Abstract
Translation of poliovirus RNA is driven by an internal ribosome entry segment (IRES) present in the 5′ noncoding region of the genomic RNA. This IRES is structured into several domains, including domain V, which contains a large lateral bulge-loop whose predicted secondary structure is unclear. The primary sequence of this bulge-loop is strongly conserved within enteroviruses and rhinoviruses: it encompasses two GNAA motifs which could participate in intrabulge base pairing or (in one case) could be presented as a GNRA tetraloop. We have begun to address the question of the significance of the sequence conservation observed among enterovirus reference strains and field isolates by using a comprehensive site-directed mutagenesis program targeted to these two GNAA motifs. Mutants were analyzed functionally in terms of (i) viability and growth kinetics in both HeLa and neuronal cell lines, (ii) structural analyses by biochemical probing of the RNA, and (iii) translation initiation efficiencies in vitro in rabbit reticulocyte lysates supplemented with HeLa or neuronal cell extracts. Phenotypic analyses showed that only viruses with both GNAA motifs destroyed were significantly affected in their growth capacities, which correlated with in vitro translation defects. The phenotypic defects were strongly exacerbated in neuronal cells, where a temperature-sensitive phenotype could be revealed at between 37 and 39.5°C. Biochemical probing of mutated domain V, compared to the wild type, demonstrated that such mutations lead to significant structural perturbations. Interestingly, revertant viruses possessed compensatory mutations which were distant from the primary mutations in terms of sequence and secondary structure, suggesting that intradomain tertiary interactions could exist within domain V of the IRES.
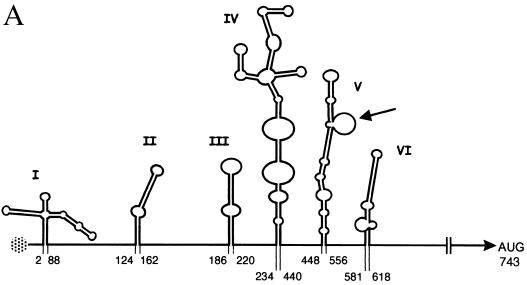
Poliovirus (PV) is often considered the prototype of the Enterovirus genus of the Picornaviridae family. As such, its genome is a single-stranded RNA molecule of positive polarity. It is now well established that the long 5′ untranslated region (5′-UTR; 742 nucleotides [nt] for PV type 1 Mahoney strain [PV1(M)]) plays different important roles in the PV life cycle. The first hundred nucleotides form a cloverleaf structure involved in RNA replication (3), acting at the initiation of negative-strand RNA synthesis (4). Similarly, on negative-strand replication intermediates, the complement of the 5′-UTR is involved in the synthesis of positive-strand viral RNA (32). Furthermore, the 5′-UTR contains an internal ribosome entry segment (IRES) which promotes translation initiation by a cap- and 5′-end-independent mechanism, following direct entry of the 40S ribosomal subunit at the 3′ end of the IRES (for a review, see reference 6). The PV IRES is approximately 450 nt long and has a predicted secondary structure comprised of five complex stem-loops (Fig. 1A), one of which has been reported to be dispensable for efficient translation at least ex vivo (9). The original predicted secondary structure of the PV 5′-UTR was initially based on thermodynamic criteria of RNA folding (23). However, this prediction was refined by taking into account physical RNA probing analyses and phylogenetic considerations, i.e., sequence conservation and covariance between different members of the Enterovirus genus (30, 34). Indeed, the IRESs of all enteroviruses known to date can be accommodated within the currently accepted predicted secondary structure shown in Fig. 1A. Furthermore, strict conservation of short blocks of sequences throughout the enterovirus IRESs has been observed (16).
FIG. 1.
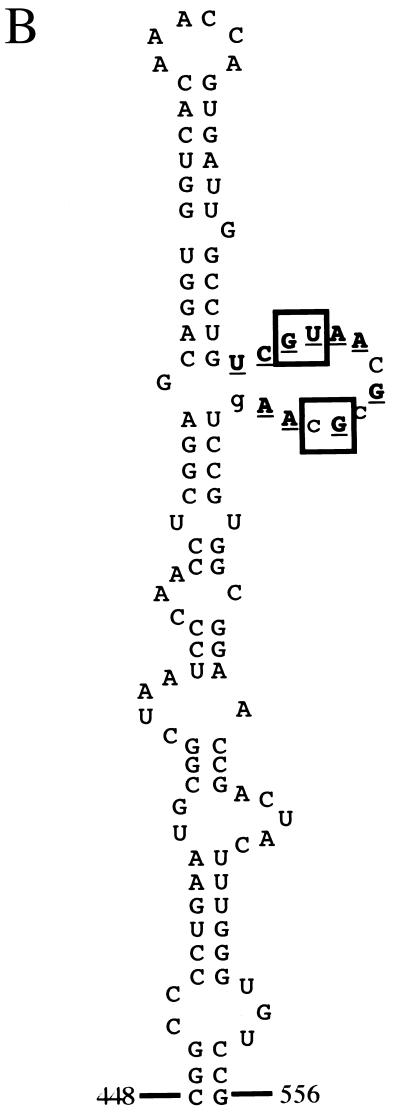
Diagram of the PV 5′-UTR. (A) Line drawing of predicted secondary structure motifs (3, 15, 30). The first and last base-paired nucleotides in each stem-loop are numbered [numbering system for PV1(M)]. Stem-loops II to VI constitute the IRES. The genome-linked protein is shown as a speckled circle, and the lateral bulge-loop of stem-loop V targeted in this study is indicated by an arrow. (B) Sequence of stem-loop V of the PV1(M) IRES. Nucleotide conservation among all members of the Enterovirus genus is shown for the lateral bulge-loop as follows: bold, underlined, uppercase letters, absolute conservation; uppercase letters, purine-pyrimidine conservation; lowercase letters, variable nucleotides. The conservation pattern was last comprehensively checked against all sequences at our disposal at the end of 1998. Nucleotides mutated in this study are boxed.
Like many other enteroviruses, PV exhibits neurotropism, and PV infection results in paralytic poliomyelitis in approximately 1% of infected humans (28). The live, attenuated vaccine strains developed by Albert Sabin for all three serotypes of PV each contains a single point mutation within the IRES which is a major determinant of attenuation (12; reviewed in reference 25). These different mutations lie in the same region of the RNA, within stem-loop V of the IRES, and result in decreased translation efficiency specifically in neuronal cells or in in vitro translation systems derived from such cells (14, 21). Additionally, a particular feature of the predicted secondary structure of stem-loop V of the IRES is that it comprises an unusually large lateral bulge (nt 511 to 524; Fig. 1A). This bulge-loop is exactly geographically juxtaposed relative to the PV1 and PV2 attenuation mutations. Furthermore, the sequence of the bulge-loop is highly conserved within all enteroviruses, including two GNAA motifs (Fig. 1B).
