Abstract
The replication-associated proteins encoded by Cucumber mosaic virus (CMV), the 1a and 2a proteins, were detected by immunogold labeling in two host species of this virus, tobacco (Nicotiana tabacum) and cucumber (Cucumis sativus). In both hosts, the 1a and 2a proteins colocalized predominantly to the vacuolar membranes, the tonoplast. While plus-strand CMV RNAs were found distributed throughout the cytoplasm by in situ hybridization, minus-strand CMV RNAs were barely detectable but were found associated with the tonoplast. In both cucumber and tobacco, 2a protein was detected at higher densities than 1a protein. The 1a and 2a proteins also showed quantitative differences with regard to tissue distributions in tobacco and cucumber. About three times as much 2a protein was detected in CMV-infected cucumber tissues as in CMV-infected tobacco tissues. In tobacco, high densities of these proteins were observed only in vascular bundle cells of minor veins. In contrast, in cucumber, high densities of 1a and 2a proteins were observed in mesophyll cells, followed by epidermis cells, with only low levels being observed in vascular bundle cells. Differences were also observed in the distributions of 2a protein and capsid protein in vascular bundle cells of the two host species. These observations may represent differences in the relative rates of tissue infection in different hosts or differences in the extent of virus replication in vascular tissues of different hosts.
Viruses utilize the cellular components and machinery of their hosts to drive their own amplification and spread through the environment. For most plant viruses containing RNA genomes, encapsidated RNA is also mRNA. Thus, following inoculation, virus particles must be disassembled and the RNAs must be translated to express protein required for virus amplification. Replication of most plant viruses with RNA genomes involves RNA-dependent RNA polymerase complexes containing both virus-encoded and plant-encoded proteins, which together form the viral replicase (1). There are one or two virus-encoded, replication-associated proteins, which are usually present at low levels within the cell and, in many instances, are detectable only during the acute replication phase. Virus replication usually occurs on specific membranes of the cell, often perturbed by vesicular formation to allow virus replication (16, 17). The specific membranes associated with virus replication have been identified for a number of viruses. There is some indication that viruses belonging to different superfamilies can form either free, smooth vesicles accumulating in the cytoplasm or membrane-associated evaginations of the cytoplasm associated with different organelles (16, 17). However, there is little relationship between the site of virus replication and the various taxonomic relationships between viruses in different genera. Thus, for example, components of the replication complexes of the alfamovirus Alfalfa mosaic virus (AMV) and the bromovirus Brome mosaic virus (BMV), both members of the family Bromoviridae, have been localized to membranes of the central vacuole (the tonoplast) (34) and the endoplasmic reticulum (ER) (21), respectively. In contrast, components of the replication complexes of the tobamovirus Tobacco mosaic virus and the pecluvirus Peanut clump virus both have been localized to the ER (8, 19), while the replication-associated proteins of the tombusviruses Cymbidium ringspot virus and Carnation Italian ringspot virus have been localized to multivesicular bodies derived from peroxisomes and to mitochondria, respectively (28).
The cucumovirus Cucumber mosaic virus (CMV) is also a member of the family Bromoviridae. Unlike most plant viruses, CMV has a very broad host range, encompassing over 1,200 species in 500 genera of 100 families of plants (9). The replicase of CMV is membrane associated and has been purified to homogeneity (14). The purified CMV replicase contains at least three proteins: the 111-kDa 1a protein and the 94-kDa 2a protein, encoded by CMV RNA 1 and RNA 2, respectively, and a plant protein of ca. 50 kDa (14). The 1a protein contains putative methyltransferase and helicase domains, and the 2a protein contains the domains specifying polymerase activity (12, 27). The nature of the plant protein remains unknown. The site of virus replication also has not been established unequivocally, although it is associated with membranes (15, 37). CMV is known to cause a number of cellular perturbations, including the formation of vesicular structures in the cytoplasm that were associated with tonoplasts and budding into the vacuole (13, 18). These vesicular structures contained fine fibrils which showed differential sensitivity to RNase digestion under conditions of high and low salt concentrations, suggesting that the RNAs might have been double-stranded RNA and thus associated with replication (13). However, such vesicles were also seen in infected tissues many weeks after infection, suggesting that these were not transient structures simply associated with CMV replication. CMV has also been reported to disrupt the internal structure of chloroplasts and, in some instances, to alter mitochondria and nuclei as well as to enhance the vesiculation of dictyosomes (18). Thus, effects related to virus replication versus pathogenicity cannot be differentiated readily. Here, we examine the question of the site of localization of the CMV replicase complexes by in situ localization of the site of accumulation of the CMV 1a and 2a replication-associated proteins in two hosts species, tobacco (Nicotiana tabacum) and cucumber (Cucumis sativus). We also examine the distribution of the 1a and 2a proteins in different tissues and compare the localization of the replication-associated proteins to the site of accumulation of plus-strand and minus-strand CMV RNAs.
MATERIALS AND METHODS
Plants, virus source, and inoculation.
Tobacco (N. tabacum cv. Samsun nn) and cucumber (C. sativus var. Venlo pickling) were used as the host plants. Three young plantlets per host were mechanically inoculated with purified CMV strain Fny (Fny-CMV) diluted to 50 ng/μl in 30 mM Na2HPO4 by gently rubbing 20 μl of the inoculum per plant onto aluminium oxide-dusted leaves. Inoculated plants were grown in a glasshouse maintained at 22 to 26°C under a 16-h daylight period.
Tissue sampling, fixation, and embedding.
The same plant samples were used for the detection of virus-encoded proteins and viral RNAs. At 6 days postinoculation (dpi), two leaf pieces (4 by 1 mm) per plant were excised from systemically infected leaves that were 2 to 2.5 cm long and did not show symptoms. Control samples were also collected from mock-inoculated plants of the same age. The individual pieces of leaf were gently vacuum infiltrated with 5 ml of 5% glutaraldehyde in piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) buffer (pH 8) and allowed to undergo fixation for 18 h at room temperature (25). Explants were rinsed twice for 1 h in the same buffer, dehydrated in ethanol, and then embedded either in Araldite or in London Resin White (LR White; medium grade). Complete polymerization of the LR White specimens was obtained by exposing the samples in sealed capsules to UV irradiation for 24 h. Ultrathin sections (silver-gold interference colors) were cut on a Riechert Ultracut E ultramicrotome, heat stretched, and mounted either on pyroxylin film nickel grids for protein detection or on gold grids for RNA detection (25).
IGL.
