Abstract
Presumably due to the capability of the hepatitis C virus (HCV) to evade the antiviral effects of alpha interferon, treatment is ineffective in more than half of chronically genotype HCV type 1 (HCV-1)-infected patients. Previous approaches to correlate the number of amino acid mutations within regions of HCV nonstructural (NS)-5A protein with virologic treatment response provided conflicting results. In the present study, we developed a new mathematical model to investigate NS5A sequences of HCV-1-infected patients. The mean number of all mutations within the complete NS5A protein was significantly higher in virologic responders compared to nonresponders (P = 0.008 and P = 0.0001 for amino acid residues predicted on the surface of the NS5A protein). Differences did not achieve statistical significance for NS5A regions that are currently assumed to be functionally relevant (e.g., the interferon sensitivity-determining region, the RNA-activated protein kinase-binding domain, etc.). Analyses of smoothed mutational frequencies showed that the number of mutations in other NS5A regions correlated with virologic response. Such a correlation was observed for both genuine and randomly generated NS5A sequences. The existence of local accumulations of mutations within genuine NS5A isolates that truly correlated with treatment response was defined by a refined test procedure. Upon considering the predicted residue accessibility, we identified the main focus of mutations correlating with treatment response to be the sequence from amino acids 2350 to 2370. Thus, evaluation of NS5A mutations in correlation with treatment response is improved by consideration of functional and predicted conformational amino acid properties. As shown by simulations with randomly generated sequences, multiple analyses of simple counts of local NS5A amino acid mutations and correlation with treatment response are insufficient. For improvement of mutational analysis, a refined specific functional data test procedure is proposed.
Chronic hepatitis C virus (HCV) infection frequently progresses to liver cirrhosis and is associated with an elevated risk for development of hepatocellular carcinoma (1, 30, 42). The only effective treatment is based on alpha interferon (IFN-α). IFN-α alone or in combination with ribavirin leads to sustained virologic response in 20 to 56% of patients with chronic hepatitis C (24, 26, 37, 53). The strongest predictive parameter for a sustained virologic response to treatment is the HCV genotype, with HCV genotype 1 (HCV-1) being the least sensitive to IFN-α-based therapy (24, 37).
For HCV as for other eukaryotic viruses, including the influenza, vaccinia, and human immunodeficiency viruses (6, 22, 27), the ability to evade the antiviral effects of IFN-α has been reported. Both the HCV envelope (E) 2 protein and the nonstructural (NS)-5A protein of HCV-1a/b isolates were shown to inhibit IFN-α-induced double-stranded RNA-activated protein kinase (PKR) in vitro (14, 51). Moreover, cellular expression of the NS5A protein from HCV-1a/b isolates confirmed inhibitory effects on IFN-α activity (13, 33, 34, 36).
In 1995 and 1996 Enomoto et al. demonstrated clinically that an increasing number of mutations within a restricted region of the HCV NS5A protein named the IFN sensitivity determining region (ISDR; amino acids [aa] 2209 to 2248 according to HCV-J [18]) was correlated with treatment response in isolates of HCV-1b-infected patients (10, 11). The importance of mutations within the NS5A ISDR of HCV-1b-infected patients was confirmed by other investigations from Japan (7, 20). In several studies from Western countries, however, the majority of HCV-1b-infected patients with sustained virologic response showed no or only a small number of mutations within the ISDR (19, 35, 44, 46, 49, 55). Furthermore, mutations within other regions of the NS5A protein (the PKR-binding domain and the variable region V3) were shown to correlate with virologic response in HCV-1-infected patients (31, 43). The methodological approach of counting the number of mutations within multiple regions of the NS5A protein for correlation with treatment response may be insufficient because it does not consider the change of functional properties by amino acid mutations and the location of the amino acids with respect to the secondary structure of the resulting protein. Furthermore, multiple tests are inappropriate when proving an overall significance for the existence of local accumulations of mutations because the probability of observing a significant correlation increases with the number of regions tested.
In the present study, we sequenced full-length NS5A genes from 45 HCV-1b-infected patients for mutational analyses in correlation with treatment outcome on the basis of viral kinetics, functional properties of amino acid mutations, and the predicted secondary structure of the NS5A protein. A new statistical model was developed for testing the significant local accumulation of mutations with specific functional data analysis procedures (47). Moreover, statistical methods were evaluated by analogous investigations on randomly generated sequences.
MATERIALS AND METHODS
Patients.
In the present study, 45 consecutive patients (31 men; mean age, 47 years [range, 20 to 72 years]) chronically infected with HCV-1b were enrolled. The diagnosis of chronic hepatitis C was based on the presence of elevated levels of aminotransferase in serum for at least 6 months and the consistent detection of serum HCV RNA. All patients were negative for hepatitis B surface antigen and antibodies to human immunodeficiency virus. Therapy was either 3 MU of recombinant IFN-α three times per week given subcutaneously for 24 to 48 weeks or 6 MU IFN-α for the initial 24 weeks, followed by 3 MU IFN-α given thrice weekly for a total of 48 weeks (cumulative doses of 216 to 648 MU). Thirty-one patients were treated in combination with ribavirin given perorally (1,000 mg/day for patients of ≤75 kg [body weight] or 1,200 mg/day for patients of >75 kg).
Quantitative measurement of HCV RNA and HCV genotyping.
Serum samples were prepared in a laminar flow bench and frozen at −80°C. Quantitative detection of serum HCV RNA from pretreatment sera and during therapy for viral kinetics (three measurements between days 2 and 18, 5 and 32, and 13 and 166, respectively) was performed by branched DNA assay (Versant Quantitative HCV RNA 3.0; Bayer Diagnostics, Emeryville, Calif.). Genotyping of HCV(48) was performed by reverse hybridization assay (INNO LiPA HCV-II; Innogenetics, Ghent, Belgium).
Amplification of HCV NS5A RNA by reverse transcription-PCR and sequence analysis.
After extraction of HCV RNA from 100 μl of pretreatment serum, cDNA was generated by using appropriate specific antisense primers. The HCV NS5A gene (codons 1973 to 2419 according to HCV-J [18]) was amplified by nested PCR in four overlapping parts with different sets of primers (Table 1). After an initial denaturation step of 95°C for 2 min, 35 cycles of 95°C for 30 s, 55°C for 50 s, and 72°C for 2 min were performed for the first- and second-round PCRs in a PE9700 thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.). The resulting amplification product was analyzed on a 1.5% agarose gel stained with ethidium bromide.
TABLE 1.
