Abstract
An important aspect of the pathophysiology of human immunodeficiency virus type 1 (HIV-1) infection is the ability of the virus to replicate in the host vigorously without a latent phase and to kill cells with a dynamic turnover of 1.8 × 109 cells/day and 10.3 × 109 virions/24 h. The transcription of HIV-1 RNA in acute infection occurs at two stages; the transcription of viral spliced mRNA occurs early, and the transcription of viral genomic RNA occurs later. The HIV-1 Tat protein is translated from the early spliced mRNA and is critical for HIV-1 genomic RNA expression. The cellular transcription factors are important for HIV-1 early spliced mRNA expression. In this study we show that virion nucleocapsid protein (NC) has a role in expression of HIV-1 early spliced mRNA. The HIV-1 NC migrates from the cytoplasm to the nucleus and accumulates in the nucleus at 18 h postinfection. Mutations on HIV-1 NC zinc fingers change the pattern of early viral spliced mRNA expression and result in a delayed expression of early viral mRNA in HIV-infected cells. This delayed HIV-1 early spliced mRNA expression occurs after proviral DNA has been integrated into the cellular genome, as shown by a quantitative integration assay. These results show that virion NC plays an important role in inducing HIV-1 early mRNA expression and contributes to the rapid viral replication that occurs during HIV-1 infection.
The human immunodeficiency virus type 1 (HIV-1) is a retrovirus which replicates in host cells with a dynamic turnover rate of 1.8 × 109 cells/day and 10.3 × 109 virions/24 h (19, 27, 34). During the acute HIV-1 infection, viral RNA is expressed in two stages, early expression of the spliced mRNA and later expression of the genomic RNA (21, 22). The Tat protein is translated from an early spliced mRNA (8, 21, 22, 25). The basal-level activity of the HIV-1 long terminal repeat (LTR) plays an important role in transcribing the HIV-1 early spliced mRNA, and the cellular general transcriptional factors appear to be major factors which regulate the LTR basal-level activity (8, 12, 25, 26, 33, 35, 38). In this report we show that the HIV-1 nucleocapsid protein (NC), a virion protein, contributes to viral spliced mRNA expression in the early stages of HIV-1 infection.
HIV-1 NC has two copies of a cysteine-histidine motif (CCHC) that is similar to the metal-binding finger domains of several proteins that interact with nucleic acids (9, 20, 29). This basic, hydrophilic protein binds genomic viral RNA in the nucleocapsid and comprises the viral core with the HIV-1 capsid protein. Each HIV-1 virion contains ∼2,000 molecules of NC (32). Virion NC enters cells with the viral genomic RNA and accumulates in the nucleus 8 h after entry (16). The early functions of NC in HIV-1 infection are reported as enhancing the first- and second-strand viral DNA synthesis and enhancing HIV-1 integrase activity in vitro (6, 11, 13, 18, 23; K. Musier-Forsyth, R. Gorelick, A. Mangla, and M. Seffernick, Proc. 2000 Meet. Retroviruses, p. 21, 2000). We have previously shown that NC binds to the HIV-1 LTR and enhances LTR-directed gene expression in vitro and in vivo (37). In this report we show that virion NC migrates from the cytoplasm to the nucleus at 10 h postinfection and that NC nuclear localization induces viral spliced mRNA expression. This role of NC comes into play after HIV-1 DNA has been integrated into the cellular genome.
MATERIALS AND METHODS
Cells and viruses.
H9 cells were grown in RPMI with 10% fetal bovine serum. Human peripheral blood mononuclear cells (PBMC) were collected from healthy blood donors and isolated with Ficoll-Hypaque gradients. The PBMC were stimulated with phytohemagglutinin and cultured in RPMI with 20% fetal bovine serum for 48 h before being used in the experiment. The viral pool of HIV-1IIIB was collected from H9/HTLV-IIIB cell culture supernatant (NIH AIDS Research & Reference Program). The HIV-1 NC mutants CCCC and SSHS and the wild-type proviruses are gifts from R. Gorelick and L. Arthur (17, 18). The viral pools of HIV-1 NC mutants CCCC and SSHS and the wild type were prepared from supernatant of 293T cell cultures after transfection (17, 18). The viruses were subjected to titer determination by a 50% tissue culture infective dose (TCID50) assay (1) on H9 cells or PBMC before they were used in the experiment.
HIV-1 acute infection.
The H9 cells and human PBMC were acutely infected with 3,000 TCID50 of each virus per 106 cells. The unattached viruses were washed out with phosphate-buffered saline. The acutely infected cells were cultured at 37°C and harvested at each time point. HIV-1 NC nuclear localization, viral early-mRNA expression, and viral DNA integration were detected with aliquots of cells which were acutely infected with wild-type and mutant viruses at equal TCID50 and harvested 4, 8, 10, 16, 18, 22, 24, 30, 32, 46, and 48 h postinfection.
Immunofluorescent staining.
The nuclear staining method was used to examine the viral protein nuclear localization (31). A total of 106 cells were acutely infected with 3 × 103 TCID50 of HIV-1IIIB, HIV-1 wild type, HIV-1 NC mutant CCCC, or HIV-1 NC mutant SSHS. Aliquots of cells were harvested at each time point and processed as described previously (31). Briefly, the cells were fixed with fixing buffer containing 2% paraformaldehyde, permeabilized on ice with buffer containing 0.2% Triton X-100, and blocked with blocking reagent (Dako, Carpinteria, Calif.) before being stained. The primary antibody SP542 was a mouse anti-human HIV-1 NCp7 monoclonal antibody which detects an epitope in the N terminus of NC (17, 24). SP542 was a gift from R. Gorelick and L. Arthur. The fluorescent staining of this antibody was specifically blocked with purified NC (HIV-1MN p7) but not with purified HIV-1 gp120 (data not shown). A normal mouse immunoglobulin G (IgG) monoclonal antibody (Sigma, St. Louis, Mo.) was used as a specificity control for SP542 and did not bind to HIV-1-infected PBMC and H9 cells or to uninfected PBMC and H9 cells in our assay conditions. The secondary antibody was an anti-mouse IgG-fluorescein isothiocyanate (Sigma). The secondary antibody alone did not bind to the infected or uninfected PBMC and H9 cells under our assay conditions. The stained cells were preserved in mounting media and examined with a fluorescence microscope (Nikon) at ×100 magnification. The same immunofluorescent-staining method was used to stain the samples which were examined under a confocal microscope, except that the second antibody was an anti-mouse IgG-Red613 (Molecular Probes, Eugene, Oreg.) which specifically labels the HIV-1 NCp7 monoclonal antibody and Sytox Green (Molecular Probes) was used to stain the nucleus. The samples were examined under the confocal microscope (Zeiss LSM 5 PASCAL) using an oil lens (63.5×) with a VARIO 2 RGB laser scanning module.
Isolation and detection of viral RNA.
