mRNA was used as an internal standard to eliminate experimental variation in transfection efficiency and in RNA isolation and its detection. Two RNA probes were used simultaneously in Northern blotting. One probe hybridized to a region common to all DHBV transcripts (nt 988 to 1665), while the second probe detected GFP mRNA. Thus, by normalizing the level of the DHBV mRNAs to the level of GFP mRNA, we could compare the level of pregenomic and subgenomic DHBV transcripts expressed from different plasmids. Upon normalization to GFP levels, it was conclusive that D2650/2671 and D686/717 synthesized less pgRNA and more subgenomic RNAs than the wild-type reference (Table 1). In these analyses, viral replication was not occurring, due to the introduction of a premature stop codon into the 5′ portion of the P gene (D686/717) or removal of the initiation codon of the C gene (D2650/2671). C and P proteins are required for packaging of pgRNA into capsids. Therefore, the lack of accumulation of pgRNA for the two variants could not be attributed to its encapsidation and reverse transcription. All subsequent analyses also were done in the absence of viral replication due to a premature termination codon in the P gene.
TABLE 1.
Analysis of deletion mutants by RNase protection assay and Northern blottinga
| DHBV | RNase protection assay
|
Northern blotting
|
|||
|---|---|---|---|---|---|
| pgb | pre-S/Sc | Splicedd | pgb | sge | |
| WT | 1.00 | 0.21 ± 0.01 | 0.30 ± 0.00 | 1.00 | 0.83 ± 0.09 |
| D686/717 | 0.02 ± 0.01 | 0.12 ± 0.07 | 1.01 ± 0.42 | 0.03 ± 0.02 | 1.38 ± 0.61 |
| D2650/2671 | 0.09 ± 0.01 | 0.15 ± 0.00 | 0.65 ± 0.14 | 0.07 ± 0.02 | 1.22 ± 0.25 |
Values are based on measurements made on poly(A)+ RNA isolated from two independent transfections. First, all DHBV RNA levels were normalized to the level of GFP mRNA. Then the value of wild-type (WT) pgRNA was set to 1.00 and all other RNA levels were accordingly adjusted, and the mean and standard deviation were calculated.
Pregenomic class of RNAs
Pre-S/S RNA.
Spliced RNA.
Subgenomic class of RNAs.
The sequences removed in the D2650/2671 and D686/717 variants lie within the intron of the spliced mRNA (Fig. 1). A possible explanation for the lack of accumulation of pgRNA was that a negative regulator of splicing had been inactivated with each deletion, resulting in efficient splicing. Consistent with this possibility, we observed a restoration in pgRNA accumulation when the deletion in D686/717 was extended to nt 840 (named D686/840) (Fig. 2, lane 4). According to this idea, splicing was inhibited in the D686/840 variant due to removal of key cis-acting sequences (such as the 3′ splice site and branch point sequence), thus leading to accumulation of pgRNA when compared to that of D686/717. The faster-migrating subgenomic RNA of D686/840 is likely the 1.8-kb S RNA.
Removal of nt 2650 to 2671 or nt 686 to 717 results in increased levels of spliced RNA.
