Abstract
Genetic recombination occurs frequently during replication of picornaviruses. To explore the intracellular site and structures involved in recombination, HeLa cells were infected with poliovirus type 1 Mahoney and type 2 Sabin. The two genomes were located by fluorescent in situ hybridization and confocal microscopy. For hybridization, type-specific fluorescent riboprobes were used to visualize the same genomic region where, in parallel, recombination was demonstrated with type-specific reverse transcription-PCR and sequencing. The hybridization analysis indicated that >85% of the replication complexes contained both type 1 and type 2 RNA sequences aligned at a lateral distance of 50 nm or less. Sequential infection of cells ruled out the possibility that the high percentage of mixed replication complexes was due to aggregation of input virus. Visualization of input genomic RNA over time showed that the viral genomes migrated to relatively few distinct, and thus presumably specific, perinuclear sites where replication started. The first recombinant RNA strands could be detected concomitantly with the onset of RNA replication. The limited number of start sites for replication may be the reason for the observed preferential formation of mixed replication complexes, each accommodating several parental RNA strands and thus allowing recombination.
Poliovirus (PV), the prototype member of the family Picornaviridae, represents in many respects the paradigm for research on other plus-strand RNA viruses. Among the fundamental insights gained by PV research is the first discovery of recombination between viral RNA genomes (30, 35). Since then, some light has been thrown on mechanistic aspects of recombination (for a review see references 1 and 57); however, it is not clear in which context of the elaborate structures comprising the viral RNA replication machinery, i.e., the replication complex (4, 6, 9, 12, 21, 22, 51, 52), recombination can occur.
PV RNA replication is preceded by translation of the incoming viral genome. Translation, initiated at the internal ribosomal entry site (IRES) (42, 43), was reported to be enhanced by binding of the cellular poly(rC) binding protein (PCBP) to the IRES (10, 11, 25) and, additionally at early stages, by an interaction of eIF4G with the IRES and the poly(A)-bound poly(A) binding protein (PABP) (50). A transition from translation to transcription was thought to be mediated by the newly synthesized viral protein 3CD, which, together with PCBP, binds to the 5′ cloverleaf of the viral mRNA (23, 24, 39). Poly(A)-bound PABP then would attach to this complex, leading (again) to a 5′-3′ interaction in the viral plus-strand RNA (29). This interaction was proposed to be a prerequisite for minus-strand RNA synthesis, the first step in genome replication (3, 29, 53). The second step in viral genome replication is plus-strand RNA synthesis, which proceeds, after multiple initiations, in the partially double-stranded replicative intermediate. The primer for RNA synthesis is VPgpUpU (41), which was found to be synthesized on a cis-acting sequence element within the 2C coding region (28, 40, 44).
Many, if not all, of these steps in the viral replication cycle seem to be linked to membranes. Translation presumably proceeds on endoplasmic reticulum (ER) membranes (21, 46, 47), and the translation products of the P2 and P3 genomic region reportedly associate with the ER (26, 55). At the same site, the protein 2BC activates the formation of vesicles (2, 7, 15, 48, 49), which build up the PV replication complex and which were found to represent ER-derived coat protein II (COPII)-induced vesicles of the anterograde membrane pathway (48). Concomitantly, the viral mRNA becomes a template for minus-strand RNA synthesis, which suggests that minus-strand synthesis could be associated with the nascent vesicular replication complex. RNA of minus-strand polarity could in fact be found in delineated membranous structures within the cell (12), but the role of membranes in minus-strand RNA synthesis remains to be defined. The mature replication complex, engaged in plus-strand RNA synthesis (20), was described to present as a cluster of vesicles, called a rosette (9). The formation of replication complexes was reported to couple translation, vesicle formation, and transcription in cis, whereby no exchange of viral protein, RNA, or membranes could be observed (21). This renders processes such as complementation and recombination enigmatic from a structural point of view and raises the question of how these processes, particularly the frequently observed recombination, can proceed.
RNA recombination is accomplished by two different mechanisms (14). Nonreplicative recombination was proposed to be mediated by ribozyme-like structures in the PV genome (27). Replicative recombination was found to occur by a template switch mechanism of the polymerase during minus-strand RNA synthesis (16, 31, 34, 45). Its frequency is 10 to 20% of recombinants among progeny strands (33), and it may take place over the entire viral genome (for reviews, see references 1 and 57). Stem-loop structures were thought to promote recombination by allowing the alignment of the parental strands for the switching process (45, 54). Recombination was considered to be important for both virus evolution and genome conservation (1), and it was found to happen in the human gut at rather high frequency after vaccination with trivalent oral polio vaccine (38).
Recombination, progressing during minus-strand RNA synthesis, requires close association between the parental donor and acceptor RNA strands of plus polarity. There are several conceivable ways in which different RNA strands could be brought into close proximity within the infected cell. First, there could be an interaction between individual replication complexes, e.g., by exchange of RNA through viral progeny RNA which, set free from a replication complex (56), would invade a second replication complex. Second, several replication complexes could merge to form a complex containing more than one parental RNA strand. Such processes were observed at late stages of virus replication when individual replication complexes were found to coalesce in a perinuclear area (12). Third, a replication complex could be built up by two or several different parental RNA strands from its very beginning and thus contain more than one replicating RNA population.
The aim of the present work was to investigate the mode of formation of presumed mixed PV replication complexes containing different parental RNA strands and thus allowing for recombination. To this end, HeLa cells were infected with two PV strains, type 1 Mahoney (M1) and type 2 Sabin (S2), and the resulting replication complexes were detected by fluorescent in situ hybridization (FISH). Type-specific fluorescent riboprobes were used to visualize the genomic region, where recombination was demonstrated with type-specific reverse transcription (RT)-PCR and sequencing. This protocol allowed the detection of parental RNA strands and recombinants without selection for viability.
After double infection, input viral genomic RNA strands were found to migrate from the cell surface to relatively few specific perinuclear sites to start RNA replication in close association. The first recombinant RNA strands were found by RT-PCR concomitantly with the onset of RNA replication, and high-resolution FISH analysis indicated that most of the replication complexes contained both PV M1 and S2 RNA sequences in close association early in infection, allowing for recombination within one (nascent) replication complex.
MATERIALS AND METHODS
Cells and virus infection.
HeLa cells were grown as monolayer cultures and infected at a multiplicity of infection of 15 PFU/cell with PV M1 and at a multiplicity of infection of 60 with PV S2 (strain P712Ch 2ab) to compensate for the different replication efficiencies. The cells were infected with either one PV type alone or with both, simultaneously or sequentially. For a sequential infection, the first virus was adsorbed to the cell at 36°C in the presence of cycloheximide (CHI) (200 μM; Sigma) for 30 min to allow for uptake and decapsidation of the virus but not for the start of replication. The cells were then washed thoroughly and infected with the second virus in the absence of CHI. To verify that input viral RNA could be detected by FISH experiments in the absence of replication, the cells were infected in the presence of 2 mM guanidine (13) to prevent viral RNA transcription. After all infection procedures, the cells were transferred to medium with 5% fetal calf serum.
In vivo labeling of viral RNA.
To quantify viral RNA, actinomycin D (AMD) (5 μg/ml; Merck, Sharp & Dohme) was added to infected cells 30 min prior to pulsing them with [5,6-3H]uridine (Amersham Pharmacia Biotech) for 10 min. Acid-precipitable radioactivity was counted in a liquid scintillation counter.