The total cellular RNA from the HIV-1-infected and mock-infected cells were isolated with the RNeasy kit (Qiagen, Valencia, Calif.) and purified with DNase following the RNeasy cleanup protocol (Qiagen).
Isolation and detection of viral DNA.
The cellular genomic DNA was isolated from the HIV-1-infected and mock-infected cells, electrophoresed in a 0.5% agarose gel, and purified with the Qiagen II gel extraction kit (Qiagen). The HIV-1-integrated DNA was examined by Alu-LTR PCR as described previously (5, 7, 15). The 2-LTR circle DNA was examined directly from the cellular genomic DNA aliquots which were not gel purified.
RT-PCR assay.
Reverse transcription-PCR (RT-PCR) was performed with the OneStep RT-PCR kit as specified by the manufacturer (Qiagen). The sequences of primers for HIV-1 spliced mRNA and cellular β-globin mRNA were as previously described (22, 28, 36). The primer pair US and ART7, which was used to detect tat, rev, and nef mRNAs, spans known splice junctions to amplify the doubly spliced mRNAs for the regulatory proteins Tat, Rev, and Nef (22). The sequences of the US and ART7 primers are 5′-TCTCTCGACGCAGGACTCGGCTTGC-3′ and 5′-TTCTATTCCTTCGGGCCTGTCG-3′, respectively (22, 36). The RT reaction was carried out at 50°C for 30 min. The initial PCR activation was performed at 95°C for 15 min and was followed by 45 cycles of denaturation at 94°C for 0.5 min, annealing at 50°C for 0.5 min, and extension at 72°C for 1 min. The final PCR products were examined on a 2% agarose gel after electrophoresis. Aliquots of cells, which were collected at each time point and were examined for HIV-1 NC localization, were examined for early viral spliced mRNA expression. The same amount of total cellular RNA from each condition was used in the RT-PCR assay.
Alu-LTR PCR and Southern hybridization assay.
Two Alu-LTR PCR assays were used to examine the HIV-1 integrated DNA in the cellular genome. The first Alu-LTR assay examines the 5′ LTR and the Alu sequence in the cellular genome, and the second Alu-LTR assay examines the 3′ LTR and the Alu sequence in the cellular genome (5, 7, 15). The assays were performed as previously described (5, 7, 15). Briefly, for the first Alu-LTR assay, a nested PCR was carried out to examine the HIV-1 LTR in the purified cellular genomic DNA. The PCR was performed by using the Taq polymerase (Gibco BRL, Rockville, Md.) and a DNA thermal cycler (Perkin-Elmer, Foster City, Calif.). The first PCR was performed with the nested primers Alu-LTR 5′ from conserved sequences of human Alu and Alu-LTR 3′ from conserved HIV-1 LTR sequences. The sequences of the primers are as follows: Alu-LTR, 5′-TCCCAGCTACTCGGGAGGCTGAGG-3′; Alu-LTR 3′, 5′-AGGCAAGCTTTATTGAGGCTTAAGC-3′. The samples were subjected to denaturation at 94°C for 4 min and then 30 cycles of denaturation at 94°C for 0.5 min, annealing at 55°C for 0.5 min, and extension at 72°C for 3 min. Following the initial PCR, a second nested PCR amplification was carried out by using an aliquot equivalent to 1/400 of the 30-cycle PCR product. The primer sequences used in this inner-set PCR are 5′-CACACACAAGGCTACTTCCCT-3′ and 5′-GCCACTCCCCAGTCCCGCCC-3′. Samples were subjected to an enzyme activation step at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 0.5 min, annealing at 55°C for 0.5 min, and extension at 72°C for 1 min. The amplified PCR products were electrophoresed on a 1.5% agarose gel. Southern hybridization followed the gel electrophoresis and used 32P-end-labeled probes which are specific to the HIV-1 LTR (7). The aliquots of cells, which were collected at each time point and were examined for HIV-1 NC localization and early viral spliced mRNA expression, were examined for provirus integration. The same amount of genomic DNA from each condition was used in the Alu-LTR PCR assay.
The second Alu-LTR PCR assay was a Taqman real-time PCR assay. The Taqman Alu-LTR PCR and 2-LTR PCR assays were performed as described previously (5). Briefly, the integrated viral DNA was detected with the four sets of standards and two sets of primers and probes. A qualification standard is derived from the known copy number of SM2 plasmid spiked with H9 cellular DNA (5). In addition to the standards which are mentioned in reference 5, we added another standard which was prepared from the gel-purified genomic DNA of OM10.1 cells (4). This standard provided a reference curve and was used for providing copy numbers derived from threshold cycles (CT value) which were ≤36 cycles (Taqman protocol, Applied Biosystems, 2001; ABI Prism 7700 user bulletin no. 2, Applied Biosystems, 2001) with 6-carboxyfluorescein (FAM). The sequences of primers and probe are as follows: Late RT forward, MH531, 5′-TGTGTGCCCGTCTGTTGTGT-3′; Late RT reverse, MH532, 5′-GAGTCCTGCGTCGAGAGAGC-3′; Late RT probe, LRT-P, 5′-(FAM)-CAGTGGCGCCCGAACAGGGA-(TAMRA)-3′; Alu forward, MH535 (below); Alu reverse, SB704, 5′-TGCTGGGATTACAGGCGTGAG-3′; and Alu probe, MH603 (below). The primer and probe concentrations were the same as described in reference 5, as were the amplification conditions. The viral DNA was detected from cellular genomic DNA in 5 × 104 cells (500 ng) in quintuplicate, and the copy number of viral DNA was read directly from the result report spreadsheet, converted from the equivalent CT value with Sequence Detector V1.7 software.
The 2-LTR circles in the specimen were detected with 2-LTR primers and probe, with a quantification standard derived from the DNA copy number of the 2-LTR circle plasmid spiked with H9 cellular DNA (5). The sequences of the primers and probe are as follows: 2-LTR circle forward, MH535, 5′-AACTAGGGAACCCACTGCTTAAG-3′; 2-LTR reverse, MH536, 5′-TCCACAGATCAAGGATATCTTGTC-3′; and 2-LTR probe, MH603, 5′-(FAM)-ACACTACTTGAAGCACTCAAGGCAAGCTTT-(TAMRA)-3′. The viral DNAs were detected from cellular genomic DNA in 5 × 104 cells (500 ng) in quintuplicate, and the copy number of viral DNA was read directly from the result report spreadsheet, converted from the equivalent CT value with Sequence Detector V1.7 software.
Densitometer tracing.
The Hewlett-Packard ScanJet IIcx was used to scan the gel photographs and photographic negatives. The density of each band was analyzed by using ImageQuant (Molecular Dynamics) software. Results are recorded as the direct reading on ImageQuant.
RESULTS
NC nuclear localization.