Immunogold labeling (IGL) was performed essentially as described previously (25). Polyclonal rabbit antisera raised against Fny-CMV 1a and 2a proteins (10) and coat protein (CP) (36) were used as the sources of immunoglobulin G (IgG), which was precipitated in saturated ammonium sulfate and diluted to 1 mg/ml in phosphate-buffered saline (PBS). Sections were pretreated with Decon 75 before incubation for 18 h at room temperature with a 1:100 dilution of IgG which had been cross-absorbed with matching protein extracts from healthy tobacco or cucumber leaf tissues. The sections were then labeled with goat anti-rabbit IgG conjugated to gold particles (15 nm; Amersham). After IGL, the sections were washed, poststained with uranyl acetate and lead citrate, and examined at 80 kV in an electron microscope (model 1200 EX; JEOL UK).
ISH.
Positive-sense and negative-sense probes for in situ hybridization (ISH) of CMV viral RNAs were generated from a derivative of pBluescript KS(+) containing a sequence equivalent to the 3′-terminal 200 nucleotides of Fny-CMV RNA 3 (10). For transcription of the positive-sense probe, the plasmid was linearized with PstI and transcribed in vitro in the presence of T7 RNA polymerase and digoxigenin-11-UTP according to the instructions of the manufacturer of the DIG RNA labeling kit (Roche Diagnostics). The negative-sense probe was synthesized from the same plasmid linearized with XhoI and transcribed with T3 RNA polymerase. Other positive-sense probes were obtained from full-length cDNA clones of Fny-CMV RNAs 1, 2, and 3 (23), linearized with PstI, labeled as described above, and hydrolyzed in alkaline carbonate buffer to reduce transcripts to an approximate size of 200 nucleotides. Nonincorporated nucleotides were removed with RNase-free gel filtration chromatography columns (P-30; Bio-Rad). ISH also was performed with two oligoprobes, either homologous or complementary to nucleotides 901 to 950 in the sequence of CMV-Fny RNA 3 (23), labeled at their 5′ ends with digoxigenin-11-dUTP.
For ISH, the protocol described by Rodriguez-Cerezo et al. (26), with some modifications, was used as standard. Briefly, grids were incubated in prehybridization buffer (50% deionized formamide, 12.5% dextran sulfate, 0.2 M NaCl, 6 mM PIPES, 1 mM EDTA, 6 mg of tRNA/ml) for 2 h at 42°C and hybridized overnight at the same temperature in the same buffer containing 1 to 5 ng of the probe/μl. For ISH with the positive-sense probe, some parameters in the standard hybridization protocol were altered in order to reduce the high level of nonspecific background. Variations made in the hybridization mixture regarded the contents of formamide (0 to 50%), tRNA (0 to 6 mg/ml), and sodium dodecyl sulfate (0 to 0.2%); the hybridization temperature (37 or 42°C); the concentration of probes (0.1 to 5 ng/μl); and the stringency and duration of washing steps (see below). A proteinase K treatment (5 mg/ml; 15 min at 25°C) and a double-stranded RNA denaturation step, which consisted of incubating the grids in prehybridization buffer at 100°C for 5 min just before hybridization, were also included. The grids were washed two times in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 30 min and two or three times in 0.1× SSC for 10 to 30 min. After the blocking step (1 h in PBS containing 1% acetylated bovine serum albumin and 0.1% Tween 20 [blocking buffer]), the grids were incubated for 4 h with a sheep antidigoxigenin antibody conjugated to 10-nm gold particles (British Biocell International), diluted 1:50 in the same blocking buffer, except that the buffer had been diluted 10-fold in PBS. The grids were then washed and stained as described for IGL.
IGL distribution and statistical analyses.
The density of gold labeling was estimated by scanning randomly selected binocular fields of view at a magnification of either ×5,000 or ×7,500 in cells belonging to different tissues. Later, the random areas scanned were confined to tonoplasts. A generalized linear model with Poisson errors and a logarithm link function was fitted to the counts from various subcellular regions (Table 1) or cell types (Tables 2 and 3). The data are expressed as a predicted mean and standard error.
TABLE 1.
Subcellular distribution of 2a protein in CMV-infected cucumber
| Subcellular location | Distribution of gold particlesa
|
Significanceb | |
|---|---|---|---|
| Predicted mean no. | SE | ||
| Tonoplast | 14.37 | 1.08 | |
| Cytoplasm | 1.85 | 0.37 | Yes |
| Plasmalemma | 0.43 | 0.17 | Yes |
| Nuclear membrane | 0.65 | 0.40 | Yes |
| Chloroplast membrane | 0.87 | 0.28 | Yes |
The distribution of gold particles in cucumber palisade mesophyll cells was determined by scanning random areas of subcellular compartments at a magnification of ×7,500 following IGL of 2a protein. The data were obtained from scanning of 10 to 12 sections. contiguous cells per section in three serial sections. The data were analyzed by using a generalized linear model with Poisson errors and a logarithm link function fitted to the counts from the different compartments.
The analysis determined that the gold particle counts obtained from the tonoplast samples were significantly higher than those obtained from the other subcellular compartments.
TABLE 2.
Distribution of CMV 1a and 2a proteins in cucumber leaf cell types
| Protein | Cell type | Distribution of gold particlesa
|
|
|---|---|---|---|
| Predicted mean no. | SE | ||
| 1a | Healthy palisade mesophyll | 0.51 | 0.18 |
| Infected upper epidermis | 7.48 | 0.86 | |
| Infected palisade mesophyll | 12.76 | 1.24 | |
| Infected spongy mesophyll | 9.31 | 1.12 | |
| Infected lower epidermis | 6.11 | 0.75 | |
| 2a | Healthy palisade mesophyll | 0.91 | 0.32 |
| Infected upper epidermis | 13.31 | 1.43 | |
| Infected palisade mesophyll | 22.68 | 2.00 | |
| Infected spongy mesophyll | 16.55 | 2.00 | |
| Infected lower epidermis | 10.86 | 1.21 | |
The distribution of counts of gold particles specific to 1a and 2a proteins (each derived from scanning of two serial sections at a magnification of ×5,000) was analyzed with regard to whether the counts were significantly different between 1a and 2a proteins for the same cell types in CMV-infected cucumber and for different cell types by using a generalized linear model with Poisson errors and a logarithm link function. There was no interaction between the two factors (cell type and protein type). Significant differences between the predicted means were calculated from a model without interactions. The counts for 2a protein were significantly higher than those for 1a protein in the various cell types. The counts for infected palisade mesophyll cells were significantly higher than those for either the other infected cell types or healthy palisade mesophyll cells. The counts for infected spongy mesophyll cells were significantly higher than those for lower epidermis cells.
TABLE 3.