Amplification of PV RNA by RT-PCR and selective amplification and sequencing of recombinants.
RNA was extracted from infected cells with phenol-chloroform, precipitated with ethanol, and reverse transcribed with random-hexamer primers and Ready-to-Go You-Prime First-Strand Beads (Amersham Pharmacia Biotech). PCR was performed with Pwo polymerase (Roche Molecular Biochemicals, Mannheim, Germany) at an annealing temperature of 55°C. After gel electrophoresis in 1.5% agarose (Gibco-Invitrogen), the PCR products were visualized with ethidium bromide in a UV illuminator. PV M1 and S2 cDNAs were amplified using the specific primers indicated in Table 1. To selectively amplify recombinants (Fig. 1) from PV M1- and S2-double-infected cells, RNA was reverse transcribed as described above and PCR was performed with the upstream primer specific for PV S2 (S up) and the downstream primer specific for PV M1 (M down), as indicated in Table 1. For sequencing, the resulting DNA band was stabbed in the gel with a micropipet and reamplified with the same primers. After gel electrophoresis, the recombinant DNA was recovered with a gel extraction kit (Qiagen), and sequencing reactions were performed with the sense S up or the antisense M down primer and analyzed in an ABI 310 Genetic Analyzer (Perkin-Elmer).
TABLE 1.
Sequences of primer sets
| Name | Nucleotide numbera | Sequence of primerb | Usec |
|---|---|---|---|
| M up | 2512-2531 | 5′-CAACACAGTCCGTGAAACGG-3′ | Amplification of PV M1 cDNA |
| M down | 3150-3130 | 5′-GTGCTGCCGACTGGTCCTTC-3′ | |
| S up | 2679-2697 | 5′-CATCCAGAGACGAACGCGA-3′ | Amplification of PV S2 cDNA |
| S down | 3152-3132 | 5′-CAGTTGAGGCTTGACCCGCT-3′ | |
| S up | 2679-2697 | 5′-CATCCAGAGACGAACGCGA-3′ | Amplification of PV S2 and PV M1 recombinant cDNA |
| M down | 3150-3130 | 5′-GTGCTGCCGACTGGTCCTTC-3′ | |
| 2687-2704 | 5′-AGGTCAAGGTCAGAGTC-3′ | Production of PV M1-specific 5′ riboprobe (5′M probe) | |
| 2800-2784 | 5′-ATTAATACGACTCACTATAGGGCACTGCAAATAGCTTAT-3′ | ||
| 3083-3099 | 5′-TCGAACGCCTATTCACA-3′ | Production of PV M1-specific 3′ riboprobe (3′M probe) | |
| 3185-3169 | 5′-ATTAATACGACTCACTATAGGGCATTTAGAGATGCTGCA-3′ | ||
| 2675-2691 | 5′-ATGTCATCCAGAGACGA-3′ | Production of PV S2-specific 5′ riboprobe (5′S probe) | |
| 2800-2784 | 5′-ATTAATACGACTCACTATAGGGCCGAAAACAATCTGCTG-3′ | ||
| 3085-3102 | 5′-GCTAATGCGTATTCCCAC-3′ | Production of PV S2-specific 3′ riboprobe (3′S probe) | |
| 3187-3171 | 5′-ATTAATACGACTCACTATAGGGCATTCAGTGAGGCAGCA-3′ | ||
| 4460-4478 | 5′-CAGTTCAAGAGCAAACACC-3′ | Production of riboprobe to detect input PV M1 and PV S2 | |
| 7440-7421 | 5′-ATTAATACGACTCACTATAGGGCTCCGAATTAAAGAAAAATT-3′ |
Numbering according to PV M1 or PV S2 plus-strand RNA.
T7 promotor sequence is in italics.
Designation of probe is in parentheses.
FIG. 1.
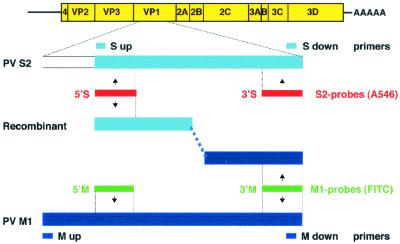
Schematic representation of primers, riboprobes, and viral sequences used. The M1 and S2 probes, tagged with different fluorochromes, and the M and S primers (Table 1) type specifically detect PV M1 or S2 sequences within parental or recombinant genomes.
Preparation of labeled PV type-specific riboprobes.
To obtain PV S2 cDNA, RNA from virus stock was extracted and reverse transcribed as described above. For PV M1, the EcoRI-linearized plasmid DNA pT7PV (kindly provided by Eckard Wimmer, Stony Brook, N.Y.) was used. Templates for riboprobe preparation were generated by PCR within the VP1 genomic region using the appropriate primers (Table 1), with the downstream primer comprising the T7 promoter (19). From the amplified DNA fragments, riboprobes of minus polarity were in vitro transcribed with T7 polymerase (Roche Molecular Biochemicals). Two riboprobes were designed to be specific for PV M1, and two were designed to be specific for PV S2 (Fig. 1). The PV M1-specific probes comprised nucleotides (nt) 2687 to 2800 and 3083 to 3185 and were designated 5′M and 3′M, respectively. For PV S2, the probes comprised nt 2675 to 2800 and 3085 to 3187 and were designated 5′S and 3′S. An ∼3,000-nt-long non-type-specific riboprobe of high sensitivity comprised nt 4460 to 7440 (Table 1). The nucleotide numbering (EMBL database) is that of the respective plus-strand RNA of PV M1 or S2. The riboprobes were labeled as described previously (19) by incorporation of UTP tagged with the fluorochrome fluorescein isothiocyanate (FITC) (ENZO, New York, N.Y.) or Alexa 546 (A546) (Molecular Probes, Eugene, Oreg.) or with digoxigenin (DIG) (Roche Molecular Biochemicals). The probe 4460-7440 was hydrolyzed to fragments of about 100 nt (19). Unincorporated tagged UTP was removed with a Micro Bio-Spin 6 column, followed by a Micro Bio-Spin 30 column (Bio-Rad, Hercules, Calif.).
Northern and Southern blot hybridization.
PV M1 or S2 suspensions were clarified by centrifugation, and the RNA was phenol-chloroform extracted, loaded onto a denaturing 0.8% agarose gel, and blotted. DNA amplified by RT-PCR from infected cells was loaded onto a 1.5% agarose gel and blotted. For hybridization, the DIG-labeled riboprobes 5′M, 3′M, 5′S, and 3′S were used as appropriate and visualized with alkaline phosphatase-coupled anti-DIG Fab fragment (ENZO).
IF and FISH.
For immunofluorescence (IF) analysis or FISH, cells were grown on glass coverslips and fixed as described previously (12) at various times postinfection (p.i.). The protocol for FISH has been described in detail previously (12, 19). For the simultaneous detection of PV M1 and S2 RNA in double-infected cells, the FITC-labeled PV M1-specific and the A546-labeled PV S2-specific riboprobes were mixed and applied. Either the 3′M and 3′S probes or all four probes were combined.
For indirect IF analysis, the monoclonal antibody (MAb) 1D3.B1 directed against PV protein 2B (20) was used, followed by a goat anti-mouse antibody coupled to Texas Red (Molecular Probes).
Microscopy and image acquisition.