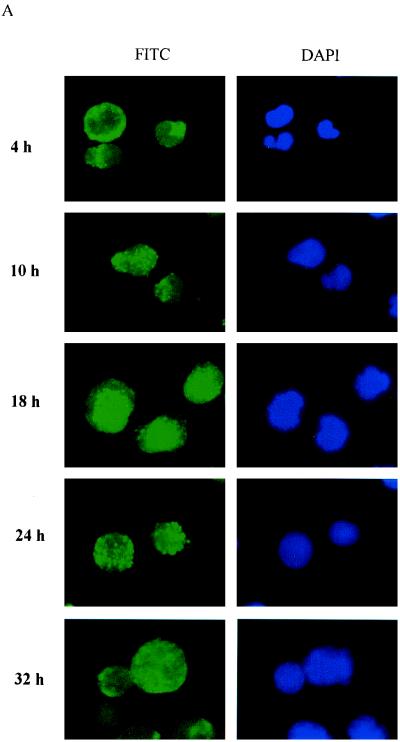
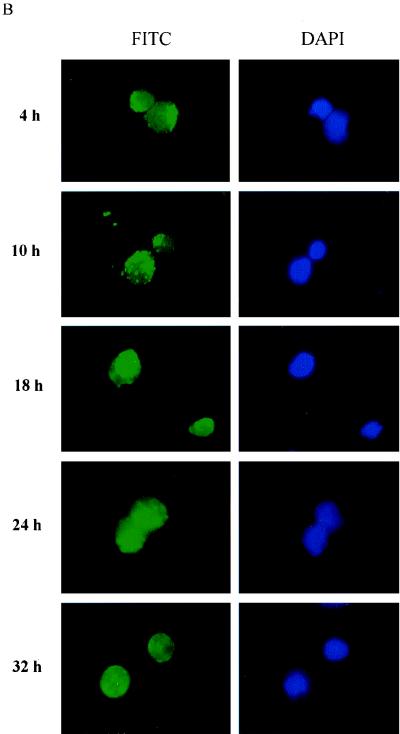
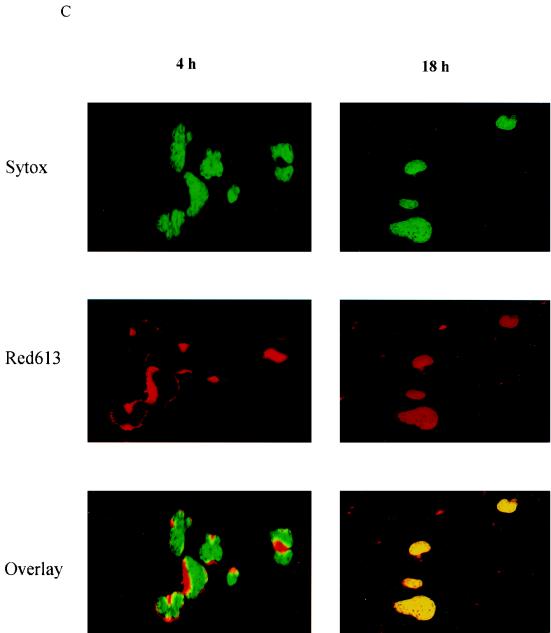
HIV-1 NC has two zinc finger motifs which are similar to the zinc fingers of many cellular DNA-binding proteins and transcriptional factors (3, 20, 29). In previous studies, virion NC has been found to migrate from the cytoplasm to the nucleus by 8 h postinfection (16). In this study, we examined the time course of virion NC nuclear localization by using immunofluorescent staining with a monoclonal antibody specific to HIV-1 NC. Our studies showed that virion NC appeared in the cytoplasm at 4 h postinfection and in the nucleus at 10 h postinfection. At 18 h postinfection, NC accumulated predominantly in the nucleus, and at 32 h post infection, it was found in both the nucleus and the cytoplasm (Fig. 1). This pattern of NC nuclear localization was observed in both human lymphatic cell line H9 cells (Fig. 1A) and in PBMC (Fig. 1B) acutely infected with HIV-1IIIB. This nuclear localization of NC was detected with a monoclonal antibody specific for HIV-1 NC, and the fluorescent staining with this antibody was blocked with purified NC (HIV-1MN p7) but not with purified HIV-1 gp120 (data not shown). We further examined the NC nuclear localization with a confocal microscope, at 4 and 18 h postinfection. At 4 h postinfection, NC signal was detected in the cytoplasm of infected cells (Fig. 1C). At 18 h postinfection, however, NC signal was clearly detected in the nucleus. The yellow signal from the laser-scanning confocal microscope (Fig. 1C) represented the combination of the NC (red staining of NC) and nuclear localization (green staining of DNA). We could count the number of cells which were infected, with NC signals being detected by a confocal microscope with a VARIO 2 RGB laser-scanning module. The NC detected in the nucleus at 18 h postinfection was in 45% of the cells counted.
FIG. 1.
NC nuclear localization by immunofluorescent staining. FITC, fluorescein isothiocyanate staining of HIV-1 NC (green); DAPI, 4′,6-diamidino-2-phenylindole staining of cellular DNA (blue). (A) NC is detected in the nucleus at 10 h postinfection and is localized predominantly in the nucleus at 18 h postinfection in H9 cells acutely infected with HIV-1IIIB. (B) NC is detected in the nucleus at 10 h postinfection and is localized predominantly in the nucleus at 18 h postinfection in PBMC acutely infected with HIV-1IIIB. (C) NC nuclear localization examined with a confocal microscope with Sytox Green staining of cellular DNA (green) and Red 613 staining of HIV-1 NC (red). At 4 h postinfection, the NC signal (red) is detected in the cytoplasm. At 18 h postinfection, the NC signal is clearly detected in the nucleus (yellow).
HIV-1 spliced mRNA expression in wild-type- and NC mutant-infected cells.
Since NC localized to the nucleus in the early stages of viral infection, we examined its effect on HIV-1 LTR-directed early viral mRNA expression. We used a strategy involving HIV-1 mutant viruses in an examination of NC nuclear localization and early viral spliced mRNA expression. These mutants are derived from HIV-1 pNL4-3, and the nucleotide sequence of the wild type is identical to previously published sequences (2, 17, 18). The CCCC mutant has an H→C mutation in the first zinc finger (CCCC/CCHC) and has nearly 100% of the viral RNA packaged into the virions as in the wild-type virus. This mutant exhibits a slow replication phenotype in cell culture compared to the wild-type HIV-1 (17). The SSHS mutant (pRB653) has C→S mutations in both zinc fingers (SSHS/SSHS). This mutant has a low level of viral RNA packaged in the virion and also shows a slow replication phenotype in cell culture compared to the wild-type HIV-1 (18). Both mutants have normal virion morphology (17, 18).