The tools necessary to precisely manipulate the PV genome have been available for over 20 years, since the development of the first full-length infectious cDNA clone (31). This development allowed site-directed mutagenesis techniques to be applied to PV, as for a DNA genome (29). Furthermore, significant technological advances during this time have rendered studies of the effects of precise substitution mutagenesis in different regions of the PV genome routine for functional analyses (39). Nevertheless, despite evidence provided by the Sabin attenuation mutations indicating that single nucleotide changes within the PV IRES can have dramatic effects, relatively few studies concerning this target have been reported to date. While such studies have ultimately revealed valuable information concerning PV IRES function, notably within stem-loop V (see, for example, reference 14), the approaches used have often been relatively unfocused and have involved multiple simultaneous substitutions (see, for example, reference 13). Furthermore, to date mutational analysis has been mainly focused on stem structures within the PV IRES, leaving putatively unpaired regions, which are more highly conserved in sequences, largely ignored.
Thus, to assess the biological function of the two GNAA motifs in the lateral bulge-loop of stem-loop V in the context of the PV IRES, the first two nucleotides of these sequences were subjected to extensive site-directed substitution mutagenesis. Either of the two GNAA motifs could be destroyed without dramatic effects on viral performance in cell cultures. However, when both motifs were destroyed, the growth of resulting viruses was impaired. Notably, such mutants were incapable of ensuring a productive infection, specifically at high temperatures and in neuronal cells, in a manner remarkably similar to what is seen with Sabin viruses. These results correlated with extremely inefficient IRES activity, as measured by in vitro translation assays. These mutants also showed major changes in RNA secondary structure, as assessed by biochemical probing.
MATERIALS AND METHODS
Bacteria and plasmid constructions.
Mutants were engineered by oligonucleotide-directed site-specific mutagenesis by using two rounds of PCR with pKK-C2 template DNA, which contains the PV1(M) cDNA with a deletion of nt 2547 to 6303 (8). For the first round of PCR, primers corresponded to nt 1 to 25 of PV1(M) cDNA (19) and antisense nt 531 to 503 or nt 503 to 531 and antisense nt 767 to 743. Thus, two overlapping mutated PCR products were generated, gel purified, and used as templates for the second round of PCR with primers corresponding to nt 1 to 25 and antisense nt 767 to 743 of PV1(M). The first PCR was carried out for 25 cycles of 45 s at 95°C, 30 s at 60°C, and 45 s at 72°C, followed by 2 min at 72°C. The second PCR consisted of 25 cycles of 45 s at 95°C, 30 s at 60°C, and 1 min at 72°C, followed by 5 min at 72°C. All PCRs were done with 50-μl volumes containing Pwo polymerase buffer, 200 μM deoxynucleoside triphosphates (dNTPs), and 2.5 U of Pwo DNA polymerase (Roche). The AgeI/MscI restriction fragments [nt 339 to 627 of PV1(M)] of the final PCR products were introduced into pKK-C2. The SalI/NheI fragments from the different pKK-C2 mutants, which had been verified by sequencing, were introduced into infectious cDNA clone pT7-PV1-D1, which is analogous to pT7-PV1-52 (26) except that nt 287 to 7055 came from the cDNA of PVR106 (31).
Domain V of wild-type or mutant IRESs [nt 448 to 556 of PV1(M)] was amplified by PCR with pT7-PV1-D1 as the template DNA. The primers were 5′-CGGCCCCTGAATGCGGC-3′ [nt 448 to 464 of PV1(M)] and 5′-CGGATCCGGACACCCAAAG-3′ [antisense nt 556 to 544 of PV1(M), with a BamHI site at the end of domain V]. PCR conditions were 1 min 20 s at 94°C; 30 cycles of 35 s at 94°C, 1 min at 55°C, and 40 s at 72°C; and 10 min at 72°C. PCRs were done with Perkin-Elmer PCR buffer, 200 μM dNTPs, 500 nM primers, and 2 to 3 U of Taq DNA polymerase (Perkin-Elmer)/100 μl. PCR products were cloned directly into vector pGEM-T Easy (Promega). The EcoRI/BamHI fragments containing domain V were transferred into vector pGEM-1 (Promega) to generate the pE series of plasmids, such that domain V started 19 nt after the end of the T7 promoter.
RNA transcription.
For transfections, purified plasmids derived from pT7-PV1-D1 were linearized with EcoRI. RNA was synthesized by using 2 U of T7 RNA polymerase (New England Biolabs)/μl in a reaction mixture containing 20 mM KH2PO4-K2HPO4 (pH 7.5), 8 mM MgCl2, 4 mM spermidine, 10 mM dithiothreitol (DTT), 1 mM each nucleoside triphosphate (NTP), and 0.5 U of RNasin (Promega)/μl for 1 h at 37°C. The quality and quantity of the RNA were evaluated by electrophoresis through agarose gels, and the RNA was used directly for transfections, without further purification.
For translation assays, purified plasmids derived from pKK-C2 were linearized with EcoRI. RNA was transcribed in vitro by using T7 RNA polymerase in a reaction mixture containing 40 mM Tris-HCl (pH 7.5), 15 mM MgCl2, 1 mM each NTP, 2 mM DTT, 0.5 U of RNasin/μl, and trace amounts of [α-32P]UTP (10 μCi/μl, 800 Ci/mmol; Amersham) to facilitate the quantification of RNA yield. RNA was purified on Sephadex G-50 spin columns (Roche), followed by ethanol precipitation.
For RNA structure probing, purified plasmids of the pE series were linearized with SalI 15 nt downstream of the 3′ end of domain V and then transcribed as for in vitro translation, except that no [α-32P]UTP was included in the reaction mixture. The quality and quantity of the RNA were evaluated by electrophoresis through agarose gels.
Cells, transfections, and infections.
HeLa cells and IMR-32 neuroblastoma cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 5 and 10% fetal calf serum (FCS), respectively.
Monolayers of HeLa cells in 35-mm plates (106 cells/plate) were used for transfection in the presence of DEAE-dextran as described previously (37). Transfection under semisolid medium (0.9% Noble agar) served to determine the specific infectivity of in vitro-transcribed RNAs and to obtain plaque-purified viruses. These viruses were amplified by picking isolated plaques, placing them in 150 μl of DMEM without serum, freezing-thawing the mixture three times, and infecting HeLa cell monolayers in 35-mm plates. The plates were incubated for 1 h at 37°C before the addition of 2 ml of DMEM supplemented with 1% FCS. Cells were harvested into medium when total cytopathic effects were observed (usually after 24 h) and were frozen-thawed three times, and the virus stock was clarified by centrifugation. Viral phenotypes were determined by plaque assays with HeLa cells at 37 and 39.5°C.
To construct one-step growth curves, monolayers of IMR-32 cells were grown in 15-mm plates for 2 to 3 days before infection with different mutants at a multiplicity of infection (MOI) of 10. After 1 h of adsorption at 37°C, 500 μl of DMEM supplemented with 1% FCS was added, and infected IMR-32 cells were incubated at 37 or 39.5°C. At different times up to 24 h postinfection, cells were scraped into the medium and collected by brief centrifugation. The medium was discarded, and the cells were washed twice with DMEM without FCS to remove extracellular virus before being resuspended in 250 μl of DMEM without serum. Cells were frozen-thawed three times, and the virus suspension was clarified by centrifugation. Virus titers were determined by plaque assays with HeLa cell monolayers at 37°C.