For conventional light microscopy, an epifluorescence microscope (Nikon E800) equipped with suitable filters and phase-contrast optics was used. To merge IF or FISH with phase-contrast pictures, the fluorescent and the phase- contrast pictures were recorded separately on Kodak TMax 400pro film under optimal exposure conditions. In addition, the same field was photographed simultaneously with both modes combined to facilitate the accurate overlay of the separate pictures by digital image processing using Adobe Photoshop software.
Optical sections were taken with a confocal laser scanning microscope (CLSM) (TCS4D; Leica Lasertechnik, Heidelberg, Germany) and recorded in a sequential-acquisition mode to avoid cross talk (“bleeding”) from one fluorochrome detection channel into the other. Background fluorescence and colocalization were determined as described earlier (48), and background was subtracted manually with Adobe Photoshop software.
RESULTS
Detection of recombination between PV M1 and S2 genomes in double-infected cells.
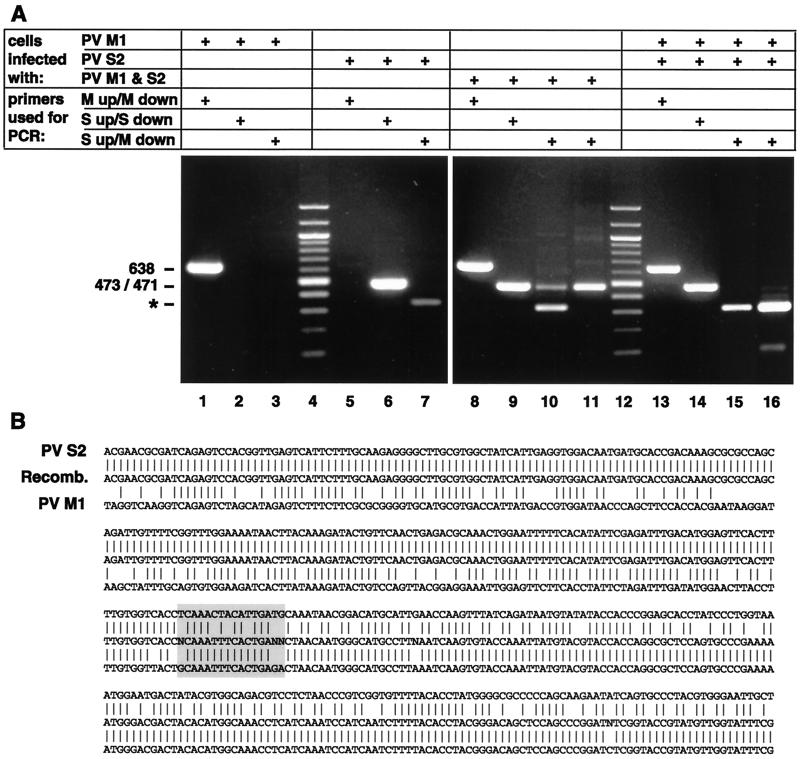
To allow the detection of recombination events during a double infection with two different PV strains, PV genomic sequences were searched to find regions of low homology. Such differing sequences were found in the VP1 region of PV M1 and S2. This enabled us to select primers (Fig. 1) to specifically amplify VP1 sequences of either PV M1 or S2. To test the specificities of the primers, HeLa cells were infected with PV M1 or S2 or with both PV types simultaneously. RNA was extracted at 3.5 h p.i., and its cDNA was amplified by PCR with the corresponding primer pairs (Table 1). Figure 2A shows that after infection with PV M1, only the M1-specific primers yielded a band of 638 nt (lanes 1 to 3), and after infection with PV S2, the S2-specific primers produced a band of 473 bp (lanes 5 to 7). The M1- and S2-specific bands were also obtained in cells infected with both PV types (lanes 8 and 9). To test whether recombination had occurred in the double-infected cells, PCR was performed with the primer combination S up and M down (Table 1). This yielded a band of 471 bp, which could be reamplified with the same primers (lanes 10 and 11). Thus, this primer pair allows the selective amplification of VP1 recombinants in a large pool of parental strands. To confirm that recombination occurred during replication in the double-infected cell and not during RT-PCR (31), RNAs extracted from either PV M1- or S2-infected cells were mixed in vitro prior to amplification (Fig. 2A, lanes 13 to 16). No (recombinant) band was obtained with the S up-M down primer pair (lane 15) even after reamplification (lane 16). In PV S2- and double-infected cells, this primer combination also consistently yielded a nonspecific band of >300 bp which was due to spurious priming of the M down primer on PV S2 cDNA (Fig. 2A, lane 7, and 3B).
FIG. 2.
(A) Specificities of primers for selective PCR after RT with random primers. In cDNAs from cells infected singly with PV M1 or S2 and in PV M1- and S2-double-infected cells, the M or S primers specifically recognize the cDNA of PV M1 (a band of 638 bp; lanes 1 and 8) or PV S2 (a band of 473 bp; lanes 6 and 9), respectively. With the S up and M down primer combination, cDNA prepared from double-infected cells allows amplification of recombinant cDNA (a band of 471 bp; lane 10). In lane 11, this band was reamplified with the same primer combination. If PV M1 and PV S2 RNAs from separately infected cells are mixed prior to RT-PCR, parental but not recombinant bands of 471 bp are found (lanes 13 to 15); lane 16 is a reamplification of the DNA shown in lane 15. The band marked with an asterisk is due to spurious priming of M down primer on PV S2 cDNA (see the text). Lanes 4 and 12 are 100-bp DNA ladders. +, present. (B) Sequence obtained from cDNA of the recombinant (Recomb.; panel A, lane 11) aligned with the sequences of PV S2 and PV M1, starting at nt 2688. The recombination locus is boxed and comprises nt 2899 to 2914 (numbering according to PV S2).
To confirm recombination, the reamplified 471-bp band obtained by PCR (Fig. 2A, lane 11) was sequenced with the S up or the M down primer. The sequencing data obtained indicated the presence of PV recombinants with a 5′S2 and a 3′M1 end (Fig. 2B). Since sequencing was done directly with the PCR product without prior cloning, the sequence had to belong to a predominant, but not necessarily homogenous, population of recombinants. The data do not allow us to narrow the recombination event more than to the region nt 2897 to 2912 (Fig. 2B).
In conclusion, intertypic replication-dependent recombination had occurred in PV M1- and S2-double-infected cells under our experimental conditions in a type-specific region of VP1. Other possible recombinants in the same or other locations have not been investigated further (see Discussion).
Specificity of riboprobes for detection of parental and recombined RNA.
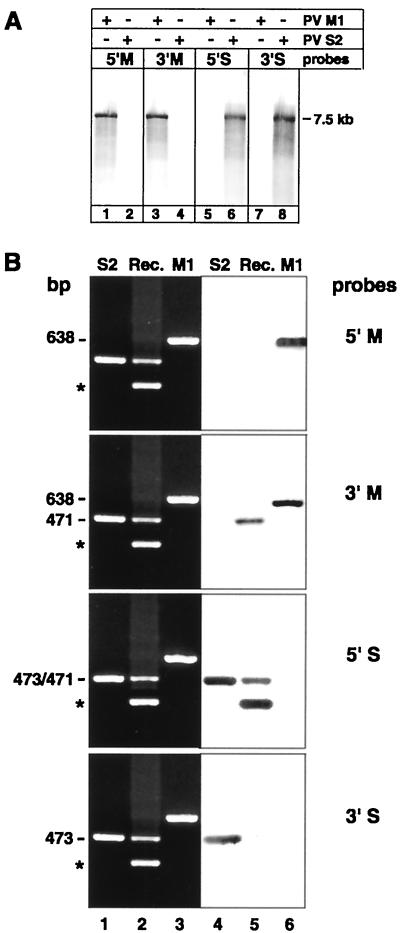
In order to detect in situ the possible intracellular site of the recombination described above, riboprobes were prepared, using appropriate sequences in the VP1 region of PV M1 and S2. To test their specificities, the DIG-labeled M and S probes were applied to Northern blots of RNA extracted from partially purified PV M1 or S2. Figure 3A shows that all four probes hybridized only to their respective target RNAs and did not nonspecifically recognize sequences in the nonhomologous viral genome.