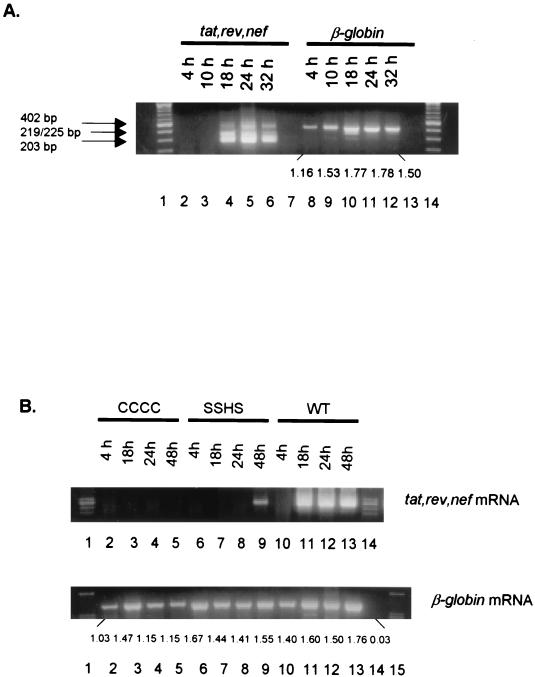
We first examined the HIV-1 spliced mRNA expression in H9 cells acutely infected with HIV-1IIIB. The HIV-1 tat, rev, and nef mRNAs were expressed in the infected cells at 18 h postinfection and were still detected at 32 h postinfection (Fig. 2A). These results are consistent with the results obtained previously (21, 22, 36). We then examined the HIV-1 spliced mRNA expression in H9 cells acutely infected with wild-type and NC mutant HIV-1. The wild-type HIV-1 expressed tat, rev, and nef mRNAs at 18 h postinfection, similar to HIV-1IIIB. The HIV-1 mutants CCCC and SSHS, which have mutations in their NC zinc finger motifs, however, have dramatically delayed tat, rev, and nef spliced mRNA expression compared to wild-type HIV-1 (Fig. 2B). The spliced mRNA could not be detected from HIV-1 mutant CCCC-infected cells at any of the time points assayed (Fig. 2B). In HIV-1 mutant SSHS-infected cells, the spliced mRNA was detected but not until 48 h postinfection (Fig. 2B). The difference in the viral mRNA detection at these time points cannot be attributed to a nonspecific effect of the wild-type infection on cells which was not observed in cells infected with the mutants. As shown in Fig. 2B, the densitometer tracing of cellular β-globin mRNA for wild-type virus infection at 18 h is 1.60, for mutant CCCC it is 1.47, and for mutant SSHS it is 1.44, indicating a similar capacity of these infected cells to transcribe a cellular mRNA at this time postinfection.
FIG. 2.
(A) Expression of HIV-1 tat, rev, and nef spliced mRNAs in H9 cells acutely infected with HIV-1IIIB. Lanes: 4 to 6, the HIV-1 tat (402 bp), rev (219/225 bp), and nef (203 bp) spliced mRNAs were expressed at 18 to 32 h postinfection; 8 to 12, β-globin mRNA expression from samples in lanes 2 to 6 (the densitometer reading of each band is shown under the gel); 7 and 13, RT-PCR DNA controls (RNA sample without the RT reaction). The aliquots of the cells in Fig. 1 were extracted for total cellular RNA, which was then digested with RNase-free DNase as previously described (36, 37). RT-PCR was performed as described in Materials and Methods. (B) Delayed expression of HIV-1 tat, rev, and nef spliced mRNAs in H9 cells acutely infected with HIV-1 NC mutants CCCC and SSHS. (Top) Lanes: 2 to 5, HIV-1 spliced mRNA was not detected in cells infected with NC mutant CCCC; 6 to 9, HIV-1 spliced mRNA of SSHS was expressed only at 48 h postinfection; 10 to 13, HIV-1 spliced mRNA of the wild type was expressed at 18 to 48 h postinfection. (Bottom) Lanes: 2 to 13, β-globin mRNA expression from samples in the top panel (the densitometer reading of each band is shown under the gel); 14, RT-PCR DNA control.
Mutant NC nuclear localization.
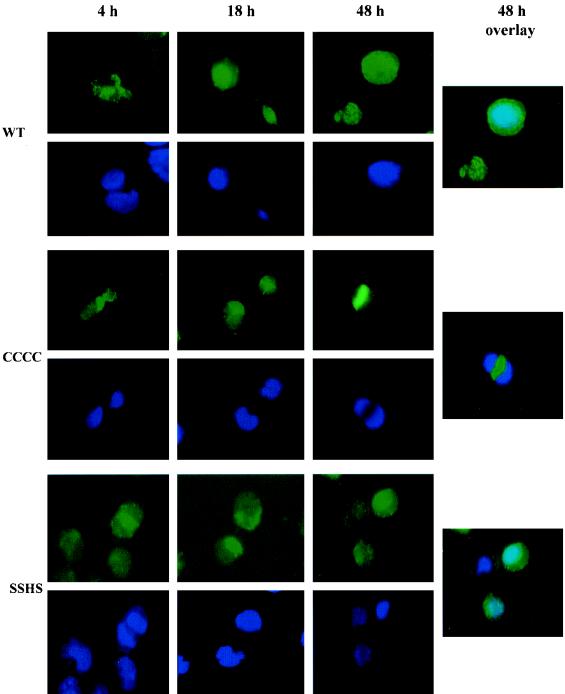
We examined the pattern of NC nuclear localization in the cells which were infected with the NC mutants to determine the relationship of NC nuclear localization and early viral mRNA expression. Consistent with the results of viral spliced mRNA expression, immunofluorescent staining showed a dramatically changed pattern of nuclear localization of mutant NC with respect to wild-type NC. The immunofluorescent staining of H9 cells showed that a majority of the mutant CCCC NC remained in the cytoplasm at 4, 18, and 48 h postinfection (Fig. 3). The strong localization of NC in the nucleus at 18 h is best shown by confocal microscopy (Fig. 1C). The mutant SSHS NC was localized in the cytoplasm at 4 and 18 h postinfection but appeared in the nucleus only very late at 48 h postinfection (Fig. 3). In comparison, the wild-type NC was localized predominantly in the nucleus at 18 h postinfection and was detected in both nucleus and cytoplasm at 48 h postinfection (Fig. 3). This pattern of NC nuclear localization was also observed in PBMC which were infected with the wild-type versus the mutant viruses (data not shown). The overlay exposure in Fig. 3 confirms that the NC from mutant CCCC was not detected in the nucleus at 48 h postinfection but that the NC from the wild-type virus and from mutant SSHS were detected in the nucleus.
FIG. 3.
Nuclear localization of NC following infection with the mutant strains. The wild-type NC was localized in the nucleus at 18 h postinfection. The mutant NC (CCCC) was detected in the cytoplasm at 18 and 48 h postinfection but not in the nucleus. The mutant NC (SSHS) was detected in the cytoplasm at 18 h postinfection and was localized in the nucleus at 48 h postinfection. The overlay pictures at 48 h postinfection show a light blue color derived from an overlay of FITC and DAPI staining, which represented the nuclear localization of wild-type and mutant NC (SSHS) at this time point. Mutant NC (CCCC) was detected in the cytoplasm at 48 h postinfection, and no light blue color was detected in the nucleus. The aliquots of cells described in the legend to Fig. 2 were processed for immunofluorescent staining as described in the legend to Fig. 1.