Blind passaging of virus stocks in IMR-32 cells was carried out as follows. Plates (35 mm) with subconfluent growth were infected at an MOI of 10 HeLa cell PFU. After adsorption for 1 h at 37°C, DMEM-2% FCS was added to a final volume of 2 ml. After 5 days, cells were scraped into the medium and frozen-thawed three times, and 15 μl (diluted to 150 μl in DMEM without FCS) was used to infect fresh plates of IMR-32 cells. This cycle was repeated three times, and the presence of virus in the harvest was verified by plaque assays with HeLa cells. These virus stocks were used to construct one-step growth curves with IMR-32 cells as described above.
To sequence virus stocks, cytoplasmic RNA extracted from infected monolayers of HeLa cells served as a template for reverse transcription-asymmetric PCR to generate single-stranded DNA. The DNA was sequenced manually by using a T7 sequencing kit (Amersham) as described previously (8). Alternatively, a double-stranded DNA product generated by reverse transcription from antisense nt 846 followed by PCR from between nt 67 and 846 was sequenced automatically by using a Big Dye Terminator kit (Amersham).
In vitro translation.
In vitro translation reactions were carried out with nuclease-treated rabbit reticulocyte lysates (RRL) (17) supplemented with HeLa or IMR-32 cell S10 extracts prepared and treated with micrococcal nuclease as described previously (8). All reactions contained 50% (vol/vol) RRL (Flexi RRL system; Promega), 417 μCi of [35S]methionine/ml (>1,000 Ci/mmol; Amersham), and 80 μM unlabeled amino acids (except for leucine and valine, which were present at 120 μM, and methionine, which was omitted). Added MgCl2 was maintained constant at 0.33 mM, whereas total added KCl was optimized on the basis of the batch of RRL used, varying from 75 to 95.5 mM. Concentrations of HeLa and IMR-32 cell extracts varied from 0 to 20% (vol/vol), and that of H100 buffer (10 mM HEPES-KOH [pH 7.5], 1 mM MgCl2, 0.1 mM EDTA, 100 mM KCl, 7 mM β-mercaptoethanol) varied from 33 to 13% (vol/vol) of the final reaction volume. Reactions were programmed with uncapped mRNAs at concentrations of 5 to 40 μg/ml, and assay reaction mixtures were incubated for 90 min at 30°C. Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis by using gels containing 23% (wt/vol) polyacrylamide. Quantification of translation products was carried out by densitometry of autoradiograms by using NIH image software, with multiple exposures of each autoradiogram to ensure that the linear response range of the film was respected.
End labeling and structure probing of RNAs.
Purified transcripts (10 μg) were dephosphorylated at their 5′ ends with 100 U of shrimp alkaline phosphatase (Roche) in a solution containing 50 mM Tris-HCl (pH 8.5) and 5 mM MgCl2 for 30 min at 37°C. Phenol-chloroform-purified RNAs (5 μg) were radiolabeled at their 5′ ends with 30 U of T4 polynucleotide kinase (Roche) in a buffer containing 50 mM Tris-HCl (pH 7.4), 10 mM MgCl2, and 5 mM DTT by using 10 μCi of [γ-32P]ATP/μg (10 μCi/μl, 3,000 mCi/mmol; Amersham) for 30 min at 37°C. Reactions were continued for 15 min after the addition of a further 30 U of T4 polynucleotide kinase and 80 nM ATP. Radiolabeled RNAs were purified from 6% acrylamide-urea gels.
Protocols for structure probing experiments were based on those of Marczinke et al. (27). Specifically, reaction mixtures contained 30,000 cpm of 5′-end-labeled RNA transcripts (corresponding to approximately 0.1 μg) and 10 μg of unlabeled yeast tRNA as a carrier. Probing with RNase T1 (Roche) was done for 20 min at 30°C with a solution consisting of 50 mM sodium cacodylate (pH 7.0) and 5 mM MgCl2 with 0, 0.1, or 0.4 U of RNase T1 in a 50-μl reaction volume. Reactions were stopped by the addition of 150 μl of ethanol, and the RNA was recovered by centrifugation. Probing with lead acetate (Merck) was done for 5 min at 30°C with a solution consisting of 20 mM HEPES, NaOH (pH 7.5), 5 mM magnesium acetate, and 50 mM potassium acetate with 0, 1, 2, or 4 mM lead acetate in a 10-μl reaction volume. Reactions were stopped by the addition of EDTA to 33 mM, and the RNA was recovered by ethanol precipitation. RNA samples were dissolved in water, mixed with an equal volume of formamide gel loading buffer (80% [vol/vol] formamide, 50 mM Tris-borate [pH 8.3], 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue), boiled for 2 min, and analyzed on 10.5% polyacrylamide-urea sequencing gels. All structure probing gels included an alkaline hydrolysis ladder of the relevant RNA as a size marker. This ladder was prepared by dissolving the dried pellet corresponding to 0.3 μg of end-labeled RNA and 10 μg of carrier tRNA in 3 μl of a solution containing 22.5 mM NaHCO3 and 2.5 mM Na2CO3 and boiling the mixture for 1 or 2.5 min.
RESULTS
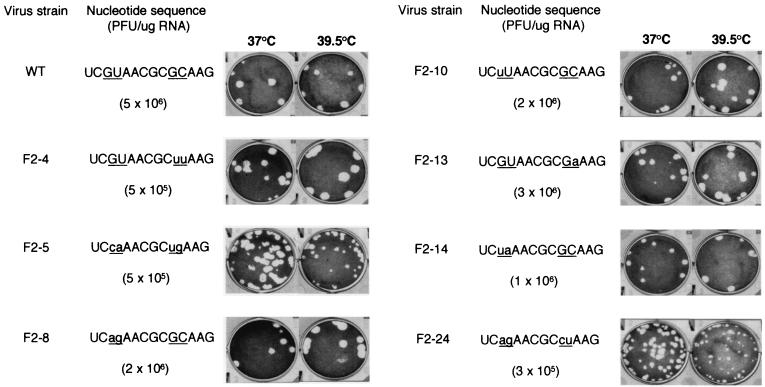
Stem-loop V of the PV IRES comprises an unusually large, highly conserved lateral bulge which encompasses two GNAA motifs (Fig. 1B). Although these sequences are not predicted to form tetraloops, such as that present in domain IV of the PV IRES, the sequence conservation pattern observed among enteroviruses would be compatible with intrabulge base pairing involving these motifs and/or possible presentation of the second motif as a tetraloop closed by a standard Watson-Crick base pair. To attempt to assess the biological function of these GNAA motifs and the biological relevance of any possible base pairing between them, they were subjected to extensive site-directed mutagenesis (Fig. 2). The first GNAA motif was destroyed by changing one (mutant F2-10) or both (mutants F2-5, F2-8, F2-14, and F2-24) of the absolutely conserved GU residues. Similarly, the second motif was eliminated by mutation of the conserved G and the variable C residues (mutants F2-4, F2-5, and F2-24). The role of the conserved AA dinucleotides was not examined in this work. Thus, for two mutants (F2-5 and F2-24), four nucleotide changes were introduced which destroyed both GNAA motifs but maintained the possibility of strong base pairing between these positions. For other mutants, such potential base pairing was at best weakened (F2-4 and F2-10) or even abolished (F2-8 and F2-14). Mutant F2-13 was constructed such that both GNAA motifs were retained, but potential base pairing was weakened.