Primers for NS5A sequencing
| Part | Primer | Name | Nucleotide sequence (5′-3′) | Positiona |
|---|---|---|---|---|
| 1 | External sense | 12s | TCCCCCACGCACTATGTGCC | 6117 |
| 1 | External antisense | 12a | TCAGTGGTCATGCCCGT | 6636 |
| 1 | Internal sense | 11s | CAGTGGATTAATGAGGA | 6213 |
| 1 | Internal antisense | 11a | TAGTGGAAATCCCCCAC | 6615 |
| 2 | External sense | 3s | GGCATGGAACATTCCCCAT | 6496 |
| 2 | External antisense | 4a | ACGGATATTTCCCTCTCATCC | 7138 |
| 2 | Internal sense | 13s | TACACCACGGGCCCCTG | 6522 |
| 2 | Internal antisense | 2a | CCGAAGCGGATCGAAAGAGTCCA | 7087 |
| 3 | External sense | 4s | ACCCCTCCCACATTACAGCAG | 6859 |
| 3 | External antisense | 8a | CCAAAGGTCTTAGTAGC | 7371 |
| 3 | Internal sense | 18s | AACCTCCTGTGGCGGCA | 7017 |
| 3 | Internal antisense | 10a | AGCTCCGCCAAGGCAGAAGA | 7350 |
| 4 | External sense | 17s | GACTACAACCCTCCACT | 7203 |
| 4 | External antisense | 15a | GGTGACGCAGCAAAGAGTTG | 7667 |
| 4 | Internal sense | 16s | GTGGTGCACGGGTGCCCATTGCC | 7257 |
| 4 | Internal antisense | 14a | CCTCCGCAGCGCATGGC | 7622 |
Nucleotide positions indicate the carboxy-terminal ends of the primers within the HCV gene according to HCV-J (18).
For direct sequencing of the different parts of NS5A, 40 μl of the second round PCR product was purified with Microcon 100 (Amicon, Witten, Germany). Sequence analysis was performed according to the manufacturer's instructions (Big Dye Deoxy Terminators; Applied Biosystems, Weiterstadt, Germany). Sequencing of the positive and the negative strands was performed by an automat (310 DNA Sequencer; Applied Biosystems). The deduced amino acid sequences of the NS5A region (codons 1973 to 2419) were compared with the NS5A sequences identified in the prototype isolates for HCV-1a (HCV-1 [8]), HCV-1b (HCV-J and HC-J4 [18, 32]), and the consensus sequence of all HCV-1b isolates (cons) investigated.
Phylogenetic and conformational analysis.
Multiple alignment of amino acid sequences, calculation of the consensus sequence (cons), and phylogenetic analyses were performed as previously described (43). Residue accessibility (buried, exposed, and unknown) within the secondary structure of the NS5A protein on the basis of the 45 full-length NS5A sequences from the present study and 25 published full-length NS5A sequences (9, 31) was predicted by the PHD and PROFacc programs of B. Rost et al. (http://maple.bioc.columbia.edu/predictprotein/) (39). The methods and validity of the prediction programs were recently reviewed by Rost and Sander (41).
Analyses of amino acid mutations according to their functional properties (conserved versus nonconserved mutations) were based on six different classes of amino acids with predicted similar properties: positive (R and K), negative (D and E), aromatic (H, F, W, and Y), polar (N, Q, S, and T), and aliphatic (A, I, L, M, V, and C) amino acids and amino acids with special conformational properties (G and P) (28).
In addition to the 45 sequences of the present study, published NS5A HCV-1b sequences were analyzed. Duverlie et al. reported full-length NS5A sequences from 11 nonresponder patients and from 8 patients who achieved a sustained virologic response (9). These sequences were retrieved from the EMBL nucleotide sequence database (accession numbers AF033358 to AF033376). NS5A isolates AF033364 and AF033365 were identical and were regarded as one isolate for sequence analyses. Nousbaum et al. published HCV-1b full-length NS5A sequences from four patients with sustained virologic responses (patients 11 and 12 were as published previously and patients 10 and 19 were from S. Polyak [unpublished data]; AF265148, AF265151, AF265145, and AF265157) and from two virologic nonresponders (patients 1 and 13; AF265142 and AF265154) (31). For analysis of the NS5A sequence from patient 19, the insertion of one nucleotide (adenine at position 6441 according to the prototype sequence HCV-J) was removed.
Kinetic modeling.
A biphasic model for the initial viral decay was used (29, 54). Estimates for the second-phase slope λ2 were obtained by a least-squares linear fit of the log viral load. Quantifications below the detection limit were included by using the Tobit approach. The resulting nonlinear minimization was performed numerically by using Matlab (MathWorks, Inc., Natick, Mass.). The quotient of pretreatment viral load and the estimated viral load 24 h after starting therapy was used as an approximation of the efficiency factor ɛ on viral production (21, 29).
Functional data analysis and statistics.
Local accumulations of mutations were analyzed on the basis of a nonparametric binomial regression model, assuming independent amino acid exchanges. This assumption was justified as a first-order approximation in the related context of sequence segmentation (5). A specific functional data analyses procedure (47) was defined to test whether differences between the mutational frequencies for responding and nonresponding patients can be explained by a constant shift only. The test was based on
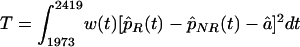 |
(1) |
where, p̂R and p̂NR are estimates of the mutational probability function for sequences from responding and nonresponding patients, respectively, and αcirc; is a frequency estimator of the global shift. p̂R and p̂NR are estimated nonparametrically by smoothing the local mutational frequencies with Gaussian kernel weights, and bandwidth b = 1.5. The asymptotic normal distribution of T was derived analytically as:
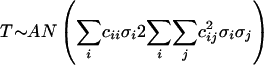 |
(2) |
where
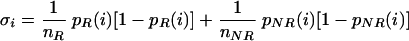 |
(3) |
and
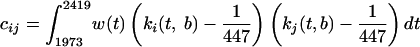 |
(4) |
with the kernel weighting scheme ki(t,b) for i = 1973,…, 2419 (i.e., amino acid positions of HCV NS5A). This distribution was used to calculate critical regions. The asymptotic results were confirmed by simulations of random sequences. Random generation of sequences was performed from independent binomially distributed pseudo random numbers with appropriately defined mutational probability functions by using Matlab.
Standard pointwise tests for smoothed mutational frequencies (bandwidth 3) of the genuine sequences and of sets of random sequences were applied. Continuous variables were compared by using the Mann-Whitney U test, and two continuous variables were analyzed by Spearman rank correlation. Unless indicated otherwise, all tests were two-tailed and P values of <0.05 were considered significant.
The clinical and biochemical characteristics of patients are expressed as the mean ± the standard deviation or median and range as appropriate.
RESULTS
NS5A amino acid sequences.
The complete NS5A region of HCV pretreatment isolates from 45 patients chronically infected with HCV-1b was investigated. Eleven patients achieved a long-time sustained virologic response (SR), with undetectable HCV RNA 18 months after termination of therapy. In 11 patients a virologic response with undetectable HCV RNA at the end of treatment, but relapse thereafter was observed (ETR). Twenty-three patients showed no virologic response to antiviral therapy (NR). For mutational analyses virologic responders (ETR and SR patients) were compared with virologic nonresponders (NR patients) to focus on effects that are correlated with IFN-α treatment. Mutations were defined always with respect to the consensus sequence of all investigated NS5A sequences. The different potentially functional regions of the NS5A protein are shown in Fig. 1.