Distribution of CMV 2a protein in infected tobacco and cucumber leaf cell types
| Plant species | Cell type | Distribution of gold particlesa
|
|
|---|---|---|---|
| Predicted mean no. | SE | ||
| Cucumber | Upper epidermis | 12.77 | 1.59 |
| Palisade mesophyll | 21.21 | 2.15 | |
| Spongy mesophyll | 16.40 | 2.26 | |
| Lower epidermis | 12.60 | 1.46 | |
| Tobacco | Upper epidermis | 3.97 | 0.61 |
| Palisade mesophyll | 6.70 | 0.88 | |
| Spongy mesophyll | 5.10 | 0.76 | |
| Lower epidermis | 3.91 | 0.57 | |
The distribution of counts of gold particles specific to 2a protein (derived from scanning of two serial sections at a magnification of ×5,000) was analyzed with regard to whether the counts were significantly different between CMV-infected cucumber and tobacco for the same cell types and for different cell types by using a generalized linear model with Poisson errors and a logarithm link function. There was no interaction between the two factors (cell type and host species). The counts for 2a protein were significantly higher in cucumber cell types than in tobacco cell types, and the counts in palisade mesophyll cells of both plant species were significantly higher than those in other cell types. Significant differences between the predicted means were calculated from a model without interactions.
To test whether the mean gold label counts for the vacuolar membrane were significantly higher than those for other parts of the plant cell, five different cell regions were compared (vacuolar membrane, cytoplasm, plasmalemma, nuclear membrane, and chloroplast membrane). To test whether the mean gold label counts in different cell types of leaves (upper epidermis, palisade mesophyll, spongy mesophyll, and lower epidermis) were different, counts from different cell types in two transections of cucumber leaves treated with different antibodies (anti-1a protein or anti-2a protein) were compared. Tests also were done to determine whether the distributions of 2a protein were significantly different in various cell types of cucumber versus tobacco. To examine the distributions of CP and 2a protein in different cell types of minor veins of tobacco versus cucumber, comparisons were made by considering only minor veins in highly infected areas of the leaf, as determined by the level of CP-specific IGL in surrounding bundle sheath (BS) cells. Two leaf samples from each host plant, with two or three minor veins per sample, were observed and analyzed.
RESULTS
Cytopathologic analysis of infected tissues.
Previous experiments showed that the accumulation of CMV 1a and 2a proteins reaches a plateau in systemically infected tissues at between 4 and 6 dpi (10). Thus, samples used for in situ localization of the viral replicase were collected at 6 dpi from newly emerged leaves. No CMV-specific symptoms were detectable at that time on the sampled plants, and typical mosaic symptoms appeared several days later. We examined early cytologic alterations in sections of tobacco samples embedded in Araldite and postfixed in osmium tetroxide, which allowed optimal resolution of cellular membrane structures. At this stage of the infection, proliferation of membranous vesicles in the cytoplasm was the only obvious effect of viral invasion observed in the samples (data not shown).
Subcellular localization of CMV replication-associated proteins.
The subcellular localization of CMV proteins in infected tissues was investigated by IGL with antibodies raised against 1a protein, 2a protein, and CP. Although immunolabeling of viral particles with anti-CP antibodies was routinely performed with Araldite-embedded specimens, positive results with anti-1a protein and anti-2a protein antibodies were obtained only with LR White-embedded material. Changes in the fixation protocol (e.g., the use of paraformaldehyde instead of glutaraldehyde as a fixative) did not affect the IGL signals under our conditions. The labeling was regarded as specific and genuine when (i) the pattern of distribution of the gold particles was repeated in the same area of cells in serial sections, (ii) the pattern was repeated in serial grids from the same specimen, and (iii) the number of gold particles was at least 2 to 3 times (and usually more than 10 times) higher than the level of nonspecific labeling in healthy tissue sections at the same subcellular locations.
As replicase complexes usually have been reported to be associated with cellular membranes, IGL signals specific for replication-associated proteins were investigated at all recognizable membrane boundaries, i.e., plasmalemma, tonoplast, chloroplast membrane, and nuclear membrane. Counts from randomly chosen areas of the cytoplasm were also included in order to consider the possible association of the replicase with mitochondria and with membranes not clearly visible in samples treated for IGL (i.e., ER and Golgi apparatus). In all the observed leaf cells of both cucumber and tobacco, labeling of both 1a protein and 2a protein was associated with the tonoplast, on which the number of gold particles counted was significantly higher than those on all other membranes (Table 1 and data not shown). As a typical example, Table 1 illustrates the quantitative analysis of the accumulation of 2a protein-specific gold particles on different membranes in cucumber palisade mesophyll cells. Only a very low background level of labeling was observed for mock-inoculated plants after IGL of 1a protein and 2a protein, on tonoplasts as well as on all other membranes (data not shown).
The distribution of 1a protein on the membrane of the central vacuole in cucumber palisade mesophyll cells is shown in Fig. 1A. At a higher magnification, colocalization of 1a protein and 2a protein in the same cellular compartment was evident when serial sections from the same specimen were labeled with anti-1a protein and anti-2a protein antibodies, respectively (Fig. 1B and C, respectively). A similar pattern of labeling, substantiating colocalization of the two replication-associated proteins, was displayed on a small immature vacuole in a cucumber palisade mesophyll cell (Fig. 1D to F). A direct comparison could also be made for the intracellular localizations of CP (Fig. 1G), 1a protein (Fig. 1H), and 2a protein (Fig. 1I) in a cucumber epidermis cell. Although the CP-specific IGL signal was widespread in the cell, a higher concentration of gold particles was observed in the area surrounding the vacuole, the same area where the replication-associated proteins colocalized (Fig. 1G to I). In tobacco leaves at the same stage of infection, immunolocalization of 1a protein and 2a protein with the corresponding antibodies confirmed the same pattern of labeling, with the accumulation of gold particles always being observed in association with the tonoplast (data not shown).
FIG. 1.
In situ localization of 1a and 2a proteins by IGL in CMV-infected cucumber leaf cells. (A) Association of 1a protein with the tonoplast of the central vacuole (V) in a transection of a palisade mesophyll cell labeled with anti-1a protein antibody. c, chloroplast. The rectangle indicates the area enlarged in panels B and C. Scale bar, 500 nm. (B and C) Serial sections from the same cell as in panel A labeled with anti-1a protein and anti-2a protein antibodies, respectively, and showing colocalization of proteins 1a and 2a in association with the tonoplast. Scale bars, 250 nm. (D) Association of 1a protein with the tonoplast of a small vacuole (V) in a transection of a palisade mesophyll cell labeled with anti-1a protein antibody. The rectangle indicates the area enlarged in panels E and F. Scale bar, 250 nm. (E and F) Serial sections from the same cell as in panel D labeled with anti-1a protein and anti-2a protein antibodies, respectively, and showing colocalization of proteins 1a and 2a in association with the tonoplast. Scale bars, 100 nm. (G) Distribution of CP throughout the cytoplasm in a transection of an epidermis cell labeled with anti-CP antibody. V, vacuole. The rectangle indicates the area enlarged in panels H and I. Scale bar, 250 nm. (H and I) Serial sections from the same cell as in panel G labeled with anti-1a protein and anti-2a protein antibodies, respectively, and showing colocalization of proteins 1a and 2a in association with the tonoplast. Scale bars, 100 nm.