As shown in Fig. 2 and determined by the RT-PCR assay, the early viral mRNA was not detected in cells infected with NC mutant CCCC at the above time points, and in HIV-1 mutant SSHS-infected cells, the spliced mRNA was detected but not until 48 h postinfection (Fig. 2B). In contrast, in the wild-type-infected cells, the viral spliced mRNA was expressed at 18 h postinfection, and these mRNAs continued to be detectable at 48 h postinfection (Fig. 2B). Mutant NC protein from CCCC and SSHS was not localized in the nucleus at 18 h postinfection, in contrast to wild-type NC, which was present in the nucleus at 18 h postinfection. Taken together, NC nuclear localization correlated with the early viral mRNA expression and a delayed NC nuclear localization resulted in a delayed early viral mRNA expression.
Viral DNA of NC mutants is integrated in the cellular genome at 18 hours postinfection.
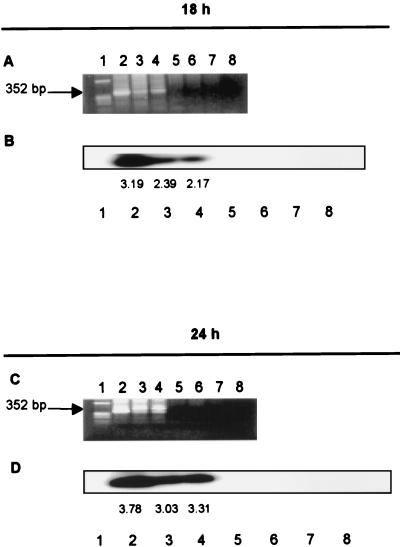
It has been previously reported that NC enhances HIV-1 reverse transcriptase activity and plays a role in provirus integration (6, 11, 18, 23; Musier-Forsyth et al., Proc. 2000 Meet. Retroviruses, 2000). We examined whether the shift in the pattern of viral spliced mRNA expression is due to a delay in provirus formation in the infected cells. We used an Alu-LTR PCR method which specifically detects integrated HIV-1 DNA but not unintegrated forms (7, 15, 30). The cellular genomic DNA was isolated and purified from the infected cells (5, 7, 15), which were infected with either wild-type or HIV-1 NC mutants. The cells infected with the NC zinc finger mutants showed a lack of tat, rev, and nef mRNA expression from 18 through 32 or 48 h postinfection. The HIV-1 proviruses, however, were detected in the cellular genomic DNA at 18 and 24 h after infection with the wild type and NC mutants (Fig. 4). The Alu-LTR PCR method did not detect the HIV-1 LTR in the cellular genomic DNA of mock-infected cells and did not amplify the plasmid-derived unintegrated form of HIV-1 DNAs, even when cellular genomic DNA was spiked with the three linearized HIV-1 plasmid DNAs (Fig. 4). The densitometer tracing of the HIV DNA bands at 24 h postinfection are 3.78 for the wild type, 3.03 for mutant CCCC, and 3.31 for mutant SSHS, indicating that similar amounts of HIV-1 DNA were integrated with these three viral infections.
FIG. 4.
Mutant HIV-1 proviruses are integrated into the cellular genome at 18 h postinfection. (A) Lanes contain a marker (lane 1), the HIV-1 LTR in genomic DNA of cells infected with wild-type (lane 2), CCCC (lane 3), and SSHS (lane 4) viruses at 18 h postinfection, and the HIV-1 LTR from linear plasmid DNA of wild-type, CCCC, and SSHS strains which were mixed with cellular DNA (lanes 5 to 7) and the HIV-1 LTR from cellular genomic DNA of cells subjected to mock infection (lane 8). (B) Southern blot analysis of samples shown in panel A. (C) Same as in panel A, except that the HIV-1 LTR was examined at 24 h postinfection. (D) Same as in panel B, except that the HIV-1 LTR was examined at 24 h postinfection. The cell aliquots described in the legend to Fig. 2 were purified for cellular genomic DNA and were amplified with Alu-LTR primers and then with nested LTR primers specific for the HIV-1 LTR (7). The results of a nested PCR are shown.
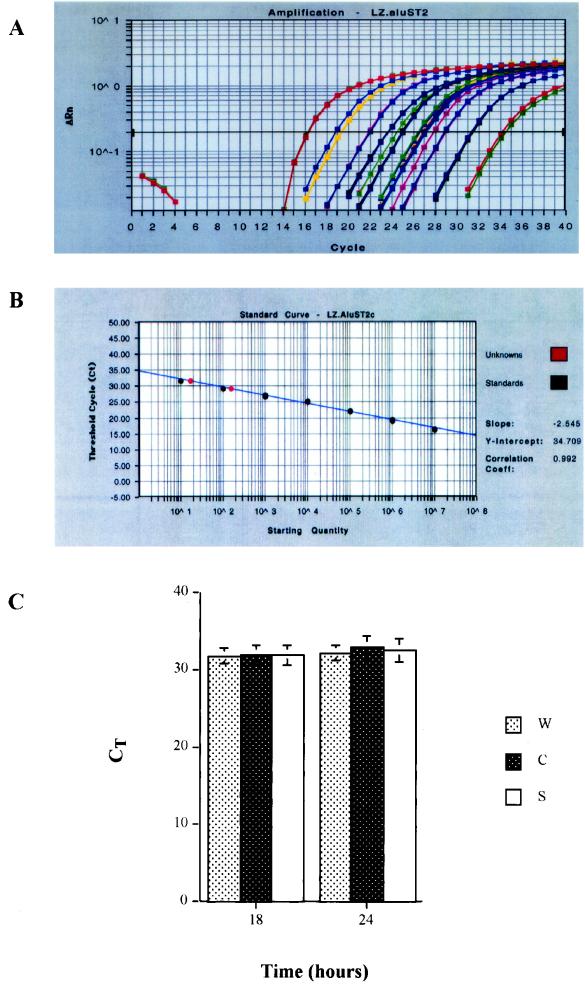
Since the detection of the amount of integrated DNA and the time of DNA integration for both mutant and wild-type viruses are important measures for assessing the role of NC in early viral spliced mRNA expression, we used a quantitative Taqman real-time Alu-PCR assay to determine the integration of viral DNA with these viruses. This assay involves Taqman PCR techniques and determines the number of copies of the HIV DNA integration in vivo (5). Using this technique, we determined the copy numbers of integrated viral DNA as well as 2-LTR circles in cell aliquots which were infected with NC mutants and wild-type virus. The integrated HIV-1 DNA and 2-LTR circle in cells infected with wild-type or mutant viruses were determined at 18 and 24 h postinfection. The copy number of the viral DNAs was read directly from the result report spreadsheet, which was converted from the equivalent threshold cycle (CT) by using Sequence Detector V1.7 software. At 18 h postinfection, the integrated DNA copy number was 143 copies in 5 × 104 cells for the wild type, 118.2 copies in 5 × 104 cells for CCCC, and 139 copies in 5 × 104 cells for SSHS (Fig. 5). There was no statistically significant difference in the copy number of integrated DNA observed in cells infected with NC mutants and the wild-type viruses (t test, P > 0.5). At 24 h postinfection, the integrated DNA copy number was slightly decreased compared to that at 18 h postinfection (t test, P > 0.5). There was no significant difference in the amount of integrated DNA detected at 24 h in cells infected with wild-type or mutant viruses (t test, P > 0.5) (Fig. 5C). The integrated DNA copy number of each virus was derived from quintuple determinations with a standard deviation of 1.26 to 1.41 ± 0.001 within the determinations. The amplification plot, the standard curve of the Alu-LTR signal, and a range of sample DNA examined by Taqman Alu-LTR PCR assay are shown (Fig. 5A and B). Under our acute infection conditions, the HIV-1 integrated DNA from infected cells could be detected from 125 to 500 ng of cellular genomic DNA in duplicate. CT values higher than or equal to 36 cycles were invalid and were not used for the calculation of copy numbers in our study.