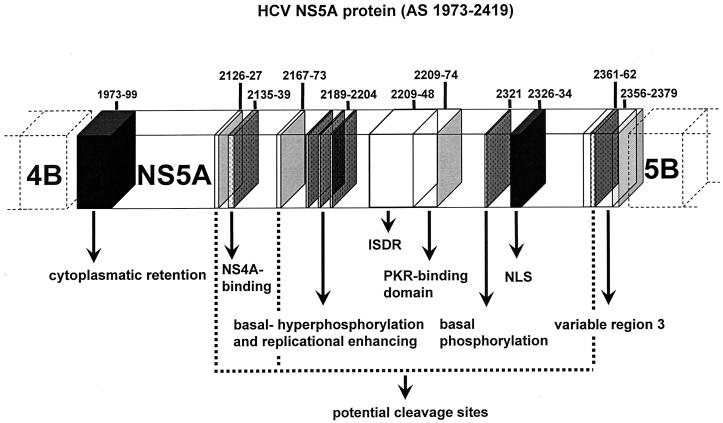
FIG. 1.
Location of different regions of interest within HCV NS5A protein. The NS5A protein (aa 1973 to 2419), the potential region for cytoplasmatic retention (aa 1973 to 1999), potential cleavage sites (aa 2126 to 2127, 2167 to 2173, and 2361 to 2362), binding to NS4A (aa 2135 to 2139), a region with potential hyperphosphorylation sites and amino acids important for replication initiation (aa 2197 to 2204), the ISDR (aa 2209 to 2248), the PKR-binding domain (aa 2209 to 2274), the HCV-1a basal phosphorylation site (aa 2321), the NLS (aa 2326 to 2334), and the V3 region (aa 2356 to 2379) are indicated according to the numbering system of HCV-1b prototype HCV-J (18).
The sequences of the complete NS5A protein (aa 1973 to 2419) of all 45 isolates are accessible in the EMBL database (accession numbers AJ507155 to AJ507199). Mutational frequencies within the different regions of the NS5A protein were compared with the treatment outcome of the different groups (SR, ETR, and NR; Table 2). The mean number of all (both conserved and nonconserved) and all nonconserved NS5A mutations in virologic responders (SR and ETR) was significantly higher than in virologic nonresponders (NR) (P = 0.008 and P = 0.014, respectively; Table 2). In addition, the number of all amino acid exchanges within the amino-terminal part of the NS5A protein correlated significantly with treatment response (P = 0.031; Table 2). However, for none of the different functionally relevant regions of the NS5A protein (the transmembrane domain, cleavage sites, NS4A association, ISDR, PKR-binding domain, nuclear localization signal [NLS], and V3 region) was a significant correlation between the number of either all amino acid mutations or all nonconserved amino acid mutations and treatment response observed (Table 2). Furthermore, hyperphosphorylation sites and amino acids shown to be important for replication initiation in the HCV replicon were highly conserved in the 45 isolates of HCV-1b-infected patients (Fig. 1 and Table 2).
TABLE 2.
Mutations in different parts of the NS5A region in 45 HCV-1b-infected patients
| NS5A region (aa range) | Treatment response to IFN-α with or without ribavirin (mean no. of mutations)a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SR (n = 11)
|
ETR (n = 11)
|
NR (n = 23)
|
Probability (SR + ETR versus NR)b
|
|||||
| All | Non-cons. | All | Non-cons. | All | Non-cons. | P (all) | P (non-cons.) | |
| Complete NS5A (1973-2419) | 20.4 (11-39) | 11.0 (5-25) | 17.2 (11-24) | 10.0 (7-15) | 14.3 (6-37) | 8.4 (5-23) | 0.008 | 0.014 |
| Amino-terminal part of NS5A (1973-2208) | 8.0 (2-13) | 3.6 (1-6) | 6.8 (3-11) | 3.5 (1-7) | 5.5 (2-11) | 2.7 (1-5) | 0.031 | 0.125 |
| Carboxy-terminal part of NS5A (2209-2419) | 12.4 (5-29) | 7.4 (3-19) | 10.4 (5-19) | 6.6 (2-12) | 8.8 (3-26) | 5.6 (2-20) | 0.084 | 0.101 |
| Transmembrane domain (1973-1999)c | 0.2 (0-2) | 0.1 (0-1) | 0.9 (0-2) | 0.4 (0-2) | 0.4 (0-2) | 0.1 (0-1) | 0.494 | 0.601 |
| Cleavage sitesd | ||||||||
| 1 (2126-2127) | 0 | 0 | 0 | 0 | 0 | 0 | ||
| 2 (2167-2173) | 0.8 (0-1) | 0.5 (0-1) | 0.6 (0-1) | 0.6 (0-1) | 0.5 (0-1) | 0.4 (0-1) | 0.092 | 0.463 |
| 3 (2361-2362) | 0.2 (0-2) | 0.2 (0-2) | 0 | 0 | 0 | 0 | ||
| NS4A association (2135-2139)e | 0.2 (0-1) | 0.1 (0-1) | 0.1 (0-1) | 0 | 0.1 (0-1) | 0 | 0.954 | |
| ISDR (2209-2248)f | 1.6 (0-8) | 1.0 (0-4) | 0.7 (0-2) | 0.6 (0-2) | 0.8 (0-5) | 0.7 (0-5) | 0.893 | 0.614 |
| PKR-binding domain (2209-2274)g | 2.9 (0-11) | 1.4 (0-4) | 1.2 (0-4) | 0.9 (0-2) | 1.6 (0-5) | 1.0 (0-5) | 0.678 | 0.563 |
| NLS (2326-2334)h | 0 | 0 | 0.2 (0-1) | 0 | 0.1 (0-1) | 0.1 (0-1) | ||
| V3 region (2356-2379)i | 3.6 (1-12) | 3.3 (1-10) | 3.6 (1-12) | 3.2 (0-9) | 2.8 (1-9) | 2.6 (1-9) | 0.228 | 0.211 |
| Hyperphosphorylation sites (2197, 2201, and 2204)j | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Replicational enhancer sites (2163, 2177, 2189, 2196, 2197, 2199, and 2204)k | 0 | 0 | 0.1 (0-1) | 0.1 (0-1) | 0 | 0 | ||
Ranges are given in parentheses. All (All) and non conserved (Non-cons.) mutations are only in comparison with the consensus sequence.
Based on comparison of the virologic responders (SR and ETR) with the virologic nonresponders (NR) by Mann-Whitney U test. Bold facing indicates statistically significant results.
The N-terminal amino acids were described as potentially responsible for anchoring of the NS5A protein to the cytoplasm (4).
Potential cleavage sites, recognition motifs for serine (aa 2167 to 2173), and caspase-like proteases (aa 2126 to 2127 and aa 2361 to 2362) were described elsewhere (25,45).