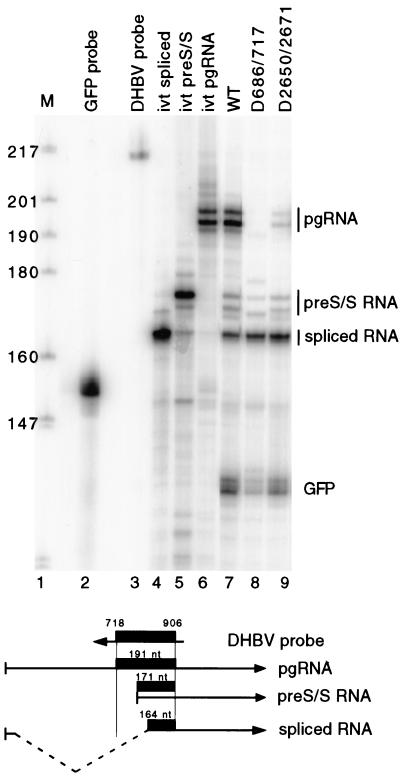
If the lack of accumulation of pgRNA for D2650/2671 and D686/717 was due to splicing, then the level of spliced RNA should increase. Unfortunately, Northern blotting cannot detect an increase in the level of spliced RNA due to its comigration with the unspliced pre-S/S mRNA. Therefore, we used an RNase protection assay to detect and measure the levels of pgRNA, spliced RNA, and unspliced pre-S/S RNA. The design of the probe is depicted in Fig. 3. A 214-nt probe was used, which is complementary to nt 718 to 906 of DHBV and contains a total of 24 nt of non-DHBV sequence at its ends. Upon annealing of the probe to the three different DHBV RNAs and digestion with RNases A and T1, protected fragments of 191, 171, and 164 nt, representing the pgRNA, pre-S/S RNA, and spliced RNA, respectively, should be generated. As described earlier, an expression vector for GFP was cotransfected with each DHBV plasmid. As an internal standard, we included a probe in the RNase protection assay to measure the level of GFP mRNA. The level of the three DHBV RNAs in each sample was normalized to the level of GFP mRNA to permit the comparison of the level of viral RNA expressed from different DHBV plasmids. To determine the identity of the protected fragments in the analysis, DNA plasmids were made that upon in vitro RNA transcription yielded RNAs similar in size to authentic viral RNAs. The in vitro-transcribed RNA standards were included in each RNase protection analysis (Fig. 3, lanes 4 to 6). The RNase protection assay was performed on the RNA expressed from the D2650/2671 and D686/717 variants and the wild-type comparison (Fig. 3, lanes 7 to 9). The measurements of the level of the three viral transcripts from two RNase protection assay analyses are presented in Table 1. Clearly, an increase in the level of spliced RNA was seen for the two variants. Overall, these data indicated that the deletion of nt 686 to 717 and deletion of nt 2650 to 2671 caused pgRNA to be spliced to a greater degree than in the wild-type reference.
FIG. 3.
RNase protection analysis of deletion variants indicates that reduction of pgRNA is due to splicing. (Top) Autoradiogram of RNase protection analysis. Lane 1, end-labeled DNA markers, MspI digest of pBR322 plasmid DNA. Sizes are indicated on the left. Lane 2, full-length GFP probe; lane 3, full-length DHBV probe; lanes 4 to 9, samples analyzed by RNase protection. Lane 4, in vitro-transcribed spliced RNA; lane 5, in vitro-transcribed pre-S/S RNA; lane 6, in vitro-transcribed pgRNA; lane 7, wild-type DHBV RNA from transfected LMH cells; lane 8, D686/717 RNA from transfected LMH cells; lane 9, D2650/2671 RNA from transfected LMH cells. Positions of protected fragments from pgRNA, pre-S/S RNA, spliced RNA, and GFP RNA are indicated on the right. (Bottom) Representation of antisense DHBV probe and the three different DHBV RNAs. The thick black line represents the probe fragment protected after RNase digestion.
The two regions containing negative regulators of splicing are predicted to base pair.
Inspection of the nucleotide sequence of the two regions containing the negative regulators of splicing indicated that they had the potential to base pair with each other. Figure 4 shows the predicted base-pairing and resultant secondary structure. The 5′ and 3′ sequences in the predicted secondary structure were named regions A and B, respectively. Interestingly, the 5′ and 3′ splice sites were within the region predicted to base pair. This observation suggested a mechanism where base pairing of regions A and B prevented splicing and permitted accumulation of pgRNA. According to this model, the secondary structure was destabilized in the presence of the D686/717 and D2650/2671 mutations. This led to a lack of accumulation of pgRNA because destabilization promoted access to pgRNA by the splicing machinery and facilitated the production of spliced RNA.
FIG. 4.