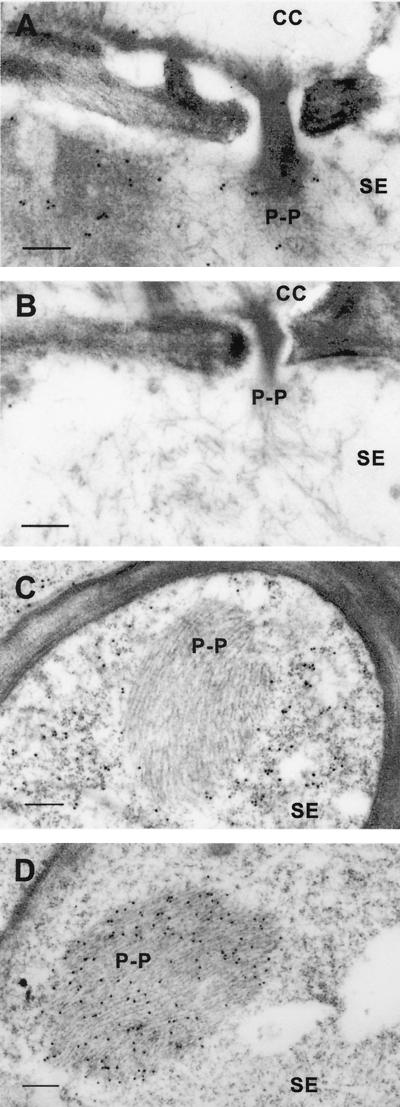
FIG.2.
IGL of filamentous PP1 in tobacco leaf cells by anti-1a protein antibody. (A) Specific labeling by anti-1a protein antibody of PP1 (P-P) in a transection of a class III vein. PP1 aggregates spread through a pore in the sieve plate joining two SE. (B) Absence of PP1 labeling by anti-2a protein antibody in a different serial section of the same cells as in panel A. (C) Absence of PP1 labeling by anti-CP antibody in a transection of a minor (class IV) vein. The CP-specific signal was widespread throughout the cytoplasm of SE. (D) Specific labeling of PP1 by anti-1a protein antibody in a transection of a minor vein of a healthy plant leaf, showing that PP1 labeling was infection independent. Scale bars, 250 nm.
At the early stage of phloem differentiation, a group of proteins known as P proteins form unique structures within sieve elements (SE). During the observation of tobacco samples for the localization of replication-associated proteins, filamentous P protein (P protein 1 [PP1]) was found to be labeled at high levels by anti-1a protein antibodies (Fig. 2A) but not by anti-2a protein antibodies (Fig. 2B) or anti-CP antibodies (Fig. 2C). This finding was the only exception found to the association described previously for 1a protein-specific immunolabeling of the vacuolar membrane. However, anti-1a protein antibodies reacted with a similar intensity to PP1 in healthy tobacco tissues (Fig. 2D). Intravascular structures of PP1 were never detected at this stage in cucumber samples (data not shown).
Distribution of CMV replication-associated proteins in leaf tissues.
The distribution of 1a protein and 2a protein in different types of cells and tissues of the two host species was analyzed by counting gold particles. When we compared the relative levels of gold labeling of the two replication-associated proteins in cucumber, 2a protein accumulated to higher levels than 1a protein in all tissues, with approximate ratios of 1.5:1 to 1.8:1 (Table 2). Higher levels of 2a protein accumulation than of 1a protein accumulation were recorded for tobacco leaf tissues (data not shown), but scores of 1a protein accumulation were not taken into account, as this protein was often barely detectable in all tobacco leaf layers. Divergent particle counts were also observed for the different leaf tissues in cucumber samples. The accumulation of 2a protein in the mesophyll layers was, on average, about twice as high as that in the epidermal layers (Tables 2 and 3), and the ratio of 1a protein in the same tissues was nearly 1.5:1 (Table 2). Inside the mesophyll, a slight preferential accumulation of 1a protein and 2a protein was observed in the palisade mesophyll versus the spongy mesophyll (Tables 2 and 3). This differential distribution of replication-associated proteins among leaf tissues also applied to tobacco samples (Table 3). Finally, a third comparison could be made with regard to the levels of accumulation of 1a protein and 2a protein in the two host species. The density of 2a protein labeling in cucumber was about three times that in tobacco for all the leaf tissues (Table 3). Higher levels of 1a protein accumulation were recorded for cucumber than for tobacco, although the levels of 1a protein accumulation in tobacco were not considered for statistical analyses.
Localization and distribution of CMV replication-associated proteins in VBs.
Several minor veins (class IV and V veins in tobacco and class VI and VII veins in cucumber) were analyzed in systemically infected leaves for the presence and distribution of CMV CP and replication-associated proteins at 6 dpi. At this stage of leaf development, minor veins were immature and the SE were densely cytoplasmic. The organelles were in place and functional, and the deposition of lignin on the cell wall was still in progress. In cucumber, where a bicollateral arrangement of the veins is observed, the differentiation between the specialized companion cells (CC) (intermediary cells [IC]) and the abaxial SE had not yet occurred (35). Although the number of vascular bundles (VBs) suitable for analysis was not sufficient for a statistically significant quantification of labeling, some differences were clearly noticeable when IGL results for tobacco and cucumber were compared (Table 4).
TABLE 4.
Distribution of 2a protein and CP in different cell types in minor veins of CMV-infected tobacco and cucumber plants
| Host species | Cell typea | Distribution of gold particles on:
|
|||
|---|---|---|---|---|---|
| 2a Proteinb
|
CPc | ||||
| Range | Avg | No. | |||
| Tobacco | BS | 19-133 | 67.2 | 14/14 | 100 |
| PP | 0-117 | 32.6 | 11/13 | 100 | |
| CC | 0-111 | 37.4 | 9/10 | 100 | |
| SE | 3-90 | 43.7 | 6/6 | 100 | |
| Cucumber | BS | 1-170 | 51.1 | 12/12 | 100 |
| Adaxial phloem | VP | 0-26 | 7.5 | 3/6 | 200-500 |
| CC | 3-37 | 15.2 | 6/6 | 200-500 | |
| SE | 0 | 0/4 | 0 | ||
| Abaxial phloem | VP | 0-12 | 4.2 | 5/6 | 10 |
| IC | 0-3 | 2.1 | 7/9 | 10 | |
| SE | 0-55 | 9.2 | 1/6 | 5-10 | |
Cell types in minor veins scored for the presence of CMV proteins (2a protein or CP).