The region from aa 2135 to 2139 was described as involved both in the association with NS4A and in the basal phosphorylation of the NS5A protein (2).
ISDR (11).
A region described to be involved in the interaction of NS5A with PKR (14).
This positively charged proline/valine-flanked region was described as a potential NLS (16).
V3 region within the carboxy-terminal part of NS5A (17).
Amino acid residues described as involved in NS5A hyperphosphorylation (50).
For 34 of 45 patients, serum samples were available for the calculation of viral kinetics during the first 12 weeks of therapy. The relative decay of HCV RNA during the first day after initiation of antiviral therapy, which approximates the efficiency factor on viral production (ɛ), and the second-phase slope (λ2) representing the degradation rate of infected hepatocytes were estimated. A higher number of mutations within the complete NS5A protein correlated with a more rapid HCV RNA decay in the second phase (one tailed P = 0.034). Furthermore, a trend toward a stronger clearance of HCV RNA in correlation with a higher number of mutations was observed (one-tailed P = 0.125).
Phylogenetic analyses showed no differences between full-length NS5A isolates from virologic responders compared to nonresponders (data not shown).
Functional data analyses.
The number of mutations did not correlate with treatment response in any of the known putative functional NS5A regions. In contrast, several studies previously reported a correlation between mutational frequencies within different NS5A regions and treatment response. The probability of observing significant correlations between the number of mutations within NS5A regions and treatment response increases with the number of regions tested. The overall significance of these tests derived from pointwise significance levels within arbitrary regions is difficult to determine.
In the present study, mutational analyses from full-length HCV-1b NS5A sequences available from patients with known treatment outcomes (n = 70; 45 patients from the present study and 25 patients from earlier studies [9, 31]) were compared with analyses of mutational frequencies for simulated sequences. The 70 NS5A sequences were divided into two groups: virologic responders (SR and ETR, n = 34) and virologic nonresponders (NR, n = 36). The simulated sequences were assigned randomly to be from virologic responders (n = 34) or from nonresponders (n = 36).
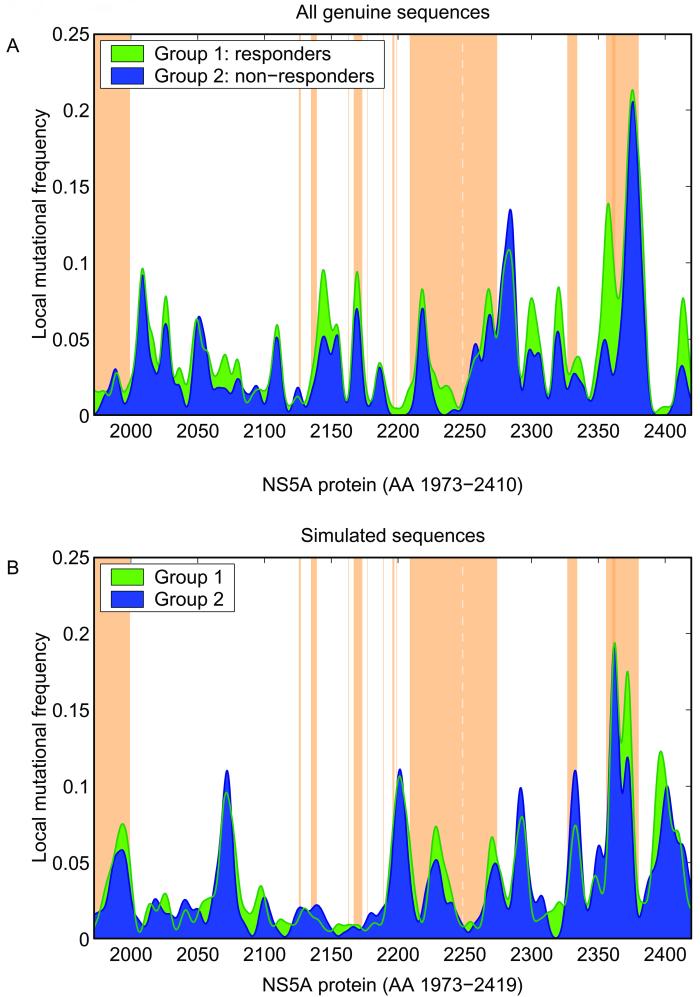
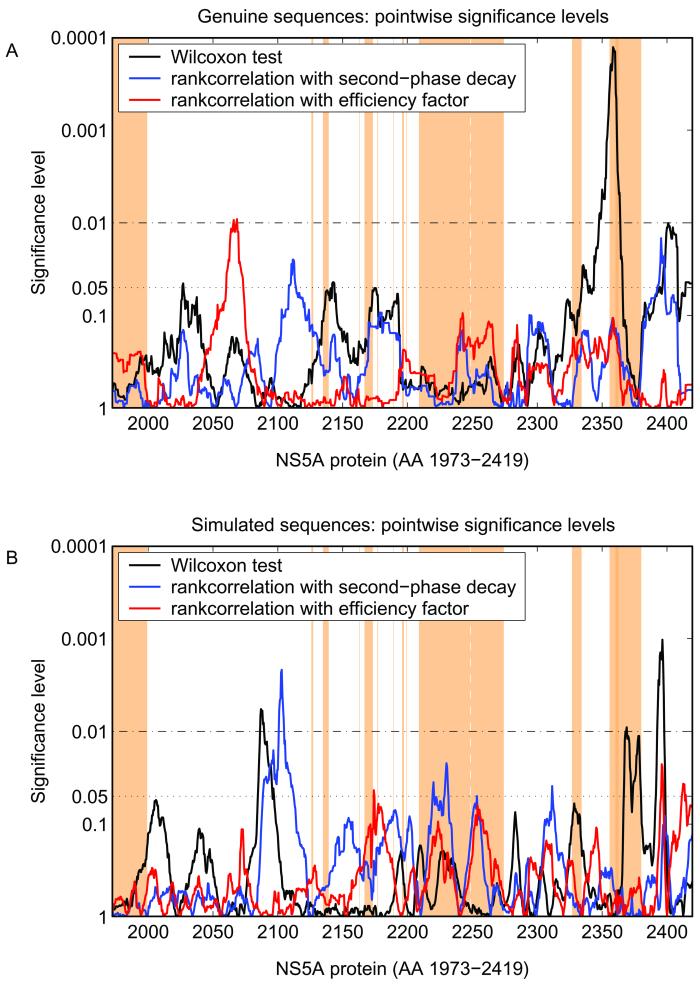
Figure 2 shows the smoothed mutational frequencies of genuine (Fig. 2A) and simulated sequences (Fig. 2B), respectively. For both genuine and simulated sequences within different NS5A regions, the number of mutations was higher for virologic responders than for nonresponders. Therefore, in a next step significance levels of the number of mutations in correlation with treatment response throughout the complete NS5A protein were calculated (Fig. 3). Different regions with a significant correlation between the number of mutations and the treatment response were observed for both genuine and simulated sequences (Fig. 3). Furthermore, regions with significant correlation between the early viral decline and treatment response were observed by screening the complete NS5A of genuine and simulated sequences (Fig. 3). Indeed, such regions could be identified in the majority (94%) of 500 simulated samples of random sequences.
