Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) is a cytoplasmic RNA virus with the unique or unusual feature of having a nucleocapsid (N) protein that is specifically transported to the nucleolus of virus-infected cells. In this communication, we show that the N protein is a phosphoprotein. Phosphoamino acid analysis of authentic and recombinant N proteins demonstrated that serine residues were exclusively phosphorylated. The pattern of phosphorylated N protein cellular distribution in comparison with that of [35S]methionine-labeled N protein suggested that phosphorylation does not influence subcellular localization of the protein. Time course studies showed that phosphorylation occurred during, or shortly after, synthesis of the N protein and that the protein remained stably phosphorylated throughout the life cycle of the virus to the extent that phosphorylated N protein was found in the mature virion. Two-dimensional electrophoresis and acid-urea gel electrophoresis showed that one species of the N protein is predominant in virus-infected cells, suggesting that multiple phosphorylated isoforms of N do not exist.
Porcine reproductive and respiratory syndrome (PRRS) is an economically important disease of pigs defined by severe respiratory disorders in piglets and widespread abortions in gestating sows and gilts. PRRS virus (PRRSV), the causative agent of PRRS, is a single-stranded, positive-sense, enveloped RNA virus classified in the family Arteriviridae, order Nidovirales (3). North American and European isolates of PRRSV represent two distinct genotypes (13, 16, 20), which have significant antigenic differences (22, 32). These observations were subsequently corroborated with the decoding of the full-length genomic sequences of both North American (1, 21, 34) and European PRRSV isolates (17). The 5′ two-thirds of the genome encodes the replicase gene that produces two polyproteins: open reading frame 1a (ORF1a) and ORF1ab, the latter of which results from a −1 frameshift during translation. The remaining one-third of the genome encodes seven structural proteins that are translated from a 3′-coterminal nested set of subgenomic mRNAs which also share a common 5′ leader sequence that is derived from the 5′ end of the genome (24, 31).
The nucleocapsid (N) protein is comprised of 123 and 128 amino acids for North American and European genotypes, respectively, and constitutes up to 40% of the protein content of the virion. This highly basic protein is also a major immunogen of the virus, with major antigenic determinants located predominantly in the central region of the protein (18, 25, 33) in an area of high surface probability (7). Mapping studies indicate that N protein conformation is readily disrupted by small deletions from the carboxy terminus and that structural integrity is essential for antigenic determinant formation (18, 33, 35). Two stretches of basic amino acids are situated in the amino terminus of the N protein. In addition to their presumed role in binding to and packaging of genomic RNA, these basic amino acids encode two potential nucleolar localization signals that may function to direct the transport of the N protein to the nucleolus (27). The biological significance of N protein nucleolar localization is not yet understood. However, in equine arteritis virus (EAV), the prototype arterivirus, continuous replication and mRNA synthesis have been observed in cells treated with leptomycin B, a chemical that disables the CRM1 export pathway and results in nuclear retention of the N protein (29). This observation suggests that replication can proceed in the absence of the N protein. Indeed, Molenkamp et al. (19) have shown that the structural proteins of EAV are dispensable for genome replication and subgenomic mRNA transcription. It is of interest, however, that the N proteins of group I, II, and III coronaviruses are also transported to the nucleolus (37), which suggests, given its evolutionary conservation, that localization of the N protein to the nucleolus may be of functional significance in the order Nidovirales. Regardless of its function in the nucleus, nucleocapsid assembly and budding of arteriviruses are cytoplasmic events, and therefore the N protein must be transported back to the cytoplasm to fulfill its role in virion assembly.
Multifunctional proteins are often regulated by phosphorylation, and this phenomenon is particularly common to positively charged, nucleic acid binding proteins that make up the nucleocapsids of such viruses as the coronavirus (12, 39), hepatitis B virus (26), hepatitis delta virus (4), influenza virus (2), rabies virus (38), herpes simplex virus (15), and parvovirus (14). Since it is well documented that phosphorylation is involved in regulating the activities of proteins at multiple levels, including nucleic acid binding, oligomerization, and nuclear transport, phosphorylation may likewise affect these properties of the PRRSV N protein.