FIG. 5.
HIV-1 DNA integration examined by the comparative CT method. (A) Amplification plot of the Taqman Alu-LTR real-time PCR assay. (B) Alu-LTR signal standard curve versus integrated viral DNA signal in the unknown specimens. Note that the CT value and the copy number are in a reverse ratio; the higher the CT number, the lower the copy number in the specimen. The integrated viral DNA signal in the specimen is represented as red dots, and the Alu-LTR standard is represented as black dots. The figure shows a titrated range of HIV-1 integrated DNA detected in the specimens (red dots) of cells infected with NC mutant virus SSHS under our acute-infection conditions. The integrated viral DNAsignal is detected in 125 to 500 ng of cellular genomic DNA in duplicate. In the mock infection control, the sample DNA with a concentration higher or lower than the above range is unable to show the Alu-LTR signal (CT, ≥40 cycles). The unknown sample with a CT value of ≥36 cycles is invalid and does not have a converted copy number by the comparative CT method (Taqman protocol, Applied Biosystems, 2001; ABI Prism 7700 user bulletin no. 2; Applied Biosystems, 2001) in this study. (C) HIV-1 DNA integration of wild-type and NC mutant viruses examined by the comparative CT method. Speckled white bar, CT value of HIV-1 integrated DNA of wild-type virus; speckled black bar, CT value of HIV-1 integrated DNA of NC mutant CCCC; solid white bar, CT value of HIV-1 integrated DNA of NC mutant SSHS. Times were 18 and 24 h postinfection. The samples were examined in quintuplicate, and the standard deviations are shown.
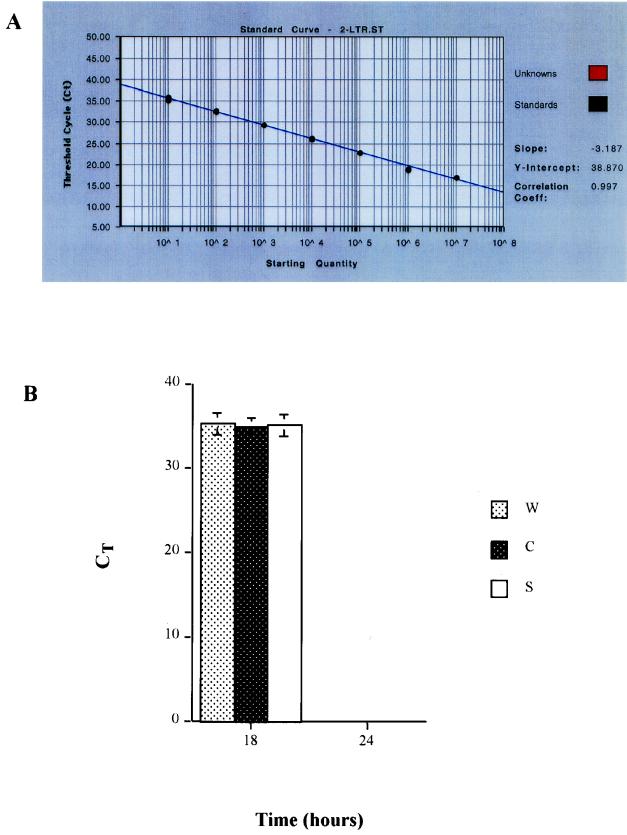
The examination of 2-LTR circle production from these viruses was performed by using the same aliquots of cellular genomic DNA used above. The formation of 2-LTR circle was nearly identical among these viruses when detected at 18 h postinfection. The copy number of 2-LTR circle was 42 copies in 5 × 104 cells in cells infected with the wild type, 47 copies in 5 × 104 cells in cells infected with NC mutant CCCC, and 43 copies in 5 × 104 cells in cells infected with NC mutant SSHS. There was no statistically significant difference in 2-LTR circle copy numbers detected in cells infected with the wild-type and NC mutant viruses (t test, P > 0.5) (Fig. 6B). The 2-LTR circle copy number of each virus was derived from quintuplicate determinations with a standard deviation of 1.05 to 1.21 ± 0.001. We were unable to detect the 2-LTR circle signal in DNA aliquots of cells infected with these viruses at 24 h postinfection (CT value, ≥36 cycles). A standard curve of 2-LTR circle signal expressed in CT value versus copy number is shown (Fig. 6A).
FIG. 6.
HIV-1 2-LTR circle examined by the comparative CT method. (A) Standard curve of 2-LTR circle signal expressed by CT value versus known copy number derived from 2-LTR circle plasmid (5). Note that the higher CT value represents a lower copy number. The valid CT numbers of the 2-LTR circle Taqman real-time PCR assay were <36 cycles. (B) HIV-1 2-LTR circle of wild-type and NC mutant viruses examined by the comparative CT method. Speckled white bar, CT value of HIV-1 2-LTR circle of wild-type virus; speckled black bar, CT value of the HIV-1 2-LTR circle of NC mutant virus CCCC; solid white bar, CT value of HIV-1 2-LTR circle of NC mutant virus SSHS. Times were 18 and 24 h postinfection. The samples were examined in quintuplicate, and the standard deviations are shown.
Taken together, these data show convincingly that at 18 and 24 h postinfection, the proviral DNA of both wild-type and NC mutant viruses had already integrated into the cellular genome, and the amount of provirus formation by the wild-type and mutant viruses was very similar by a Taqman real-time quantitative PCR assay. The mutation in the NC of these mutants shows a significant effect on viral RNA expression determined by the in vivo studies, and this effect is not explained by a difference in proviral integration. The two Alu-LTR PCR assays confirmed that the HIV-1 DNA of wild-type and mutant viruses is already present in the cellular genome at 18 h postinfection, and the copy number of integrated viral DNA is equivalent between the wild-type and mutant viruses. These results show that the mutations in the NC zinc finger motifs affect LTR-directed early spliced mRNA expression and that this effect of NC occurs after the HIV-1 DNA is integrated into the cellular genome.