FIG. 3.
(A) Northern blot hybridization with the DIG-labeled M (lanes 1 to 4) or S (lanes 5 to 8) riboprobe on PV M1 or S2 7.5-kb-genome-length RNA extracted from corresponding virus stocks shows type specificity of probes. The position of the 7.5-kb marker RNA is indicated. +, present; −, absent. (B) Southern blot hybridization with the DIG-labeled type-specific riboprobes on cDNA from single- or double-infected cells. Ethidium bromide-stained gels (lanes 1 to 3) of RT-PCR-amplified PV S2 (lane 1), recombinant PV S2-M1 (Rec.; lane 2), and PV M1 RNA (lane 3) are juxtaposed to their corresponding blots hybridized with the DIG-labeled M and S probes (lanes 4 to 6). The probes recognize PV M1 cDNA of 638 bp or PV S2 cDNA of 473 bp at their 5′ and 3′ termini. The 471-bp cDNA of the PV S2-M1 recombinant is recognized by the 5′S and the 3′M probes but not the 5′M and the 3′S probes. The asterisks indicate nonspecific bands as in Fig. 2.
The same probes were also hybridized to DNA obtained by RT-PCR from PV M1-, S2-, or double-infected cells. Figure 3B shows, in ethidium bromide-stained gels, the presence and locations of the respective bands and, in the juxtaposed blot, that the 5′M1 and 3′M1 probes specifically recognize PV M1 (lane 6) and the S2 probes specifically recognize PV S2 sequences (lane 4). The recombinant 471-bp band, obtained from double-infected cells, was recognized by the 5′S and the 3′M probes (lane 5), which is in agreement with the sequencing data (Fig. 2B), indicating recombination between the corresponding 5′S2 and 3′M1 sequences. The nonspecific >300-bp band (Fig. 3B and 2A, lane 7) was recognized by the 5′S probe and not by the 3′S probe, which indicates that it contained PV S2 sequences upstream of the binding site of the 3′S probe and was produced by false priming of the M down primer on the PV S2 template.
Thus, hybridization on blotted cDNA and RNA confirmed the type specificities of the riboprobes for their respective target sequences within the VP1 genomic region, intended for the in situ localization.
Detection of mixed PV M1- and S2 RNA-containing replication complexes as possible recombination sites.
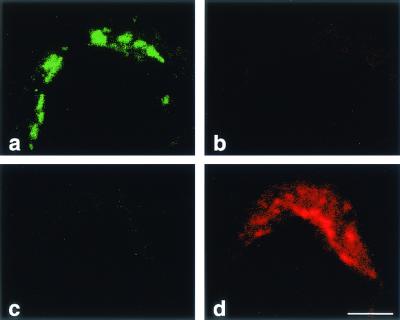
Replicative recombination between RNA strands requires their close spatial association during replication, presumably in the same replication complex. To detect possible recombination sites within double-infected cells, PV M1 and S2 and their recombinant RNA were located by type-specific FISH in the CLSM. Type specificity necessitated the use of comparatively short probes of about 100 nt (Table 1), so in many experiments, two probes of the same type specificity were combined to enhance the sensitivity of the FISH. To confirm the specificities of the probes for FISH, the four probes were mixed and FISH was performed on cells infected with either PV M1 or S2. Figure 4 shows that the FITC-labeled PV M1 probes and the A546-labeled S2 probes hybridized type specifically (Fig. 4a and d) with no cross-reactivity detectable (Fig. 4b and c), even with digital overamplification of the background signal (not shown). The same FISH results were obtained by conventional light microscopy (not shown).
FIG. 4.
Type specificity of the fluorescent riboprobes used for FISH was evaluated. Cells were infected with either PV M1 (a and b) or PV S2 (c and d) and hybridized with a mixture of all M and S probes. (a and c) Pictures taken in the green-detecting channel; (b and d) pictures taken in the red channel. (a) FITC-tagged 5′ and 3′M probes hybridized to PV M1 RNA. (d) A546-tagged 5′ and 3′S probes hybridized to PV S2 RNA. (b) PV M1 RNA-containing structures were not recognized by the S probes. (c) PV S2 was not recognized by the M probes. In addition, there is no visible cross talk from the green to the red channel (a and b) and the red to the green channel (d and c). Bar, 5 μm.
Hybridization in PV M1- and S2-double-infected cells with the same four probes showed that a large proportion of the M1 and S2 RNA strands colocalized. Since viral RNA at that stage of the infectious cycle is contained in replication complexes, the FISH results mean that, surprisingly, the majority of the replication complexes contained plus-strand RNA sequences of both PV types (Fig. 5a to c). In this experiment, all four riboprobes (Fig. 1) were used, and therefore, mixed replication complexes could have contained both parental and recombinant RNA or merely multiplying recombinants which had arisen elsewhere.
FIG.5.