Predicted secondary structure of pgRNA. (A) Predicted base pairing between regions A and B. Nucleotide coordinates are indicated. Splice donor and acceptor sites are indicated with arrows. Extent of deletion in D2650/2671 and D686/717 clones is shown. (B) Location of regions A and B on pgRNA. Region A is 75 nt from the 5′ end of pgRNA. Region B is 1,030 nt 3′ of region A.
Regions A and B base pair to positively affect the accumulation of DHBV pgRNA by suppressing its splicing.
To test the role of base pairing between regions A and B, we analyzed a set of variants, referred to as the 2-series, in which base pairing was disrupted and then restored (Fig. 5). Six base-pairing partners were changed within region A (A2 variant) or region B (B2 variant). A third variant, named A2/B2, contained both the A2 and B2 substitutions and was restored in its potential to base pair. LMH cells were transfected with plasmids that expressed each of the variants, and poly(A)+ RNA was isolated 3 days later. Northern blotting indicated that both the A2 and B2 variants accumulated less pgRNA than the wild-type standard (Fig. 5 and Table 2). Accompanying the decrease in pgRNA levels was an increase in the level of subgenomic RNAs. More importantly, the double mutant A2/B2 was restored for accumulation of pgRNA, albeit partially. In addition, the level of subgenomic RNAs for the A2/B2 variant was less than that of either of the single mutants, but greater than that of the wild type. This result indicated that base pairing between regions A and B contributed to the accumulation of pgRNA.
FIG. 5.
Disruption of base pairing between regions A and B results in accumulation of lower levels of pgRNA; restoration of base pairing increases the accumulation of pgRNA. (A) The A2 and B2 variants have 6 nt substituted. (B) Northern blotting of 2-series variants. Poly(A)+ RNA from transfected LMH cells was analyzed. Lane 1, wild-type standard; lane 2, B2 variant; lane 3, A2/B2 variant; lane 4, A2 variant. Positions of pgRNA (PG), subgenomic RNA (SG), and GFP RNA are indicated on the left. The blot was hybridized with plus-strand-specific probes that detect DHBV nt 988 to 1665 and GFP mRNA.
TABLE 2.
Analysis of 2-series variants by RNase protection assay and Northern blottinga
| DHBV | RNase protection assay
|
Northern blotting
|
|||
|---|---|---|---|---|---|
| pgb | pre-S/Sc | Splicedd | pgb | sge | |
| WT | 1.00 | 0.10 ± 0.04 | 0.45 ± 0.08 | 1.00 | 0.69 ± 0.04 |
| B2 | 0.09 ± 0.01 | 0.12 ± 0.04 | 0.96 ± 0.04 | 0.08 ± 0.02 | 1.25 ± 0.01 |
| A2 | 0.07 ± 0.02 | 0.12 ± 0.05 | 1.01 ± 0.04 | 0.07 ± 0.02 | 1.31 ± 0.07 |
| A2/B2 | 0.68 ± 0.13 | 0.39 ± 0.14 | 0.55 ± 0.05 | 0.73 ± 0.06 | 1.10 ± 0.10 |
Values are based on measurements made on poly(A)+ RNA isolated from independent transfections, with two for the wild type (WT) and B2, and four for A2 and A2/B2. First, all DHBV RNA levels were normalized to the level of GFP mRNA. Then, the value of WT pgRNA was set to 1.00 and all other RNA levels were accordingly adjusted, and then the mean and standard deviation were calculated.
Pregenomic class of RNAs.
pre-S/S RNA.
Spliced RNA.
Subgenomic class of RNAs.