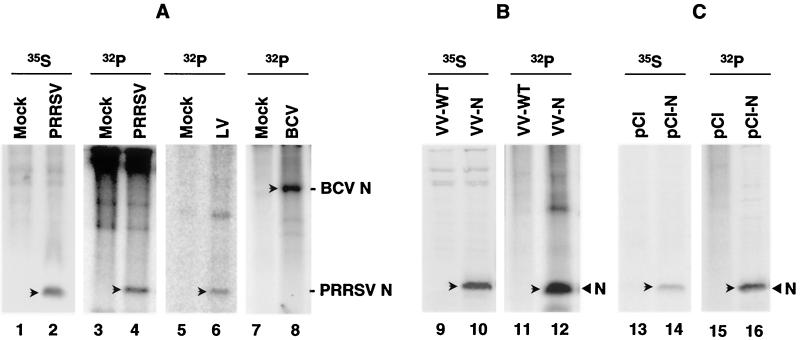
In order to determine whether the PRRSV N protein is phosphorylated, MARC-145 cells (a subclone of MA104 monkey kidney cells [11]) infected with the PA8 strain of PRRSV were metabolically labeled with [32Pi]orthophosphate from 32 to 38 h postinfection (p.i.). Cell lysates were prepared using radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate [DOC], 150 mM NaCl, 50 mM Tris-HCl [pH 7.4], 10 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 2 mM phenylmethylsulfonyl fluoride) and subjected to immunoprecipitation with a mixture of N-specific monoclonal antibodies (MAbs) (33). A 15-kDa 32Pi-labeled protein was precipitated from virus-infected cell lysates (Fig. 1A, lane 4). This protein corresponded to the [35S]methionine-labeled N protein (lane 2), indicating that the PRRSV N protein was a phosphoprotein. Since the N protein amino acid similarity between the two PRRSV genotypes is only 58%, it was of interest to examine whether the Lelystad virus (European genotype) N protein was a phosphoprotein. As shown in Fig. 1A, lane 6, the Lelystad virus N protein was specifically labeled by 32Pi. The efficacy of 32Pi metabolic labeling was verified using the bovine coronavirus (BCV) N protein, a known phosphoprotein (39). As demonstrated in Fig. 1A, lane 8, a 55-kDa phosphorylated protein was immunoprecipitated from BCV-infected cell lysates by using an N-specific MAb. These data demonstrate that the PRRSV N protein is modified by phosphorylation and that N protein phosphorylation is a common feature of PRRSV, irrespective of genotype. Moreover, since the EAV N protein has been shown to be phosphorylated (41), this suggests that phosphorylation of N may be of functional importance for the family Arteriviridae.
FIG. 1.
Phosphorylation of the PRRSV N protein. (A) N protein phosphorylation in virus-infected cells. MARC-145 cells were infected with the PA8 strain (North American genotype) or Lelystad strain (European genotype) of PRRSV and labeled at 32 h p.i. for 6 h with either 50 μCi of [35S]methionine/ml or 300 μCi of [32Pi]orthophosphate/ml. For BCV, Mardin-Darby bovine kidney cells were infected and labeled at 24 h p.i. for 6 h with 32Pi. Cell lysates were immunoprecipitated with a mixture of the N-specific MAbs for PA8 virus or with MAb NS99 for Lelystad virus. The BCV N protein was precipitated using MAb MD8-3. The precipitates were separated by SDS-12% PAGE followed by autoradiography. Lanes 1, 3, 5, and 7, uninfected; lanes 2 and 4, PA8 infected; lane 6, Lelystad virus infected; lane 8, BCV infected. (B) N protein phosphorylation in recombinant vaccinia virus-infected cells. MARC-145 cells were infected with either wild-type vaccinia virus (VV-WT) or recombinant vaccinia virus expressing the PRRSV N protein (VV-N) and radiolabeled at 12 h p.i. for 6 h with [35S]methionine or 32Pi. Cell lysates were immunoprecipitated as describe above. Lanes 9 and 11, wild-type vaccinia virus-infected cells; lanes 10 and 12, recombinant vaccinia virus-infected cells. (C) N protein phosphorylation in N gene-transfected cells. COS-1 cells were transfected with plasmids pCI-Neo or pCI-Neo-N containing the N coding sequence and labeled at 36 h posttransfection for 12 h with either [35S]methionine or 32Pi. Lanes 13 and 15, pCI-Neo transfected; lanes 14 and 16, pCI-Neo-N transfected. Arrows denote the N protein of BCV or PRRSV.
To investigate whether N protein phosphorylation required the presence of other PRRSV constituents, phosphorylation of the N protein expressed from a recombinant vaccinia virus (VV-N) was examined. The N protein immunoprecipitated from recombinant vaccinia virus-infected cell lysates was found to be phosphorylated (Fig. 1B, lane 12), thereby indicating that N protein phosphorylation does not require the presence of other structural or nonstructural components of PRRSV. To address the possible involvement of vaccinia virus-specific functions in this process, the N gene was subcloned into the eukaryotic expression vector pCI-Neo (Invitrogen) and used to transfect COS-1 cells. The transfected cells were radiolabeled with either [35S]methionine or 32Pi from 36 to 48 h posttransfection, and cell lysates were immunoprecipitated with a mixture of N-specific MAbs. A 15-kDa phosphorylated protein was identified in the N gene-transfected cells (Fig. 1C, lane 16), indicating that N protein phosphorylation was not mediated by a vaccinia virus-encoded kinase, but rather by a cellular kinase. As we were unable to identify any autophosphorylation activity associated with the N protein coupled to Sepharose beads by in vitro kinase assay, we concluded that N protein phosphorylation was mediated by a cellular kinase(s).