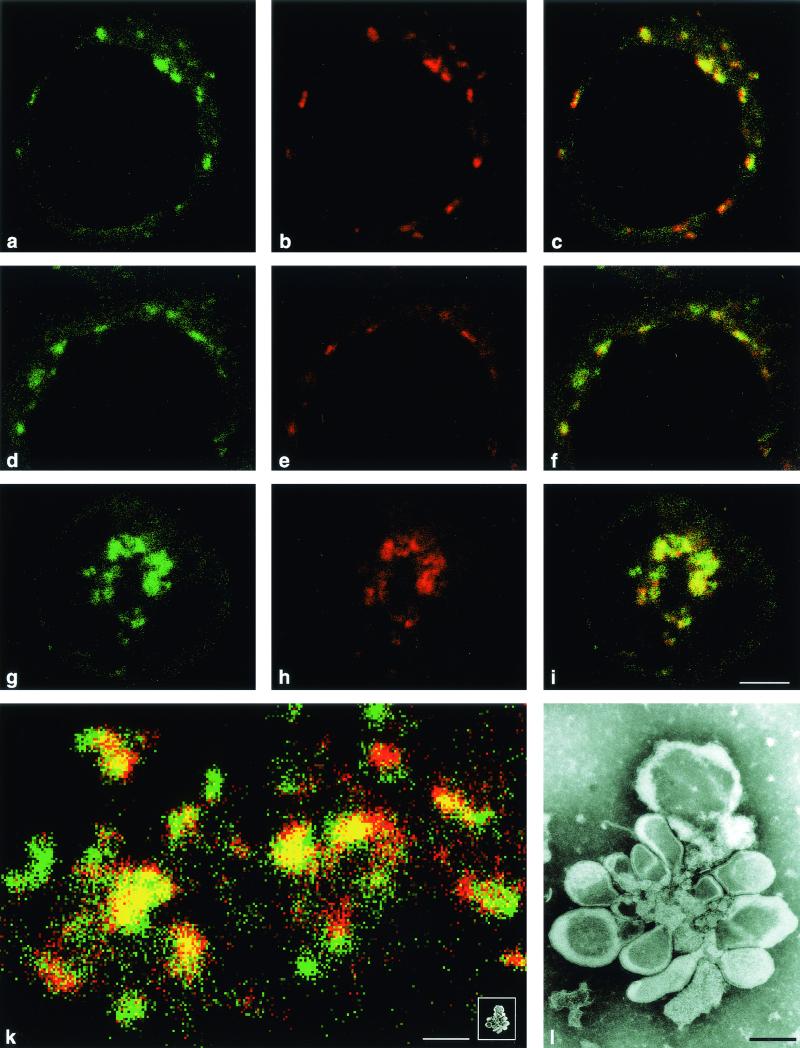
Type-specific detection of different viral genomic sequences in cells double infected with PV M1 and S2 at 3.5 h p.i. (a to c) FISH was performed with all four riboprobes, i.e., the FITC-tagged 5′M and 3′M probes and A546-tagged 5′S and 3′S probes. PV M1 RNA is visualized in the green channel (a), and PV S2 RNA is visualized in the red channel (b). An overlay of both pictures (c) shows largely colocalization (yellow). (d to i) FISH performed with 3′M and 3′S probes indicates colocalization (yellow) if PV M1 and S2 RNAs are present on two different strands (parental and/or recombinant) but not if only one upgrowing recombinant is present (Fig. 1). PV M1 (green) (d and g) and PV S2 (red) (e and h) (overlaid in panels f and i) was detected at early (d to f) and later (g to i) stages of infection (for definitions of early and late, see the text). The majority of the replication complexes contain two different types of RNA. Bar, 5 μm. (k) Several replication complexes are shown at higher magnification in a double-infected cell hybridized with all four riboprobes. Most replication complexes contain both types of RNA closely associated (yellow pixels) in their central parts. (Inset) The same replication complex shown in the electron micrograph in panel l scaled to the same magnification as the confocal micrograph. Bar, 1 μm. (l) Electron micrograph of an isolated replication complex, called a rosette (9). Bar, 0.1 μm.
To avoid the detection of replication complexes containing a multiplying recombinant only (Fig. 1) and to test for presumably recombination-competent replication complexes containing PV M1 and S2 sequences on two separate (parental and/or already recombinant) RNA strands, FISH with only the 3′M (FITC) and 3′S (A546) probes was performed. Due to a slight asynchrony in the viral replication, cells with individual perinuclear replication complexes (considered to be cells at an earlier stage in the infectious cycle) and cells with a more central and condensed location of the replication complexes (cells at a later stage [12]) can be seen in preparations at 3.5 h p.i. FISH with the 3′M and 3′S probes (Fig. 5d to i) showed that the majority of the replication complexes in earlier and later stages exhibited colocalization of both M1 and S2 signals and thus contained RNA with M1 and S2 sequences on separate strands, which would allow recombination. Quantification of the replication complexes showed that at early and late stages at least 85% of all replication complexes carried both signals (Table 2).
TABLE 2.
Percentages of replication complexes containing PV M1 and PV S2 sequences
| Fluorescent riboprobesa | Stage in replication cycleb | % Replication complexes containing sequences of:
|
||
|---|---|---|---|---|
| M1 only | S2 only | M1 and S2 | ||
| 5′M, 3′M, 5′S, 3′S | Early; n = 229 | 10 | 3 | 87c |
| Late; n = 115 | 5.2 | 0.8 | 94c | |
| 3′M, 3′S | Early; n = 87 | 15 | 0 | 85d |
| Late; n = 129 | 9.3 | 2.3 | 88.4d | |
See Fig. 1.
For definitions of early and late, see Results. n, number of replication complexes evaluated.
A mixed replication complex may contain M1 and S2 RNA and/or recombinant RNA.
A mixed replication complex contains both M1 and S2 RNA sequences downstream of the recombination locus on separate parental and/or separate recombinant RNA strands.
Analysis of CLSM pictures at high magnification revealed the spatial arrangement and the colocalization of PV M1 or S2 sequences in several individual replication complexes (Fig. 5k). Sequences PV M1 and S2 were found to colocalize predominantly in the central parts of the complexes. To score as colocalization in the CLSM protocol used, the red and green signals must be recorded within the same 50-nm-diameter pixel. Therefore, the two signals, and hence the corresponding target RNA sequences, have to be associated at a distance of 50 nm or less.
To illustrate the dimensions of the replication complexes and the replicating RNA in Fig. 5k, Fig. 5l shows an electron micrograph of an isolated PV replication complex (a rosette [6, 9]). The rosette, with a long axis of ∼1 μm, harbors replicating PV RNA with a stretched out length of 2.7 μm (37). For comparison, the same rosette is reproduced to scale in the inset to Fig. 5k.
Our data indicate that most of the replication complexes are built up by RNA strands differing in sequence at the same positions in VP1. Early in the infectious cycle, they might be the infecting parental RNA strands; later, they might conceivably be parental and/or recombinant strands. The mixed replication complexes, therefore, could continuously give rise to intertypic recombination.
Tracing individual input viral genomes from cell entry to the site of replication complex formation.
The experiments presented above suggest the formation of replication complexes containing two different (parental) RNA molecules. The statistical likelihood for two parental RNA strands to encounter and build up a mixed replication complex, however, seems quite low, given the small size of a replication complex compared to that of a cell. The chance for the formation of mixed replication complexes would be higher if preferential sites for the formation of replication complexes existed. To test for such possible sites, we traced, by FISH, input viral genomes on their way from virus adsorption to the formation of replication complexes.
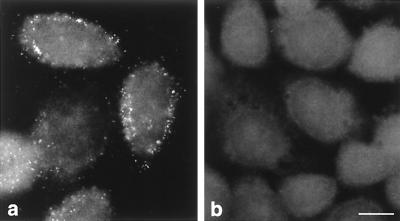
To establish that input PV genomes can be detected with the FISH protocol used, cells were infected with PV M1, blocked with guanidine-HCl to prevent RNA replication, and subjected to FISH with a plus-strand-detecting riboprobe ∼3,000 nt long. The length of the riboprobe allowed us to achieve the necessary sensitivity for input RNA detection but abolished type specificity. A discrete granular signal (Fig. 6a), clearly above the background fluorescence seen in mock-infected cells (Fig. 6b), was observed and is considered to be indicative of input viral RNA.
FIG. 6.
Validation of the protocol to detect input viral RNA by FISH with a conventional microscope. (a) Cells were infected with PV M1 in the presence of 2 mM guanidine-HCl to prevent viral RNA replication and hybridized with a FITC-labeled probe of 2,980 nt at 2 h p.i. The discrete dotted signals thus represent input viral RNA. (b) Uninfected cells are devoid of signal. Bar, 10 μm.