DISCUSSION
Our data show that wild-type HIV-1 NC accumulated predominantly in the nucleus at 18 h postinfection. Mutations in NC zinc finger domains impaired NC nuclear localization and also retarded HIV-1 early spliced mRNA expression. This change in HIV-1 early spliced mRNA expression occurs after the HIV-1 provirus integration into the cellular genome. Densitometer readings of the HIV-1 DNA bands reveal that similar amounts of HIV-1 DNA are integrated at 24 h postinfection by both wild-type and mutant viruses, so that the profound differences in early spliced mRNA expression are not explainable by a marked difference in provirus integration. Our data do not reveal the mechanism whereby NC regulates early viral spliced mRNA expression, but they clearly indicate that an HIV-1 virion protein is contributing to HIV-1 early-gene transcription. It is important to distinguish the specific roles of NC in enhancing RT, enhancing proviral integration, and regulating early spliced mRNA expression since the interaction of NC with LTR occurs in all three processes (6, 11, 18, 37; Musier-Forsyth et al., Proc. 2000 Meet. Retroviruses, 2000). When combined with the data in our previous paper (37) showing that HIV-1 NC binds to HIV-1 LTR and enhances transcription from the LTR, our data presented here show that following nuclear localization and provirus integration, NC induces HIV-1 early spliced mRNA expression in HIV-1-infected cells.
We used an immunofluorescence assay to detect NC localization following viral entry and examined the localization of wild-type and mutant NC in a system where HIV infects the host cells. Our results with light fluorescent and confocal microscopy show that the virion NC migrates from the cytoplasm to the nucleus at 10 h postinfection and accumulates predominantly in the nucleus at 18 h postinfection. At 18 h postinfection, approximately 45% of cells show NC in the nucleus. At 24 and 32 h postinfection, the NC signal is detected in both the nucleus and the cytoplasm. The NC signal detected 4 to 18 h postinfection appears to represent the pathway of virion NC migration to the nucleus during this time, based on immunofluorescent staining at different time points following the viral infection. The NC signal detected at 24 and 32 h, however, may represent two conditions. First, de novo-synthesized NC was detected in the form of Gag polyproteins present in the cytoplasm at this time. Second, the virion NC shuttled back to the cytoplasm. Considering the results of previous studies by other researchers, the first condition appears to be more likely since NC is detected in 45% of cells at 18 h postinfection. It is known that mature NC follows a pathway different from Gag polyprotein in the HIV-1 infection. It has been previously shown that virion NC migrates toward the nucleus and localizes in the nucleus following viral entry (16). Gag polyprotein migrates toward the cytoplasmic membrane and buds off from the host cell in HIV infection (14).
The localization of the NC from the two mutants was also examined in our study. The immunofluorescent-staining assay showed that mutant NCs entered the cytoplasm with a similar pattern to the wild type but had a significant delay in the nuclear expression compared to the wild type. Mutant NC of CCCC virus remained in the cytoplasm at 48 h postinfection, and mutant NC of SSHS virus remained in the cytoplasm at 18 h postinfection and was detected in the nucleus only at 48 h postinfection. In contrast, the wild-type NC accumulated predominantly in the nucleus at 18 h postinfection; 48 h postinfection, the signal was detected in both the cytoplasm and nucleus (Fig. 3). This is best shown in the confocal microscopy overlay picture of NC localization at 18 h postinfection (Fig. 1C). The delay in nuclear localization of the mutant NCs correlates with a delayed expression of early viral spliced mRNA (Fig. 2B). In addition, the two mutant viruses have delayed viral replication in viral multiplication assays (17, 18). Our study shows that a delayed early viral spliced mRNA expression could be a factor in the slow replication of these viruses. A delay in early RNA transcription could produce a delay in the production of genomic RNA.
The NC mutants CCCC and SSHS have point mutations in their zinc finger motifs. The CCCC mutant has the first CCHC zinc finger mutated to the steroid hormone receptor CCCC, and the SSHS mutant has the CCHC of both zinc fingers mutated to SSHS which is severely defective in zinc binding (17, 18; Musier-Forsyth et al., Proc. 2000 Meet. Retroviruses, 2000). Although these mutations in NC have different effects on the efficiency of HIV-1 first- and second-strand DNA synthesis examined by in vitro RT and tRNA primer unwinding and annealing assays (18; Musier-Forsyth et al., Proc. 2000 Meet. Retroviruses, 2000), the HIV-1 DNA is easily detected in cells infected with these mutants (17, 18). In agreement with the viral culture studies, we found that the proviruses of the two NC mutants were detected in the cellular genome at 18 h postinfection, similar to that of the wild-type virus. At 24 h, quantitation by densitometer tracings reveals that very similar amounts of integrated DNA are present with the wild-type and mutant infections and the profound differences in early mRNA expression are not likely to be due to a difference in the amount of integrated provirus at 24 h. Previous studies have suggested that the sequences surrounding the NC zinc fingers are important for HIV-1 NC nucleic acid binding (9, 23). Our results suggest that mutations that change the type of finger rather than preventing the binding of zinc (17, 18; Musier-Forsyth et al., Proc. 2000 Meet. Retroviruses, 2000) abolish the pattern of NC nuclear localization and early viral mRNA expression but do not prevent provirus integration.
Since it is important to establish that the role of NC in early RNA expression is not explained by a difference in proviral DNA integration with wild-type and mutant viruses, we used two Alu-LTR PCR assays to examine the integration of HIV-1 DNA among these viruses. The first Alu-LTR PCR assay uses a set of primers which detect the HIV-1 DNA integration from the 5′ LTR (7). The second Alu-LTR PCR assay, the Taqman PCR, uses probes and primers which detect the HIV-1 DNA integration from the 3′ LTR (5). Both assays show that HIV-1 DNA was detected in the cellular genome at 18 and 24 h postinfection of both wild-type and NC mutant viruses, but the wild-type virus expresses viral RNA and the NC mutants exhibit a delayed viral RNA expression or a reduction in the amount of mRNA produced so that it is not detected by the PCR assay. The two assays are based on different PCR mechanisms; therefore, one cannot directly compare the band density from one PCR assay with the copy number from the other. Nevertheless, each assay has its own merit and has examined viral DNA integration in the 5′ to Alu sequence and in the 3′ to Alu sequence from cellular DNA following infection with the NC mutant and wild-type viruses. In comparison, the Alu-PCR assay with the nested PCR technique is simpler and more economical and can detect the viral DNA integration in clinical samples (7, 15). The Taqman PCR assay emphasizes the specificity of detection of a target sequence and detects the viral DNA integration well if the 3′ LTR and Alu sequences are within the 250-bp range. Furthermore, the Taqman real-time PCR assay provides a copy number for HIV-1 DNA integrated per cell and showed that the wild type and mutants have similar amounts of integration at 18 and 24 h postinfection. The 2-LTR circle assay also showed that similar amounts of HIV-1 DNA are being formed in the nucleus but that this is not integrated. These controls strongly support our finding that the HIV-1 NC is playing a role in HIV-1 early mRNA expression. Taken together, our results demonstrate a functional role of NC in inducing early viral mRNA expression and that this function of NC occurs after the viral DNA integration.