FIG. 2.
Effects of site-directed mutagenesis of the two GNAA motifs in the lateral bulge-loop of PV1(M) stem-loop V on virus viability. The sequence of the entire loop is given with mutation sites underlined. For each mutant, changes from the wild-type (WT) sequence are indicated by lowercase letters. The specific infectivity of in vitro-derived transcripts (PFU per microgram of RNA) was determined by transfection of HeLa cell monolayers, which were then overlaid with semisolid medium. Plaque phenotypes of recovered viruses were determined after infection of HeLa cell monolayers for 48 h at the indicated temperatures.
Effects of mutations in PV IRES stem-loop V GNAA motifs on viral phenotypes.
All of the mutants tested were viable upon transfection of HeLa cell monolayers, although a 1-log drop in specific infectivity was observed when the second GNAA motif was destroyed (Fig. 2). In all instances, the viruses recovered had retained the mutations introduced (data not shown). Most of these viruses grew as well as the wild-type virus in HeLa cells, as exemplified by their plaque phenotypes (Fig. 2); in addition, certain mutants actually performed slightly better than the wild type at elevated temperatures (for example, mutants F2-4 and F2-8). Only mutants F2-5 and, in particular, F2-24 evidenced mild growth defects which were exacerbated at elevated temperatures. However, even for these mutants, virus stocks showed the same titers at 39.5°C as at 37°C (data not shown).
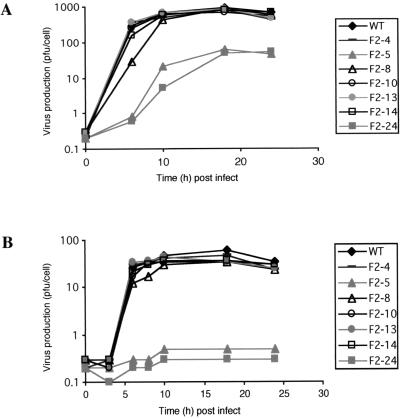
On the other hand, the latter two mutants were severely handicapped in human neuronal cells, those of neuroblastoma cell line IMR-32, as evidenced by their one-step growth curves (Fig. 3). At 37°C, wild-type virus produced over 10-fold more infectious virions per cell than did F2-5 and F2-24 mutant viruses. The defective nature of these mutants was particularly marked at elevated temperature, where they did not produce any detectable infectious virions. This finding was in sharp contrast to the findings for most other mutant viruses, which grew as well as the wild-type virus in these cells. Only mutant F2-8 seemed to have a slightly slow viral cycle, as indicated by the slight lag before the maximum titer was reached. This finding was particularly noticeable at lower temperatures (Fig. 3).
FIG. 3.
One-step growth curves for PV1(M) IRES mutants in neuronal cells. IMR-32 cell monolayers were infected with various mutants at an MOI of 10 and incubated at 37°C (A) or 39.5°C (B). At the indicated times postinfection (post infect), intracellular virus was harvested and quantified (PFU per cell) by titration on HeLa cells. WT, wild type.
Correction of growth defects in neuronal cells correlates with the presence of suppressor mutations.
Mutants F2-5 and F2-24 differed from all of the others by several criteria. As mentioned above, they were the only mutants where both GNAA motifs were destroyed by the introduction of four mutations. Furthermore, certain of their mutations were unique in the context of this study. Thus, the lack of growth of mutants F2-5 and F2-24 in neuronal cells could have been due to one of several explanations. Therefore, it was of interest to determine the nature of any potential revertant viruses capable of growing in IMR-32 cells. To this end, multiple independent stocks of F2-5 and F2-24 produced in HeLa cells were blind passaged three times in IMR-32 cells as described in Materials and Methods.
In each instance where a virus was recovered, growth capacities compared to those of the wild-type and parental mutant viruses were determined with both IMR-32 and HeLa cells (Table 1). Revertant viruses could be classified into two groups in terms of virus production in IMR-32 cells. Members of the first group (e.g., R5-1.4 and R24-11) performed as well as or better than the wild-type virus. Members of the second group (e.g., R5-1.1 and R24-9.1) still produced significantly more virus than the parental mutant virus but two- to fourfold less than the wild-type virus. With HeLa cells, three plaque phenotypes could be distinguished: wild-type large plaques, parental mutant-like medium plaques, and plaques significantly smaller than those of the parental mutant. No correlation could be established between growth properties in the two cell types.
TABLE 1.
Phenotypic and genotypic properties of revertant viruses obtained upon blind passaging of mutants in nonpermissive cells
| Virus | Reversionsa | Plaque phenotypeb | Virus production in IMR-32 cellsc |
|---|---|---|---|
| Wild type | None | lp | 500 |
| F2-5 | None | mp | 30 |
| R5-1.1 | None | sp | 150 |
| R5-1.4 | U476→A | lp | 600 |
| R5-2.2 | None | mp | 180 |
| R5-3 | U529→A | lp | 870 |
| R5-4 | C512→G ± U529→A | lp | 670 |
| R5-7 | U476→A | mp | 640 |
| F2-24 | None | mp | 30 |
| R24-9.1 | C520→U | mp | 190 |
| R24-11 | C471→U | mp | 1100 |
| R24-13 | C517→U | lp | 140 |
| R24-14.2 | U529→A | sp | 600 |
| R24-14.5 | C517→U ± U529→A | lp | 600 |
| R24-15 | None | sp | 260 |
Sequence differences between revertant viruses (R5 and R24) and their parents (F2-5 and F2-24, respectively) were determined from nt 67 to 615. ±, mixed population, encompassing both viruses with a U and an A at nt 529.
Determined with HeLa cells. Viruses were scored as large, medium, or small plaques (lp, mp, or sp, respectively) after incubation for 48 h at 37°C.
Expressed as the number of intracellular infectious virions (measured by titration on HeLa cells) produced per IMR-32 cell over 24 h at 37°C following infection at an MOI of 10.
The genomes of these viruses were sequenced throughout the IRES (Table 1). In only one instance was a primary site reversion observed, within the second GNAA motif (R24-9.1). However, this reversion did not recreate the second GNAA motif but rather recapitulated the sequence of mutant F2-4. Both parents gave rise to reversion events elsewhere within the lateral bulge (R5-4, R24-13, and R24-14.5). Interestingly, for such revertants, at least those derived from F2-24, good virus production in neuronal cells seemed to depend on the presence of a second mutation, a U-to-A change at nt 529 within the proximal part of stem-loop V (compare R24-13 to R24-14.5). Indeed, the presence of this mutation alone correlated with a wild-type level of virus production in neuronal cells (R24-14.2). This same U-to-A reversion was seen for the F2-5 parental virus (R5-3) and again correlated with a high level of virus production in neuronal cells. Intruigingly, the plaques on HeLa cells were large or small, depending on the parental context. It is noteworthy that several different reversion events were evidenced in the proximal part of stem V. Finally, revertant viruses were obtained that carried no reversion mutations in the IRES. Although these revertants showed relatively low levels of virus production in neuronal cells, at least two of them (R5-1.1 and R24-15) could be clearly distinguished from their parents on the basis of very small plaques on HeLa cells (Table 1).