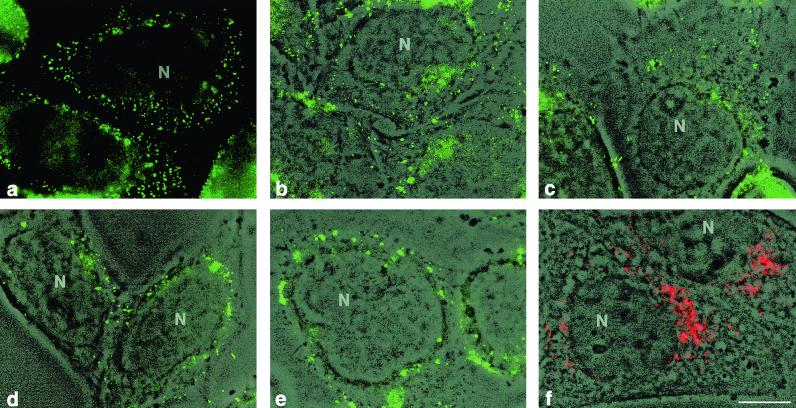
In PV M1- and S2-double-infected cells, input viral genomes were distributed over the cell at 0.5 h p.i. (Fig. 7a). To locate the FISH signals with respect to the entire cell, phase-contrast and FISH pictures of the same microscopic field were overlaid. Figure 7b shows the locations of the FISH signals over the cytoplasm, as well as at the cell periphery. At 1.0 h p.i., the signals were found throughout the cytoplasm of the cell, but less at its surface (Fig. 7c). At 1.5 h p.i., the number of signals per cell was decreased, and they appeared as larger clumps around the nucleus (Fig. 7d). About an hour later, the first PV vesicles forming viral replication complexes became visible in a perinuclear region by IF analysis with anti-2B MAb, previously established as a vesicle marker (21, 48) (Fig. 7f). Their location was compatible with the perinuclear location of the FISH signal (Fig. 7e). The number of replication complexes at 2.5 h p.i. appeared definitely lower than the number of input RNA signals at 0.5 h p.i. (Fig. 7a). Optical sections obtained by confocal microscopy of the same specimens confirmed the location of the nascent replication complexes adjacent to the nucleus (not shown; similar pictures are presented in Fig. 5a to f).
FIG. 7.
Conventional micrographs of cells infected with PV M1 and S2. (a to e) FISH was performed with a FITC-labeled probe of 2,980 nt at different times p.i. (b to e) The images were superimposed on the corresponding phase-contrast images. Viral RNA is found at the cell periphery and in the cytoplasm at 0.5 (a and b) and 1 (c) h p.i. and in a juxtanuclear region at 1.5 h p.i. (d). The signals in panels a to d represent input viral RNA. Replication complexes start to form around 2.5 h p.i. (e), as judged from the appearance of PV vesicles, visualized in a parallel specimen by phase-contrast-IF with anti-2B MAb (red) (f). Bar, 10 μm.
In conclusion, the location of the RNA signal during the early phases of the infection suggests that the viral input RNA migrates to a perinuclear region, where translation and, subsequently, RNA replication in a replication complex starts at relatively few specific sites.
Temporal correlation of replication and recombination.
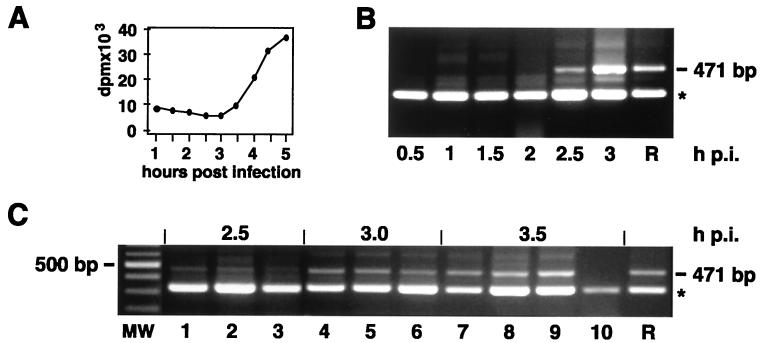
The data presented indicate that mixed replication complexes are common in double-infected cells. We wanted to know whether recombination, in our system, occurred at early stages of replication complex formation, i.e., when the first replication complexes were emerging at their presumably specific sites. To correlate the kinetics of viral RNA synthesis with the emergence of recombinants, incorporation of a radioactive precursor into viral RNA in cells double infected with PV M1 and S2 (Fig. 8A) was compared to the appearance of the 471-bp recombinant cDNA band after RT-PCR (Fig. 8B). From 3 h p.i. onward, AMD-resistant RNA synthesis could be measured above background, which is relatively high in this cell system (Fig. 8A). The first recombinants could be detected at 2.5 h p.i. (Fig. 8B), which clearly puts the earliest recombination event at the very beginning of the viral growth cycle, when the first replication complexes were formed.
FIG. 8.
Viral RNA replication and appearance of recombinants during the viral growth cycle in double-infected cells. (A) Kinetics of PV M1 and S2 RNA synthesis in the presence of AMD, as measured by incorporation of [3H]uridine. (B) Recombinants (471-bp band) are detected by type-specific PCR with the primer pair S up and M down from 2.5 h p.i. onward. (C) Appearance of recombinants in sequentially or simultaneously infected cells. Recombinants (471-bp band) were detected by type-specific PCR at the indicated times p.i. Lanes 1, 4, and 7, cells were infected with PV M1 in the presence of CHI, washed, and then infected with PV S2; lanes 2, 5, and 8, cells were first infected with PV S2 and then with PV M1 as described above; lanes 3, 6, and 9, cells were mock infected in the presence of CHI, washed, and then simultaneously infected with PV M1 and S2; lane 10, cells were infected simultaneously with PV M1 and S2 and kept under CHI to prevent virus replication, which resulted in a lower intensity of the nonspecific band. R, recombinant; ∗, nonspecific band as in Fig. 2; MW, molecular size ladder.
In the experiments described above, cells were infected by adding PV M1 and S2 simultaneously. This could have resulted in mixed virus aggregates, which would facilitate recombination and mimic the proposed preferential sites of replication complex formation. To exclude this possibility, cells were infected sequentially in order to prevent virus aggregation at the adsorption and penetration step. However, even a small temporal advantage (10 min) of the first virus led to considerable inhibition of the second virus as monitored by type-specific FISH at early time points (not shown). Therefore, the first virus was adsorbed to the cells in the presence of CHI. Thirty minutes later, the drug and unadsorbed virus were washed out and the second virus was added, which resulted in sequential adsorption and uptake of the two virus strains but simultaneous onset of replication. Figure 8C shows that recombinants, as measured by RT-PCR, appeared at the same time p.i. (2.5 h), regardless of whether adsorption with the parental PV strains was sequential (lanes 1, 2, 4, 5, 7, and 8) or simultaneous (lanes 3, 6, and 9). Consistent with the findings in Fig. 8B, recombinant RNA could not be found without ongoing RNA replication, i.e., earlier than 2.5 h p.i. (not shown) or in cells kept under CHI (Fig. 8C, lane 10).
These findings make aggregation of the viral inoculum an unlikely factor for the early occurrence of recombination and the high percentage of mixed replication complexes observed in our experiments. They rather support the view that preferential perinuclear sites exist where several RNA genomes together can form a replication complex which contains more than one replicating genome and thus allows for recombination.
DISCUSSION
In the present study, we addressed the question of how recombination between different viral RNA genomes may take place in the infected cell. The process of recombination implies the close association of two parental RNA strands, and thus, we asked how viral genomes might find each other. How might this happen early in infection, when the genomes are still spread out in a comparatively large cell and have not yet accumulated in replication complexes? On the other hand, the observation that these complexes are built up in cis and are tightly closed (21) suggests that genetic exchange of replicating RNA enclosed in replication complexes would again be impeded.