Total number of gold particles per cell after 2a protein-specific IGL. Data were derived from 4 to 14 independent counts per cell type. No., number of cells containing gold particles/total number of cells analyzed.
Number of gold particles per cell after CP-specific IGL. A representative area corresponding to the field of view at a magnification of ×7,500 was chosen for particle scoring in each cell. Scores for BS cells were arbitrarily set at 100; other scores were calculated proportionally.
In tobacco, virus localization at the level of the minor veins, visualized by CP-specific IGL, showed a uniform extent of infection in BS cells, CC, and phloem parenchyma (PP) cells. The number of gold particles in BS cells after CP-specific IGL was used as the reference for an approximate comparison of virus accumulation in other VB cells (Table 4). Virus infection was never detected inside the xylem (data not shown). The most intense labeling in the whole leaf by both anti-1a protein and anti-2a protein antibodies was found in BS cells, CC, and PP cells inside the minor veins. In all these cells, vacuoles formed a complex structure common to both healthy and infected cells and appeared as a system of several vesicles in transverse sections (Fig. 3A). Gold labeling specific for 1a protein and 2a protein colocalized on the tonoplast, with the highest concentration in the cytoplasmic strands between individual vacuolar vesicles (Fig. 3B and C) and often in association with an amorphous electron-dense material of an unknown nature (Fig. 3D). Several dozen and up to 120 gold particles were usually counted around vacuoles inside a single CC or PP cell. The counts of 2a protein labeling are shown in Table 4. The 1a protein-specific gold particle counts, although present to a lower extent (see above), followed a similar pattern. Even the immature SE were sites of virus infection, with high levels of labeling produced by anti-CP antibodies (Fig. 2C and Table 4). In immature SE, where vesiculated vacuoles were also present, labeling with both anti-2a protein antibodies (Fig. 4A and Table 4) and anti-1a protein antibodies (data not shown) was observed at the same density as in CC and PP cell.
FIG. 3.
In situ localization of 1a and 2a proteins by IGL in a minor vein of a CMV-infected tobacco leaf. (A) Section of a class V immature vein of tobacco, used to identify the different cell types examined for the presence of CMV replication-associated proteins. BS, bundle sheath; X, xylem; XP, xylem parenchyma; PP, phloem parenchyma. The rectangle indicates the location of the area shown in panels B to D. Scale bar, 2 μm. (B to D) Serial sections from the same minor vein as in panel A labeled with anti-1a protein antibody (B) and anti-2a protein antibody (C and D). Proteins 1a and 2a colocalized in association with the tonoplast, and dense aggregates of gold particles were observed between individual vacuolar vesicles. Arrowheads indicate amorphous electron-dense areas specifically labeled by anti-2a protein antibody. Scale bars: B and D, 250 nm; C, 500 nm.
FIG. 4.
In situ localization of 2a protein by IGL in an immature SE of a CMV-infected leaf. (A) Localization of 2a protein to the tonoplast of vacuolar vesicles (V) in a transection of an SE in an immature minor vein of tobacco labeled with anti-2a protein antibody. PP, phloem parenchyma. Scale bar, 250 nm. (B) Localization of 2a protein to the tonoplast of a small vacuole (V) in a transection of an SE in an immature minor vein of cucumber labeled with anti-2a protein antibody. Scale bar, 100 nm.
In cucumber, complex vesiculated structures were found in vacuoles in BS cells, and again dense 1a protein- and 2a protein-specific labeling was found on these structures associated with the tonoplast. Inside VBs, CP-specific IGL indicated a nonhomogeneous distribution of CMV particles in different cell types, with the highest level of labeling being found in the CC and vascular parenchyma (VP) cells bordering the mature adaxial SE and the lowest level of labeling being found in the immature abaxial IC-SE complexes (Table 4). A virus-specific IGL signal was not detected in the mature adaxial SE (Table 4). An intermediate level of virus-specific IGL was found in surrounding BS cells (Table 4). The distribution of the IGL signals specific to the replication-associated proteins in the vascular cells resembled the virus localization to some extent (Table 4 and data not shown). Immunolabeling showed a limited accumulation of 1a protein and 2a protein in CC in the adaxial position, less frequent labeling in VP cells in the adaxial position, and a rather low level of IGL in the abaxial VP cells and IC (Table 4). In the only instance where 2a protein was detected substantially in a nondifferentiated SE cell, the signal again was associated with the tonoplast of a small round vacuole (Table 4 and Fig. 4B).
ISH analysis of CMV RNA.
Tobacco and cucumber sections from the same tissue samples as those used for IGL were treated for ISH to determine whether both plus-strand and nascent minus-strand viral RNAs were present in the same sites where the 1a and 2a proteins accumulated. A number of different riboprobes and oligoprobes were used in order to achieve the best sensitivity and specificity of the technique. Except for probes generated from the 3′-terminal 200 nucleotides of RNA 3, the other probes all produced high levels of nonspecific background for both healthy and infected tissues. Thus, the RNA 3-specific terminal probes were used in both negative-sense and positive-sense orientations in all subsequent experiments.
CMV plus-strand RNAs were detected throughout the cytoplasm of infected cucumber cells at a density comparable to that of CP-specific IGL (Fig. 5A). The observation that plus-strand RNA-specific signals were detected at high levels on paracrystalline arrays of viral particles (Fig. 5A) indicates that the ISH technique was unable to distinguish between free and encapsidated CMV RNAs. Thus, the accumulation of plus-strand RNA-specific signals on the tonoplast was likely to be correlated with the high level of CP-specific immunolabeling also observed at the same compartment (compare Fig. 5B and Fig. 1G). Similar results were obtained with infected tobacco sections, whereas no plus-strand RNA-specific labeling was detected on healthy plant sections (data not shown).
FIG. 5.