Defective mutants have an altered RNA structure.
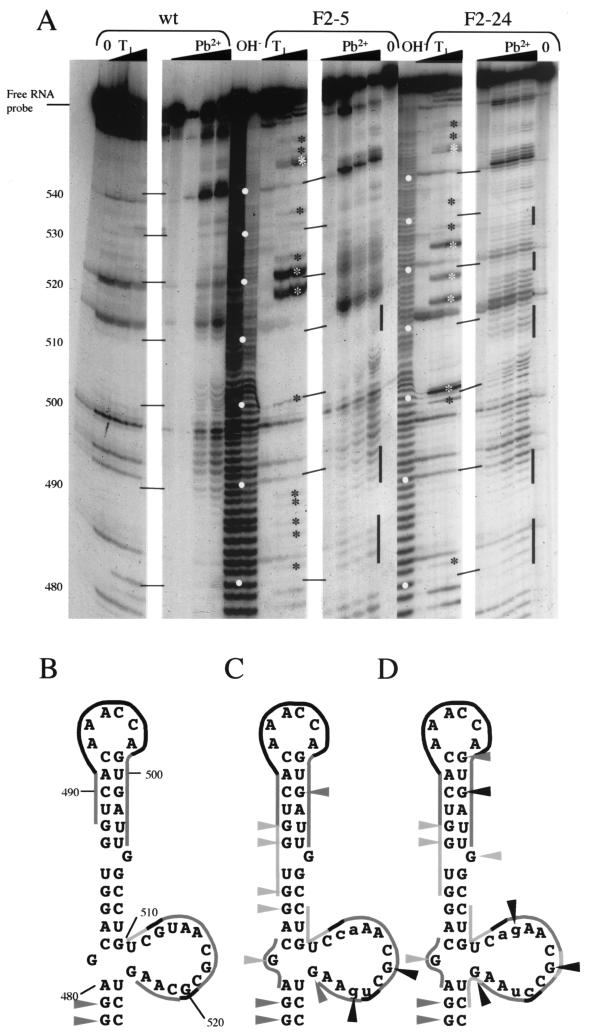
To examine directly the possibility of mutagenesis-induced RNA structural modifications, biochemical probing experiments were carried out (Fig. 4). In addition to RNAs of defective mutants F2-5 and F2-24, that of the quasi-wild-type-phenotype mutant F2-8 was examined. Two different agents were used: RNase T1, which cleaves unpaired G residues, and lead acetate, which cleaves RNA in single-stranded regions. The results of lead acetate probing carried out on the wild-type domain V RNA were generally in good agreement with the currently accepted predicted secondary structure model (Fig. 4A, wt, and 4B). In the region from nt 478 to 527, two signals were obtained: the first corresponded to the apex of domain V (approximately nt 490 to 500), and the second corresponded to the lateral bulge-loop. However, the number and intensity of lead-induced hits in the latter were surprisingly low. Furthermore, the apparent RNase T1 signals observed in this lateral bulge-loop were nonspecific (Fig. 4A, second lane, no enzyme present), indicating that the G residues were not accessible to RNase T1, possibly due to steric hindrance. Indeed, throughout the top of domain V, the only specific RNase T1 signals seen corresponded to nt 478 and 479.
FIG. 4.
Structure probing of 5′-end-labeled mutant RNAs. (A) In vitro transcripts corresponding to stem-loop V of PV1(M) RNA (wt) or mutant derivatives (F2-5 and F2-24) were subjected to limited RNA hydrolysis. Reactions were performed such that each molecule would receive only a single hit (as evidenced by the fact that >90% of the radiolabeled probe remained intact [free RNA probe]) either with RNase T1 (T1 lanes,from left to right: 0, 0.1, and 0.4 U) or with lead acetate (Pb2+ lanes, from left to right: 0, 1, 2, and 4 mM [final concentrations]). Control reactions consisted of either pure RNA (0 lanes) or alkaline hydrolysis ladders (OH− lanes). The autoradiogram of the 10.5% acrylamide-urea gel is shown (the first 30 nt were deliberately cropped, as no differences in profiles between wt and mutant RNAs were seen). Open circles in the OH− lanes indicate intervals of 10 nt, as indicated on the left. Differences between the mutant profiles and the wild-type profiles are indicated by asterisks for RNase T1 and black bars for lead acetate. (B, C, and D) Different probing profiles summarized for the wild type and mutants F2-5 and F2-24, respectively (mutations are indicated by lowercase letters). The predicted secondary structure of nt 478 to 527 of wild-type PV1(M) RNA (top of domain V of the IRES) is shown (30). RNase T1 hits are shown as triangles; lines highlight lead acetate-sensitive regions. The intensity of the symbols is proportional to the intensity of the signals obtained in the probing experiments.
No differences were observed between mutant F2-8 and the wild type, except for a predictable displacement of a nonspecific RNase T1 signal from nt 513 to 514 (data not shown). In contrast, notable differences from the wild type were obtained upon probing of F2-5 and F2-24 mutant RNAs, both quantitatively and qualitatively (Fig. 4A). The F2-5 mutant showed specific RNase T1 signals in the lateral bulge-loop, not only at the mutation site but also at the unchanged G518 and G524 residues (Fig. 4C). The signals corresponding to G518 and G521 were of an extremely high intensity. Furthermore, for this mutant, RNase T1 signals were observed throughout the top of domain V (Fig. 4A and C). Finally, RNase T1 hits were seen at the base of the stem, at G552 and particularly G548-G549 (Fig. 4A). In terms of the lead acetate profile, the stretches evidenced for the wild-type and F2-8 RNAs were extended for the F2-5 mutant RNA. Additionally, a new short lead acetate-sensitive region could be identified facing the lateral bulge-loop (nt 480 to 482). These results attest to the fact that while the global RNA secondary structure may be conserved, it is at least significantly destabilized in mutant F2-5. Further evidence for structural destabilization was provided by probing of mutant F2-24 RNA (Fig. 4D). The lead acetate profile was globally similar to that of mutant F2-5 RNA rather than wild-type RNA. Again, additional RNase T1 signals seen within the bulge-loop were not attributable to the mutations introduced; there was a particularly strong signal at G524 (Fig. 4A and D). Furthermore, as for F2-5, RNase T1 hits were observed throughout domain V, with the addition of a clear signal at G499 and a significantly stronger signal at G501. Such changes in the RNase T1 profile at a significant distance from the actual mutation site (in terms of sequence or predicted secondary structure) are particularly interesting in that they suggest as-yet-unpredicted long-range RNA-RNA interactions.
Mutant IRESs are defective in translation efficiencies.