Our objective was to find the intracellular site where recombination occurs. To locate RNA of PV serotypes 1 or 2 in the infected cell, fluorescent type-specific riboprobes were used, and the presence of the donor and acceptor plus strands, PV M1 and S2, in a topologically close setting was considered to be indicative of a possible site of recombination.
Recombination was reported to occur, in principle, over the entire viral genome (31, 32), although reduced in a possibly temperature-dependent way in the P1 genomic region (18). For our work, we had chosen an RNA stretch, within VP1 of the P1 genomic region, sufficiently differing in sequence between PV M1 and S2 to allow for type-specific RNA localization by FISH. Luckily, in this region, recombination occurred reproducibly, as shown by type-specific PCR and hybridization of type-specific probes to and sequencing of the PCR products. These assays are not dependent on the viability of the recombinants, and this might have helped to detect recombination in a region where it was reported to be low (18). Surprisingly, we found recombination to occur at a predominant site within the tested stretch of about 450 nt. This might support the view that RNA structures which favor template switching exist (45, 54). Conceivably, other recombinants may develop outside of or within the VP1 region investigated, notably a possible reversed recombinant having an M1 5′ and an S2 3′ end. However, for the RNA localization experiments reported here, it was important to demonstrate that recombination as such may occur in the RNA region tested, and thus, other possible recombinants than the one characterized in Fig. 1, 2, and 3 were not explored.
Using parental PV strains not carrying markers which restrict replication, the conditions under which recombination occurred were not restrictive for the multiplication of either of the two viral partners. As expected, recombination was detected only if the parental strands were allowed to replicate in double-infected cells. This was shown by mixing PV M1 and S2 RNAs just before RT-PCR (Fig. 2A) or inhibiting replication in double-infected cells with CHI (Fig. 8C, lane 10), which did not give rise to recombinants. Furthermore, in time course experiments, recombinants were found only after the onset of viral replication (compare Fig. 8A and B). Before that time, only parental RNA genomes, but no recombinants, could be detected (not shown). These findings also argue against an RT-PCR artifact in the detection of the recombinants.
By locating RNA sequences of both PV serotypes by FISH, we found a surprisingly high percentage (>85%) of individual replication complexes containing both recombination partners, PV M1 and S2 RNAs, early in infection. Thus, exchange of RNA between different replication complexes is not required for recombination, and structural constraints for the polymerase to switch from a donor to an acceptor RNA strand seem to be low in mixed replication complexes.
The presence of a large percentage of mixed replication complexes and the emergence of recombinants concomitantly with the onset of replication prompted us to investigate when and where the parental RNA strands would come together. Therefore, the early steps of replication complex formation were followed with a highly sensitive fluorescent non-type-specific ribopobe. We were able to detect input viral RNA genomes and follow their migration up to the site where viral RNA replication started. After emerging as large numbers of extremely small granules at the cell periphery, the input RNA moved centripetally toward the nucleus. After having reached the perinuclear region, the RNA was found to locate to larger but fewer spots. Concomitantly, the first PV vesicle clusters appeared, as demonstrated by IF with a MAb against protein 2B, a marker protein for the PV vesicles (5, 20, 48). Such early vesicle clusters represent actively RNA-synthesizing replication complexes, as was demonstrated previously (8).
Input viral RNA, in migrating to the nuclear periphery to start replication, showed a clear decrease in the number of FISH signals. This could be due to either degradation of genomes, leaving only a few replication-competent RNA strands, or to nesting of several strands at few but specific sites. Rapid degradation of decapsidated viral RNA does not seem likely, since FISH signals persisted in cells with guanidine-blocked viral replication until 2 h p.i. (Fig. 6) and were found up to 7 h p.i. (not shown).
Our data indicate that (few) preferential start sites exist where PV replication is initiated, i.e., where minus-strand RNA synthesis is launched. These start sites may well correspond to our previously described delineated membrane-associated and minus-strand RNA-containing structures (“germ centers”) ∼1 μm in diameter (12). They were found to persist in number and size throughout the viral replication cycle, becoming more and more embedded in a progressively growing area with predominantly plus-strand RNA. Whether the actual coming together of different genomes takes place only at the start sites of replication or also during the migration of the genomes from the cell periphery to the perinuclear area is unknown. However, experiments with sequential infection ruled out aggregation of input virus, and the FISH data (Fig. 7) can be interpreted to mean that individual viral genomes are separated until later in their migration across the cytoplasm. We favor the interpretation that the genomes come together at a few preferential start sites for replication complex formation over the view that nascent replication complexes could attract additional viral genomes. The latter possibility seems less likely, since the formation of a PV replication complex was found to proceed in cis (21).
The possible existence of limiting replication sites was discussed in earlier work (17), in which competition for such putative sites was considered to be an explanation for the interference of two viruses in double-infected cells. The view that preferential start sites for viral RNA replication exist is also supported by previous observations on the location of COPII-mediated vesicle formation in PV-infected and uninfected cells (48). In uninfected cells, the COPII proteins induce the vesicles of the anterograde membrane pathway and are distributed throughout the cytoplasm (reference 48 and references therein). In contrast, in PV-infected cells, the COPII proteins are involved in the formation of and are present on PV vesicles and the COPII proteins were found in a perinuclear location (48). However, the mechanism by which PV chooses its start site for RNA replication is unknown and may include translational or transcriptional processes and the presence of indispensable cellular factors.
A limited number of preferential start sites for viral replication clearly would facilitate the formation of mixed replication complexes. Analysis of the spatial relationship between RNA strands in mixed replication complexes indicated a close arrangement of the genomic segments visualized by our type-specific riboprobes. To score as colocalization, different RNA strands must have a lateral distance of <50 nm, i.e., less than the pixel size used in our confocal microscopy protocol. Due to the small diameter of an RNA strand, even more than two RNA strands may be lying in parallel across a 50-nm-diameter area. The actual colinear alignment of those segments of the parental RNA strands involved in the switching process was proposed to be mediated by distinct secondary structures (45, 54). In addition, it was recently suggested (36) that viral RNA synthesis takes place on the surfaces of two-dimensional viral polymerase lattices forming small tubules connected to the surfaces of PV vesicles. The sharing of such lattices by parental RNA strands could favor their recombination (36).
Recombination takes place early in the infectious cycle (31). We report here that viral input genomes migrate to preferential replication start sites where, by the cooperation of two or more different genomes, replication complexes are formed, of which the majority contain a mixed RNA population and thus can autonomously provoke genetic rearrangement.
Acknowledgments
We are indebted to Vadim I. Agol, Institute for Poliomyelitis, Moscow, Russia, for valuable discussions, suggestions, and critical reading of the manuscript and to Lukas Landmann, Institute of Anatomy, Basel, Switzerland, for help and hospitality at the confocal microscope. We thank Eckard Wimmer, Stony Brook, N.Y., for providing the full-length PV M1 plasmid pT7PV and Andrea Glaser and Rainer Gosert for helpful discussions.
This work was supported by grant 31-055397.98 from the Swiss National Science Foundation.