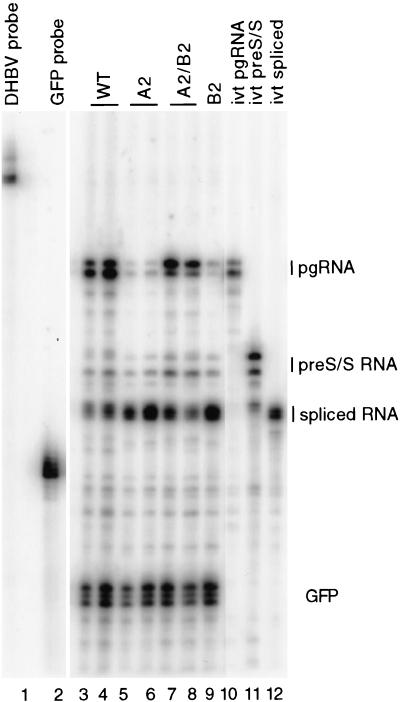
Next, we performed an RNase protection analysis on the variants of the 2-series (Fig. 6). As expected, the level of pgRNA for the A2 and B2 mutants decreased relative to that of wild type, and the double mutant, A2/B2, was partially restored in its accumulation of pgRNA (Table 2). The spliced RNA accumulated to higher levels for the A2 and B2 variants than the wild type, while the A2/B2 mutant accumulated levels that were closer to the level seen in the wild type (Table 2). This analysis indicated that regions A and B base pair to positively affect the accumulation of DHBV pgRNA by suppressing its splicing.
FIG. 6.
The A/B secondary structure suppresses splicing to positively affect accumulation of pgRNA. Autoradiogram results of an RNase protection assay of the 2-series variants are shown. Poly(A)+ RNA was isolated from transfected LMH cells and analyzed. Lane 1, full-length DHBV probe; lane 2, full-length GFP probe; lanes 3 and 4, wild-type DHBV RNA; lanes 5 and 6, independently isolated molecular clones of A2 variant; lanes 7 and 8, independently isolated molecular clones of A2/B2 variant; lane 9, B2 variant; lane 10, in vitro-transcribed pgRNA; lane 11, in vitro-transcribed pre-S/S RNA; lane 12, in vitro-transcribed spliced RNA. Positions of protected fragments from pgRNA, pre-S/S RNA, spliced RNA, and GFP RNA are indicated on the right.
To further evaluate the role of base pairing between regions A and B, three additional series of substitution variants were analyzed (the 3-series, 4-series, and 6-series) (Fig. 7). Each variant had multiple nucleotides changed to disrupt base pairing potential. In general, the Northern blot analyses of these variants were consistent with the idea that regions A and B base pair to suppress splicing. For all three series of variants, mutations in region B resulted in decreased accumulation of pgRNA and an increase in the level of subgenomic RNAs (Tables 3 and 4). Similarly for all three region A variants, a decrease in the accumulation of pgRNA was seen. But, only for one of the region A variants, A6, was an increase in the level of subgenomic RNAs seen (Table 4). The other two region A mutants (A3 and A4) supported the synthesis of less subgenomic RNAs (Tables 3 and 4). In all three series of variants, the restoration of base pairing between regions A and B lead to increased accumulation of pgRNA relative to the individual single mutations, although the magnitude of the increases was not as large as was seen for the A2/B2 variant. In general, these analyses were consistent with the conclusion that regions A and B base pair to regulate splicing, although the A3 and A4 variants had a more complex phenotype, suggesting that other processes were affected.
FIG. 7.
Northern blot analysis of 3-, 4-, and 6-series variants. Poly(A)+ RNA from transfected LMH cells was analyzed. (A) Sequence of the individual variants of the 3-, 4-, and 6-series. (B) Northern blot of 3-series variants. Lane 1, wild type; lane 2, B3 variant; lane 3, A3/B3 variant; lane 4, A3 variant. (C) Northern blot of 4-series variants. Lane 1, wild type; lane 2, B4 variant; lane 3, A4/B4 variant; lane 4, A4 variant. (D) Northern blot of 6-series variants. Lane 1, wild type; lane 2, A6 variant; lane 3, A6/B6 variant; lane 4, B6 variant. Positions of pgRNA (PG), subgenomic RNA (SG), and GFP RNA are indicated on the left. The blot was hybridized with plus-strand-specific probes that detect DHBV nt 988 to 1665 and GFP mRNA.