Localization of viral RNA by ISH in vascular cells of veins of CMV-infected leaves. (A) Detection of CMV plus-strand RNA associated with paracrystalline arrays of virus particles (arrowheads) in a transection of a cucumber phloem parenchyma cell in a class IV or V vein. The plus-strand RNA was detected throughout the cytoplasm. In the inset, a larger aggregate of virions in a different phloem parenchyma cell is shown. Scale bars, 250 nm. (B) Detection of CMV plus-strand RNA in a transection of a tobacco CC of a minor vein. The CMV RNA-specific hybridization signal was widely dispersed in the cytoplasm, but clusters of gold particles were associated with the tonoplast around vacuolar vesicles (V). Scale bar, 250 nm. (C and D) Serial transections of a CMV-infected cucumber VB labeled with anti-2a protein antibody (C) and with a CMV minus-strand RNA-specific probe (D) and showing two small vacuoles, V1 and V2, in a phloem parenchyma cell. The 2a protein (C) and minus-strand RNA (arrowheads in D) were associated with the tonoplast of V1 and in between V1 and V2. Scale bars, 100 nm. The RNA probes were either complementary (A and B) or homologous (D) to the 3′-terminal 200 nt of CMV RNA 3.
Hybridization with a positive-sense probe to detect minus-strand viral RNA at the putative site of CMV replication resulted in poor labeling signals on all the leaf tissues of both host plant species. Although several parameters in the hybridization method were changed to increase the specificity of signals, an effective, although minimal, improvement was observed only when proteinase K treatment and denaturation steps were included in the protocol. The procedures for hybridization and washing were set to provide the highest stringency (see Materials and Methods). Under these conditions, although the preservation of tissues after ISH treatment was reduced, making the analysis of the cellular structures rather difficult, three samples were obtained from cucumber sections in which minus-strand RNA-specific signals accumulated at higher levels than the nonspecific background labeling of tonoplasts. Figure 5C and D show serial sections of a small vacuole in a VP cell labeled, respectively, with anti-2a protein antibody and the minus-strand RNA-specific riboprobe. The signal of the replication-associated protein typically was found associated with the tonoplast, while the hybridization signal showed labeling suggestive of a similar distribution (Fig. 5D). Under these conditions, no accumulation of minus-strand RNA-specific ISH labeling was observed on healthy plant sections (data not shown).
DISCUSSION
As has been observed for other plus-strand RNA viruses, CMV RNA replication occurs in close association with intracellular membranes (15, 37). Earlier studies on the cytopathic effect of CMV and Tomato aspermy virus, another member of the genus Cucumovirus, in infected host tissues (13, 18) showed alterations of the vacuole. The presence of vesicles associated with the tonoplast protruding internally and the presence of double-stranded RNA in those vesicles suggested the tonoplast to be the site of cucumovirus replication (13). However, in our experience, such structures are not present at the acute stage of virus replication in cucumber, tobacco (this study), or Nicotiana clevelandii (C. N. Mayers, I. M. Roberts, and P. Palukaitis, unpublished observations). In this study, we used electron microscopy coupled with serologic and in situ RNA detection techniques to establish the intracellular localization of the CMV replicase and the site of CMV RNA replication. The results presented here demonstrate that the CMV replication-associated proteins colocalize to the tonoplast in all leaf tissues of both tobacco and cucumber (Fig. 1, 3, and 4 and Tables 1 and 2). Moreover, while only low levels of negative-sense CMV RNAs were detectable, they were associated with tonoplasts containing 2a protein (Fig. 5).
The sites of accumulation of the replication-associated proteins and minus-strand RNA have been identified for two other members of the family Bromoviridae, the bromovirus BMV and the alfamovirus AMV (7, 21, 34). BMV minus-strand RNA and the replication-associated proteins 1a and 2a colocalized with vesicles originating from the ER, mostly concentrated in the perinuclear region in infected barley protoplasts, as analyzed by immunofluorescence confocal microscopy (21). BMV 1a and 2a proteins and newly synthesized RNAs also colocalized with electron-dense bodies widespread in the cytoplasm and probably of the same origin in barley leaf sections after detection by IGL (7). AMV minus-strand RNA and the replication-associated P1 and P2 proteins were detected in association with the tonoplast in cowpea protoplasts by confocal microscopy, although P2 was found both at the tonoplast and in the cytoplasm (34). Hence, AMV and CMV replicase proteins colocalize at the same subcellular compartment, a feature possibly shared by the ilarvirus Tobacco streak virus (18). The accumulation of the CMV 1a and 2a proteins in tobacco and cucumber leaf sections was always restricted to the tonoplast and the areas closely surrounding the vacuoles, and the specific IGL signal was never observed in the cytoplasm. However, Western blotting of fractionated extracts from CMV-infected tissue showed the presence of free, histidine-tagged 2a protein, presumably in the cytoplasm (11). Thus, as with AMV, there may be a pool of the CMV 2a protein that, under certain conditions, is not associated with the 1a protein.
In contrast to the situation with 2a protein, fractions of extracts from CMV-infected tissues showed 1a protein to be solely membrane associated (11). For BMV, 1a protein itself was shown to direct interactions with the membranes, targeting the replication complex replicase to the ER (3, 5). Using transgenic tobacco plants expressing the CMV 1a protein (2), we attempted to assess the ability of the CMV 1a protein to associate with the tonoplast in the absence of the 2a protein, but we were not able to detect any 1a protein-specific IGL signals. This result was probably due to the low concentration of transgenically expressed 1a protein, which was present at a level that could support the replication of coinoculated RNA 2 and RNA 3 but could not be detected by immunoblot analysis (2).
The potential of the IGL technique to yield quantitative results was also used to determine how replication-associated proteins accumulated throughout the systemically infected leaf. We found that 2a protein accumulated in systemically infected leaves of tobacco and cucumber at levels significantly higher than 1a protein, an observation that was consistently repeated in all leaf tissues. This finding is in agreement with the results of previous quantification studies using immunoblotting, where the Fny-CMV 2a protein was found to accumulate in systemically infected zucchini squash leaves at levels up to 10 times higher than the 1a protein at 6 dpi (10).
The labeling of tobacco PP1 with anti-1a protein antibody (Fig. 2) or with any other plant virus protein-specific antisera has never been reported, to our knowledge. No obvious similarities were found in a comparison of the amino acid sequences of the Fny-CMV 1a protein (22) and two PP1 isolates from the phloem of Cucurbita maxima, the only such PP1 sequences available in the protein databases (4). Thus, either tobacco PP1 has a protein sequence very different from that of C. maxima PP1 or 1a protein-specific antiserum was able to react with a neotope(s) not identifiable on the primary sequence. Besides being part of the CMV replicase, 1a protein has been shown to have a role in the rate of systemic movement (10), whereas PP1 is involved in sealing damaged SE and in long-distance signaling through the phloem (32). Thus, the suggestion that 1a protein may in part mimic a role of PP1 in some hosts is intriguing, although there is little supporting evidence.