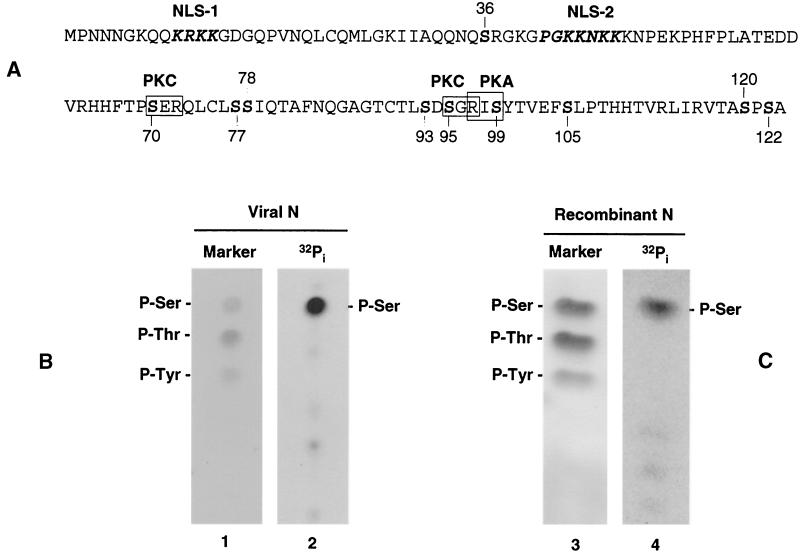
The N protein of PA8 contains 1 tyrosine, 9 threonine, and 10 serine residues that can potentially function as targets for phosphorylation (Fig. 2A). To determine the identity of the phosphorylated amino acid(s), phosphoamino acid analysis was carried out. The 32Pi-labeled N protein immunoprecipitated from either PRRSV- or VV-N-infected cells was separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to a polyvinylidene difluoride membrane in CAPS buffer (pH 10.4; Sigma). The region of the membrane containing the N protein was excised and hydrolyzed in 6 N HCl at 110°C for 2 h. The hydrolysate was lyophilized and dissolved in water containing 0.5 μg each of phosphoserine, phosphothreonine, and phosphotyrosine standards (Sigma). The samples were subsequently spotted onto silica gel plates (Whatman flexible-backed TLC plate; Fisher Scientific) and electrophoresed in one dimension in 0.5% pyridine-5% acetic acid (pH 3.5) for 150 min at 400 V with cooling at 10°C. The amino acid standards were visualized by ninhydrin staining (Fig. 2B and C, lanes 1 and 3). The same plates were then exposed to a phosphorimager to obtain radiographic images of the labeled amino acid(s). The results demonstrated that the N protein prepared from PRRSV-infected cells was phosphorylated exclusively on serine residues (Fig. 2B, lane 2), and no evidence was obtained to suggest that either threonines or tyrosines were phosphorylated. Additional experiments were performed using the recombinant N protein purified from recombinant vaccinia virus-infected cells, and identical results were obtained, such that the liberated phosphoamino acid corresponded to phosphoserine (Fig. 2C, lane 4), thereby confirming that the PRRSV N protein is a serine phosphoprotein.
FIG. 2.
(A) Amino acid sequence and the predicted functional motifs of the PRRSV N protein. Nuclear localization signals are indicated in bold italics (27), and the protein kinase motifs for protein kinase C (PKC) (S-X-R) and PKA (R-X-S) are outlined with boxes. Numbers indicate the positions of serine residues. (B and C) Identification of phosphoamino acids of the N protein. MARC-145 cells infected with PRRSV (B) or recombinant vaccinia virus expressing the PRRSV N protein (C) were radiolabeled with 300 μCi of 32Pi/ml and subjected to immunoprecipitation with an N-specific MAb mixture. The precipitates were resolved by SDS-PAGE and transferred to polyvinylidene difluoride membrane. N protein bands were excised and hydrolyzed with 6N HCl for 2 h at 110°C. Amino acids were separated by one-dimensional electrophoresis on cellulose plates in 0.5% pyridine-5% acetic acid [pH 3.5] for 150 min at 400 V with cooling at 10°C. Unlabeled phosphoamino acid standards were visualized by staining with 0.25% ninhydrin (lanes 1 and 3), and the 32Pi-labeled amino acids were visualized by autoradiography (lanes 2 and 4).
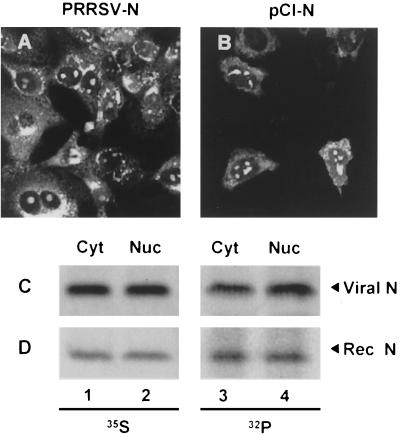
Rowland et al. (27) have demonstrated that, in addition to its cytoplasmic distribution, the N protein of PRRSV is found in the nucleolus of virus-infected cells. As phosphorylation is commonly used to regulate the subcellular localization of proteins, we wanted to examine whether phosphorylation of the N protein had an effect on its cellular compartmentalization. To confirm nucleolar localization of the N protein derived from the PA8 strain, confocal microscopy of virus-infected and N gene-transfected cells was performed. In PRRSV-infected cells, we monitored N protein distribution over time and found that N protein staining varied from intensely nucleolar during the early stages of infection (6 to 20 h p.i.) to exclusively cytoplasmic (30 to 48 h p.i.) during the later stages of infection (data not shown). Distribution of the N protein in PRRSV-infected cells at 25 h p.i. is illustrated in Fig. 3A. This image is representative of the various cellular compartments within which the N protein is found to localize over time. These include the intensely stained regions in the endoplasmic reticulum (ER), perinuclear region, and nucleolus as well as the diffusely stained cytoplasm. Although the same intense nucleolar staining of the N protein was consistently observed in the N gene-transfected cells, we did not see marked accumulation of the N protein in the ER, but rather the distribution of N was more diffuse in the cytoplasm (Fig. 3B). This difference in cytoplasmic distribution between recombinant and viral N protein may be due to the fact that replication complex formation and virus assembly do not take place in N gene-transfected cells and these processes are likely what concentrate the N protein in the ER