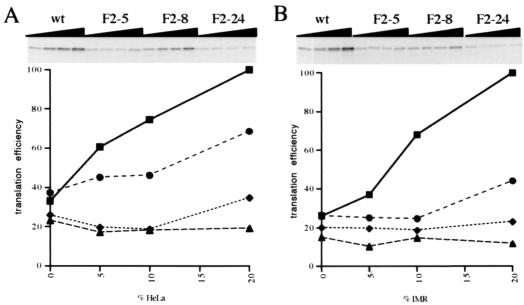
Since it seemed likely that the growth defects and altered RNA probing profiles seen for F2-5 and F2-24 could be accommodated by a hypothesis of impaired translation initiation, in vitro translation analyses were carried out. It is known that PV translation initiation efficiency and fidelity are significantly enhanced by supplementation of the RRL system with cytoplasmic extracts from PV-permissive cells (10) (Fig. 5, wt). Preliminary experiments in which RNA concentrations were varied and reactions included 18% HeLa cell extracts indicated that translation efficiencies indeed correlated with overall growth properties (data not shown), whether they were defects (mutants F2-5 and F2-24) or enhanced growth (mutant F2-4). In addition, differences in translation efficiencies were exacerbated at low RNA concentrations.
FIG. 5.
Translation efficiencies for PV1(M) IRES mutants in vitro. RRL-based translation reactions were programmed with 5 μg of uncapped in vitro transcripts synthesized from pKK-C2 derivatives/ml (see Materials and Methods). Reaction mixtures contained 95.5 mM added KCl (final concentration) and were supplemented with increasing concentrations (0, 5, 10, and 20%, by volume, from left to right) of S10 extracts from HeLa (A) or IMR-32 (B) cells. The different RNAs are indicated above the autoradiograms of the dried sodium dodecyl sulfate-23% polyacrylamide gels. The results of densitometric quantifications are shown below the autoradiograms (▪, wt; ♦, F2-5; •, F2-8; ▴, F2-24). Translation efficiency is expressed as a percentage of that of wild-type RNA in reaction mixtures supplemented with 20% (by volume) S10 extracts.
Thus, further experiments were carried out at a low and constant RNA concentration, with variable amounts of added cell extracts (Fig. 5). Such an approach should allow intrinsically inefficient IRESs to be distinguished from those whose responses to added cell extracts are specifically altered, i.e., those affected in their requirements for noncanonical translation factors. To examine the possibility that the latter effects were cell type specific, translation reaction mixtures were supplemented with extracts from either HeLa cells (Fig. 5A) or IMR-32 cells (Fig. 5B). As expected, the translation of wild-type RNA was significantly stimulated upon inclusion of S10 extracts in the translation reaction mixtures, up to approximately threefold at the highest dose tested. F2-8 RNA translation was less stimulated by the presence of S10 extracts, notably when those from neuronal cells were used. Indeed, unlike the wild-type RNA, this RNA responded only to the highest dose of neuronal cell extracts tested (Fig. 5B). In contrast, translation of the defective mutant F2-5 and F2-24 RNAs was completely insensitive to the presence of neuronal cell extracts and showed little or no response to the inclusion of HeLa cell extracts in translation assays (Fig. 5). Thus, apparent differences in the growth capacities of these mutants in HeLa cells versus IMR-32 cells are not reflected by differences in translation profiles, depending on the source of S10 extracts. However, it is likely that the in vitro translation approach used does not faithfully reproduce the cellular environment. In this context, it should be noted that a maximum of 20% of the translation extract consisted of S10 extract and that this quantity was not sufficient for wild-type RNA to reach a plateau of translation efficiency.
Overall, the translation defects exhibited in in vitro translation assays correlated with the phenotypic properties of the different mutant viruses. Thus, mutants F2-5 and F2-24, which have a slow cycle in HeLa cells and grow very poorly in neuronal cells, show very inefficient translation in vitro and show little or no response to the presence of noncanonical translation factors. The relatively low level of translation of F2-8 reflects the fact that the in vitro translation assays were carried out at 30°C and this virus seems to display a certain degree of cold sensitivity.
DISCUSSION
Generally, the sequence of putatively unpaired regions throughout the IRES is conserved among all members of the enterovirus-rhinovirus group of picornaviruses (16). Thus, it has been proposed that such motifs play critical roles in IRES function, for example, through RNA-RNA tertiary interactions or protein-RNA interactions. Such hypotheses could be investigated by a site-specific mutagenesis approach coupled to functional analyses of the resulting mutants. In this study, we chose the highly conserved lateral bulge of stem-loop V of the PV IRES as a specific target sequence, based on several criteria: (i) this loop is unusually large and as such would not be expected to be totally unpaired; (ii) it encompasses a GNAA motif which could be presented as a tetraloop, although such a structure has not been predicted to date; and (iii) it could be expected to harbor determinants of pathogenesis, based on its close proximity to nucleotides known to be implicated in neurovirulence (14, 25).
Our results clearly show that maintenance of the sequence conservation observed among natural isolates is not required for virus viability. In fact, it seems that a wide range of sequence variation would be tolerated in this region. Indeed, even transversions are accepted at the three absolutely conserved residues targeted in this study. This is also true for other residues within this same bulge-loop (T. A. A. Pöyry, A. M. Borman, K. M. Kean, and R. J. Jackson, unpublished data). Thus, the question of GNRA tetraloop presentation could be denied. Furthermore, we found no evidence for the importance of potential intraloop base pairing between the two GNAA motifs. However, results derived from the study of mutants F2-5 and F2-24 provided evidence for the existence or introduction of intradomain tertiary RNA-RNA interactions.
Biochemical probing analyses showed that, overall, the wild-type lateral bulge is less open than one would expect for an unpaired loop, based on the absence of specific RNase T1 hits and the low intensity of the lead acetate profile compared to those of the apical loop (nt 492 to 498). This finding is in agreement with the results of a previous enzymatic probing study of RNAs derived from reversion mutations of PV mutants, which provided evidence that this bulge is, in fact, relatively unexposed (36). Similarly, the top of stem-loop V is more accessible to lead ions than would be expected for a tightly base-paired stem. One could postulate that this finding reflects long-range intradomain tertiary RNA-RNA interactions. Concomitant destabilization of both the top and the bottom of stem V, along with the bulge itself, observed upon mutation within the bulge would be compatible with this hypothesis. Further evidence in support of such an idea was provided by the phenotypic revertants obtained upon blind passaging of mutants F2-5 and F2-24 on nonpermissive neuronal cells, since such revertants frequently harbored mutations in the proximal part of stem V. Experiments are in progress to formally prove that these are indeed functional compensatory suppressor reversions.