TABLE 3.
Analysis of 3-series variants by RNase protection assay and Northern blottinga
| RNase protection assay
|
Northern blotting
|
|||
|---|---|---|---|---|
| pgb | Splicedc | pgb | sgd | |
| WT | 1.00 | 0.36 ± 0.04 | 1.00 | 0.69 ± 0.07 |
| B3 | 0.16 ± 0.05 | 0.82 ± 0.28 | 0.14 ± 0.02 | 1.07 ± 0.24 |
| A3 | 0.14 ± 0.08 | 0.26 ± 0.15 | 0.09 ± 0.04 | 0.55 ± 0.22 |
| A3/B3 | 0.36 ± 0.10 | 0.16 ± 0.07 | 0.25 ± 0.04 | 0.37 ± 0.07 |
Values are based on measurements made on poly(A)+ RNA isolated from independent transfections, with five for wild type (WT), four for A3/B3, and three for B3 and A3. First, all DHBV RNA levels were normalized to the level of GFP mRNA. Then, the value of WT pgRNA was set to 1.00 and all other RNA levels were accordingly adjusted, and then the mean and standard deviation were calculated.
Pregenomic class of RNAs.
Spliced RNA.
Subgenomic class of RNAs.
TABLE 4.
Northern blot analysis of 4- and 6-series variantsa
| DHBV | pgb | sgc |
|---|---|---|
| WT | 1.00 | 0.90 ± 0.21 |
| B4 | 0.05 ± 0.01 | 1.63 ± 0.04 |
| A4 | 0.18 ± 0.03 | 0.74 ± 0.06 |
| A4/B4 | 0.37 ± 0.04 | 1.13 ± 0.17 |
| B6 | 0.04 ± 0.02 | 1.81 ± 0.06 |
| A6 | 0.06 ± 0.01 | 1.63 ± 0.11 |
| A6/B6 | 0.37 ± 0.04 | 1.96 ± 0.14 |
Values are based on measurements made on poly(A)+ RNA isolated from independent transfections, with three for wild type (WT) and B6, and two for B4, A4, A4/B4, A6, and A6/B6. First, all DHBV RNA levels were normalized to the level of GFP mRNA. Then, the value of WT pgRNA was set to 1.00 and all other RNA levels were accordingly adjusted, and then the mean and standard deviation were calculated.
Pregenomic class of RNA.
Subgenomic class of RNA.
To better understand the phenotypes of the 3-series variants, we performed the RNase protection assay on poly(A)+ RNA isolated from three independent transfections (Table 3). The A3 and B3 variants accumulated less pgRNA than the wild-type standard. The B3 mutant accumulated more spliced RNA than the wild type, while the A3 variant made less spliced RNA. The A3/B3 double mutant accumulated more pgRNA than either of the single mutants but less than the wild type. The A3/B3 mutant accumulated less spliced RNA than both of the single mutants and the wild type. Overall for the 3-series variants, the RNase protection analysis and Northern blotting analysis displayed similar trends. In general, the results were consistent with the conclusion that base pairing between regions A and B positively affects pgRNA accumulation by suppressing its splicing.
DISCUSSION
At the onset of our studies, the mechanism for simultaneous accumulation of spliced and unspliced versions of the DHBV pgRNA was not understood. Through our studies we have uncovered a major, if not sole, component of that mechanism. An RNA secondary structure that contains the 5′ and 3′ splice sites suppresses splicing to permit accumulation of pgRNA. The analysis of the 2-series variants (Fig. 5 and 6; Table 2) provides compelling evidence for this conclusion. A novel aspect of this mechanism is that both the splice donor and acceptor sequences are simultaneously inactivated in a single process to suppress splicing. Inspection of the nucleotide sequence of other avian hepadnaviruses reveals the potential for the formation of similar secondary structures (data not shown). Whether splicing of mammalian hepadnavirus RNA is regulated in a similar fashion remains to be determined. Computer-aided secondary structure analysis was inconclusive. Lastly, whether other reverse-transcribing viruses use a similar mechanism to permit accumulation of unspliced RNA is an intriguing question whose answer is currently not known.