and perinuclear regions of the cell, as has been documented for EAV (19, 30). To examine whether phosphorylated N protein was specifically localized in one particular compartment of the cell, fractionation experiments were conducted. Virus-infected or N gene-transfected cells were labeled with either [35S]methionine or 32Pi and separated into cytoplasmic and nuclear fractions according to the protocol of Jameel et al. (6). Cell monolayers were washed twice with phosphate-buffered saline and then scraped into 1 ml of phosphate-buffered saline. After low-speed centrifugation, cell pellets equivalent to a 60-mm-diameter plate were resuspended in 0.5 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 140 mM NaCl, 5 mM iodoacetamide, 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride) and incubated on ice for 1 h. Lysates were centrifuged at 14,000 rpm for 30 min (Micromax; International Equipment Co., Needham Heights, Mass.). The supernatant (cytoplasmic fraction) was removed to a fresh tube, and 50 μl of a 10× DOC-SDS solution (10% sodium DOC, 1% SDS) was added to it. The pellet was washed once in 0.5 ml of lysis buffer as described above and resuspended in 0.5 ml of RIPA buffer (nuclear fraction). Both fractions were immunoprecipitated with an N-specific MAb mixture. In accordance with the immunofluorescence data shown in Fig. 3A and B, approximately equal amounts of the [35S]-labeled N protein was found to be associated with the cytoplasmic and nuclear fractions (Fig. 3C and D, lanes 1 and 2), whether or not other viral constituents were present. An equivalent distribution was observed with the 32Pi-labeled N protein, such that equal amounts of N were associated with both the cytoplasmic and nuclear fractions (Fig. 3C and D, lanes 3 and 4). As a control, β-galactosidase extracts prepared from cells transfected with pCMV-Sport-β-Gal (Invitrogen) were fractionated and the cellular distribution of this resident cytoplasmic reporter protein was assessed colorimetrically. The results from the β-galactosidase assay confirmed the authenticity of the cytoplasmic and nuclear fractions prepared using this fractionation method (data not shown). We concluded, therefore, that phosphorylation does not appear to limit or confine the N protein to a particular region of the cell, such as the nucleolus, suggesting that N protein nucleolar localization is not likely to be regulated by phosphorylation.
FIG. 3.
Subcellular distribution of the N protein. PRRSV-infected (25 h p.i.) (A) or N gene-transfected (B) cells were fixed with 4% formaldehyde and permeabilized with 0.1% NP-40. The cells were stained with an N-specific MAb mixture followed by Alexa Fluor 488 goat anti-mouse antibody and examined by laser scanning confocal microscopy. (C and D). Subcellular fractionation of the N protein. PRRSV (C) or recombinant vaccinia virus-infected (D) cells were radiolabeled with either [35S]methionine (lanes 1 and 2) or 32Pi (lanes 3 and 4) and separated into cytoplasmic (lanes 1 and 3) and nuclear (lanes 2 and 4) fractions followed by immunoprecipitation with an N-specific MAb mixture and SDS-PAGE.
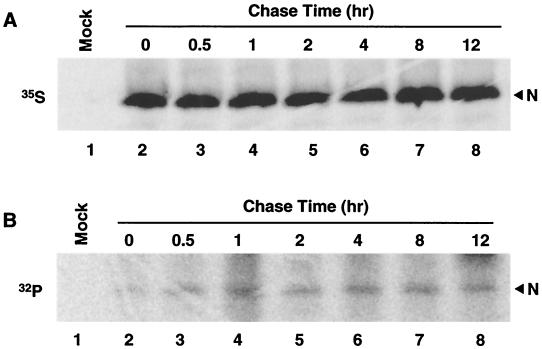
To examine the kinetics of N protein phosphorylation, virus-infected cells were starved for 1 h at 32 h p.i. and pulse-labeled with 500 μCi of 32Pi/ml for 30 min. The cells were either harvested immediately or chased for 0.5, 1, 2, 4, 8, and 12 h and then subjected to immunoprecipitation with N-specific MAbs. As shown in Fig. 4B, a single species of 32Pi-labeled protein was identified, which was stable for at least 12 h following its synthesis. The 32Pi-labeled protein was visible immediately following the 30-min pulse, which suggests that the N protein is rapidly phosphorylated following translation. The amount of [35S]methionine- and 32Pi-labeled N protein remained constant throughout the chase period (Fig. 4A and B, respectively), demonstrating that the N protein was not subject to degradation over time and that phosphorylation was stable.
FIG. 4.
Pulse-chase analysis of the PRRSV N protein. Following 1 h of starvation in deficient medium, PRRSV-infected cells were pulse-labeled at 32 h p.i. for 30 min in medium supplemented with either 150 μCi of [35S]methionine/ml (A) or 500 μCi of 32Pi/ml (B) and then chased for the indicated time intervals. At various time points postlabeling, cells were harvested and the lysates were reacted with an N-specific MAb mixture. The immunoprecipitated proteins were separated by SDS-12% PAGE and visualized by autoradiography. Mock-infected cells were harvested immediately following the pulse-label (lane 1). The chase times (in hours) are displayed above the panels.