An alternative explanation of our results is that it is not the wild-type RNA which encompasses long-range intradomain RNA-RNA interactions, but rather that such interactions have been generated in mutants F2-5 and F2-24 and that they are deleterious. For example, the F2-24 sequence CGCC520 is complementary to GGCG533, and an interaction between these elements would interfere with the formation of stems at nt 468 to 470 and 533 to 535 and stems at nt 474 to 480 and 525 to 531. The lead acetate profile of F2-24 in the bulge region supports this hypothesis, indicating pairing in the region of nt 517 to 520 (Fig. 4); however, no expected concomitant modification in the structure probing profile was seen in the region of nt 468 to 480 (data not shown). Similarly, the results concerning revertant viruses are somewhat ambiguous. While R24-9.1, R24-13, and R24-14.5 could be taken as providing evidence that the CGCC520 sequence is incompatible with virus growth in neuronal cells, revertants R24-11 and R24-14.2 showed that this is clearly not the case, and these revertants do not involve the sequence elements cited above. Further experiments are required to discriminate categorically between the existence of intradomain tertiary RNA-RNA interactions in wild-type PV1 or the generation of deleterious structural motifs in certain mutants.
Interestingly, the mutations catalogued for the phenotypic revertants were almost exclusively C-to-U transitions or U-to-A transversions. In particular, the frequency of transversions was unexpectedly high relative to previous reports concerning nucleotide substitutions by the PV polymerase (20). This finding could be indicative of selective RNA polymerase error bias, such as that seen for the human immunodeficiency virus reverse transcriptase, which is prone to G-to-A hypermutations (38). Indeed, firm evidence of this phenomenon at specific sites elsewhere in the PV genome has been found (K. M. Kean, L. Balaci, and A. E. Gorbalenya, unpublished observations). Alternatively, the nature of the reversions could reflect RNA structure constraints. In effect, as U476 and U529 face each other across stem V, U-to-A transversions at either of these positions would strengthen the stem via the formation of a canonical A-U base pair, like that found in all strains of PV other than PV1(M). Thus, the revertant data could be interpreted as reflecting compensatory secondary structure reorganization within domain V of the PV IRES. However, we do not believe this to be the case. Such an explanation would not correlate well with the structural perturbations evidenced by probing of mutant RNAs or with the marked temperature dependence of phenotypic defects. Instead, globally, our results bear witness to tertiary rather than secondary structure modifications. Thus, we propose a model in which the lateral bulge-loop is the motor element of structural condensation of domain V, through intradomain tertiary RNA-RNA interactions.
For the hepatitis C virus (a flavivirus) IRES, convincing tertiary structure models have been proposed based on experimental data. It has been shown that this IRES adopts a specific ion-dependent tertiary fold (18). The extended nature of the RNA helices has been confirmed by electron microscopy studies, which show that the hepatitis C virus IRES does not form a globular structure (5, 35). In contrast, the tertiary folds proposed to date for the PV IRES are theoretical (1, 22). Although it is anticipated that data from electron microscopy studies will soon be available for this IRES (5), the resolution of such analyses precludes a comprehension of higher-order structure at the nucleotide level. Thus, genetic approaches such as that described here will remain powerful tools for providing insight into the finer details of picornavirus IRES tertiary structure. Such information will be invaluable for the exploitation of the potential of IRESs as possible targets for antiviral therapies or for the rational conception of attenuated viruses which could serve as vaccines (see below).
Phenotypic revertants of F2-5 and F2-24 were obtained that did not carry additional mutations anywhere in the IRES. It is tempting to speculate that these viruses would harbor mutations in the 2A protease gene. Indeed, it was previously reported that amino acid changes in 2A were able to compensate for domain V disruptions, at least ex vivo (24). Notably, one of the mutations which gave rise to reversions in 2A was the first nucleotide of the bulge under study here. Interestingly, the compensatory effect of 2A changes was found to be cell specific (33).
Our results based on virus viability in cell culture systems raises the question of whether the observed sequence conservation within the lateral bulge-loop in stem V of enterovirus and rhinovirus IRESs is fortuitous or whether this loop plays a role specifically in pathogenesis in the animal host. The former hypothesis is untenable, since evolution would tend toward diversity within a group of related sequences provided that function was not lost and that sequence change was not inordinately difficult. High percentages of positions throughout the IRESs of all known enteroviruses and rhinoviruses are variable (16), showing that there is no intrinsic reason for random sequence conservation in this region of the genome. Evidence that the lateral bulge-loop in stem V of the PV IRES could play a role in pathogenesis was provided by the two mutants which were specifically defective in neuronal cells. Although they formed small plaque on HeLa cells, mutants F2-5 and F2-24 actually produced the same numbers of virions per cell as the wild-type virus in these cells and were merely slow growing (data not shown). However, in neuronal cells, they produced less than 10% of the wild-type number of virions per cell at 37°C, and as the temperature was increased, they actually became incapable of ensuring a productive infection (Fig. 3). This situation is reminiscent of that seen with the attenuated Sabin strain of PV1, which produces 10-fold less virus than the wild-type strain per HeLa cell but does not achieve even 1/100 the wild-type virion production per neuronal cell (2). Similarly, mutations in stem V, just below the lateral bulge, also result in a neuronal cell-specific reduction of virus yield (14). Our results highlight the participation of this entire region of RNA in PV neurovirulence.
The vaccine strain of PV1 is relatively stably attenuated, compared to those of PV2 and particularly PV3, as assessed by the excretion of neurovirulent revertant viruses of the different serotypes by vaccinees (11) and the relative frequency of vaccine-associated disease. These findings probably result from the presence of multiple attenuating determinants scattered throughout the PV1 genome. In contrast, only two or three mutations contribute to the attenuation phenotypes of the PV2 and PV3 vaccine strains, including the transition substitutions in domain V of the IRES, which undergo rapid reversion in the gut of vaccinees (reviewed in reference 7). The sequences of mutants F2-5 and F2-24 differ from that of the wild type either by two transitions plus two transversions or by four transversions. Thus, such viruses could be considered more attractive potential vaccine candidates than the existing strains, with reduced capacities of reversion to neurovirulence during multiplication in the human gut. Of particular interest is the possibility that the same mutations would result in the attenuation of other viruses closely related to PV1. Experiments designed to address this question are currently in progress.
Acknowledgments
Cécile E. Malnou and Tuija A. A. Pöyry contributed equally to this work.
We thank Andy Borman for help and encouragement throughout this work and Ian Brierley for sharing detailed RNA structure mapping protocols with us. We are also grateful to Eric Westhof and Alexander Gorbalenya for stimulating and insightful discussions. Finally, we thank Catherine Gibbs (Cambridge, England) and Josiane Blin and Alex Anselme-Vatin (Paris, France) for infrastructure support.
This work was supported in part by a grant (BIO4-CT95-0045) from the European Commission Framework IV Biotechnology Program to R.J.J. (and others), with additional support to R.J.J. from the Wellcome Trust. K.M.K. is grateful for support from the MENRT (PRFMMIP and Réseau National Hépatite C), the AFM, the ANRS, and the Pasteur Institute (Programme de Recherche Clinique and Contrat d'Incitation à la Recherche en Vue d'Applications). C.E.M. is funded by Bourse de Doctorat d'Ingénieur from the Centre National de la Recherche Scientifique.