FIG. 2.
Smoothed mutational frequencies of all full-length NS5A sequences from HCV-1b-infected patients (A) and of a typical sample of simulated sequences randomly assigned to derive from virologic responders and nonresponders (B). Virologic responders (SR and ETR) are indicated in green, and virologic nonresponders are indicated in blue. The brown-shaded areas indicate the potential functional regions within the NS5A protein according to the data shown in Fig. 1.
FIG. 3.
Pointwise significance levels of mutations in correlation with early viral kinetics (efficiency factor ɛ and second-phase decay λ2) and of mutational frequencies in correlation with treatment response of full-length NS5A sequences from HCV-1b-infected patients (A) and of simulated sequences from Fig. 2B randomly assigned to belong to the genuine viral kinetic data (B). The black line indicates the significance levels from the correlation of mutational frequencies with treatment response. The blue and red lines indicate the significance levels from the correlation of the number of mutations with the efficiency factor and second-phase decay, respectively. Dotted horizontal lines indicate the 0.05 and 0.01 significance levels. The brown-shaded areas show the potential functional regions within the NS5A protein according to Fig. 1.
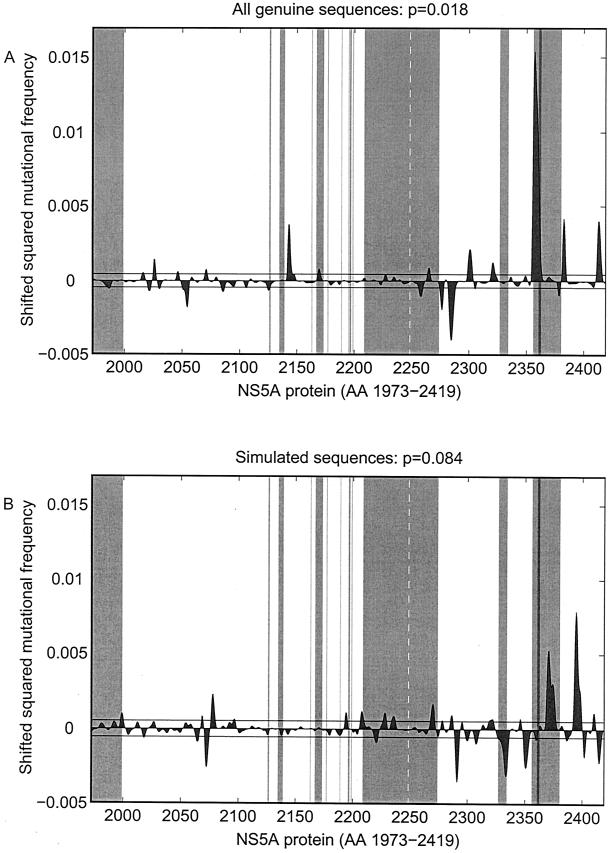
A refined test procedure was introduced to assess an overall significance level for the existence of local accumulations of mutations truly correlated with treatment response. Genuine NS5A sequence analyses of shifted squared differences of smoothed mutational frequencies from virologic responders and virologic nonresponders showed a significant local accumulation of mutations (P = 0.018) with a main focus upstream of the V3 region (aa 2350 to 2370; Fig. 4A). No such local accumulation of mutations was detected in 94% of 500 random samples of simulated sequences (Fig. 4B).
FIG. 4.
Shifted squared differences (shown with the original sign [±]) of smoothed mutational frequencies from virologic responders and virologic nonresponders based on the full-length NS5A sequences from HCV-1b-infected patients (A) and on the simulated sequences from Fig. 2B (B). Significant local differences are present if the gray-shaded areas under the curve are larger than the area within the black horizontal lines. The gray-shaded vertical bars show the potential functional regions within the NS5A protein determined according to the data presented in Fig. 1.
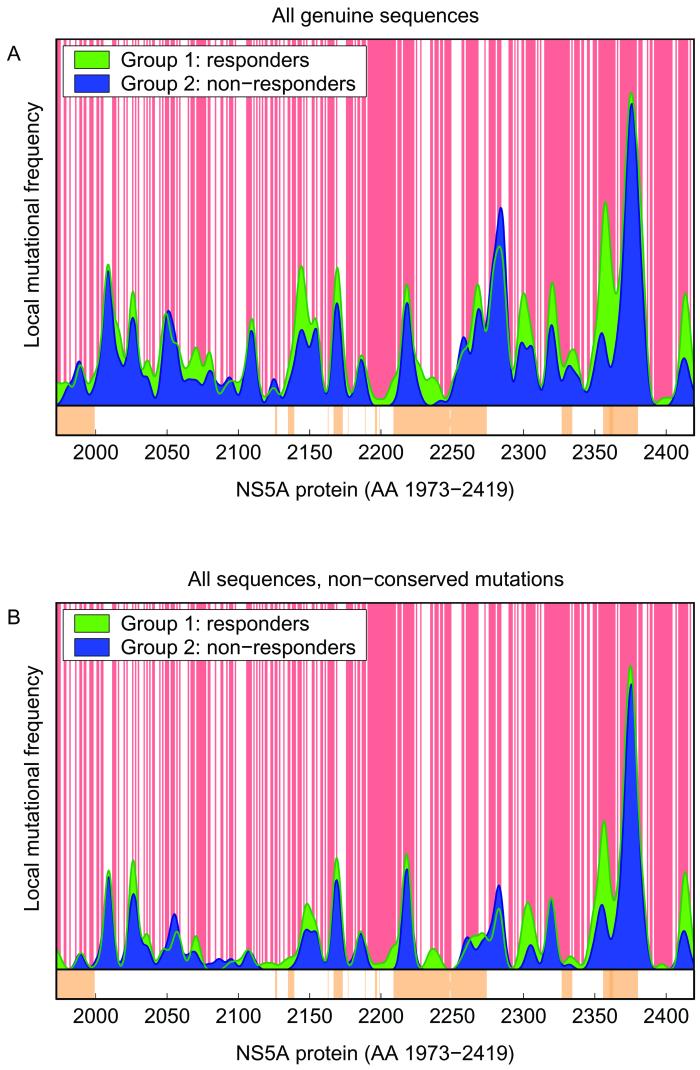
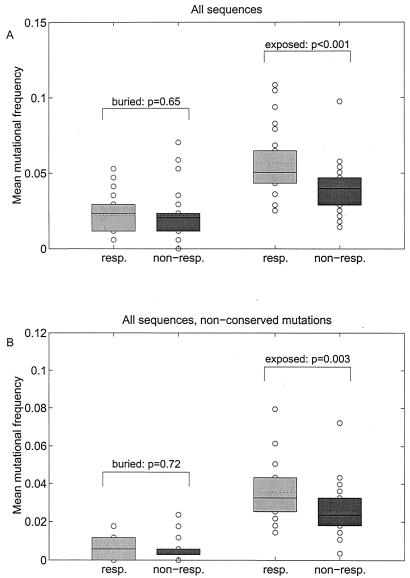
Furthermore, the functional properties of amino acids and also the predicted localization of amino acid mutations within the secondary structure of the resulting protein were considered. Figure 5A shows the smoothed mutational frequencies throughout the complete NS5A protein based on the predicted localization of amino acids within the core (buried) or at the surface (exposed) of the NS5A protein. This analysis was also performed for nonconserved amino acid mutations only (Fig. 5B). The mean number of mutations within the complete NS5A protein for predicted buried amino acids was not different between virologic responders and virologic nonresponders (P = 0.646). For predicted exposed amino acids, however, the number of mutations for virologic responders was significantly higher than for nonresponders (P = 0.0001, Fig. 6A). For analysis of nonconserved NS5A mutations only, no significant difference was detected between virologic responders and nonresponders for predicted buried amino acids (P = 0.725). For predicted exposed amino acids, the number of mutations was significantly higher in virologic responders than in nonresponders (P = 0.003; Fig. 6B).