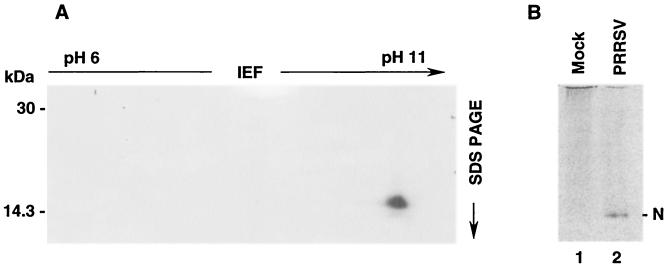
To determine whether multiple isoforms of the N protein exist, two-dimensional isoelectric focusing was carried out. [35S]methionine-labeled N protein was purified from virus-infected cells by immunoprecipitation and subsequently concentrated into a low salt buffer (25 mM Tris-HCl [pH 7.4]) using a Microcon filter device (model YM-10; Millipore). Precast Immobiline dry strips (pH 6 to 11, linear gradient; Amersham Biosciences) were rehydrated in 8 M urea, 0.5% Triton X-100, 0.5% carrier ampholyte (pH 6 to 11), and 0.2% (wt/vol) dithiothreitol for 16 h. The samples were applied to the anodic end of the strip and electrophoresed for a total of 25,000 V-h at 20°C. For the second-dimension separation, the equilibrated strips were placed on top of a 15% polyacrylamide gel in the usual position of the stacking gel, and PAGE was performed in the Tris-glycine buffer system followed by autoradiography. Only a single species of N was discernible in the two-dimensional separation, and this species migrated with a basic pI well within the range of the predicted pI for the N protein of 10.08 (Fig. 5A). These results suggested that one form of the N protein was predominant in virus-infected cells. To further evaluate whether the N protein could be resolved into multiple isomers, [35S]methionine-labeled N protein was subjected to acid-urea gel electrophoresis. Acid-urea separation has typically been employed to evaluate the processing of basic proteins into their multiple isoforms (23). Again, only a single species of the N protein was identified using this separation method (Fig. 5B, lane 2). Therefore, we did not obtain any evidence to indicate that differentially phosphorylated isoforms of the N protein exist. Taken together, our results suggest that N protein phosphorylation is unlikely to be transient but rather is uniform and stable.
FIG. 5.
Two-dimensional analysis and acid-urea gel electrophoresis of the PRRSV N protein. (A) For two-dimensional analysis, the N protein was purified by immunoprecipitation from virus-infected cells radiolabeled with 50 μCi of [35S]methionine/ml. For the first dimension, the samples were applied to precast Immobiline dry strips (pH 6 to 11, linear pH gradient) and focused at 20°C for a total of 25,000 V-h. For the second-dimension separation, the equilibrated strips were placed on top of an SDS-15% polyacrylamide gel and electrophoresed at 15 mA for the first 15 min and 30 mA for a total of 5 h. The gel was dried and autoradiographed. (B) Analysis of the PRRSV N protein by acid-urea gel electrophoresis. MARC-145 cells were infected with PRRSV and radiolabeled with 50 μCi of [35S]methionine/ml between 32 and 38 h p.i. Cell lysates were prepared and subjected to immunoprecipitation. The proteins were eluted from Sepharose beads in acid-urea sample buffer (5% acetic acid, 9 M urea, 0.002% methyl green) by heating samples at 80°C for 5 min. After prerunning gels in reverse polarity for 2 h at 150 V in 5% acetic acid, samples were loaded and electrophoresed at a constant voltage of 150 V for 1.5 h in a 15% polyacrylamide gel containing 6.4 M urea and 5% acetic acid. The gel was dried and autoradiographed. Lane 1, mock infected; lane 2, PRRSV infected.
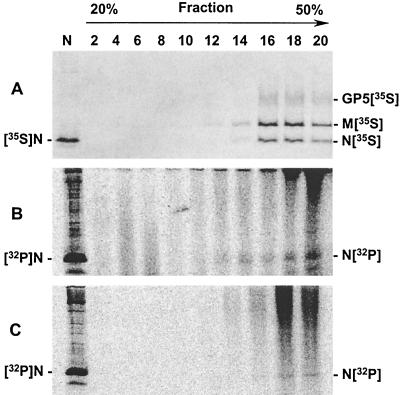
Given that the addition of phosphate groups to the N protein appeared to be a stable modification, it was therefore expected that the N protein in the virion would be phosphorylated. To examine the virion N protein, PRRSV-infected cells were radiolabeled at 32 h p.i. for 16 h with either 50 μCi of [35S]methionine/ml or 300 μCi of 32Pi/ml and the culture supernatants were collected. After cell debris was removed, the virions were pelleted through a 20% sucrose cushion. The virus pellet was then resuspended in Tris-EDTA buffer containing 100 mM NaCl and separated on a 20-to-50% (wt/vol) sucrose gradient at 50,000 rpm for 5 h in an SW55Ti rotor (Beckman). The purification procedure was performed in parallel, such that one sample contained 32Pi-labeled virus and the other sample contained [35S]methionine-labeled virus. A total of 20 fractions were collected from the top to the bottom of the gradient, and aliquots from even-numbered fractions were resolved by SDS-PAGE under reducing conditions. Three major viral proteins were readily identified in fractions 16 to 20 of the [35S]methionine-labeled virion preparation (Fig. 6A). The fastest-migrating band represented the N protein, and the next two higher-molecular-weight bands represented the M and GP5 proteins, respectively. In the case of the 32Pi-labeled virion preparation, a 15-kDa protein was identified in fractions 14 to 20 (Fig. 6B), and this protein migrated at the same rate as the immunoprecipitated N protein (Fig. 6B, lane N). To confirm the specificity of the phosphorylated band identified in Fig. 6B, the same fractions were subjected to immunoprecipitation with a mixture of N-specific MAbs. Immunoprecipitation of the N protein from individual fractions containing the intact virions was performed by adding a final concentration of 0.5% SDS, 1% Triton X-100, and 1% sodium DOC to the reaction, which served to disrupt the viral envelope, thus making the N protein accessible to MAbs. The identical bands were immunoprecipitated from these fractions (Fig. 6C), demonstrating that the 32Pi-labeled band identified in Fig. 6B was in fact the N protein. We concluded, therefore, that the virion-associated N protein is phosphorylated.