We think the A/B secondary structure is directly affecting splicing rather than acting as a requirement for pgRNA nuclear export and indirectly affecting splicing. We have reached this conclusion because the pgRNA expressed from the variant D686/840, which has region B completely removed, accumulates in the cytoplasm as well as wild-type pgRNA (data not shown). On a related note, the mechanism by which DHBV pgRNA is exported from the nucleus has not been reported. It is likely there is one or more export elements in DHBV pgRNA similar to those described in the mammalian hepadnaviruses (26).
Our findings concerning the function of the A/B secondary structure were not limited to LMH cells. A decrease in accumulation of pgRNA for the A2 and B2 variants and a restoration of pgRNA accumulation for the A2/B2 mutant were seen in Northern blotting of poly(A)+ RNA isolated from Huh7 and HepG2 cells (data not shown). In addition, our findings do not appear to be a peculiar feature of expression of DHBV RNA from 1.5-mer plasmids. To mimic the authentic transcription template, we transfected LMH cells with in vitro-synthesized circular DHBV monomers of the D686/717, 2-, and 3-series variants. Northern blot analysis of the in vitro-synthesized cccDNA revealed patterns and trends similar to the analysis with the 1.5-mer plasmids (data not shown).
Region A is within the terminally redundant portions of pgRNA. Thus, pgRNA has a 5′ and 3′ copy of region A. In the majority of our analyses, using 1.5-mer plasmids, only the 5′ copy of region A was mutated. Clearly, the 5′copy of region A can base pair with region B to suppress splicing. A significant increase in splicing is seen when only the 5′ copy of region A is mutated. This indicates that the 3′ copy of region A containing the wild-type sequence does not base pair with region B to efficiently suppress splicing. Whether the 3′ copy of region A can base pair with region B to suppress splicing at a low level was not directly addressed in our study and awaits further experimentation.
In general, transcription and splicing are thought to be coupled (19). In these situations, it is believed that a 5′ splice site is recognized and bound by the U1 snRNP almost as soon as it is synthesized by the polymerase. Given this possibility, U1 snRNP would bind the 5′ splice site found in region A of our model as it emerges from the polymerase. Consequently, U1 snRNP binding would prevent region A from base pairing with region B, as our model predicts. Some considerations to address this possibility include the following: (i) sequences in DHBV may not be optimal for efficient interaction with components of the spliceosome; (ii) a local secondary structure near the 5′ end of pgRNA, epsilon (Fig. 1), may form in the nascent transcript to prevent U1 snRNP binding; and (iii) transcription and splicing are not necessarily coupled during DHBV transcription. Ultimately, additional experiments are needed to better understand the relationship between transcription and splicing for DHBV.
Other examples of RNA secondary structures suppressing splicing have been described. Many of these cases involve cellular genes that undergo alternative splicing. Typically, a local secondary structure involving either the 5′ or 3′ splice site contributes to the suppression of splicing of a particular exon by occluding the splicing machinery. The chicken beta-tropomyosin gene (15), the microtubule-associated tau protein gene (24), the immunoglobulin M heavy chain gene (25), the mammalian hnRNP A1 gene (2), the murine neuronal cell adhesion molecule gene (8), and the human growth hormone gene (9) are examples. Regulation of splicing of DHBV pgRNA is different from these cases because both the 5′ and 3′ splice sites are simultaneously base paired in the same stem.