Plant viruses move systemically in the phloem and, after unloading into the mesophyll from the class III vein network, invade newly formed tissues, moving locally from cell to cell through the plasmodesmata (PD) (24). Virus spread into the mesophyll precedes the infection of both the epidermal layers and the VBs (24). Thus, the higher level of accumulation of replication-associated proteins in the cucumber mesophyll (Fig. 3) may be a consequence of early entry of the virus in that tissue. However, the timing of viral invasion can only partially explain the differential distribution of replication-associated proteins in cucumber, where tissues infected later showed lower levels of 1a protein and 2a protein accumulation (see below). In tobacco leaves, in contrast, the highest density of labeling was found in VB cells, where virus entry followed the invasion of the surrounding mesophyll (Table 4). Furthermore, 1a protein in tobacco was detected almost exclusively in the SE, CC, and PP cells (Fig. 3B and data not shown), indicating a preferential targeting of the replicase to the vacuoles in VB cells. The lower levels of accumulation of 2a protein and, even more so, of 1a protein in tobacco, in comparison with the levels seen in cucumber epidermis and mesophyll cells, can be explained, at least in part, by the different volumes occupied by the vacuoles in young developing leaves. In tobacco cells, the vacuoles were already greatly extended and the remaining cytoplasm was limited to thin strands appressed against the cell borders, while in cucumber cells, the single vacuole or more vacuoles represented 50% or less of the total cell volume. Thus, low densities of IGL labeling at this stage of leaf development may reflect an effect of the dispersion of the replication-associated complexes on a larger tonoplast surface.
In this study, we analyzed the localization of CMV gene products in two host species, tobacco and cucumber, which use, respectively, an apoplastic pathway and a symplastic pathway for phloem loading (20, 31). In both loading systems, though, trafficking of viruses from cell to cell inside the VBs seems to occur symplastically through the PD (29). From the mesophyll, infection progresses into the minor veins, usually following the route from the BS via the PP to CC and SE (6, 20). In several instances, boundaries between BS and VP, VP and CC, and CC and SE have been shown to represent barriers preventing or delaying viral cell-to-cell diffusion inside the VBs (6). We observed that tobacco was thoroughly infected by CMV, showing no barriers to viral diffusion at the minor vein level; in addition, the levels of accumulation of replicase proteins in these VBs were very high in all the cell types of the VBs. However, unlike what was seen for tobacco, infection by CMV in the VBs of cucumber was not as uniform and reflected the bicollateral structure of its minor veins, in that VP cells and CC in the adaxial positions were infected at a high level, while in the abaxial positions, the level of infection was significantly lower (Table 4). These findings can be explained by considering that the adaxial (internal) phloem in minor veins of Cucurbita pepo matures rapidly and may be specialized in the import of photoassimilates (33), although in Cucumis melo, the adaxial phloem has been demonstrated to be able neither to export nor to import photoassimilates (30). Therefore, the ability of the adaxial phloem to provide early access to the virus moving long distance via the vascular system needs further investigation, especially since it is generally accepted that phloem unloading of both macromolecules and viruses does not occur in minor veins (24). The alternative hypothesis, that the accumulation of high levels of virus in the adaxial phloem may be due to cell-to-cell movement from the mesophyll, is not consistent with the ultrastructure of the minor veins in cucurbit leaves. In these minor veins, numerous PD establish intensive symplastic connections between IC and BS cells in the abaxial phloem, whereas only small numbers of PD are observed in the adaxial phloem at the BS-CC and BS-VP boundaries (30, 33).
The accumulation of 1a and 2a proteins in minor veins of cucumber only partially resembled the distribution of the virus. Although CC and VP cells in the adaxial positions were highly infected, the actual levels of accumulation of 1a and 2a proteins in these cells were lower than expected (compared with the IGL signal in the BS) (Table 4). Taken together, our data suggest that host factors affect the level of accumulation of CMV replicase proteins other than just via viral cell-to-cell movement in leaf tissues. Some of these factors may be merely physical. Examples of these include the tonoplast extension and its effect on replicase dispersion and the lack of vacuoles in many vascular cells in cucumber minor veins, which could make the cells themselves unsuitable for viral replicase accumulation. Moreover, these factors act in a host-specific manner and in a tissue-specific manner, as shown by the almost opposite distributions displayed by 1a and 2a proteins in tobacco and cucumber. In tobacco, the preferential sites of accumulation of replication-associated proteins were inside the veins, while in cucumber, the replication-associated proteins accumulated at the highest levels in mesophyll cells, secondarily in epidermis cells, and only at lower levels in vein cells. Identification of these host factors will prove an interesting challenge.
Acknowledgments
We greatly appreciate the advice and assistance of Lynn Broadfoot and Christine Hackett at Biomathematics and Statistics Scotland (BioSS) with regard to the statistical analyses of the data.
This work was supported in part by a Marie Curie Fellowship of the European Commission Programme Human Potential, under contract number HPMF-CT2000-00632, granted to F.C. and by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department.
REFERENCES
- 1.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Canto, T., and P. Palukaitis. 1998. Transgenically expressed cucumber mosaic virus RNA 1 simultaneously complements replication of cucumber mosaic virus RNAs 2 and 3 and confers resistance to systemic infection. Virology 250:325-336. [DOI] [PubMed] [Google Scholar]
- 3.Chen, J., and P. Ahlquist. 2000. Brome mosaic virus polymerase-like protein 2a is directed to the endoplasmic reticulum by helicase-like viral protein 1a. J. Virol. 74:4310-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark, A. M., K. R. Jacobsen, D. E. Bostwick, J. M. Dannehoffer, M. I. Skaggs, and G. A. Thompson. 1997. Molecular characterization of a phloem-specific gene encoding the filament protein, phloem protein 1 (PP1), from Cucurbita maxima. Plant J. 12:49-61. [DOI] [PubMed] [Google Scholar]
- 5.den Boon, J. A., J. Chen, and P. Ahlquist. 2001. Identification of sequences in brome mosaic virus replicase protein 1a that mediate association with endoplasmic reticulum membranes. J. Virol. 75:12370-12381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, B. 1998. Intercellular protein trafficking through plasmodesmata. Plant Mol. Biol. 38:279-310. [PubMed] [Google Scholar]
- 7.Dohi, K., M. Mori, I. Furusawa, K. Mise, and T. Okuno. 2001. Brome mosaic virus replicase proteins localize with the movement protein at infection-specific cytoplasmic inclusions in infected barley leaf cells. Arch. Virol. 146:1607-1615. [DOI] [PubMed] [Google Scholar]
- 8.Dunoyer, P., C. Ritzenthaler, O. Hemmer, P. Michler, and C. Fritsch. 2002. Intracellular localization of the peanut clump virus replication complex in tobacco BY-2 protoplasts containing green fluorescent protein-labeled endoplasmic reticulum or Golgi apparatus. J. Virol. 76:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwardson, J. R., and R. G. Christie. 1991. Cucumoviruses, p. 293-319. In J. R. Edwardson and R. G. Christie (ed.), CRC handbook of viruses infecting legumes. CRC Press, Inc., Boca Raton, Fla.