FIG. 6.
Phosphorylation of the virion N protein. Virus-infected cells were labeled between 36 and 48 h p.i., and the culture supernatants were harvested. The supernatants were preclarified to remove cellular debris before concentrating the virus through a 20% sucrose cushion for 2 h at 25,000 rpm in an SW28 rotor (XL-90 ultracentrifuge; Beckman Instruments, Inc.). Virus pellets obtained from the 20% sucrose cushion were resuspended in Tris-EDTA buffer and further purified in a 20-to-50% continuous sucrose gradient at 50,000 rpm (SW55Ti) for 5 h. Fractions were taken from the top of the gradients and aliquots from alternate fractions were directly analyzed without immunoprecipitation (A and B) or immunoprecipitated using N-specific MAbs (C), followed by SDS-PAGE under reducing conditions and autoradiography. (A) [35S]methionine-labeled virions; (B and C) 32Pi-labeled virions. GP5[35S], [35S]methionine-labeled GP5 glycoprotein; M[35S], [35S]methionine-labeled membrane-associated protein; N[35S], [35S]methionine-labeled nucleocapsid protein; N[32P], [32P]phosphate-labeled nucleocapsid protein.
In this communication, we have demonstrated that the PRRSV N protein is a phosphoprotein. Given that EAV and coronavirus nucleocapsid proteins are also phosphoproteins (41, 12), phosphorylation of the nucleocapsid protein appears to be a common feature of nidoviruses. Evolutionary conservation of this chemical modification suggests that phosphorylation of the N protein may be of significant biological importance for the virus and, therefore, it warranted further investigation. Phosphorylation of the N protein occurs not only in PRRSV-infected cells but also in N gene-transfected cells, where the N protein is synthesized in the absence of other viral components. Therefore, phosphorylation of the N protein is not an artifact and cellular kinases are involved in this process. Phosphoamino acid analysis was performed on authentic and recombinant N proteins, and in each case the liberated phosphorylated amino acid corresponded to phosphoserine. Given that analysis of a phosphoprotein purified by immunoprecipitation would represent the average state of phosphorylation, it is therefore unlikely that the N protein would be transiently phosphorylated on either threonine or tyrosine residues. Since identification of phosphorylated serine residues through proteolytic digestion of the 32Pi-labeled N protein was not possible due to the small size of the resultant digestion products, individual serine residues were mutated to alanine and examined for their phosphorylation properties (data not shown). Results from these experiments revealed that the N protein was phosphorylated on more than one residue and, thus, an alternative approach is currently being taken in order to thoroughly delineate the location of the phosphorylated serine residues.
Since the PRRSV N protein is both a cytoplasmic and nuclear-associated phosphoprotein, phosphorylation does not appear to be the mechanism determining N protein subcellular localization, unless of course transient hyperphosphorylated forms of the N protein exist. This, however, is an unlikely scenario because results from the pulse-chase and two-dimensional electrophoresis and acid-urea electrophoresis experiments suggest that the N protein population is rather homogeneous, with one species of N existing. Because the amount of 32Pi-labeled N protein remained constant throughout the chase period, this implies that phosphorylation of N is relatively stable and that cycling between phosphorylated and unphosphorylated species seems improbable. We examined purified virus particles to determine whether the virion N protein, following egress from the cell, was phosphorylated. As was expected, given the stable nature of N protein phosphorylation, the virion N protein was shown to be phosphorylated. Therefore, N appears to be phosphorylated throughout the extent of the virus life cycle. We did not, however, explore the possibility of N protein dephosphorylation occurring during uncoating, as it does in the case of the mouse hepatitis coronavirus (MHV) N protein (8).