FIG.5.
Smoothed mutational frequencies of full-length NS5A sequences from HCV-1b-infected patients with respect to the localization of amino acid residues within the secondary structure of the NS5A protein. Red-shaded areas indicate exposed amino acid residues, and white areas indicate buried amino acid residues or unknown localization. (A and B) Smoothed mutational frequencies based on all amino acid mutations (A) and based on nonconserved amino acid mutations only (B). Virologic responders (SR and ETR) are indicated in green, and virologic nonresponders are indicated in blue. The brown-shaded areas at the bottom indicate the potential functional regions within the NS5A protein according to the data presented in Fig. 1.
FIG. 6.
Box plots of mean mutational frequencies of full-length NS5A sequences from HCV-1b-infected patients based on all mutations (A) and on all nonconserved mutations (B). Horizontal joint and dotted lines indicate median and mean, respectively. The term “buried” indicates analysis for amino acids that are predicted to be located within the buried core of the NS5A protein or with unknown localization; the term “exposed” indicates analysis for amino acids that are predicted to be located on the surface of the NS5A protein. Abbreviations: resp., virologic responders to antiviral therapy (SR and ETR); non-resp., virologic nonresponders to antiviral therapy (NR).
Local analysis based on the predicted residue accessibility (buried and exposed) showed a significant local accumulation of amino acid mutations (P = 0.038). The main focus on aa 2350 to 2370 upstream of the V3 region was confirmed.
DISCUSSION
Four or more mutations within the ISDR of the HCV NS5A protein have been clinically correlated with sensitivity to IFN-α-based treatment (7, 11, 44, 46, 49, 52). However, in the majority of patients from western countries with a virologic response, fewer than four mutations within the ISDR are present, and sensitivity or resistance to antiviral therapy cannot be explained by the ISDR hypothesis (44, 46, 49, 55). Expression in cell culture systems generally confirmed IFN-α resistance mechanisms of the NS5A protein (13, 33, 34, 36). However, resistance to IFN-α was independent of the presence or absence of mutations within the ISDR. Therefore, other regions within the NS5A protein outside the ISDR may be involved in IFN resistance mechanisms. Mutations in other regions of the NS5A protein (PKR-binding domain, V3 region, carboxy-terminal part) have been described as associated with virologic response (9, 15, 31, 43).
In the present study, full-length NS5A sequences of 45 patients infected with HCV-1b were investigated for a possible correlation of mutational frequencies with treatment response. Patients were treated with IFN-α with or without ribavirin, and the response to antiviral therapy was carefully characterized. Furthermore, the initial virologic response to antiviral therapy was assessed by first- and second-phase kinetic analysis (29, 54). Since putative NS5A treatment resistance mechanisms have been characterized as directed against IFN-dependent pathways, patients with a virologic treatment response (SR and ETR patients) were compared to virologic nonresponders (NR patients).
The number of mutations throughout the complete NS5A protein correlated positively with the virologic response to treatment (P = 0.008). Furthermore, independent of different treatment regimens (3 to 6 MU of IFN-α with or without the addition of ribavirin), a more rapid second-phase viral decline correlated significantly with multiple mutations within the NS5A protein (one-tailed P = 0.034). However, in none of the known presumably functional regions within the NS5A protein (transmembrane domain, cleavage sites, NS4A association, ISDR, PKR-binding domain, NLS, and V3 region) was any significant correlation between mutations and treatment response observed.
Therefore, in a next step, analyses in other regions of the NS5A protein were performed based on 70 full-length NS5A sequences (31). Screening multiple regions of the complete NS5A by pointwise significance levels showed several regions with significant correlations between the mutational frequencies and treatment response and significant correlations between the mutational frequencies and the early viral decline for genuine sequences (Fig. 3A). Because the significance levels of multiple and correlated tests are difficult to interpret, random sequences were generated on the basis of a newly developed nonparametric binomial model and randomly assigned to be from virologic responders or nonresponders (Fig. 3B). Indeed, regions with pointwise significant correlations between mutational frequencies and virologic response could be identified in 94% of 500 simulated samples of random sequences. These findings are neither restricted to specific model assumptions nor to the assumed mutational probability function. Hence, comparing results from genuine sequences with those from randomly generated sequences can help to avoid misinterpretations of statistical results in general and especially showed that the identification of local accumulations of mutations from pointwise significance levels as in previous studies is problematic.
For further analyses of NS5A isolates, the functional consequences of amino acid exchanges, as well as predictions of the localization of amino acid residues in the core or on the surface of the resulting NS5A protein were studied. Amino acids were grouped according to functional properties to differentiate between conserved and nonconserved amino acid mutations (28). For conformational analysis, the amino acid residue accessibility within the secondary structure of the NS5A protein was predicted (39, 40). Exchange of amino acids with identical functional and conformational properties (conserved), as well as exchange of amino acids within the core of the resulting NS5A protein, should be of minor relevance, whereas nonconserved amino acid mutations and especially mutations for amino acids located on the predicted surface of the NS5A protein may be of functional importance. Analysis of the genuine NS5A isolates of the 70 HCV-1b-infected patients for predicted buried amino acid residues showed no differences between the virologic responders and nonresponders for the number of all (conserved and nonconserved) mutations, as well as all nonconserved mutations, in correlation with the treatment response. However, the number of all mutations and all nonconserved mutations was significantly higher for virologic responders compared to that for nonresponders for amino acids predicted to be located at the surface of the resulting NS5A protein (P = 0.0001 and P = 0.0029, respectively).