REFERENCES
- 1.Agol, V. I. 1991. The 5′-untranslated region of picornaviral genomes. Adv. Virus Res. 40:103-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I., S. G. Drozdov, T. A. Ivannikova, M. S. Kolesnikova, M. B. Korolev, and E. A. Tolskaya. 1989. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 63:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino, R., G. E. Rieckhof, and D. Baltimore. 1991. A functional ribonucleoprotein complex forms around the 5′end of poliovirus RNA. Cell 63:369-380. [DOI] [PubMed] [Google Scholar]
- 4.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beales, L. P., D. J. Rowlands, and A. Holzenburg. 2001. The internal ribosome entry site (IRES) of hepatitis C virus visualized by electron microscopy. RNA 7:661-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA: translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 7.Blondel, B., G. Duncan, T. Couderc, F. Delpeyroux, N. Pavio, and F. Colbère-Garapin. 1998. Molecular aspects of poliovirus biology with a special focus on the interactions with nerve cells. J. Neurovirol. 4:1-26. [DOI] [PubMed] [Google Scholar]
- 8.Borman, A. M., F. G. Déliat, and K. M. Kean. 1994. Sequences within the poliovirus internal ribosome entry segment control viral RNA synthesis. EMBO J. 13:3149-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dildine, S. L., and B. L. Semler. 1989. The deletion of 41 proximal nucleotides reverts a poliovirus mutant containing a temperature-sensitive lesion in the 5′ noncoding region of genomic RNA. J. Virol. 63:847-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysates. J. Virol. 50:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, G., N. T. Begg, N. Cammack, and P. D. Minor. 1990. Virus excretion and mutation by infants following primary vaccination with live oral poliovaccine from two sources. J. Med. Virol. 32:92-95. [DOI] [PubMed] [Google Scholar]
- 12.Evans, D. M. A., G. Dunn, P. D. Minor, G. C. Schild, A. J. Cann, G. Stanway, J. W. Almond, K. Currey, and J. V. Maizel. 1985. Increased neurovirulence associated with a single nucleotide change in a noncoding region of the Sabin type 3 poliovirus genome. Nature 314:548-550. [DOI] [PubMed] [Google Scholar]
- 13.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haller, A. A., S. R. Stewart, and B. L. Semler. 1996. Attenuation stem-loop lesions in the 5′ noncoding region of poliovirus RNA: neuronal cell-specific translation defects. J. Virol. 70:1467-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson, R. J., M. T. Howell, and A. Kaminski. 1990. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem. Sci. 15:477-483. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, R. J., S. L. Hunt, C. L. Gibbs, and A. Kaminski. 1994. Internal initiation of translation of picornavirus RNAs. Mol. Biol. Rep. 19:147-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson, R. J., and T. Hunt. 1983. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 96:50-74. [DOI] [PubMed] [Google Scholar]
- 18.Kieft, J. S., K. Zhou, R. Jubin, M. G. Murray, J. Y. N. Lau, and J. A. Doudna. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292:513-529. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura, N., B. L. Semler, P. G. Rothberg, G. R. Larsen, C. J. Adler, A. J. Dorner, E. A. Emini, R. Hanecak, J. J. Lee, S. van der Werf, C. W. Anderson, and E. Wimmer. 1981. Primary structure, gene organization and polypeptide expression of poliovirus RNA. Nature 291:547-553. [DOI] [PubMed] [Google Scholar]
- 20.Kuge, S., N. Kawamura, and A. Nomoto. 1989. Strong inclination toward transition mutations in nucleotide substitutions by poliovirus replicase. J. Mol. Biol. 207:175-182. [DOI] [PubMed] [Google Scholar]
- 21.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le, S. Y., and J. V. Maizel. 1998. Evolution of a common structural core in the internal ribosome entry sites of picornavirus. Virus Genes 16:25-38. [DOI] [PubMed] [Google Scholar]
- 23.Le, S. Y., and M. Zuker. 1990. Common structures of the 5′ non-coding RNA in enteroviruses and rhinoviruses. J. Mol. Biol. 216:729-741. [DOI] [PubMed] [Google Scholar]
- 24.Macadam, A. J., G. Ferguson, T. Fleming, D. M. Stone, J. W. Almond, and P. D. Minor. 1994. Role for poliovirus protease 2A in cap independent translation. EMBO J. 13:924-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macadam, A. J., D. M. Stone, J. W. Almond, and P. D. Minor. 1994. The 5′ noncoding region and virulence of poliovirus vaccine strains. Trends Microbiol. 2:449-454. [DOI] [PubMed] [Google Scholar]
- 26.Marc, D., G. Drugeon, A.-L. Haenni, M. Girard, and S. van der Werf. 1989. Role of myristoylation of poliovirus capsid protein VP4 as determined by site-directed mutagenesis of its N-terminal sequence. EMBO J. 8:2661-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marczinke, B., R. Fischer, M. Vidakovic, A. J. Bloys, and I. Brierley. 1998. Secondary structure and mutational analysis of the ribosomal frameshift signal of Rous sarcoma virus. J. Mol. Biol. 284:205-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minor, P. D. 1997. Poliovirus, p. 555-574. In N. Nathanson (ed.), Viral pathogenesis. Lippincott-Raven, Philadelphia, Pa.
- 29.Oostra, B. A., R. Harvey, B. K. Ely, A. F. Markham, and A. E. Smith. 1983. Transforming activity of polyoma virus middle-T antigen probed by site-directed mutagenesis. Nature 304:456-459. [DOI] [PubMed] [Google Scholar]
- 30.Pilipenko, E. V., V. M. Blinov, L. I. Romanova, A. N. Sinyakov, S. V. Maslova, and V. I. Agol. 1989. Conserved structural domains in the 5′-untranslated region of picornaviral genomes: an analysis of the segment controlling translation and neurovirulence. Virology 168:201-209. [DOI] [PubMed] [Google Scholar]
- 31.Racaniello, V. R., and D. Baltimore. 1981. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 214:916-919. [DOI] [PubMed] [Google Scholar]
- 32.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowe, A., G. L. Ferguson, P. D. Minor, and A. J. Macadam. 2000. Coding changes in the poliovirus protease 2A compensate for 5′NCR domain V disruption in a cell-specific manner. Virology 269:284-293. [DOI] [PubMed] [Google Scholar]
- 34.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 35.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 36.Stewart, S. R., and B. L. Semler. 1999. Pyrimidine-rich region mutations compensate for a stem-loop V lesion in the 5′ noncoding region of poliovirus genomic RNA. Virology 264:385-397. [DOI] [PubMed] [Google Scholar]
- 37.Teterina, N. L., K. M. Kean, A. E. Gorbalenya, V. I. Agol, and M. Girard. 1992. Analysis of the functional significance of amino acid residues in the putative NTP-binding pattern of the poliovirus 2C protein. J. Gen. Virol. 73:1977-1986. [DOI] [PubMed] [Google Scholar]
- 38.Vartanian, J.-P., A. Meyerhans, B. Åsjö, and S. Wain-Hobson. 1991. Selection, recombination, and G→A hypermutation of human immunodeficiency virus type 1 genomes. J. Virol. 65:1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wimmer, E. 1993. Genetics of poliovirus. Annu. Rev. Genet. 27:353-436. [DOI] [PubMed] [Google Scholar]