Phosphorylation is an important posttranslational modification that has been shown to modulate a variety of macromolecular events in virus biology. As with the PRRSV N protein, numerous plant and animal virus nucleocapsid proteins are phosphorylated, and in many cases this chemical modification serves to stabilize distinct conformations of the protein (40). Although we have not addressed the biological function of N protein phosphorylation in this study, the consequence of viral capsid protein phosphorylation in general is well documented and typically falls into one of three categories: modulation of nucleic acid binding activity, regulation of nuclear localization, or mediation of protein-protein interactions including oligomerization. Phosphorylation modulates the RNA binding activity of many viral proteins, and instances of both positive and negative regulation have been cited in the literature. For example, in the human T-cell leukemia virus type 2, phosphorylation of the Rex protein enhances RNA binding activity (5), whereas for the hepatitis B virus core protein and the rabies virus nucleoprotein, phosphorylation leads to decreased RNA binding activity (9, 38). In the case of MHV, dephosphorylation of the N protein occurs upon virus entry into neutral endosomes and is required for subsequent uncoating and release of the viral RNA (8). Therefore, phosphorylated MHV N appears to exert a much higher affinity for genomic RNA than does the unphosphorylated N protein. Since phosphorylation of the N protein is conserved across the nidoviruses, it is quite possible that a similar mechanism of phosphorylation-regulated RNA binding may occur with the PRRSV N protein during uncoating. Alternatively, phosphorylation may regulate dimer formation or oligomerization, as has been speculated in the case of hepatitis C virus, where phosphorylation appears to modulate core protein dimer formation (28). In fact, the PRRSV N protein exists as a dimer in the mature virion (36) and, therefore, it is possible that phosphorylation may be involved in dimerization and subsequent oligomerization of the N protein for capsid assembly. Phosphorylation has also been shown to regulate nuclear localization of viral proteins. In the case of hepatitis B virus, phosphorylation regulates the core protein nuclear localization by inducing a conformational change that exposes a nuclear localization signal in the carboxy terminus of the core protein, which allows core binding to the nuclear pore complex through the importin-mediated pathway (10). This seems unlikely, however, for the PRRSV N protein, since our experimental data suggest that the phosphorylated N protein is distributed in all cellular compartments and thus phosphorylation does not appear to restrict its localization. Considering these aspects, the mechanism by which phosphorylation of the N protein plays a role in the PRRSV life cycle is currently under investigation.
Acknowledgments
This study was supported by funds from the Ontario Ministry of Agriculture, Food and Rural Affairs and from Ontario Pork.
We are grateful to P. Dobos for his help with phosphoamino acid analysis and critical reading of the manuscript. S.K.W. is a recipient of the Ontario Graduate Scholarship.
REFERENCES
- 1.Allende, R., T. L. Lewis, Z. Lu, D. L. Rock, G. F. Kutish, A. Ali, A. R. Doster, and F. A. Osorio. 1999. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J. Gen. Virol. 80:307-315. [DOI] [PubMed] [Google Scholar]
- 2.Arrese, M., and A. Portela. 1996. Serine 3 is critical for phosphorylation at the N-terminal end of the nucleoprotein of influenza virus A/Victoria/3/75. J. Virol. 70:3385-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 4.Chang, M. F., S. C. Baker, L. H. Soe, T. Kamahora, J. G. Keck, S. Makino, S. Govindarajan, and M. M. Lai. 1988. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J. Virol. 62:2403-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Green, P. L., M. T. Yip, Y. Xie, and I. S. Chen. 1992. Phosphorylation regulates RNA binding by the human T-cell leukemia virus Rex protein. J. Virol. 66:4325-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jameel, S., M. Zafrullah, M. H. Ozdener, and S. K. Panda. 1996. Expression in animal cells and characterization of the hepatitis E virus structural proteins. J. Virol. 70:207-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janin, J., and S. Wodak. 1978. Conformation of amino acid side-chains in proteins. J. Mol. Biol. 125:357-386. [DOI] [PubMed] [Google Scholar]
- 8.Kalicharran, K., D. Mohandas, G. Wilson, and S. Dales. 1996. Regulation of the initiation of coronavirus JHM infection in primary oligodendrocytes and L-2 fibroblasts. Virology 225:33-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kann, M., and W. H. Gerlich. 1994. Effect of core protein phosphorylation by protein kinase C on encapsidation of RNA within core particles of hepatitis B virus. J. Virol. 68:7993-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kann, M., B. Sodeik, A. Vlachou, W. H. Gerlich, and A. Helenius. 1999. Phosphorylation-dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol. 145:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim, H. S., J. Kwang, I. J. Yoon, H. S. Joo, and M. L. Frey. 1993. Enhanced replication of porcine reproductive and respiratory syndrome (PRRS) virus in a homogeneous subpopulation of MA-104 cell line. Arch. Virol. 133:477-483. [DOI] [PubMed] [Google Scholar]
- 12.Laude, H., and P. S. Masters. 1995. The coronavirus nucleocapsid protein, p. 141-163. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 13.Mardassi, H., S. Mounir, and S. Dea. 1994. Identification of major differences in the nucleocapsid protein genes of a Quebec strain and European strains of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 75:681-685. [DOI] [PubMed] [Google Scholar]