Our analysis does not determine whether the A/B secondary structure has additional positive roles in the accumulation of pgRNA. None of the variants in which base pairing was restored accumulated pgRNA to wild-type levels. The reason for this is not clear, but several possibilities come to mind. For example, a specific sequence within the secondary structure could be required for pgRNA accumulation, which could indicate an interaction with one or more proteins. Alternatively, the lack of complete restoration could reflect a lower thermostability of the A/B secondary structures for the restoration variants. However, computer-aided thermostability predictions do not support this idea (data not shown). Lastly, if the sequence within either region A or B makes contributions to other processes involved in pgRNA accumulation, then complete restoration might not be expected. In fact, the pet element, a cis-acting element necessary for pgRNA accumulation, is located close to, if not overlapping, region A. It is proposed that pet prevents transcription termination during the first pass of RNA transcription through a termination region on the cccDNA template (1, 5, 12). Further studies will be necessary to determine whether region A and pet overlap.
Our analysis was in the absence of viral DNA synthesis. Therefore, the lack of accumulation of variant pgRNAs cannot be attributed to their encapsidation and replication. In addition, nonsense-mediated decay of variant RNAs does not account for their lack of accumulation. Derivatives of the B2 and B3 variants that did not contain a premature stop codon in the 5′ portion of the P gene also failed to accumulate pgRNA (data not shown).
Our analysis suggests a model in which two isomers of pgRNA coexist in the nucleus. One population of pgRNA has regions A and B base paired and ultimately accumulates in the cytoplasm as pgRNA. The second population of pgRNA does not have A base paired with B and is directed into a splicing pathway resulting in its accumulation as spliced RNA in the cytoplasm. Whether an equilibrium exists between the two pools and whether the proportion of RNA in the two populations can be dynamically altered is not known. Although the role of pgRNA in the viral life cycle is clearly understood, the function of the spliced RNA is not clear. Based on its structure, it is proposed to be an mRNA for the L protein (18). If accurate, then increased splicing of pgRNA could lead to increased expression of L protein. L protein is required for virion morphogenesis and, at least with DHBV, L protein negatively regulates the accumulation of cccDNA (22). Another possible role for splicing in the viral life cycle would be to decrease the level of pgRNA to modulate the level of viral replication.
Region A and part of the RNA encapsidation signal, epsilon, overlap. The epsilon is comprised of a local RNA secondary structure. Therefore, the pgRNA cannot simultaneously contain the epsilon and A/B structures. Additionally, a third conformational isomer, one that undergoes translation and likely does not contain either the epsilon or the A/B structure, should exist. Consequently, one or more mechanisms to regulate the cytoplasmic proportions of the various pgRNA conformational isomers is likely to exist.
Acknowledgments
We thank Jeff Ross and Jesse Summers for helpful discussions during the course of this work. We thank Jeff Habig, Ning Liu, Kristin Ostrow, Jeff Ross, and Jesse Summers for critical review of the manuscript.
This work was supported by NIH grants R29 GM50263, P01 CA22443, P30 CA07175, P30 CA14520, and T32 CA09135 and by ACS grant JFRA-651.