The existence of local accumulations of mutations truly correlating with the treatment response was analyzed by a new specific functional data analysis procedure. Significant local differences of mutational frequencies were detected for the complete NS5A protein (P = 0.018), as well as on the basis of predicted buried and exposed amino acid residues (P = 0.038), between virologic responders and nonresponders with a main focus upstream of the V3 region (aa 2350 to 2370) of the NS5A protein. At present, the functional relevance of this region is unknown. In a recent study, mutations in two variable regions within the carboxy-terminal part of the NS5A protein (a region downstream the PKR-binding domain and the V3 region) were characterized as correlated with the treatment outcome (31). Therefore, together with the results of the present study, the V3 region and flanking sequences may be involved in IFN-α resistance. The potential function of this NS5A region should be investigated in further studies, e.g., in the new HCV replicon model.
In conclusion, we have shown with the help of randomly generated sequences that simple analysis of counts of mutations in different NS5A regions is an insufficient approach for identifying local accumulations of mutations correlated with treatment response. A new nonparametric binomial model was developed to overcome this problem. In addition to the present study of HCV isolates, this model and the proposed analytical method have potential implications for similar investigations in other viruses (i.e., human immunodeficiency virus, hepatitis B virus) (12, 38).
The detection of significant mutational differences in regions within the NS5A protein of virologic responders and nonresponders is improved by consideration of functional properties of amino acid exchanges and the predicted accessibility of amino acid residues within the secondary structure of the resulting protein. Based on the investigations of nonconserved and predicted exposed amino acid mutations, one region (aa 2350 to 2370) within NS5A in particular was identified that showed significant differences between virologic responders and nonresponders to antiviral therapy.
Acknowledgments
We are grateful to Andreas Heger for computational support and to Steve Polyak for critically reading the manuscript and helpful discussions.
REFERENCES
- 1.Alter, M. J., H. S. Margolis, K. Krawczynski, F. N. Judson, A. Mares, W. J. Alexander, P. Y. Hu, J. K. Miller, M. A. Gerber, and R. E. Sampliner, et al. 1992. The natural history of community-acquired hepatitis C in the United States. N. Engl. J. Med. 327:1899-1905. [DOI] [PubMed] [Google Scholar]
- 2.Asabe, S. I., Y. Tanji, S. Satoh, T. Kaneko, K. Kimura, and K. Shimotohno. 1997. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J. Virol. 71:790-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1975. [DOI] [PubMed] [Google Scholar]
- 4.Brass, V., E. Bieck, R. Montserret, B. Wolk, J. A. Hellings, H. E. Blum, F. Penin, and D. Moradpour. 2001. An aminoterminal amphipathic alpha-helix mediates membrane association of the hepatitis C virus nonstructural protein 5A. J. Biol. Chem. 277:8130-8139. [DOI] [PubMed] [Google Scholar]
- 5.Braun, J. V., and H. G. Müller. 1998. Statistical methods for DNA sequence segmentation. Stat. Sci. 13:142-146. [Google Scholar]
- 6.Carroll, K., O. Elroy-Stein, B. Moss, and R. Jagus. 1993. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J. Biol. Chem. 268:12837-12842. [PubMed] [Google Scholar]
- 7.Chayama, K., A. Tsubota, M. Kobayashi, K. Okamoto, M. Hashimoto, Y. Miyano, H. Koike, M. Kobayashi, I. Koida, Y. Arase, S. Saitoh, Y. Suzuki, N. Murashima, K. Ikeda, and H. Kumada. 1997. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology 25:745-749. [DOI] [PubMed] [Google Scholar]
- 8.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, A. Medina-Selby, P. J. Barr, A. J. Weiner, and D. W. Bradley. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duverlie, G., H. Khorsi, S. Castelain, O. Jaillon, J. Izopet, F. Lunel, F. Eb, F. Penin, and C. Wychowski. 1998. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J. Gen. Virol. 79:1373-1381. [DOI] [PubMed] [Google Scholar]
- 10.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, N. Izumi, F. Marumo, and C. Sato. 1995. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J. Clin. Investig. 96:224-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enomoto, N., I. Sakuma, Y. Asahina, M. Kurosaki, T. Murakami, C. Yamamoto, Y. Ogura, N. Izumi, F. Marumo, and C. Sato. 1996. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N. Engl. J. Med. 334:77-81. [DOI] [PubMed] [Google Scholar]
- 12.Erhardt, A., U. Reineke, D. Blondin, W. H. Gerlich, O. Adams, T. Heintges, C. Niederau, and D. Häussinger. 2000. Mutations of the core promoter and response to interferon treatment in chronic replicative hepatitis B. Hepatology 31:716-725. [DOI] [PubMed] [Google Scholar]
- 13.François, C., G. Duverlie, D. Rebouillat, H. Khorsi, S. Castelain, H. E. Blum, A. Gatignol, C. Wychowski, D. Moradpour, and E. F. Meurs. 2000. Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis. J. Virol. 74:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gale, M., Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze. 1998. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol. Cell. Biol. 18:5208-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerotto, M., F. Dal Pero, P. Pontisso, F. Noventa, A. Gatta, and A. Alberti. 2000. Two PKR inhibitor HCV proteins correlate with early but not sustained response to interferon. Gastroenterology 119:1649-1655. [DOI] [PubMed] [Google Scholar]
- 16.Ide, Y., L. Zhang, M. Chen, G. Inchauspe, C. Bahl, Y. Sasaguri, and R. Padmanabhan. 1996. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene 182:203-211. [DOI] [PubMed] [Google Scholar]
- 17.Inchauspe, G., S. Zebedee, D. H. Lee, M. Sugitani, M. Nasoff, and A. M. Prince. 1991. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc. Natl. Acad. Sci. USA 88:10292-10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kato, N., M. Hijikata, Y. Ootsuma, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 87:9524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khorsi, H., S. Castelain, A. Wyseur, J. Izopet, V. Canva, A. Rombout, D. Capron, J. P. Capron, F. Lunel, L. Stuyver, and G. Duverlie. 1997. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alfa therapy in French patients. J. Hepatol. 27:72-77. [DOI] [PubMed] [Google Scholar]
- 20.Kurosaki, M., N. Enomoto, T. Murakami, I. Sakuma, Y. Asahina, C. Yamamoto, T. Ikeda, S. Tozuka, N. Izumi, F. Marumo, and C. Sato. 1997. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology 25:750-753. [DOI] [PubMed] [Google Scholar]
- 21.Lam, N. P., A. U. Neumann, D. R. Gretch, T. E. Wiley, A. S. Perelson, and T. J. Layden. 1997. Dose-dependent acute clearance of hepatitis C genotype 1 virus with interferon alpha. Hepatology 26:226-231. [DOI] [PubMed] [Google Scholar]