- 14.Maroto, B., J. C. Ramirez, and J. M. Almendral. 2000. Phosphorylation status of the parvovirus minute virus of mice particle: mapping and biological relevance of the major phosphorylation sites. J. Virol. 74:10892-10902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McNabb, D. S., and R. J. Courtney. 1992. Posttranslational modification and subcellular localization of the p12 capsid protein of herpes simplex virus type 1. J. Virol. 66:4839-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng, X.-J., P. S. Paul, P. G. Halbur, and M. A. Lum. 1995. Phylogenetic analysis of the putative M (ORF 6) and N (ORF 7) genes of porcine reproductive and respiratory syndrome virus (PRRSV): implication for the existence of two genotypes of PRRSV in the USA and Europe. Arch. Virol. 140:745-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meulenberg, J. J. M., M. M. Hulst, E. J. de Meijer, P. J. M. Moonen, A. den Besten, E. P. De Kluyver, G. Wensvoort, and R. J. M. Moormann. 1993. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology 192:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meulenberg, J. J. M., A. P. van Nieuwstadt, A. van Essen-Zandbergen, J. N. A. Bos-de-Ruijter, J. P. M. Langeveld, and R. H. Meloen. 1998. Localization and fine mapping of antigenic sites on the nucleocapsid protein of porcine reproductive and respiratory syndrome virus with monoclonal antibodies. Virology 252:106-114. [DOI] [PubMed] [Google Scholar]
- 19.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 20.Murtaugh, M. P., M. R. Elam, and L. T. Kakach. 1995. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch. Virol. 140:1451-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nelson, C. J., M. P. Murtaugh, and K. S. Faaberg. 1999. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J. Virol. 73:270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson, E. A., J. Christopher-Hennings, T. Drew, G. Wensvoort, J. E. Collins, and D. A. Benfield. 1993. Differentiation of U.S. and European isolates of porcine reproductive and respiratory syndrome virus by monoclonal antibodies. J. Clin. Microbiol. 31:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panyim, S., and R. Chalkley. 1969. High resolution acrylamide gel electrophoresis of histones. Arch. Biochem. Biophys. 130:337-346. [DOI] [PubMed] [Google Scholar]
- 24.Pasternak, A. O., E. van den Born, W. J. Spaan, and E. J. Snijder. 2001. Sequence requirements for RNA strand transfer during Nidovirus discontinuous subgenomic RNA synthesis. EMBO J. 20:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez, M. J., J. Sarraseca, J. Garcia, A. Sanz, J. Plana-Duran, and J. I. Casal. 1997. Epitope mapping of the nucleocapsid protein of European and North American isolates of porcine reproductive and respiratory syndrome virus. J. Gen. Virol. 78:2269-2278. [DOI] [PubMed] [Google Scholar]
- 26.Roossinck, M. J., and A. Siddiqui. 1987. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J. Virol. 61:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rowland, R. R., R. Kervin, C. Kuckleburg, A. Sperlich, and D. A. Benfield. 1999. The localization of porcine reproductive and respiratory syndrome virus nucleocapsid protein to the nucleolus of infected cells and identification of a potential nucleolar localization signal sequence. Virus Res. 64:1-12. [DOI] [PubMed] [Google Scholar]
- 28.Shih, C. M., C. M. Chen, S. Y. Chen, and Y. H. Lee. 1995. Modulation of the trans-suppression activity of hepatitis C virus core protein by phosphorylation. J. Virol. 69:1160-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tijms, M. A., Y. van der Meer, and E. J. Snijder. 2002. Nuclear localization of non-structural protein 1 and nucleocapsid protein of equine arteritis virus. J. Gen. Virol. 83:795-800. [DOI] [PubMed] [Google Scholar]
- 30.van der Meer, Y., H. van Tol, J. Krijnse Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 72:6689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Marle, G., J. C. Dobbe, A. P. Gultyaev, W. Luytjes, W. J. Spaan, and E. J. Snijder. 1999. Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc. Natl. Acad. Sci. USA 96:12056-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wensvoort, G., E. P. de Kluyver, E. A. Luijtze, A. den Besten, L. Harris, J. E. Collins, W. T. Christianson, and D. Chladek. 1992. Antigenic comparison of Lelystad virus and swine infertility and respiratory syndrome (SIRS) virus. J. Vet. Diagn. Investig. 4:134-138. [DOI] [PubMed] [Google Scholar]
- 33.Wootton, S. K., E. A. Nelson, and D. Yoo. 1998. Antigenic structure of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus. Clin. Diagn. Lab. Immunol. 5:773-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wootton, S., D. Yoo, and D. Rogan. 2000. Full-length sequence of a Canadian porcine reproductive and respiratory syndrome virus (PRRSV) isolate. Arch. Virol. 145:2297-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wootton, S., G. Koljesar, L. Yang, K. J. Yoon, and D. Yoo. 2001. Antigenic importance of the carboxy-terminal beta-strand of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. Clin. Diagn. Lab. Immunol. 8:598-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wootton, S., and D. Yoo. 2001. Homotypic interactions of the nucleocapsid protein of porcine reproductive and respiratory syndrome virus (PRRSV). Adv. Exp. Med. Biol. 494:627-632. [DOI] [PubMed] [Google Scholar]
- 37.Wurm, T., H. Chen, T. Hodgson, P. Britton, G. Brooks, and, J. A. Hiscox. 2001. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J. Virol. 75:9345-9356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, J., H. Koprowski, B. Dietzschold, and Z. F. Fu. 1999. Phosphorylation of rabies virus nucleoprotein regulates viral RNA transcription and replication by modulating leader RNA encapsidation. J. Virol. 73:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo, D., G. J. Cox, and I.-D. Yoo. 1992. Phosphorylation of the nucleocapsid protein of bovine coronavirus expressed with a recombinant baculovirus vector. J. Microbiol. Biotechnol. 2:122-128. [Google Scholar]
- 40.Yu, M., and J. Summers. 1994. Phosphorylation of the duck hepatitis B virus capsid protein associated with conformational changes in the C terminus. J. Virol. 68:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeegers, J. J., B. A. Van der Zeijst, and M. C. Horzinek. 1976. The structural proteins of equine arteritis virus. Virology 73:200-205. [DOI] [PubMed] [Google Scholar]