REFERENCES
- 1.Beckel-Mitchener, A., and J. Summers. 1997. A novel transcriptional element in circular DNA monomers of the duck hepatitis B virus. J. Virol. 71:7917-7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchette, M., and B. Chabot. 1997. A highly stable duplex structure sequesters the 5′ splice site region of hnRNP A1 alternative exon 7B. RNA 4:405-419. [PMC free article] [PubMed] [Google Scholar]
- 3.Boris-Lawrie, K., T. M. Roberts, and S. Hull. 2001. Retroviral RNA elements integrate components of post-transcriptional gene expression. Life Sci. 69:2697-2709. [DOI] [PubMed] [Google Scholar]
- 4.Buscher, M., W. Reiser, H. Will, and H. Schaller. 1985. Transcripts and the putative RNA pregenome of duck hepatitis B virus: implications for reverse transcription. Cell 40:717-724. [DOI] [PubMed] [Google Scholar]
- 5.Chang, C., R. C. Hirsch, and D. Ganem. 1995. Sequences in the preC region of duck hepatitis B virus affect pregenomic RNA accumulation. Virology 207:549-554. [DOI] [PubMed] [Google Scholar]
- 6.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. USA 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Condreay, L. D., C. E. Aldrich, L. Coates, W. S. Mason, and T. T. Wu. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cote, J., and B. Chabot. 1997. Natural base-pairing interactions between 5′ splice site and branch site sequences affect mammalian 5′ splice site selection. RNA 11:1248-1261. [PMC free article] [PubMed] [Google Scholar]
- 9.Estes, P. A., N. E. Cooke, and S. A. Liebhaber. 1992. A native RNA secondary structure controls alternative splice-site selection and generates two human growth hormone isoforms. J. Biol. Chem. 267:14902-14908. [PubMed] [Google Scholar]
- 10.Ganem, D., and R. J. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In B.N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 11.Goff, S. P. 2001. Retroviridae: the retroviruses and their replication, p. 1871-1939. In B.N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Virology, 4th ed. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 12.Huang, M., and J. Summers. 1994. pet, a small sequence distal to the pregenome cap site, is required for expression of the duck hepatitis B virus pregenome. J. Virol. 68:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katz, R. A., and A. M. Skalka. 1990. Control of retroviral RNA splicing through maintenance of suboptimal processing signals. Mol. Cell. Biol. 10:696-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiss-Laszlo, Z., and T. Hohn. 1996. Pararetro- and retrovirus RNA: splicing and the control of nuclear export. Trends Microbiol. 4:480-485. [DOI] [PubMed] [Google Scholar]
- 15.Libri, D., A. Piseri, and M. Y. Fiszman. 1991. Tissue-specific splicing in vivo of the beta-tropomyosin gene: dependence on an RNA secondary structure. Science 252:1842-1845. [DOI] [PubMed] [Google Scholar]
- 16.Mason, W. S., G. Seal, and J. Summers. 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 36:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McNally, M. T., R. R Gontarek, and K. Beemon. 1991. Characterization of Rous sarcoma virus intronic sequences that negatively regulate splicing. Virology 185:99-108. [DOI] [PubMed] [Google Scholar]
- 18.Obert, S., B. Zachmann-Brand, E. Deindl, W. Tucker, R. Bartenschlager, and H. Schaller. 1996. A splice hepadnavirus RNA that is essential for virus replication. EMBO J. 15:2565-2574. [PMC free article] [PubMed] [Google Scholar]
- 19.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 20.Schneider, R., D. Fernholz, G. Wildner, and H. Will. 1991. Mechanism, kinetics, and role of duck hepatitis B virus e-antigen expression in vivo. Virology 182:503-512. [DOI] [PubMed] [Google Scholar]
- 21.Sprengel, R., C. Kuhn, H. Will, and H. Schaller. 1985. Comparative sequence analysis of duck and human hepatitis B virus. J. Med. Virol. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 22.Summers, J., P. M. Smith, M. J. Huang, and M. S. Yu. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J. Virol. 65:1310-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuttleman, J. S., C. Pourcel, and J. Summers. 1986. Formation of the pool of covalently closed circular viral DNA in hepadnavirus-infected cells. Cell 47:451-460. [DOI] [PubMed] [Google Scholar]
- 24.Varani, L., M. Hasegawa, M. G. Spillantini, M. J. Smith, J. R. Murrell, B. Ghetti, A. Klug, M. Goedert, and G. Varani. 1999. Structure of tau exon 10 splicing regulatory element RNA and destabilization by mutations of frontotemporal dementia and parkinsonism linked to chromosome 17. Proc. Natl. Acad. Sci. USA 96:8229-8234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watakabe, A., K. Inoue, H. Sakamoto, and Y. Shimura. 1989. A secondary structure at the 3′ splice site affects the in vitro splicing reaction of mouse immunoglobulin mu chain pre-mRNAs. Nucleic Acids Res. 17:8159-8169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen, T. S. B. 1998. Posttranscriptional regulation of gene expression in hepadnaviruses. Semin. Virol. 8:319-326. [Google Scholar]