Acknowledgments
This work was supported in part by grants from NIH (AI028691) and the amfAR Foundation (02717-28-RG). J.Z. is a recipient of an amfAR Foundation grant.
We gratefully acknowledge the generosity of R. Gorelick of NIH in providing the HIV-1 mutants containing mutations in the zinc finger regions of NC and for critically reading the manuscript prior to submission. We thank J. Sodroski for critically reading the manuscript and providing very helpful suggestions. We thank F. Bushman for providing technical support and reagents of the Taqman PCR assay used in measuring HIV-1 DNA integration. We also thank L. Bhagat, K. Bryant, P. Sharma, Z. Jin, C. De Melo, I. Ghiran, H. Wang, K. Sugiyama, and H. Vo for generous advice, laboratory assistance, and reagents.
REFERENCES
- 1.ACTG Technical Advisory Committee. 1994. Virus stock infectivity titration, p. RES1-RES5. In F. B. Hollinger (ed.), ACTG virology manual for HIV laboratories. National Institutes of Health, Bethesda, Md.
- 2.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bess, J. W., Jr., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type I, and other retroviruses. J. Virol. 66:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butera, S. T., V. L. Perez, B. Y. Wu, G. J. Nabel, and T. M. Folks. 1991. Oscillation of the human immunodeficiency virus surface receptor is regulated by the state of viral activation in a CD4+ cell model of chronic infection. J. Virol. 65:4645-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, S. L., M. S. T. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 6.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coffin, J. M. 1996. Retroviridae, p. 763-843. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, Philadelphia, Pa.
- 9.Dannull, J., A. Surovoy, G. Jung, and K. Moelling. 1994. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO. J. 13:1525-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 11.Druillennec, S., A. Caneparo, H. de Rocquigny, and B. P. Roques. 1999. Evidence of interactions between the nucleocapsid protein NCp7 and the reverse transcriptase of HIV-1. J. Biol. Chem. 274:11283-11288. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg, M. B., D. Baltimore, and A. D. Frankel. 1991. The role of Tat in the human immunodeficiency virus life cycle indicates a primary effect on transcriptional elongation. Proc. Natl. Acad. Sci. USA 88:4045-4049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feng, Y. X., T. D. Copeland, L. E. Henderson, R. J. Gorelick, W. J. Bosche, J. G. Levin, and A. Rein. 1996. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc. Natl. Acad. Sci. USA 93:7577-7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 15.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. M. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614-1622. [DOI] [PubMed] [Google Scholar]
- 16.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 17.Gorelick, R. J., T. D. Gagliardi, W. J. Bosche, T. A. Wiltrout, L. V. Coren, D. J. Chabot, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 1999. Strict conservation of the retroviral nucleocapsid protein zinc finger is strongly influenced by its role in viral infection processes: characterization of HIV-1 particles containing mutant nucleocapsid zinc-coordinating sequences. Virology 256:92-104. [DOI] [PubMed] [Google Scholar]
- 18.Guo, J., T. Wu, J. Anderson, B. F. Kane, D. G. Johnson, R. J. Gorelick, L. E. Henderson, and J. G. Levin. 2000. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J. Virol. 74:8980-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123-126. [DOI] [PubMed] [Google Scholar]
- 20.Kadonaga, J. T., K. R. Carner, F. R. Masiarz, and R. Tjian. 1987. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell 51:1079-1090. [DOI] [PubMed] [Google Scholar]
- 21.Kim, S. Y., R. Byrn, J. Groopman, and D. Baltimore. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708-3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klotman, M. E., S. Kim, A. Buchbinder, A. DeRoss, D. Baltimore, and F. Wong-Staal. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc. Natl. Acad. Sci. USA 88:5011-5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lener, D., V. Tanchou, B. P. Roques, S. F. Le Grice, and J. L. Darlix. 1998. Involvement of HIV-1 nucleocapsid protein in the recruitment of reverse transcriptase into nucleoprotein complexes formed in vitro. J. Biol. Chem. 273:33781-33786. [DOI] [PubMed] [Google Scholar]
- 24.Liu, B., R. Dai, C. J. Tian, L. Dawson, R. Gorelick, and X. F. Yu. 1999. Interaction of the human immunodeficiency virus type 1 nucleocapsid with actin. J. Virol. 73:2901-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciw, P. A. 1996. Human immunodeficiency viruses and their replication, p. 845-916. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Raven Press, Philadelphia, Pa.
- 26.Noriega, R. T. Hay, D. Harrich, R. B. Gaynor, J. L. Virelizier, and F. Arenzanaseisdedos. 1995. Absolute dependence on kB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO. J. 14:1552-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582-1586. [DOI] [PubMed] [Google Scholar]
- 28.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 29.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 30.Sonza, S., A. Maerz, N. Deacon, J. Meanger, J. Mills, and S. Crowe. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spector, R. D., L. D. Goldman, and L. A. Leinwand. 1998. Subcellular localization of genes and their products, p. 105.1-105.4. In R. D. Spector, L. D. Goldman, and L. A. Leinwand (ed.), Cells; a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Turner, B. G., and M. F. Summers. 1999. Structural biology of HIV. J. Mol. Biol. 285:1-32. [DOI] [PubMed] [Google Scholar]
- 33.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 34.Wei, X. P., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117-122. [DOI] [PubMed] [Google Scholar]
- 35.Xiao, H., J. T. Lis, and K. T. Jeang. 1997. Promoter activity of Tat at steps subsequent to TATA-binding protein recruitment. Mol. Cell. Biol. 17:6898-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, J. L., H. Choe, B. J. Dezube, M. Farzan, P. L. Sharma, X. C. Zhou, L. B. Chen, M. Ono, S. Gillies, Y. Wu, J. G. Sodroski, and C. S. Crumpacker. 1998. The bis-azo compound FP-21399 inhibits HIV-1 replication by preventing viral entry. Virology 244:530-541. [DOI] [PubMed] [Google Scholar]
- 37.Zhang, J. L., P. L. Sharma, and C. S. Crumpacker. 2000. Enhancement of the basal-level activity of HIV-1 long terminal repeat by HIV-1 nucleocapsid protein. Virology 268:251-263. [DOI] [PubMed] [Google Scholar]
- 38.Zhou, Q., and P. A. Sharp. 1996. Tat-SF1: cofactor for stimulation of transcriptional elongation by HIV-1 Tat. Science 274:605-610. [DOI] [PubMed] [Google Scholar]