REFERENCES
- 1.Agol, V. I. 1997. Recombination and other genomic rearrangements in picornaviruses. Semin. Virol. 8:1-9. [Google Scholar]
- 2.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 3.Barton, D. J., B. J. ODonnell, and J. B. Flanegan. 2001. 5 ′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz, K., D. Egger, and L. Pasamontes. 1987. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology 160:220-226. [DOI] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, and T. Pfister. 1994. Characteristics of the poliovirus replication complex. Arch. Virol. Suppl. 9:147-157. [DOI] [PubMed] [Google Scholar]
- 6.Bienz, K., D. Egger, T. Pfister, and M. Troxler. 1992. Structural and functional characterization of the poliovirus replication complex. J. Virol. 66:2740-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1983. Intracellular distribution of poliovirus proteins and the induction of virus-specific cytoplasmic structures. Virology 131:39-48. [DOI] [PubMed] [Google Scholar]
- 8.Bienz, K., D. Egger, Y. Rasser, and W. Bossart. 1980. Kinetics and location of poliovirus macromolecular synthesis in correlation to virus-induced cytopathology. Virology 100:390-399. [DOI] [PubMed] [Google Scholar]
- 9.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bolten, R., D. Egger, R. Gosert, G. Schaub, L. Landmann, and K. Bienz. 1998. Intracellular localization of poliovirus plus- and minus-strand RNA visualized by strand-specific fluorescent in situ hybridization. J. Virol. 72:8578-8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caliguiri, L. A., and I. Tamm. 1968. Action of guanidine on the replication of poliovirus RNA. Virology 35:408-417. [DOI] [PubMed] [Google Scholar]
- 14.Chetverin, A. B., H. V. Chetverina, A. A. Demidenko, and V. I. Ugarov. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 16.Cooper, P. D. 1977. Genetics of picornaviruses. Comp. Virol. 9:133-207. [Google Scholar]
- 17.Cords, C. E., and J. J. Holland. 1964. Interference between enteroviruses and conditions effecting its reversal. Virology 22:226-234. [DOI] [PubMed] [Google Scholar]
- 18.Duggal, R., and E. Wimmer. 1999. Genetic recombination of poliovirus in vitro and in vivo: temperature-dependent alteration of crossover sites. Virology 258:30-41. [DOI] [PubMed] [Google Scholar]
- 19.Egger, D., R. Bolten, C. Rahner, and K. Bienz. 1999. Fluorochrome-labeled RNA as a sensitive, strand-specific probe for direct fluorescence in situ hybridization. Histochem. Cell Biol. 111:319-324. [DOI] [PubMed] [Google Scholar]
- 20.Egger, D., L. Pasamontes, R. Bolten, V. Boyko, and K. Bienz. 1996. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J. Virol. 70:8675-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 74:6570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etchison, D., and E. Ehrenfeld. 1981. Comparison of replication complexes synthesizing poliovirus RNA. Virology 111:33-46. [DOI] [PubMed] [Google Scholar]
- 23.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(RC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Gene Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 26.Giachetti, C., S. S. Hwang, and B. L. Semler. 1992. cis-acting lesions targeted to the hydrophobic domain of a poliovirus membrane protein involved in RNA replication. J. Virol. 66:6045-6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gmyl, A. P., E. V. Belousov, S. V. Maslova, E. V. Khitrina, A. B. Chetverin, and V. I. Agol. 1999. Nonreplicative RNA recombination in poliovirus. J. Virol. 73:8958-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodfellow, I., Y. Chaudhry, A. Richardson, J. Meredith, J. W. Almond, W. Barclay, and D. J. Evans. 2000. Identification of a cis-acting replication element within the poliovirus coding region. J. Virol. 74:4590-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirst, G. K. 1962. Genetic recombination with Newcastle disease virus, polioviruses, and influenza. Cold Spring Harbor Symp. Quant. Biol. 27:303-309. [DOI] [PubMed] [Google Scholar]
- 31.Jarvis, T. C., and K. Kirkegaard. 1992. Poliovirus RNA recombination: mechanistic studies in the absence of selection. EMBO J. 11:3135-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King, A. M. 1988. Preferred sites of recombination in poliovirus RNA: an analysis of 40 intertypic cross-over sequences. Nucleic Acids Res. 16:11705-11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.King, A. M. Q. 1988. Genetic recombination in positive strand RNA viruses, p. 149-165. In E. Domingo, J. J. Holland, and P. Ahlquist (ed.), RNA genetics. CRC Press, Boca Raton, Fla.
- 34.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ledinko, N. 1963. Genetic recombination with poliovirus type 1 studies of crosses between a normal horse serum-resistant mutant and several guanidine-resistant mutants of the same strain. Virology 20:107-119. [DOI] [PubMed] [Google Scholar]
- 36.Lyle, J. M., E. Bullitt, K. Bienz, and K. Kirkegaard. 2002. Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296:2218-2222. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, J., R. E. Lundquist, and J. V. Maizel, Jr. 1978. Structural studies of the RNA component of the poliovirus replication complex. II. Characterization by electron microscopy and autoradiography. Virology 85:445-455. [DOI] [PubMed] [Google Scholar]
- 38.Minor, P. D., A. John, M. Ferguson, and J. P. Icenogle. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J. Gen. Virol. 67:693-706. [DOI] [PubMed] [Google Scholar]
- 39.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 40.Paul, A. V., E. Rieder, D. W. Kim, J. H. vanBoom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul, A. V., J. H. vanBoom, D. Filippov, and E. Wimmer. 1998. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature 393:280-284. [DOI] [PubMed] [Google Scholar]
- 42.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 44.Rieder, E., A. V. Paul, D. W. Kim, J. H. vanBoom, and E. Wimmer. 2000. Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J. Virol. 74:10371-10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanova, L. I., V. M. Blinov, E. A. Tolskaya, E. G. Viktorova, M. S. Kolesnikova, E. A. Guseva, and V. I. Agol. 1986. The primary structure of crossover regions of intertypic poliovirus recombinants: a model of recombination between RNA genomes. Virology 155:202-213. [DOI] [PubMed] [Google Scholar]
- 46.Roumiantzeff, M., J. V. Maizel, Jr., and D. F. Summers. 1971. Comparison of polysomal structures of uninfected and poliovirus infected HeLa cells. Virology 44:239-248. [DOI] [PubMed] [Google Scholar]
- 47.Roumiantzeff, M., D. F. Summers, and J. V. Maizel, Jr. 1971. In vitro protein synthetic activity of membrane-bound poliovirus polyribosomes. Virology 44:249-258. [DOI] [PubMed] [Google Scholar]
- 48.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. J. Hong, H. P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 75:9808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suhy, D. A., T. H. Giddings, and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interactions with eIF4G stimulates picornavirus IRES-dependent translations. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda, N., R. J. Kuhn, C. F. Yang, T. Takegami, and E. Wimmer. 1986. Initiation of poliovirus plus-strand RNA synthesis in a membrane complex of infected HeLa cells. J. Virol. 60:43-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takegami, T., R. J. Kuhn, C. W. Anderson, and E. Wimmer. 1983. Membrane-dependent uridylylation of the genome-linked protein VPg of poliovirus. Proc. Natl. Acad. Sci. USA 80:7447-7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 55.Towner, J. S., M. M. Mazanet, and B. L. Semler. 1998. Rescue of defective poliovirus RNA replication by 3AB-containing precursor polyproteins. J. Virol. 72:7191-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Troxler, M., D. Egger, T. Pfister, and K. Bienz. 1992. Intracellular localization of poliovirus RNA by in situ hybridization at the ultrastructural level using single-stranded riboprobes. Virology 191:687-697. [DOI] [PubMed] [Google Scholar]
- 57.Wimmer, E., C. U. T. Hellen, and X. Cao. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]