- 22.Lee, T. G., J. Tomita, A. G. Hovanessian, and M. G. Katze. 1990. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc. Natl. Acad. Sci. USA 87:6208-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 25.Markland, W., R. A. Petrillo, M. Fitzgibbon, T. Fox, R. McCarrick, T. McQuaid, J. R. Fulghum, W. Chen, M. A. Fleming, J. A. Thomson, and S. P. Chambers. 1997. Purification and characterization of the NS3 serine protease domain of hepatitis C virus expressed in Saccharomyces cerevisiae. J. Gen. Virol. 78:39-43. [DOI] [PubMed] [Google Scholar]
- 26.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alpha-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 27.McMillan, N. A., R. F. Chun, D. P. Siderovski, J. Galabru, W. M. Toone, C. E. Samuel, T. W. Mak, A. G. Hovanessian, K. T. Jeang, and B. R. Williams. 1995. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology 213:413-424. [DOI] [PubMed] [Google Scholar]
- 28.Mirny, L. A., and E. I. Shakhnovich. 1999. Universally conserved positions in protein folds: reading evolutionary signals about stability, folding kinetics and function. J. Mol. Biol. 291:177-196. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-a therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 30.Niederau, C., S. Lange, T. Heintges, A. Erhardt, M. Buschkamp, D. Hürter, M. Nawrocki, L. Kruska, F. Hensel, W. Petry, and D. Häussinger. 1998. Prognosis of chronic hepatitis C: results of a large prospective cohort study. Hepatology 28:1687-1695. [DOI] [PubMed] [Google Scholar]
- 31.Nousbaum, J., S. J. Polyak, S. C. Ray, D. G. Sullivan, A. M. Larson, R. L. Carithers, Jr., and D. R. Gretch. 2000. Prospective characterization of full-length hepatitis C virus NS5A quasispecies during induction and combination antiviral therapy. J. Virol. 74:9028-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto, H., M. Kojima, S. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, D. A. Peterson, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2-year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 33.Paterson, M., C. D. Laxton, H. C. Thomas, A. M. Ackrill, and G. R. Foster. 1999. Hepatitis C virus NS5A protein inhibits interferon antiviral activity, but the effects do not correlate with clinical response. Gastroenterology 117:1187-1197. [DOI] [PubMed] [Google Scholar]
- 34.Podevin, P., A. Sabile, R. Gajardo, N. Delhem, A. Abadie, P. Y. Lozach, L. Beretta, and C. Brechot. 2001. Expression of hepatitis C virus NS5A natural mutants in a hepatocytic cell line inhibits the antiviral effect of interferon in a PKR-independent manner. Hepatology 33:1503-1511. [DOI] [PubMed] [Google Scholar]
- 35.Polyak, S. J., S. McArdle, S. L. Liu, D. G. Sullivan, M. Chung, W. T. Hofgartner, R. L. Carithers, Jr., B. J. McMahon, J. I. Mullins, L. Corey, and D. R. Gretch. 1998. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J. Virol. 72:4288-4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polyak, S. J., D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch. 1999. Characterization of the effects of hepatitis C virus nonstructural 5A protein expression in human cell lines and on interferon-sensitive virus replication. Hepatology 29:1262-1271. [DOI] [PubMed] [Google Scholar]
- 37.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, J. Albrecht, et al. 1998. Randomised trial of interferon α2b plus ribavirin for 48 weeks or for 24 weeks versus interferon α2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 38.Ross, L., M. Johnson, R. DeMasi, Q. Liao, N. Graham, M. Shaefer, and M. St. Clair. 2000. Viral genetic heterogeneity in HIV-1-infected individuals is associated with increasing use of HAART and higher viremia. AIDS 14:813-819. [DOI] [PubMed] [Google Scholar]
- 39.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-77. [DOI] [PubMed] [Google Scholar]
- 40.Rost, B., and C. Sander. 1994. Conservation and prediction of solvent accessibility in protein families. Proteins 20:216-226. [DOI] [PubMed] [Google Scholar]
- 41.Rost, B., and C. Sander. 2000. Third generation prediction of protein structures. Methods Mol. Biol. 143:71-95. [DOI] [PubMed] [Google Scholar]
- 42.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q. L. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarrazin, C., T. Berg, J. H. Lee, B. Rüster, B. Kronenberger, W. K. Roth, and S. Zeuzem. 2000. Mutations in the protein kinase-binding domain of the NS5A protein in patients infected with hepatitis C virus type 1a are associated with treatment response. J. Infect. Dis. 181:432-441. [DOI] [PubMed] [Google Scholar]
- 44.Sarrazin, C., T. Berg, J. H. Lee, G. Teuber, C. F. Dietrich, W. K. Roth, and S. Zeuzem. 1999. Improved correlation between multiple mutations within the NS5A region and virological response in European patients chronically infected with hepatitis C virus type 1b undergoing combination therapy. J. Hepatol. 30:1004-1013. [DOI] [PubMed] [Google Scholar]
- 45.Satoh, S., M. Hirota, T. Noguchi, M. Hijikata, H. Handa, and K. Shimotohno. 2000. Cleavage of hepatitis C virus nonstructural protein 5A by a caspase-like protease(s) in mammalian cells. Virology 270:476-487. [DOI] [PubMed] [Google Scholar]
- 46.Sáiz, J. C., F. X. López-Labrador, S. Ampurdanés, J. Dopazo, X. Forns, J. M. Sánchez-Tapias, and J. Rodés. 1998. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J. Infect. Dis. 177:839-847. [DOI] [PubMed] [Google Scholar]
- 47.Silverman, B. W. 1996. Smoothed functional principal components analysis by choice of norm. Ann. Statist. 24:1-24. [Google Scholar]
- 48.Simmonds, P., E. C. Holmes, T. A. Cha, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 49.Squadrito, G., F. Leone, M. Sartori, B. Nalpas, P. Berthelot, G. Raimondo, S. Pol, and C. Brechot. 1997. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology 113:567-572. [DOI] [PubMed] [Google Scholar]
- 50.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Taylor, D. R., S. T. Shi, P. R. Romano, G. N. Barber, and M. M. C. Lai. 1999. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science 285:107-110. [DOI] [PubMed] [Google Scholar]
- 52.Witherell, G. W., and P. Beineke. 2001. Statistical analysis of combined substitutions in nonstructural 5A region of hepatitis C virus and interferon response. J. Med. Virol. 63:8-16. [PubMed] [Google Scholar]
- 53.Zeuzem, S., S. V. Feinman, J. Rasenack, E. J. Heathcote, M. Y. Lai, E. Gane, J. O'Grady, J. Reichen, M. Diago, A. Lin, J. Hoffman, and M. J. Brunda. 2000. Peginterferon α-2a in patients with chronic hepatitis C. N. Engl. J. Med. 343:1666-1672. [DOI] [PubMed] [Google Scholar]
- 54.Zeuzem, S., E. Herrmann, J. H. Lee, J. Fricke, A. U. Neumann, M. Modi, G. Colucci, and W. K. Roth. 2001. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon α2a. Gastroenterology 120:1438-1447. [DOI] [PubMed] [Google Scholar]
- 55.Zeuzem, S., J. H. Lee, and W. K. Roth. 1997. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon α. Hepatology 25:740-744. [DOI] [PubMed] [Google Scholar]