- 10.Gal-On, A., I. Kaplan, M. J. Roossinck, and P. Palukaitis. 1994. The kinetics of infection of zucchini squash by cucumber mosaic virus indicate a function for RNA 1 in virus movement. Virology 205:280-289. [DOI] [PubMed] [Google Scholar]
- 11.Gal-On, A., T. Canto, and P. Palukaitis. 2000. Characterization of genetically modified cucumber mosaic virus expressing histidine-tagged 1a and 2a proteins. Arch. Virol. 145:37-50. [DOI] [PubMed] [Google Scholar]
- 12.Habili, N., and R. H. Symons. 1989. Evolutionary relationship between luteoviruses and other RNA plant viruses based on sequence motifs in their putative RNA polymerases and nucleic acid helicases. Nucleic Acids Res. 17:9543-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hatta, T., and R. I. B. Francki. 1981. Cytopathic structures associated with tonoplasts of plant cells infected with cucumber mosaic and tomato aspermy viruses. J. Gen. Virol. 53:343-346. [Google Scholar]
- 14.Hayes, R. J., and K. W. Buck. 1990. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63:363-368. [DOI] [PubMed] [Google Scholar]
- 15.Jaspars, E. M. J., D. S. Gill, and R. H. Symons. 1986. Viral RNA synthesis by a particulate fraction from cucumber seedlings infected with cucumber mosaic virus. Virology 144:410-425. [DOI] [PubMed] [Google Scholar]
- 16.Lesemann, D.-E. 1991. Specific cytological alterations in virus-infected plant cells, p. 147-159. In K. Mendgen and D.-E. Lesemann (ed.), Electron microscopy of plant pathogens. Springer-Verlag KG, Berlin, Germany.
- 17.Lesemann, D.-E. 1999. Virus specific cytological effects in infected plant cells. Phyton (Horn) 39:41-45. [Google Scholar]
- 18.Martelli, G. P., and M. Russo. 1985. Virus-host relationships: symptomatological and ultrastructural aspects, p. 163-205. In R. I. B. Francki (ed.), Polyhedral virions with tripartite genomes. Plenum Press, New York, N.Y.
- 19.Mas, P., and R. N. Beachy. 1999. Replication of tobacco mosaic virus on endoplasmic reticulum and role of the cytoskeleton and virus movement protein in intracellular distribution of viral RNA. J. Cell Biol. 147:945-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oparka, K. J., and S. Santa Cruz. 2000. The great escape: phloem transport and unloading of macromolecules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51:323-347. [DOI] [PubMed] [Google Scholar]
- 21.Restrepo-Hartwig, M. A., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rizzo, T. M., and P. Palukaitis. 1989. Nucleotide sequence and evolutionary relationships of cucumber mosaic virus (CMV) strains: CMV RNA 1. J. Gen. Virol. 70:1-11. [DOI] [PubMed] [Google Scholar]
- 23.Rizzo, T. M., and P. Palukaitis. 1990. Construction of full-length cDNA clones of cucumber mosaic virus RNA 1, 2, and 3: generation of infectious RNA transcripts. Mol. Gen. Genet. 222:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Roberts, A. G., S. Santa Cruz, I. M. Roberts, D. A. M. Prior, R. Turgeon, and K. J. Oparka. 1997. Phloem unloading in sink leaves of Nicotiana benthamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9:1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts, I. M. 1994. Factors affecting the immunogold labelling of plant virus antigens in thin sections. J. Virol. Methods 50:155-166. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Cerezo, E., K. Findlay, J. G. Shaw, G. P. Lomonossoff, S. G. Qiu, P. Linstead, M. Shanks, and C. Risco. 1997. The coat and cylindrical inclusion proteins of a potyvirus are associated with connections between plant cells. Virology 236:296-306. [DOI] [PubMed] [Google Scholar]
- 27.Rozanov, M. N., E. V. Koonin, and A. E. Gorbalenya. 1992. Conservation of the putative methyltransferase domain: a hallmark of the ′Sindbis-like' supergroup of positive-strand RNA viruses. J. Gen. Virol. 73:2129-2134. [DOI] [PubMed] [Google Scholar]
- 28.Rubino, L., and M. Russo. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431-437. [DOI] [PubMed] [Google Scholar]
- 29.Santa Cruz, S. 1999. Phloem transport of viruses and macromolecules—what goes in must come out. Trends Microbiol. 6:237-241. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz, K., B. Cuypers, and M. Moll. 1987. Pathway of assimilate transfer between mesophyll cells and minor veins in leaves in Cucumis melo L. Planta 171:19-29. [DOI] [PubMed] [Google Scholar]
- 31.Schulz, A. 1998. Phloem. Structure related to function. Prog. Bot. 59:431-475. [Google Scholar]
- 32.Thompson, G. A., and A. Schulz. 1999. Macromolecular trafficking in the phloem. Trends Plant Sci. 4:354-360. [DOI] [PubMed] [Google Scholar]
- 33.Turgeon, R., J., A. Webb, and R. F. Evert. 1975. Ultrastructure of minor veins in Cucurbita pepo leaves. Protoplasma 83:217-232. [Google Scholar]
- 34.van der Heijden, M. W., J. E. Carette, P. J. Reinhoud, A. Haegi, and J. F. Bol. 2001. Alfalfa mosaic virus replicase proteins P1 and P2 interact and colocalize at the vacuolar membrane. J. Virol. 75:1879-1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Volk, G. M., R. Turgeon, and D. U. Beebe. 1996. Secondary plasmodesmata formation in the minor-vein phloem of Cucumis melo L. and Cucurbita pepo L. Planta 199:425-432. [Google Scholar]
- 36.Wahyuni, W. S., R. G. Dietzgen, K. Hanada, and R. I. B. Francki. 1992. Serological and biological variation between and within subgroup I and II strains of cucumber mosaic virus. Plant Pathol. 41:282-297. [Google Scholar]
- 37.Young, N. D., P. Palukaitis, and M. Zaitlin. 1987. Characterization of multimeric forms of cucumber mosaic virus satellite RNA, p. 243-252. In C. J. Arntzen and C. Ryan (ed.), Proceedings of UCLA symposium. Molecular strategies for crop protection. Alan R. Liss, Inc., New York, N.Y.